Abstract
Cytochrome c oxidase (CcO) is the terminal enzyme in the electron transfer chain in the inner membrane of mitochondria. It contains four metal redox centers, two of which, CuB and heme a3, form the binuclear center (BNC), where dioxygen is reduced to water. Crystal structures of CcO in various forms have been reported, from which ligand-binding states of the BNC and conformations of the protein matrix surrounding it have been deduced to elucidate the mechanism by which the oxygen reduction chemistry is coupled to proton translocation. However, metal centers in proteins can be susceptible to X-ray-induced radiation damage, raising questions about the reliability of conclusions drawn from these studies. Here, we used microspectroscopy-coupled X-ray crystallography to interrogate how the structural integrity of bovine CcO in the fully oxidized state (O) is modulated by synchrotron radiation. Spectroscopic data showed that, upon X-ray exposure, O was converted to a hybrid O∗ state where all the four metal centers were reduced, but the protein matrix was trapped in the genuine O conformation and the ligands in the BNC remained intact. Annealing the O∗ crystal above the glass transition temperature induced relaxation of the O∗ structure to a new R∗ structure, wherein the protein matrix converted to the fully reduced R conformation with the exception of helix X, which partly remained in the O conformation because of incomplete dissociation of the ligands from the BNC. We conclude from these data that reevaluation of reported CcO structures obtained with synchrotron light sources is merited.
Keywords: synchrotron radiation, bioenergetics, optical absorption, crystallography, metastable states
Abbreviations: APS, Advanced Photon Source; BNC, binuclear center; bCcO, bovine heart CcO; CC, closed conformer; CcO, cytochrome c oxidase; DOE, Department of Energy; heme a and heme a3, heme A moieties in bCcO; O∗, a hybrid derivative of resting oxidized bCcO resulting from X-ray-induced metal reduction; OC, open conformer; PDB, Protein Data Bank; R∗, a bCcO derivative produced by annealing the O∗ species; SSRL, Stanford Synchrotron Radiation Lightsource; TtCcO, CcOs from Thermus thermophilus
Cytochrome c oxidase (CcO) is the terminal enzyme of the electron transport chain in the inner mitochondrial membrane in eukaryotes and the plasma membrane in prokaryotes (1, 2, 3). It catalyzes the four-electron reduction of dioxygen to two water molecules and harnesses the redox energy to pump four protons across the membrane against an electrochemical proton concentration gradient (2, 4).
Crystal structures of bovine heart CcO (bCcO) show that it is a homodimer (5) with each monomer comprised of 13 subunits and four redox sites: CuA, heme a, CuB, and heme a3 (Fig. 1A). CuB and heme a3 constitute the so-called binuclear center (BNC), where dioxygen is reduced to water. During the reduction reaction, four electrons are sequentially passed from cytochrome c via CuA and heme a to the dioxygen bound to heme a3 in the BNC; at the same time, four protons (the “substrate protons”) are taken up from the negative side of the mitochondrial membrane to generate two water molecules. Coupled with the dioxygen reduction chemistry, four additional protons (the “pumped protons”) are translocated from the negative to the positive side of the mitochondrial membrane (2).
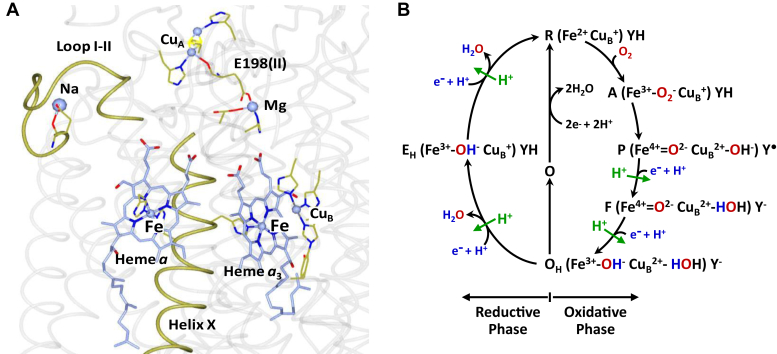
Figure 1.
Structure of the metal centers in subunits I and II. In bCcO (A) and the proposed biphasic sequential mechanism (B). A, shows the locations of the four redox centers, CuA, heme a, CuB, and heme a3, with respect to helix X and loop I–II in bCcO. The structure is from Protein Data Bank (ID: 7TIE). The Mg atom and the E198 from subunit II that links Mg to CuA are also shown as indicated. B, shows the proposed dioxygen reduction mechanism depicting the correlation of the changes in the oxidation and ligation states of the BNC with the entry of four electrons and four substrate protons (in blue) into the BNC and the translocation of four pumped protons (in green) (see the full description in the main text). If OH produced at the end of the oxidative phase is allowed to relax to the resting O state, its reduction to R does not support proton translocation. bCcO, bovine heart CcO; BNC, binuclear center.
Time-resolved studies showed that the oxygen reduction reaction follows a biphasic mechanism (Fig. 1B) (6, 7, 8, 9, 10). In the oxidative phase, the fully reduced enzyme (R) binds O2 to form the O2 complex (A), which then converts to the fully oxidized enzyme (OH) via the P and F intermediates, during which two electrons and two substrate protons (blue) are loaded into the BNC and two pumped protons (green) are translocated through the protein matrix to the positive side of the membrane. In the ensuing reductive phase, OH converts back to the R state via the putative EH intermediate, during which two additional electrons and two substrate protons (blue) are transferred into the BNC; at the same time, two water molecules are released, and two pump protons (green) are translocated. All the oxygen intermediates, except EH, have been identified spectroscopically (8, 9, 10, 11).
The OH intermediate produced at the end of the oxidative phase is redox equivalent to the resting oxidized O state, but if OH is allowed to relax to O, its subsequent reduction to R is unable to drive proton translocation (6), indicating that OH is structurally distinct from the resting O state. To identify the molecular origin enabling the OH state, but not the O state, to translocate protons, it is important to fully understand the identity of the ligand(s) in the BNC and the conformations of the two heme groups, as well as the protein environment surrounding them in the two states. The crystal structures of bCcO, as well as several bacterial CcOs, in their resting O state have been reported (12, 13, 14, 15). However, the identity of the ligand(s) in the BNC remains elusive. In bCcO (12), as well as in Paracoccus denitrificans CcO (14), the ligand was assigned as a peroxide bridging the heme a3 iron and CuB, but this assignment has been questioned (16, 17). Similar ambiguity exists in other bacterial CcOs, where the ligand density was modeled by a water coordinated to heme a3 and a hydroxide ion coordinated to CuB (13) or a single hydroxide ion, or water, bridging the two metal centers (18, 19).
The characterization of the ligand identity in the BNC of CcO is complicated by X-ray-induced radiation damage (16, 20, 21). X-ray-induced radiation damage to protein crystals has been extensively studied (22, 23). An upper dose limit of 30,000 kGy for typical proteins has been established by Owen et al. (24). This dose limit is much lower for metalloproteins, ∼3 kGy, as metal centers in proteins are susceptible to photoinduced reduction and/or ligand dissociation (21, 25, 26, 27, 28). Radiation-induced metal reduction in the oxidized forms of CcO crystals has been recognized based on optical spectra acquired following X-ray exposure (12, 18, 29, 30, 31), which raises an important question: do the structures solved by synchrotron light sources reflect the genuine forms of CcO? To address this question, here we sought to use microspectroscopy-coupled X-ray crystallography at the BL9-2 beamline at Stanford Synchrotron Radiation Lightsource (SSRL) to systematically interrogate the effects of synchrotron radiation on the structural properties of bCcO in the fully oxidized O state. As a comparison, a set of O and R structures were obtained at Sector 31 of the Advanced Photon Source (APS).
Results and discussion
X-ray-induced reduction of redox centers
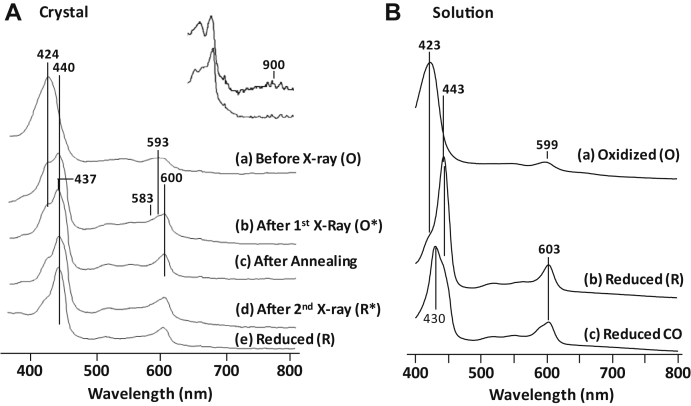
We crystalized bCcO in the fully oxidized state, O, and acquired its diffraction data at 100 K with an X-ray exposure of ∼161 kGy at the BL9-2 beamline at SSRL. The structure was refined to a resolution of 2.35 Å (Protein Data Bank [PDB] ID: 7TIH, Table S1). It is similar to a higher resolution structure obtained at the APS (PDB ID: 7TIE, Table S1) and other reported structures of fully oxidized bCcO obtained with synchrotron light sources (32, 33). Prior to the diffraction measurement, an optical absorption spectrum of the crystal was acquired in situ with a microspectrometer equipped at the beamline. The spectrum has a Soret and visible band at 424 and ∼593 nm, respectively (Fig. 2A, spectrum a). It is in good agreement with that of the O state obtained in free solution (Fig. 2B, spectrum a, see the supporting information for the assignment of the bands). However, following the diffraction measurement, the crystal exhibited a new spectrum with (i) the appearance of a Soret band at 440 nm, at the expense of the 424 nm band (Fig. 2A, spectrum b), (ii) the development of a sharp visible band at ∼600 nm, and the concomitant reduction of the broad 593 nm band, and (iii) the growth of a small shoulder at ∼583 nm and two weak bands in the 500 to 560 nm region. We assign the spectral component with the 440 and 600 nm bands to a low-spin ferrous heme a, whereas the remaining component with the 424 and 593 nm bands to a low-spin ferrous heme a3 with a sixth ligand coordinated to its iron (34, 35). The latter assignment is supported by the similarity of the visible bands to those of CO-bound reduced bCcO, where the ferrous heme a3 is coordinated by the CO ligand (Fig. 2B, spectrum c). Associated with the changes in the heme-associated bands, the broad band at ∼900 nm, which is characteristic of the cupric CuA, disappeared (see the upper inset in Fig. 2A), indicating that CuA is reduced. Taken together, the spectral data suggest that all the four redox centers in bCcO are completely reduced by the X-ray beam, but the ligation state of the BNC is distinct from that in the fully reduced R state (spectrum b in Fig. 2), which is free of ligands. Structural comparison indicates that the protein structure is analogous to the authentic O structure obtained with serial femtosecond crystallography (PDB ID: 6NMP), where radiation damage is minimized (36), instead of the R structure (PDB ID: 7THU, Table S1), indicating that X-ray exposure during the diffraction measurement induces complete reduction of the four redox centers, but the protein matrix is locked in the fully oxidized O state. This hybrid state is referred to as O∗ hereinafter.
Figure 2.
Absorption spectra of the fully oxidized bCcO crystal studied. In this work (A) versus those of bCcO obtained in free solution (B). A, spectra a and b were obtained before and after the first X-ray exposure, respectively; spectra c and d were obtained from the same crystal following annealing before and after the second X-ray exposure, respectively. Spectrum e is a reference spectrum of a fully reduced ligand-free crystal. The upper right inset displays an expanded view of spectrum a (top) and b (bottom) showing the presence of the broad band at ∼900 nm associated with CuA in the cupric state in spectrum a, which is absent in spectrum b. bCcO, bovine heart CcO.
Annealing-induced protein conformational relaxation
We proposed that the O∗ state is stabilized at the cryogenic temperature because of limited protein motion. To allow the relaxation of the protein matrix, we annealed the O∗ crystal to above the protein glass transition temperature (∼200 K) for 2 s and then lowered the temperature back down to 100 K and acquired a new set of diffraction data at the same irradiated spot. In parallel, we obtained the in-crystal spectra before and after the diffraction measurement to identify potential changes in the redox and/or ligand coordination states of the metal centers.
As shown in spectrum c in Figure 2A, after annealing, the Soret band at 440 nm became somewhat sharper and shifted slightly to 437 nm, whereas the 424 nm band was slightly weakened. At the same time, the visible band at 600 nm was sharpened and the shoulders at 583 and 593 nm were diminished. These spectral features suggest the development of a new ligand-free ferrous heme a3 component and small adjustment in the heme a conformation. Following the second diffraction measurement, only minor modifications in the spectrum were identified (spectrum d), which possibly reflect small fine-tuning in the protein matrix surrounding the redox centers. The data indicate that the second X-ray exposure did not lead to additional reduction of the redox centers, consistent with the view that the redox centers had been fully reduced by the first X-ray exposure. In general, the spectral data indicate that the annealing following the first X-ray exposure induced partial dissociation of the heme a3 ligand as well as the relaxation of the heme a conformation and the protein matrix surrounding the redox centers. The fact that spectrum c and d are distinct from that of the fully reduced R species (spectrum e) indicates that the annealing treatment is insufficient to drive the full conversion of O∗ to R.
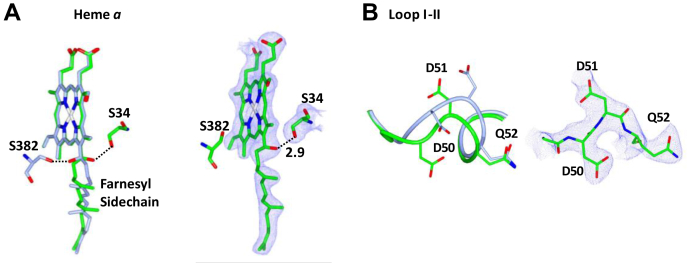
The structure of bCcO acquired following the annealing was solved to a resolution of 2.45 Å (PDB ID: 7TII, Table S1). This unique structure, referred to as R∗ hereinafter, is significantly different from the O∗ structure obtained prior to the annealing, in particular, in three structural regions, heme a, loop I–II, and helix X, where large differences were identified in the resting oxidized O versus the fully reduced R structures (Figs. S1 and S2). In the R∗ structure, the farnesyl side chain of heme a rotated by 120° with respect to that in the O∗ structure, such that the OH group forms a new H-bond with S34 instead of S382 (Fig. 3A, green versus blue structure), a hallmark of the reduced heme a (36). Likewise, loop I–II in the R∗ structure was best modeled with the conformation characteristic of the R structure, whereas that in the O∗ structure was best modeled with the conformation characteristic of the O structure (Fig. 3B, green versus blue structure). These data indicate that the annealing induces structural transitions in heme a and loop I–II from their oxidized conformations to their reduced conformations.
Figure 3.
Electron density maps. Heme a (A) and loop I–II (B) in the R∗ state. The left panels of (A) and (B) show the structures of R∗ (green) overlaid with O∗ (blue) to highlight the annealing-induced structural transition. The 2FO–FC maps associated with the two structural elements in the R∗ state, shown in the right panels, are contoured at 1.25 σ and 1.5 σ, respectively. It is noted that there were no positive and negative features identifiable in the associated FO–FC density maps countered at 3.0 σ (not shown).
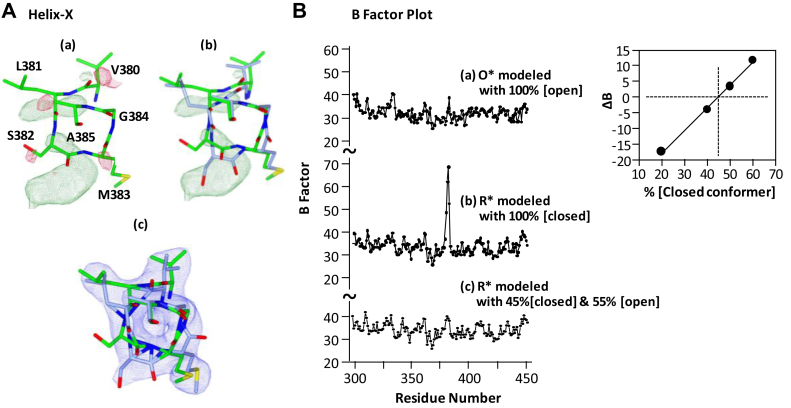
In contrast to heme a and loop I–II, the annealing-induced structural transition in the helix X region was more complicated. Helix X is a key structural element of bCcO, as it hosts the aforementioned S382 residue, as well as the proximal ligands of heme a and heme a3 (H378 and H376, respectively). Our previous studies showed that the conformation of helix X is sensitive to the ligand-binding state of heme a3, regardless of its oxidation state (21) (Fig. S2). In the ligand-free ferrous state, it adopts a normal α-helical structure (referred to as the closed conformer [CC] hereinafter), but when a ligand binds to heme a3, the helical structure of the [380–385] fragment is disrupted (21) (referred to as the open conformer [OC], hereinafter). The [380–385] fragment in the O∗ structure could be modeled with the OC without detectable features in the FO–FC difference map, indicating the presence of a ligand coordinated to the ferrous heme a3. In contrast, when the same fragment in the R∗ structure was modeled with the CC, unusually high B-factor values were detected in the 382-residue region (Fig. 4B, trace b); in addition, in the FO–FC difference map, significant positive features coinciding with those expected for the OC were identified (Fig. 4A, panel a), suggesting the coexistence of the CC and OC. This scenario is in good agreement with the spectral data shown in Figure 2A, indicating partial dissociation of the heme a3 ligand upon annealing. It is important to note that annealing-induced ligand dissociation following X-ray-induced reduction is not unprecedented. It has been documented in bacterial CcOs (18, 30) and in several ferric hemeprotein systems (27, 37), which share a common feature that the heme iron ligands (either a water or a hydroxide) are constrained in the heme pockets when the metal centers were reduced and were unable to dissociate unless the temperature of the crystals was raised above the protein glass transition temperature.
Figure 4.
Electrondensity mapsandB-factor plotsof helix X in theR∗state. A, the FO–FC map obtained by modeling the R∗ structure with the closed conformer (green sticks) shown in (A) indicates that the electron density associated with helix X cannot be fully accounted by the pure closed conformer. The map is countered at 3.0 σ; the positive and negative features are shown in green and red, respectively. The residual positive features (green) were reproduced in (B) to demonstrate that they coincide well with the open conformer (blue sticks), indicating the coexist of the two conformers. The 2FO–FC map (countered at 1.25 σ) shown in (C) was obtained with the R∗ structure modeled with a mixed open conformer (55%) and closed conformer (45%); the absence of significant features in the FO–FC map (contoured at 3.0 σ, not shown) supports the reliability of the modeling. B, the average backbone B-factors (in Å2) of the R∗ structure obtained by modeling with the closed conformer (B) or with a mixed open (55%) and closed (45%) conformer (C). The comparable B-factor plot of the O∗ structure modeled with the open conformer is shown in (A) as a reference. The right inset shows a plot of ΔB as a function of the percent contribution of the closed conformer derived from the B-factor analysis described in the main text. The crossed dashed lines indicate that a mixed 55% open conformer and 45% closed conformer leads to ΔB = 0, where the averaged B-factors of the two conformers are comparable.
To determine the relative contributions of the two conformers, we employed a B-factor analysis method (38). The R∗ structure was modeled with x[OC] + (1 − x)[CC], where the parameter “x” represents the fractional occupancy of the OC. We varied “x” and recorded ΔB, defined as the average backbone B-factor of the CC minus that of the OC in the [379–386] residue range. The average B-factors of the two conformers are predicted to be comparable; hence, ΔB is expected to be zero. Based on this analysis, we found that the R∗ structure was best modeled with a mixture of a 55% OC and a 45% CC (see the right inset in Fig. 4B). This structure assignment is supported by the observation that there are no detectable features in the FO–FC difference map when the R∗ structure was modeled with the mixed conformer (structure c, Fig. 4A), and that the B-factor plot of the final refined structure with the mixed conformer (trace c, Fig. 4B) is comparable with that of the O∗ structure (trace a).
Ligand-binding state of the BNC
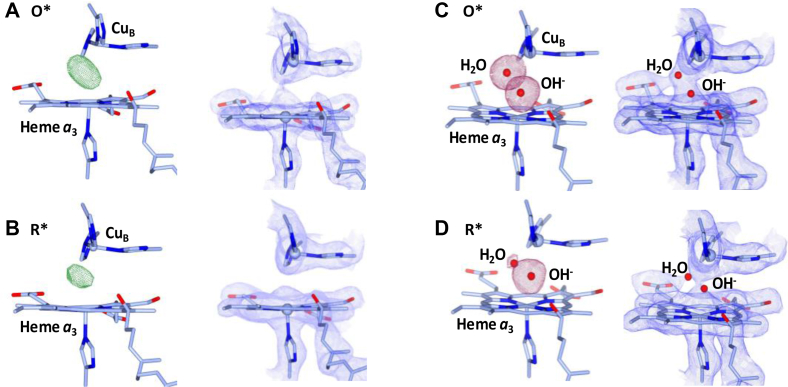
To determine how the protein conformational relaxation correlates with the ligand-binding state of the BNC, we examined the electron density in the BNC of the O∗ and R∗ states. Strong ligand density was identified in the FO–FC difference map in the O∗ state when the ligand(s) were not modeled (Fig. 5A). In the R∗ state, the electron density was significantly reduced (Fig. 5B), but not entirely eliminated, consistent with the view that the ligand(s) are only partially dissociated from the BNC.
Figure 5.
Ligand-binding state of the BNC in the O∗ versus R∗. A and B, show the FO–FC maps (left panels) and 2FO–FC maps (right panels), contoured at 7.00 and 1.50 σ, respectively, of the O∗ and R∗ states without the ligand modeled. The green density shown in the FO–FC maps highlight the unmodeled ligands in the BNC. C and D, show the polder maps of the ligands in the BNC of the O∗ and R∗ states, respectively (contoured at 4.5 σ), and the corresponding 2FO–FC electron density maps with the ligands modeled with a hydroxide ion and a water molecule in the BNC (contoured at 1.00 σ). The ligand occupancies are estimated to be 86 and 80%, respectively, for O∗ and 63 and 35%, respectively, for R∗. No positive or negative features are identifiable in the associated FO–FC density maps contoured at 3.0 σ (not shown), supporting the reliability of the modeling. BNC, binuclear center.
What is the identity of the ligand(s) in the BNC of the O∗ and R∗ states? As the O∗ and R∗ states are formed by X-ray exposure to the O state, [O → O∗ → R∗], it is important to first consider the structure of the O state. It has been proposed that the heme a3 and CuB in the resting oxidized O state are bridged by a peroxide, with the [Fe3+–O−–O−–CuB2+] configuration (12, 39). However, it is unclear how the peroxide is formed in the BNC and what prevents the peroxide from spontaneous O–O bond scission. Moreover, modeling the ligand density with an unrestrained O–O bond leads to a structure with an excessively long O–O bond distance (1.75–1.9 Å) (12, 39). Here, we hypothesize that, in the resting O state, heme a3 and CuB are coordinated by a hydroxide ion and a water molecule, respectively [Fe3+–OH−…H2O–CuB2+], as that originally proposed for the structure of OH based on spectroscopic studies conducted in free solutions (9) and that proposed for a comparable O structure of a bacterial aa3 type of CcO (13). In addition, we assume that X-ray-induced metal reduction does not affect the nature of the ligands, whereas annealing allows partial dissociation of the ligands from the BNC. Accordingly, we modeled the ligand density in O∗ and R∗ with OH− and H2O coordinated to heme a3 and CuB, respectively.
As shown in Figure 5C, the ligand density in the O∗ state can be nicely accounted for by a OH− ion that is coordinated to heme a3 and a H2O molecule that is 2.35 Å away from CuB. Although with the current resolution, H2O and OH− are indistinguishable, the presence of two oxygen-containing ligands in the BNC is evident (see the polder map (40) on the left panel). With two other water molecules in the vicinity of the BNC as an internal standard (assuming both water molecules have full occupancy) (Fig. S3), the occupancy of the OH− and H2O ligands was estimated to be 86 and 80%, respectively (Table S2). Similar results were obtained on a comparable oxidized enzyme (PDB ID: 7TIE, Fig. S4) obtained at the APS (although absorption spectra before and after X-ray exposure were not obtained). Likewise, the ligand density in the R∗ structure can be nicely accounted by OH− and H2O, with the occupancy of the OH− ligand slightly reduced to 63%, whereas that of the H2O ligand dramatically reduced to 35% (Fig. 5D, left panel). The 2FO–FC maps resulting from the modeling are shown in the right panels of Figure 5, C and D; the absence of detectable features in the corresponding FO–FC difference maps supports the reliability of the fitting.
The O–O distance between the OH− and the H2O ligands in the O∗ structure is 1.95 Å, somewhat shorter than a typical H-bond. It is conceivable that the reduction of the metal centers in the BNC in the O∗ state leads to a high-energy configuration, [Fe2+–OH−...H2O–CuB1+], where the two oxygen ligands are pushed together. It is notable that short O–O distances between the two oxygen ligands in the BNC of the oxidized forms of CcOs from Thermus thermophilus (TtCcO) have been reported. Specifically, crystallographic studies of caa3 TtCcO carried out by Lyons et al. (41) showed that the ligand density was best modeled with a water molecule on heme a3 and a hydroxide on CuB with an O–O separation of 2.3 Å. Similarly, density functional theory calculations of ba3 TtCcO carried out by Du et al. (42) showed that when the ligands were modeled with a hydroxide coordinated to heme a3 and a water coordinated to CuB, the O–O distance was 2.45 Å.
Conclusions
X-ray-induced reduction of metal centers can be problematic in structural determination of metalloproteins. Here, we demonstrated that X-ray exposure to the oxidized bCcO crystal during the diffraction measurements led to an O∗ structure, where the four redox centers were fully reduced, but its protein matrix was locked in the resting O structure at the cryogenic temperature employed. Although the reduction of the metal centers did not affect the polypeptide structure, the hydroxide and water ligands in the BNC were pushed together by the reduced heme a3 iron and CuB. Annealing of the O∗ crystal to above the glass transition temperature enabled the relaxation of the protein to a new R∗ structure, where the polypeptide adopted the fully reduced R structure, except that the helix X partially remained in the O conformation as the ligands in the BNC were only partially dissociated. The incomplete transition of the O∗ structure to the fully reduced R structure upon annealing suggests a high-energy barrier for ligand dissociation from the BNC during the O → R transition in the oxygen reduction cycle.
Experimental procedures
Sample preparation
CcO was isolated and purified from bovine hearts by standard procedures (43, 44). The protein was further purified by concentrating it in an Amicon apparatus with a 200 kD molecular weight cutoff filter, where it was crystallized as microcrystals. The microcrystals were harvested and redissolved in pH 6.8 phosphate buffer (40 mM) containing 0.2% decylmaltoside to generate the crystallization protein stock. This as-prepared resting oxidized enzyme has a Soret absorption peak at 424 nm, characteristic of the “fast” enzyme (45, 46) (Fig. 2). The crystals were grown in pH 6.8 phosphate buffer (40 mM) with 0.2% decylmaltoside and 2.5% PEG4000, using an under-oil batch method. Nanocrystal seeds were included in the crystallization solution to ensure the reproducibly of the crystals. To produce the seeding solution, large crystals were crashed and sonicated in the mother solution. The large plate-like crystals, with a typical size of 400 × 400 × 80 μm, were harvested and cryoprotected by 45% ethylene glycol using a stepwise soaking protocol. The crystals were then flash frozen in liquid nitrogen.
Diffraction and optical absorption measurements
The before and after annealing diffraction data were obtained at a cryogenic temperature (100 K) at the beamline BL9-2 at the SSRL. The X-ray beam size was 100 × 60 μm, and the photon flux was 4.3 × 1010 (photons/s). We first acquired an absorption spectrum of the fully oxidized crystal to ensure its integrity. We then collected the first set of diffraction data to determine its structure. After the diffraction measurement, we acquired another absorption spectrum to examine X-ray-induced changes to the redox centers. We next blocked the cold nitrogen stream, which maintained the temperature of the crystal at 100 K, for 2 s until the temperature of the crystal was raised above the protein glass transition temperature (∼200 K), to promote the relaxation of the protein matrix. We then acquired a new set of diffraction data to determine the structural impact of the annealing. We acquired the absorption spectra of the crystal before and after the second diffraction measurement to examine the annealing-induced structural changes and the impact of the second X-ray exposure.
All the absorption spectra were obtained in situ with a microspectrometer equipped at the beamline BL9-2. The light beam, coinciding with the X-ray beam, was directed to the sample along the shortest axis of the plate-like crystal, to ensure that the whole spectrum in the 400 to 1000 nm region could be measured without saturation problems. The spot size of the light beam (∼50 × 50 μm) was much smaller than that of the X-ray beam (∼100 × 60 μm), which ensured that only the X-ray-irradiated area was probed.
The X-ray diffraction data of the resting oxidized O and fully reduced R crystals were acquired at the APS at the Argonne National Laboratory as references. The X-ray beam size was 110 × 80 μm, the photon flux was 3.6 × 1012 (photons/s). The fully reduced R crystal was prepared by soaking the O crystal with dithionite under anaerobic conditions. The complete reduction of the crystal was confirmed by optical absorption measurements.
Diffraction data analysis
The diffraction images were indexed, integrated, and scaled with XDS (47) and AIMLESS (48). Molecular replacement was conducted with Phaser (49, 50) through the CCP4i graphic interface (51). Further model building was performed using COOT (52). Structure refinements were performed using Refmac5 (51, 53, 54). Data processing and refinement statistics are summarized in Table S1. The structural models were displayed with CCP4MG (55).
Data availability
The atomic parameters and structure factors (PDB IDs: 7TIH, 7TII, 7TIE, and 7THU) have been deposited in the PDB (https://www.rcsb.org/). All other data are contained within the article and can be shared upon request.
Supporting information
This article contains supporting information (21, 34, 45, 46).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76F00515. The SSRL Structural Molecular Biology Program is support by the DOE Office of Biological and Environmental Research and the National Institutes of Health, National Institute of General Medical Sciences (grant no.: P30GM133894). The reference structural data were collected by the Lilly Research Laboratories Collaborative Access Team beamline staff at Sector 31 of the Advanced Photon Source (APS). This research used resources of the APS, a US DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract number DE-AC02-06CH11357. Use of the Lilly Research Laboratories Collaborative Access Team beamline at Sector 31 of the APS was provided by Eli Lilly Company, which operates the facility.
Author contributions
I. I., S.-R. Y., and D. L. R. conceptualization; I. I. validation; I. I., S.-R. Y., and D. L. R. formal analysis; I. I., S.-R. Y., and D. L. R. writing–review & editing; I. I., S. R., and A. C. funding acquisition.
Funding and additional information
This study was supported by the National Institutes of Health National Institute of General Medical Sciences grants R01 GM126297 (to D. L. R. and S.-R. Y.), R21 GM127944 (to D. L. R.), and R01 GM115773 (to S.-R. Y.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Edited by Ruma Banerjee
Contributor Information
Syun-Ru Yeh, Email: syun-ru.yeh@einsteinmed.edu.
Denis L. Rousseau, Email: denis.rousseau@einsteinmed.edu.
Supporting information
References
- 1.Sousa F.L., Alves R.J., Ribeiro M.A., Pereira-Leal J.B., Teixeira M., Pereira M.M. The superfamily of heme-copper oxygen reductases: Types and evolutionary considerations. Biochim. Biophys. Acta. 2012;1817:629–637. doi: 10.1016/j.bbabio.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 2.Belevich I., Verkhovsky M.I. Molecular mechanism of proton translocation by cytochrome c oxidase. Antioxid. Redox Signal. 2008;10:1–29. doi: 10.1089/ars.2007.1705. [DOI] [PubMed] [Google Scholar]
- 3.Wikstrom M., Sharma V. Proton pumping by cytochrome c oxidase - a 40year anniversary. Biochim. Biophys. Acta Bioenerg. 2018;1859:692–698. doi: 10.1016/j.bbabio.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Yoshikawa S., Shimada A. Reaction mechanism of cytochrome C oxidase. Chem. Rev. 2015;115:1936–1989. doi: 10.1021/cr500266a. [DOI] [PubMed] [Google Scholar]
- 5.Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 6.Verkhovsky M.I., Jasaitis A., Verkhovskaya M.L., Morgan J.E., Wikstrom M. Proton translocation by cytochrome c oxidase. Nature. 1999;400:480–483. doi: 10.1038/22813. [DOI] [PubMed] [Google Scholar]
- 7.Rousseau D.L. Bioenergetics. Two phases of proton translocation. Nature. 1999;400:412–413. doi: 10.1038/22664. [DOI] [PubMed] [Google Scholar]
- 8.Babcock G.T., Varotsis C. Discrete steps in dioxygen activation--the cytochrome oxidase/O2 reaction. J. Bioenerg. Biomembr. 1993;25:71–80. doi: 10.1007/BF00762849. [DOI] [PubMed] [Google Scholar]
- 9.Han S., Takahashi S., Rousseau D.L. Time dependence of the catalytic intermediates in cytochrome c oxidase. J. Biol. Chem. 2000;275:1910–1919. doi: 10.1074/jbc.275.3.1910. [DOI] [PubMed] [Google Scholar]
- 10.Kitagawa T., Ogura T. Time-resolved resonance Raman investigation of oxygen reduction mechanism of bovine cytochrome c oxidase. J. Bioenerg. Biomembr. 1998;30:71–79. doi: 10.1023/a:1020511612194. [DOI] [PubMed] [Google Scholar]
- 11.Varotsis C., Zhang Y., Appelman E.H., Babcock G.T. Resolution of the reaction sequence during the reduction of O2 by cytochrome oxidase. Proc. Natl. Acad. Sci. U. S. A. 1993;90:237–241. doi: 10.1073/pnas.90.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aoyama H., Muramoto K., Shinzawa-Itoh K., Hirata K., Yamashita E., Tsukihara T., Ogura T., Yoshikawa S. A peroxide bridge between Fe and Cu ions in the O2 reduction site of fully oxidized cytochrome c oxidase could suppress the proton pump. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2165–2169. doi: 10.1073/pnas.0806391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin L., Hiser C., Mulichak A., Garavito R.M., Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koepke J., Olkhova E., Angerer H., Muller H., Peng G., Michel H. High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: New insights into the active site and the proton transfer pathways. Biochim. Biophys. Acta. 2009;1787:635–645. doi: 10.1016/j.bbabio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Tiefenbrunn T., Liu W., Chen Y., Katritch V., Stout C.D., Fee J.A., Cherezov V. High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wikstrom M. Active site intermediates in the reduction of O(2) by cytochrome oxidase, and their derivatives. Biochim. Biophys. Acta. 2012;1817:468–475. doi: 10.1016/j.bbabio.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Kim E., Chufan E.E., Kamaraj K., Karlin K.D. Synthetic models for heme-copper oxidases. Chem. Rev. 2004;104:1077–1133. doi: 10.1021/cr0206162. [DOI] [PubMed] [Google Scholar]
- 18.Liu B., Chen Y., Doukov T., Soltis S.M., Stout C.D., Fee J.A. Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from thermus thermophilus: Structure of the reduced form of the enzyme. Biochemistry. 2009;48:820–826. doi: 10.1021/bi801759a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson R., Safari C., Dods R., Nango E., Tanaka R., Yamashita A., Nakane T., Tono K., Joti Y., Bath P., Dunevall E., Bosman R., Nureki O., Iwata S., Neutze R., et al. Serial femtosecond crystallography structure of cytochrome c oxidase at room temperature. Sci. Rep. 2017;7:4518. doi: 10.1038/s41598-017-04817-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaila V.R., Oksanen E., Goldman A., Bloch D.A., Verkhovsky M.I., Sundholm D., Wikstrom M. A combined quantum chemical and crystallographic study on the oxidized binuclear center of cytochrome c oxidase. Biochim. Biophys. Acta. 2011;1807:769–778. doi: 10.1016/j.bbabio.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Ishigami I., Zatsepin N.A., Hikita M., Conrad C.E., Nelson G., Coe J.D., Basu S., Grant T.D., Seaberg M.H., Sierra R.G., Hunter M.S., Fromme P., Fromme R., Yeh S.-R., Rousseau D.L. Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature. Proc. Natl. Acad. Sci. U. S. A. 2017;114:8011–8016. doi: 10.1073/pnas.1705628114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garman E.F. Radiation damage in macromolecular crystallography: What is it and why should we care? Acta Crystallogr. Section D, Biol. Crystallogr. 2010;66:339–351. doi: 10.1107/S0907444910008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garman E.F., Owen R.L. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallogr. Section D, Biol. Crystallogr. 2006;62:32–47. doi: 10.1107/S0907444905034207. [DOI] [PubMed] [Google Scholar]
- 24.Owen R.L., Rudino-Pinera E., Garman E.F. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4912–4917. doi: 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beitlich T., Kuhnel K., Schulze-Briese C., Shoeman R.L., Schlichting I. Cryoradiolytic reduction of crystalline heme proteins: Analysis by UV-vis spectroscopy and X-ray crystallography. J. Synchrotron Radiat. 2007;14:11–23. doi: 10.1107/S0909049506049806. [DOI] [PubMed] [Google Scholar]
- 26.Yi J., Orville A.M., Skinner J.M., Skinner M.J., Richter-Addo G.B. Synchrotron X-ray-induced photoreduction of ferric myoglobin nitrite crystals gives the ferrous derivative with retention of the O-bonded nitrite ligand. Biochemistry. 2010;49:5969–5971. doi: 10.1021/bi100801g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denisov I.G., Victoria D.C., Sligar S.G. Cryoradiolytic reduction of heme proteins: Maximizing dose dependent yield. Radiat. Phys. Chem. 2007;76:714–721. doi: 10.1016/j.radphyschem.2006.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfanzagl V., Beale J.H., Michlits H., Schmidt D., Gabler T., Obinger C., Djinovic-Carugo K., Hofbauer S. X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner. J. Biol. Chem. 2020;295:13488–13501. doi: 10.1074/jbc.RA120.014087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueno G., Shimada A., Yamashita E., Hasegawa K., Kumasaka T., Shinzawa-Itoh K., Yoshikawa S., Tsukihara T., Yamamoto M. Low-dose X-ray structure analysis of cytochrome c oxidase utilizing high-energy X-rays. J. Synchrotron Radiat. 2019;26:912–921. doi: 10.1107/S1600577519006805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Qin L., Ferguson-Miller S. Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1284–1289. doi: 10.1073/pnas.1012846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferguson-Miller S., Hiser C., Liu J. Gating and regulation of the cytochrome c oxidase proton pump. Biochim. Biophys. Acta. 2012;1817:489–494. doi: 10.1016/j.bbabio.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshikawa S., Shinzawa-Itoh K., Nakashima R., Yaono R., Yamashita E., Inoue N., Yao M., Fei M.J., Libeu C.P., Mizushima T., Yamaguchi H., Tomizaki T., Tsukihara T. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 33.Yano N., Muramoto K., Shimada A., Takemura S., Baba J., Fujisawa H., Mochizuki M., Shinzawa-Itoh K., Yamashita E., Tsukihara T., Yoshikawa S. The Mg2+-containing water cluster of mammalian cytochrome c oxidase collects four pumping proton equivalents in each catalytic cycle. J. Biol. Chem. 2016;291:23882–23894. doi: 10.1074/jbc.M115.711770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanneste W.H. The stoichiometry and absorption spectra of components a and a-3 in cytochrome c oxidase. Biochemistry. 1966;5:838–848. doi: 10.1021/bi00867a005. [DOI] [PubMed] [Google Scholar]
- 35.Argade P.V., Ching Y.C., Rousseau D.L. Resonance Raman spectral isolation of the a and a3 chromophores in cytochrome oxidase. Biophysical J. 1986;50:613–620. doi: 10.1016/S0006-3495(86)83500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishigami I., Lewis-Ballester A., Echelmeier A., Brehm G., Zatsepin N., Grant T., Jesse Coe J., Lisova S., Nelson G., Zhang S., Dobson Z., Boutet S., Sierra R., Batyuk A., Fromme P., et al. Snapshot of an oxygen intermediate in the catalytic reaction of cytochrome c oxidase. Proc. Nat. Acad. Sci. U. S. A. 2019;116:3572–3577. doi: 10.1073/pnas.1814526116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engler N., Ostermann A., Gassmann A., Lamb D.C., Prusakov V.E., Schott J., Schweitzer-Stenner R., Parak F.G. Protein dynamics in an intermediate state of myoglobin: Optical absorption, resonance Raman spectroscopy, and x-ray structure analysis. Biophys. J. 2000;78:2081–2092. doi: 10.1016/S0006-3495(00)76755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimada A., Etoh Y., Kitoh-Fujisawa R., Sasaki A., Shinzawa-Itoh K., Hiromoto T., Yamashita E., Muramoto K., Tsukihara T., Yoshikawa S. X-ray structures of catalytic intermediates of cytochrome c oxidase provide insights into its O2 activation and unidirectional proton-pump mechanisms. J. Biol. Chem. 2020;295:5818–5833. doi: 10.1074/jbc.RA119.009596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirata K., Shinzawa-Itoh K., Yano N., Takemura S., Kato K., Hatanaka M., Muramoto K., Kawahara T., Tsukihara T., Yamashita E., Tono K., Ueno G., Hikima T., Murakami H., Inubushi Y., et al. Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL. Nat. Methods. 2014;11:734–736. doi: 10.1038/nmeth.2962. [DOI] [PubMed] [Google Scholar]
- 40.Liebschner D., Afonine P.V., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., Adams P.D. Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 2017;73:148–157. doi: 10.1107/S2059798316018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyons J.A., Aragao D., Slattery O., Pisliakov A.V., Soulimane T., Caffrey M. Structural insights into electron transfer in caa3-type cytochrome oxidase. Nature. 2012;487:514–518. doi: 10.1038/nature11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han Du W.G., McRee D., Gotz A.W., Noodleman L. A water molecule residing in the Fea3(3+)...CuB(2+) Dinuclear center of the resting oxidized as-isolated cytochrome c oxidase: A density functional study. Inorg. Chem. 2020;59:8906–8915. doi: 10.1021/acs.inorgchem.0c00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshikawa S., Choc M.G., O'Toole M.C., Caughey W.S. An infrared study of CO binding to heart cytochrome c oxidase and hemoglobin A. Implications re O2 reactions. J. Biol. Chem. 1977;252:5498–5508. [PubMed] [Google Scholar]
- 44.Mochizuki M., Aoyama H., Shinzawa-Itoh K., Usui T., Tsukihara T., Yoshikawa S. Quantitative reevaluation of the redox active sites of crystalline bovine heart cytochrome c oxidase. J. Biol. Chem. 1999;274:33403–33411. doi: 10.1074/jbc.274.47.33403. [DOI] [PubMed] [Google Scholar]
- 45.Baker G.M., Noguchi M., Palmer G. The reaction of cytochrome oxidase with cyanide. Preparation of the rapidly reacting form and its conversion to the slowly reacting form. J. Biol. Chem. 1987;262:595–604. [PubMed] [Google Scholar]
- 46.Moody A.J., Cooper C.E., Rich P.R. Characterisation of 'fast' and 'slow' forms of bovine heart cytochrome-c oxidase. Biochim. Biophys. Acta. 1991;1059:189–207. doi: 10.1016/s0005-2728(05)80204-x. [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;D69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCoy A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallographica. Section D, Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Section D, Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallographica. Section D, Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Section D, Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Section D, Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 55.McNicholas S., Potterton E., Wilson K.S. Presenting your structures: The CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 2011;D67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic parameters and structure factors (PDB IDs: 7TIH, 7TII, 7TIE, and 7THU) have been deposited in the PDB (https://www.rcsb.org/). All other data are contained within the article and can be shared upon request.