Summary
Somatic activating variants in PIK3CA, the gene that encodes the p110α catalytic subunit of phosphatidylinositol 3-kinase (PI3K), have been previously detected in ∼80% of lymphatic malformations (LMs).1,2 We report the presence of somatic activating variants in BRAF in individuals with LMs that do not possess pathogenic PIK3CA variants. The BRAF substitution p.Val600Glu (c.1799T>A), one of the most common driver mutations in cancer, was detected in multiple individuals with LMs. Histology revealed abnormal lymphatic channels with immunopositivity for BRAFV600E in endothelial cells that was otherwise indistinguishable from PIK3CA-positive LM. The finding that BRAF variants contribute to low-flow LMs increases the complexity of prior models associating low-flow vascular malformations (LM and venous malformations) with mutations in the PI3K-AKT-MTOR and high-flow vascular malformations (arteriovenous malformations) with mutations in the RAS-mitogen-activated protein kinase (MAPK) pathway.3 In addition, this work highlights the importance of genetic diagnosis prior to initiating medical therapy as more studies examine therapeutics for individuals with vascular malformations.
Keywords: lymphatic malformation, PIK3CA, BRAF, post-zygotic, mosaicism, clinical diagnostics, ddPCR, VANSeq, endothelium, vascular
Somatic activating PIK3CA mutations are the only known genetic cause of isolated lymphatic malformations. We detected somatic BRAF p.Val600Glu variants in lymphatic malformations lacking PIK3CA mutations. BRAF mutant cells were localized to lymphatic endothelium, supporting the importance of endothelial cells in driving formation of vascular malformations.
Main text
Disorganized morphogenesis of arteries, veins, capillaries, and lymphatic vessels results in vascular malformations, a relatively common congenital malformation associated with significant morbidity.4 Vascular malformations are classified into high-flow lesions, which include arteriovenous malformations (AVMs), and low-flow lesions, which include venous malformations (VeMs) and lymphatic malformations (LMs). Individuals with vascular malformations typically have no family history, because most are caused by post-zygotic (mosaic) activating mutations in oncogenes within the phosphatidylinositol 3-kinase (PI3K)-AKT and RAS-mitogen-activated protein kinase (MAPK) pathways.1, 2, 3 Treatments for vascular malformations are primarily invasive and include sclerotherapy, embolization, and open surgical resection,4 but the identification of specific activating mutations in well-known oncogenic signaling pathways has led to trials examining the efficacy of targeted medical therapies.5, 6, 7, 8, 9, 10, 11
Previous work has shown that approximately 80% of isolated LMs have somatic pathogenic variants in PIK3CA,1,2,12 the gene that encodes for the catalytic subunit of PI3K, a component of the PI3K-AKT pathway.13 Although mutations in other genes (including NRAS, KRAS, CBL, ARAF, and EPHB4) have been identified in complex lymphatic anomalies, such as diffuse lymphangiomatosis and Gorham-Stout disease,14 PIK3CA is the only gene associated with isolated LMs to date. The vast majority (>90%) of LM-associated pathogenic variants occur at one of three locations,2 referred to as “hotspots”: c.1624G>A (p.Glu542Lys), c.1633G>A (p.Glu545Lys), and c.3140A>G (p.His1047Arg), all of which result in PI3K hyperactivation.15,16 The fraction of DNA molecules that possess the pathogenic PIK3CA variant (referred to as the variant allele fraction [VAF]) within LM tissue is typically very low (<10%),2 and it has been hypothesized that a fraction of LMs without a detected PIK3CA variant in fact do carry a PIK3CA variant that was “missed” due to low-level mosaicism. It is also possible that additional genes play a role. Here, we report the identification of somatic BRAF mutations in LMs without a detected PIK3CA variant.
LM tissue from 106 individuals was screened for the three PIK3CA (GenBank: NM_006218.4) hotspots (p.Glu542Lys, p.Glu545Lys, and p.His1047Arg) as well as the less common but amplicon-overlapping p.His1047Leu substitution using droplet digital polymerase chain reaction (ddPCR) assays and molecular inversion probes, as previously reported (Supplemental methods).2 Following this screening, 22 individuals remained without a detected PIK3CA variant. Fifteen of these individuals had sufficient DNA (14 lesion-derived and 1 cyst fluid) for further testing, which was sent for high-depth targeted sequencing using a 44-gene panel, referred to as VANseq (vascular anomaly sequencing) (see Supplemental methods and Table S1) throughout the rest of this paper.
VANseq identified variants in 6/15 individuals (Table 1). One individual (LR18-536) had a non-hotspot PIK3CA variant, c.1035T>A (p.Asn345Lys) that could not have been detected by hotspot allele-specific ddPCR screening. This variant is absent from the Genome Aggregation Database (gnomAD), is predicted to be damaging by several in silico tools, and has been previously reported in numerous individuals with cancer as well as one individual with congenital lipomatous overgrowth, vascular malformations, epidermal nevis, spinal/skeletal anomalies/scoliosis (CLOVES) syndrome.1,17,18 Functional studies have demonstrated that this substitution results in PI3K pathway hyperactivity.15,16 Although not previously reported in association with isolated LMs, we interpreted this variant as being pathogenic19 and confirmed the presence of the variant in additional samples from that individual using ddPCR. There was variation in VAF from undetectable to 1.4% within lesion samples (Table 1), as we have previously described.2
Table 1.
Somatic variants in LM detected by VANseq and confirmed by ddPCR
|
VANseq reads |
ddPCR droplets |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subject | Agea | Sex | Variant | Sample | VAF (%) | Var | WT | VAFb(%) | Var | WT |
| LR18-536 | 2 years | F | PIK3CA p.Asn345Lys | lesion A | 1.3 | 21 | 1,669 | 1.1 | 144 | 13,401 |
| lesion B | – | – | – | 0.2 | 23 | 10,709 | ||||
| lesion C | – | – | – | NEG | 5 | 12,251 | ||||
| lesion D | – | – | – | 0.2 | 22 | 12,569 | ||||
| lesion E | – | – | – | 1.4 | 151 | 10,827 | ||||
| lesion F | – | – | – | 0.5 | 88 | 23,161 | ||||
| lesion G | – | – | – | 0.9 | 74 | 9,877 | ||||
| skin | – | – | – | 0.3 | 30 | 9,480 | ||||
| salivary gland | – | – | – | NEG | 0 | 31,662 | ||||
| LR16-278 | 2 years | F | PIK3CA p.Glu545Lys | lesion | 0.7 | 12 | 1,709 | 0.5 | 48 | 12,295 |
| LR16-264 | 3 years | F | PIK3CA p.Glu545Lys | lesion | 10.6 | 7 | 59c | 4.8 | 437 | 9,551 |
| LR17-322 | 1 year | M | BRAF p.Val600Glu | lesion | 2.1 | 34 | 1,618 | 1.7 | 165 | 10,555 |
| skin | – | – | – | NEG | 0 | 10,374 | ||||
| LR19-346 | 5 months | F | BRAF p.Val600Glu | lesion, deep | 0.6 | 7 | 1,143 | 1.2 | 69 | 5,872 |
| lesion, inferior | – | – | – | 0.9 | 56 | 6,389 | ||||
| lesion, superior | – | – | – | NEG | 2 | 9,584 | ||||
| lesion, no location | – | – | – | 3.6 | 91 | 2,479 | ||||
| skin | – | – | – | NEG | 0 | 4,304 | ||||
| fat | – | – | – | NEG | 1 | 5,340 | ||||
| muscle | – | – | – | NEG | 0 | 6,129 | ||||
| LR19-443 | 1 month | M | BRAF p.Val600Glu | cyst fluid, Ad | – | – | – | 0.2 | 29 | 13,055 |
| cyst fluid, Bd | 0.3 | 4 | 1,458 | 0.1 | 10 | 9,661 | ||||
| cyst fluid, Cd | – | – | – | 0.3 | 31 | 13,292 | ||||
| cyst fluid pellet | – | – | – | NEG | 15 | 30,798 | ||||
ddPCR, droplet digital polymerase chain reaction; NEG, no variant detected; VAF, variant allele fraction; Var, variant; WT, wild type.
Age at time of tissue or cyst fluid attainment.
ddPCR VAF calculated using droplet concentrations and only reported for samples in which sample variant concentration was statistically different from WT control variant concentration based on 95% total error confidence intervals.
Lower than typical coverage.
Cell-free DNA was assayed from cyst fluid samples.
VANseq detected a hotspot PIK3CA variant (p.Glu545Lys) in two individuals (LR16-278 and LR16-264) who had previously been screened for this allele by ddPCR.2 We re-examined prior data from both cases. LR16-278’s prior ddPCR had six variants and 1,055 reference droplets but did not meet our positive criteria, as the 95% confidence interval overlapped with wild-type samples (Supplemental methods). The initial ddPCR run for LR16-264 had one variant and 5,305 reference droplets, but subsequent testing from the original stock DNA dilution was unambiguously positive by VANseq and ddPCR (VAFs of 10.6% and 4.8%, respectively). Although provenance testing was not possible to prove it, we suspect this resulted from a sample swap during the original screening. Poor sample quality could also be a factor, as LR16-265 had lower than typical coverage on VANseq (Table 1). These examples highlight difficulties in using tiered screening assays, which increase the likelihood of sample swaps, and also demonstrate consideration for repeat testing when the diagnostic pre-test probability is high.20 Previous publications from our lab and others have highlighted the utility of repeat testing when the diagnostic pre-test probability is high.2,21 We are confident that the pathogenic variant has now been identified for both of these individuals.
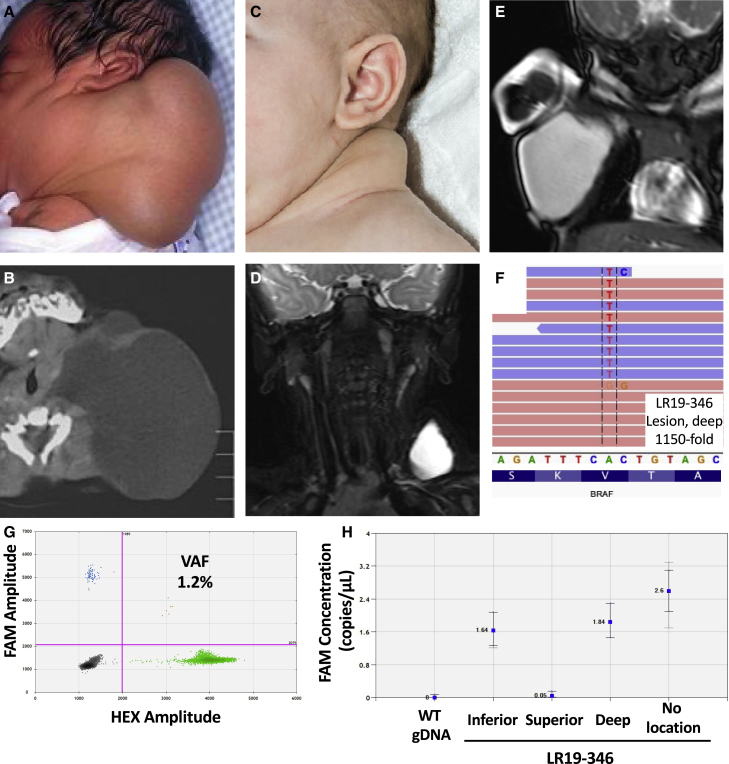
VANseq identified a pathogenic BRAF variant in 3 of the 15 LMs without a detected PIK3CA variant (LR17-322, LR19-346, and LR19-443). All three possessed the same variant (GenBank: NM_004333.6:c.1799T>A, resulting in p.Val600Glu), which was confirmed by ddPCR in multiple independent tissues, when available (Table 1; Figures 1F–1H). Three additional LMs without a detected PIK3CA variant (LR17-134, LR17-319, and LR18-572) possessed three or more reads supporting the BRAF p.Val600Glu substitution but were not confirmed by ddPCR so were not classified as being BRAF positive (Table S2). The presence of three or four alternate base calls out of 1,200–1,500 reads is comparable to the inherent error rate of next-generation sequencing (NGS).22 The discrepancy between the VANseq and ddPCR results for these three samples reflects the lower error rate of ddPCR and highlights the challenges in accurate detection of variants with extremely low allele frequency.
Figure 1.
Clinical features of BRAF-mutated LM and confirmation of genetic diagnosis
(A–G) Clinical photos of LR17-322 (A) and LR19-346 (C), showing posterior neck LMs. Corresponding computed tomography (CT) (LR17–322; B) and MRI (LR19-346, D; LR19-443, E) images demonstrate macrocystic lesions with minimal septations of the posterior lateral neck and axilla. Integrated Genomics Viewer image for LR19-346 demonstrates somatic BRAF p.Val600Glu variant (F), confirmed on droplet digital PCR (G).
(H) Variant concentration image from Quantasoft shows variability in mutation prevalence between samples (H).
Note: (A) and (B) were previously published prior to identification of this individual’s genetic variant.43
All three individuals with BRAF p.Val600Glu substitutions had macrocystic LMs diagnosed at birth (Figure 1). LR17-322 had a large, macrocystic lesion of the posterior neck, de Serres stage 1, that resolved spontaneously over the first few months of life (Figures 1A and 1B). Surgery was performed at 1 year of age to remove remaining LM and redundant skin. LR19-346’s LM was also isolated to the neck, de Serres stage 1, and was resolving with just observation until an upper respiratory infection induced swelling and the decision was made to remove it surgically (Figures 1C and 1D). LR19-443 had a large macrocystic LM of the axilla that was treated with sclerotherapy at 1 month of age (Figure 1E). This individual did not have surgery, but cell free DNA (cfDNA) from aspirated cyst fluid was available for genetic diagnosis. All individuals did well after intervention with no evidence of recurrence and no further procedures or therapy.
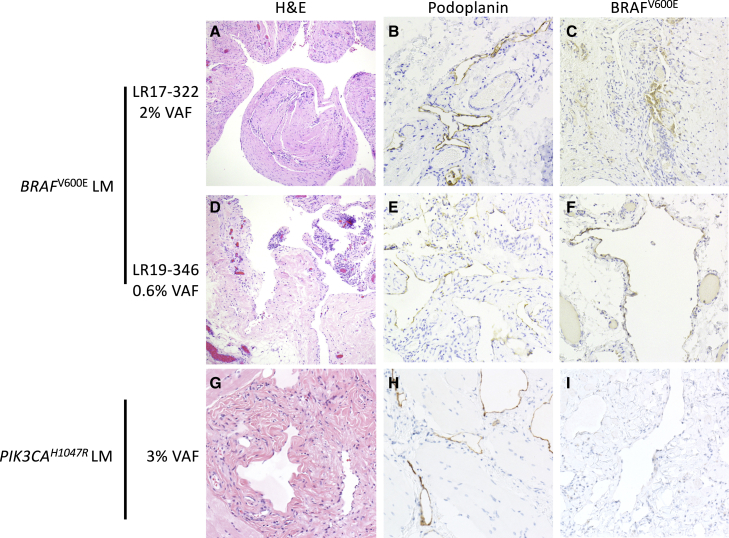
Histopathological examination of tissues from two BRAF p.Val600Glu containing LMs showed numerous dilated cystic channels with bland, flattened epithelium that was immunopositive for podoplanin, a marker of lymphatic endothelial cells (Figure 2).23 There were no distinguishing histopathological features between BRAF and PIK3CA mutant LMs. The extremely low VAFs of the BRAF p.Val600Glu substitutions (0.3%–2%) indicate that most cells within the malformation do not possess the BRAF substitution.2 We hypothesized that BRAF mutant cells would be primarily located within the lymphatic endothelial cells, as has previously been shown in LMs with PIK3CA mutations.24, 25, 26, 27 To test this, we used a BRAF p.Val600Glu-specific monoclonal antibody (VE1).28 BRAF p.Val600Glu immunostaining was present in cyst-lining endothelial cells in LR17-322 and LR19-346, but not in other cells within the lesion (Figure 2). We detected no BRAF p.Val600Glu staining in two other LM samples bearing p.Glu545Lys and p.His1047Arg PIK3CA substitutions (Figure 2; data not shown), demonstrating specificity. These results both confirm the presence of the BRAF substitutions within these lesions and demonstrate their localization to lymphatic endothelial cells.
Figure 2.
Histology and immunohistochemistry of PIK3CA and BRAF mutated LMs
LM tissue from two individuals with BRAF p.Val600Glu substitutions (A–F) and one individual with PIK3CA p.His1047Arg substitution.
(G–I). H&E stains (A), (D), and (G) show dilated cystic channels with bland, flattened epithelium. (B), (E), and (H) show presence of podoplanin (a.k.a. D2-40) immunoreactivity in endothelial cells. Panels on the right show BRAF p.Val600Glu immunoreactivity (VE1 staining) in endothelial cells in BRAF mutant LM (C and F), but not in PIK3CA mutant LM (I).
When these results are combined with our previous reports,2,29 a more complete picture of allelic and locus heterogeneity within isolated LMs appears (Table 2). PIK3CA variants were found in 88% of the 101 individuals in our cohort, 92% of which occurred at one of the three PIK3CA hotspots. BRAF p.Val600Glu variants were found in 3% of individuals with isolated LMs—a small proportion but a clinically important finding, as responses to targeted drug therapies may differ. For example, some BRAF inhibitors produce paradoxical activation of the MAPK pathway and corresponding cellular proliferation in tumors possessing oncogenic mutations in RAS or upstream receptors.30,31 The application of VANseq to our cohort of 101 individuals with isolated LMs brought our overall diagnostic rate from ∼80% to over 90%, and currently, only 9/101 individuals with adequate DNA now remain without a genetic diagnosis.2
Table 2.
The genetic spectrum of LM, including BRAF p.Val600Glu
|
PIK3CA |
BRAF |
NEGa | |||||
|---|---|---|---|---|---|---|---|
| p.His1047Arg | p.Glu545Lys | p.Glu542Lys | Other | Total | p.Val600Glu | ||
| Zenner et al.2 | 22 | 18 | 18 | 6 | 64 | – | – |
| Zenner et al.29 | 7 | 9 | 5 | 0 | 21 | – | – |
| Current study | 1 | 2 | 0 | 1 | 4 | 3 | 9 |
| Total, n = 101b | 30 | 29 | 23 | 7 | 89 (88.1%) | 3 (3.0%) | 9 (8.9%) |
All negative samples for the first two studies were included in this study if adequate sample was available for VANseq testing.
Total includes only individuals with detected mutations or sufficient DNA to undergo VANseq testing.
BRAF is one of the most frequently mutated genes in cancer with a predilection for melanoma, thyroid cancer, colon cancer, and non-small cell lung cancer. p.Val600Glu is the most common oncogenic BRAF substitution, accounting for >90% of BRAF mutations.32 Non-mosaic constitutional missense and in-frame deletions in BRAF have been reported in RASopathies (e.g., Cardiofaciocutaneous syndrome, Noonan syndrome, and Noonan syndrome with multiple lentigines),33 but the p.Val600Glu substitution has never been reported in these diseases. This is likely due to the fact that the BRAF p.Val600Glu substitution is not compatible with embryonic survival except in the mosaic state (i.e., the Happle hypothesis).34 This conclusion is supported by the embryonic lethality seen in constitutional expression of BRAF p.Val600Glu in mouse embryos.35 Somatic BRAF p.Val600Glu variants have previously been reported to cause AVMs, though activating mutations in KRAS and MAP2K1 are more common causes.3,36,37 The precise mechanisms by which somatic BRAF p.Val600Glu substitutions cause LMs in some cases and AVMs in others likely has to do with the timing and location of the post-zygotic mutation. Additional studies are needed to examine this further. Although activating mutations in oncogenes raise concern for an increased risk of cancer, PIK3CA-related overgrowth syndromes have a low risk18 and BRAF p.Val600Glu variants are detected in >80% of benign melanocytic nevi, indicating that the single mutation is insufficient to produce melanoma.38
All three individuals with BRAF p.Val600Glu substitutions in our study had similar clinical phenotypes—large, macrocystic lesions of the neck or body that resolved spontaneously or were treated very early in life. Under the surgical staging system for LMs (de Serres staging), these three individuals would be classified as having stage 1 lesions (unilateral and below the hyoid).39 Stage 1 lesions make up only ∼31% of total LMs in recent studies,2,39,40 suggesting that the LMs with BRAF mutations may represent a milder phenotype than LMs with PIK3CA mutations. Although our cohort of LMs with BRAF mutations (n = 3) is too small for genotype-phenotype correlations, we speculate that there may be enrichment for BRAF mutations in individuals with milder, non-surgical LMs, as genetic diagnosis in most LMs to date has required surgically resected tissue. Additional studies of more LMs with BRAF variants, perhaps using non-invasive diagnostic methods, such as cyst-fluid-based cfDNA,29 will be needed to provide a more balanced view of the genetic spectrum among LMs. The presence of pathogenic BRAF variants within the cyst fluid of macrocystic LMs is consistent with our previous study identifying pathogenic PIK3CA variants within this compartment.29 Further studies are needed to assess the relative yield of cyst fluid versus tissue as a diagnostic analyte.
Endothelial cells play a key role in the pathogenesis of vascular malformations, and isolation of endothelial cells from these lesions enriches the detection of somatic variants.24, 25, 26, 27 Prior work in AVMs has shown KRAS-mutation-specific staining of endothelial cells,41 but this has not previously been possible for LMs, as there is no PIK3CA-mutant-specific antibody. The presence of BRAFV600E staining in lymphatic endothelial cells within the lesions supports the hypothesis that cell-non-autonomous effects, such as signaling to or recruitment of wild-type cells to the lesion, contribute to the formation of LMs. Cell-non-autonomous effects have been previously suggested to cause cartilage overgrowth in AVMs, but additional studies will be needed to examine this further.42
In conclusion, we demonstrate that a somatic activating pathogenic BRAF variant (c.1799T>A, [p.Val600Glu]) is present in 3% of our cohort of individuals with isolated lymphatic malformations. Screening isolated LMs for the three PIK3CA hotspots is an efficient and cost-effective approach but will potentially miss clinically important non-hotspot PIK3CA and BRAF variation. Our use of VANseq, a high-depth, full-gene sequencing panel, increased the positivity rate for our cohort of LM from ∼80% to >90%. In addition, our results suggest the need for studies to examine the efficacy of BRAF inhibition in the treatment of lymphatic malformations.
Data and code availability
The published article includes all data generated or analyzed during this study.
Acknowledgments
We thank the participants and their families. This study was funded by the US National Institutes of Health under National Heart, Lung, and Blood Institute (NHLBI) grants F32HL147398(to K.Z.) and R01 HL130996(to J.T.B.), as well as a Burroughs Wellcome Career Award for Medical Scientists 1014700 (to J.T.B.) and a Seattle Children’s Hospital Guild Association Funding Focus Award (to J.A.P.). We also acknowledge the Seattle Children’s Vascular Anomalies Program and especially the interventional radiology team for their support in sample collection, as well as Dr. Raj Kapur for his assistance with pathology. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the funding sources.
Declaration of interests
R.A.B. is a co-founder of EigenHealth, Inc; a consultant to SpiWay, LLC; and holds a financial interest of ownership equity with Wavely Diagnostics, Inc. The remaining authors declare no competing interests.
Published: April 14, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2022.100101.
Web resources
Catalogue of Somatic Mutations in Cancer, https://cancer.sanger.ac.uk/cosmic.
Seattle Children’s Hospital Lab Test Catalogue, Vascular Anomaly Sequencing Panel (VANSeq) https://seattlechildrenslab.testcatalog.org/show/LAB1920-1.
Supplemental information
References
- 1.Luks V.L., Kamitaki N., Vivero M.P., Uller W., Rab R., Bovee J.V., Rialon K.L., Guevara C.J., Alomari A.I., Greene A.K., et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J. Pediatr. 2015;166:1048–1054. doi: 10.1016/j.jpeds.2014.12.069. e1041-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zenner K., Cheng C.V., Jensen D.M., Timms A.E., Shivaram G., Bly R., et al. Genotype correlates with clinical severity in PIK3CA-associated lymphatic malformations. JCI Insight. 2019;4 doi: 10.1172/jci.insight.129884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Olabi L., Polubothu S., Dowsett K., Andrews K.A., Stadnik P., Joseph A.P., Knox R., Pittman A., Clark G., Baird W., et al. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J. Clin. Invest. 2018;128:1496–1508. doi: 10.1172/JCI98589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padia R., Bly R., Bull C., Geddis A.E., Perkins J. Medical management of vascular anomalies. Curr. Treat Options Pediatr. 2018;4:221–236. doi: 10.1007/s40746-018-0130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams D.M., Trenor C.C., 3rd, Hammill A.M., Vinks A.A., Patel M.N., Chaudry G., Wentzel M.S., Mobberley-Schuman P.S., Campbell L.M., Brookbank C., et al. Efficacy and safety of sirolimus in the treatment of complicated vascular anomalies. Pediatrics. 2016;137:e20153257. doi: 10.1542/peds.2015-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards E.A., Phelps A.S., Cooke D., Frieden I.J., Zapala M.A., Fullerton H.J., et al. Monitoring arteriovenous malformation response to genotype-targeted therapy. Pediatrics. 2020;146 doi: 10.1542/peds.2019-3206. [DOI] [PubMed] [Google Scholar]
- 7.Hammer J., Seront E., Duez S., Dupont S., Van Damme A., Schmitz S., Hoyoux C., Chopinet C., Clapuyt P., Hammer F., et al. Sirolimus is efficacious in treatment for extensive and/or complex slow-flow vascular malformations: a monocentric prospective phase II study. Orphanet J. Rare Dis. 2018;13:191. doi: 10.1186/s13023-018-0934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lekwuttikarn R., Lim Y.H., Admani S., Choate K.A., Teng J.M.C. Genotype-guided medical treatment of an arteriovenous malformation in a child. JAMA Dermatol. 2019;155:256–257. doi: 10.1001/jamadermatol.2018.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker V.E.R., Keppler-Noreuil K.M., Faivre L., Luu M., Oden N.L., De Silva L., Sapp J.C., Andrews K., Bardou M., Chen K.Y., et al. Safety and efficacy of low-dose sirolimus in the PIK3CA-related overgrowth spectrum. Genet. Med. 2019;21:1189–1198. doi: 10.1038/s41436-018-0297-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Triana P., Dore M., Cerezo V.N., Cervantes M., Sanchez A.V., Ferrero M.M., Gonzalez M.D., Lopez-Gutierrez J.C. Sirolimus in the treatment of vascular anomalies. Eur. J. Pediatr. Surg. 2017;27:86–90. doi: 10.1055/s-0036-1593383. [DOI] [PubMed] [Google Scholar]
- 11.Venot Q., Blanc T., Rabia S.H., Berteloot L., Ladraa S., Duong J.P., Blanc E., Johnson S.C., Hoguin C., Boccara O., et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature. 2018;558:540–546. doi: 10.1038/s41586-018-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brouillard P., Schlogel M.J., Homayun Sepehr N., Helaers R., Queisser A., Fastre E., Boutry S., Schmitz S., Clapuyt P., Hammer F., et al. Non-hotspot PIK3CA mutations are more frequent in CLOVES than in common or combined lymphatic malformations. Orphanet J. Rare Dis. 2021;16:267. doi: 10.1186/s13023-021-01898-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fruman D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C., Abraham R.T. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makinen T., Boon L.M., Vikkula M., Alitalo K. Lymphatic malformations: genetics, mechanisms and therapeutic strategies. Circ. Res. 2021;129:136–154. doi: 10.1161/CIRCRESAHA.121.318142. [DOI] [PubMed] [Google Scholar]
- 15.Dogruluk T., Tsang Y.H., Espitia M., Chen F., Chen T., Chong Z., Appadurai V., Dogruluk A., Eterovic A.K., Bonnen P.E., et al. Identification of variant-specific functions of PIK3CA by rapid phenotyping of rare mutations. Cancer Res. 2015;75:5341–5354. doi: 10.1158/0008-5472.CAN-15-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gymnopoulos M., Elsliger M.A., Vogt P.K. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl. Acad. Sci. U S A. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O'Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gripp K.W., Baker L., Kandula V., Conard K., Scavina M., Napoli J.A., Griffin G.C., Thacker M., Knox R.G., Clark G.R., et al. Nephroblastomatosis or Wilms tumor in a fourth patient with a somatic PIK3CA mutation. Am. J. Med. Genet. A. 2016;170:2559–2569. doi: 10.1002/ajmg.a.37758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akobeng A.K. Understanding diagnostic tests 2: likelihood ratios, pre- and post-test probabilities and their use in clinical practice. Acta Paediatr. 2007;96:487–491. doi: 10.1111/j.1651-2227.2006.00179.x. [DOI] [PubMed] [Google Scholar]
- 21.Djemie T., Weckhuysen S., von Spiczak S., Carvill G.L., Jaehn J., Anttonen A.K., Brilstra E., Caglayan H.S., de Kovel C.G., Depienne C., et al. Pitfalls in genetic testing: the story of missed SCN1A mutations. Mol. Genet. Genomic Med. 2016;4:457–464. doi: 10.1002/mgg3.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoler N., Nekrutenko A. Sequencing error profiles of Illumina sequencing instruments. NAR Genom Bioinform. 2021;3 doi: 10.1093/nargab/lqab019. lqab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukunaga M. Expression of D2-40 in lymphatic endothelium of normal tissues and in vascular tumours. Histopathology. 2005;46:396–402. doi: 10.1111/j.1365-2559.2005.02098.x. [DOI] [PubMed] [Google Scholar]
- 24.Blesinger H., Kaulfus S., Aung T., Schwoch S., Prantl L., Rosler J., Wilting J., Becker J. PIK3CA mutations are specifically localized to lymphatic endothelial cells of lymphatic malformations. PLoS One. 2018;13:e0200343. doi: 10.1371/journal.pone.0200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boscolo E., Coma S., Luks V.L., Greene A.K., Klagsbrun M., Warman M.L., Bischoff J. AKT hyper-phosphorylation associated with PI3K mutations in lymphatic endothelial cells from a patient with lymphatic malformation. Angiogenesis. 2015;18:151–162. doi: 10.1007/s10456-014-9453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glaser K., Dickie P., Neilson D., Osborn A., Dickie B.H. Linkage of metabolic defects to activated PIK3CA alleles in endothelial cells derived from lymphatic malformation. Lymphat. 2018;16:43–55. doi: 10.1089/lrb.2017.0033. [DOI] [PubMed] [Google Scholar]
- 27.Osborn A.J., Dickie P., Neilson D.E., Glaser K., Lynch K.A., Gupta A., Dickie B.H. Activating PIK3CA alleles and lymphangiogenic phenotype of lymphatic endothelial cells isolated from lymphatic malformations. Hum. Mol. Genet. 2015;24:926–938. doi: 10.1093/hmg/ddu505. [DOI] [PubMed] [Google Scholar]
- 28.Ritterhouse L.L., Barletta J.A. BRAF V600E mutation-specific antibody: a review. Semin. Diagn. Pathol. 2015;32:400–408. doi: 10.1053/j.semdp.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Zenner K., Jensen D.M., Cook T.T., Dmyterko V., Bly R.A., Ganti S., Mirzaa G.M., Dobyns W.B., Perkins J.A., Bennett J.T. Cell-free DNA as a diagnostic analyte for molecular diagnosis of vascular malformations. Genet. Med. 2021;23:123–130. doi: 10.1038/s41436-020-00943-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cichowski K., Janne P.A. Drug discovery: inhibitors that activate. Nature. 2010;464:358–359. doi: 10.1038/464358a. [DOI] [PubMed] [Google Scholar]
- 31.Hatzivassiliou G., Song K., Yen I., Brandhuber B.J., Anderson D.J., Alvarado R., Ludlam M.J., Stokoe D., Gloor S.L., Vigers G., et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 32.Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L., et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarkozy A., Carta C., Moretti S., Zampino G., Digilio M.C., Pantaleoni F., Scioletti A.P., Esposito G., Cordeddu V., Lepri F., et al. Germline BRAF mutations in Noonan, LEOPARD, and cardiofaciocutaneous syndromes: molecular diversity and associated phenotypic spectrum. Hum. Mutat. 2009;30:695–702. doi: 10.1002/humu.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Happle R. Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J. Am. Acad. Dermatol. 1987;16:899–906. doi: 10.1016/s0190-9622(87)80249-9. [DOI] [PubMed] [Google Scholar]
- 35.Mercer K., Giblett S., Green S., Lloyd D., DaRocha Dias S., Plumb M., Marais R., Pritchard C. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res. 2005;65:11493–11500. doi: 10.1158/0008-5472.CAN-05-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goss J.A., Huang A.Y., Smith E., Konczyk D.J., Smits P.J., Sudduth C.L., Stapleton C., Patel A., Alexandrescu S., Warman M.L., et al. Somatic mutations in intracranial arteriovenous malformations. PLoS One. 2019;14:e0226852. doi: 10.1371/journal.pone.0226852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong T., Yan Y., Li J., Radovanovic I., Ma X., Shao Y.W., Yu J., Ma Y., Zhang P., Ling F., et al. High prevalence of KRAS/BRAF somatic mutations in brain and spinal cord arteriovenous malformations. Brain. 2019;142:23–34. doi: 10.1093/brain/awy307. [DOI] [PubMed] [Google Scholar]
- 38.Pollock P.M., Harper U.L., Hansen K.S., Yudt L.M., Stark M., Robbins C.M., Moses T.Y., Hostetter G., Wagner U., Kakareka J., et al. High frequency of BRAF mutations in nevi. Nat. Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 39.de Serres L.M., Sie K.C., Richardson M.A. Lymphatic malformations of the head and neck. A proposal for staging. Arch. Otolaryngol. Head Neck Surg. 1995;121:577–582. doi: 10.1001/archotol.1995.01890050065012. [DOI] [PubMed] [Google Scholar]
- 40.Bonilla-Velez J., Whitlock K.B., Ganti S., Zenner K., Cheng C.V., Jensen D.M., Pham M.M., Mitchell R.M., Dobyns W., Bly R.A., et al. Acetylsalicylic acid suppression of the PI3K pathway as a novel medical therapy for head and neck lymphatic malformations. Int. J. Pediatr. Otorhinolaryngol. 2021;151:110869. doi: 10.1016/j.ijporl.2021.110869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oka M., Kushamae M., Aoki T., Yamaguchi T., Kitazato K., Abekura Y., Kawamata T., Mizutani T., Miyamoto S., Takagi Y. KRAS G12D or G12V mutation in human brain arteriovenous malformations. World Neurosurg. 2019;126:e1365–e1373. doi: 10.1016/j.wneu.2019.03.105. [DOI] [PubMed] [Google Scholar]
- 42.Konczyk D.J., Goss J.A., Smits P.J., Sudduth C.L., Al-Ibraheemi A., Greene A.K. Arteriovenous malformation MAP2K1 mutation causes local cartilage overgrowth by a cell-non autonomous mechanism. Sci. Rep. 2020;10:4428. doi: 10.1038/s41598-020-61444-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perkins J.A., Maniglia C., Magit A., Sidhu M., Manning S.C., Chen E.Y. Clinical and radiographic findings in children with spontaneous lymphatic malformation regression. Otolaryngol. Head Neck Surg. 2008;138:772–777. doi: 10.1016/j.otohns.2008.02.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data generated or analyzed during this study.