Abstract
Plant extracellular vesicles (EVs) play critical roles in the cross-kingdom trafficking of molecules from hosts to interacting microbes, most notably in plant defense responses. However, the isolation of pure, intact EVs from plants remains challenging. A variety of methods have been utilized to isolate plant EVs from apoplastic washing fluid (AWF). Here, we compare published plant EV isolation methods, and provide our recommended method for the isolation and purification of plant EVs. This method includes a detailed protocol for clean AWF collection from Arabidopsis thaliana leaves, followed by EV isolation via differential centrifugation. To further separate and purify specific subclasses of EVs from heterogeneous vesicle populations, density gradient ultracentrifugation and immunoaffinity capture are then utilized. We found that immunoaffinity capture is the most precise method for specific EV subclass isolation when suitable specific EV biomarkers and their corresponding antibodies are available. Overall, this study provides a guide for the selection and optimization of EV isolation methods for desired downstream applications.
Keywords: Cell-to-cell communication, extracellular vesicles, isolation methods, plants Interacting with pathogens
INTRODUCTION
Cell-to-cell communication between plants and pathogens requires the secretion and delivery of molecular signals into extracellular environments and their subsequent transport into interacting organisms. This process is critical for both plant defense and pathogen virulence (Kimura et al., 2001; Mahlapuu et al., 2016; Toruno et al., 2016). Recent studies have demonstrated that RNAs, including regulatory small RNAs (sRNAs), are able to move between pathogens and their hosts and regulate biological processes in recipient cells (Knip et al., 2014; Zhang et al., 2016b; Cai et al., 2018; Cai et al., 2019b; Huang et al., 2019). Previously, the mechanisms underlying sRNA movement through multiple barriers and into the opposing host or fungal cells were largely unknown. However, recent studies have shown that extracellular vesicles (EVs) can traffic sRNAs from plants to their pathogens (Cai et al., 2018). Plant EVs have also generated further interest because of their numerous functions in bioactive molecule exchange and cell-to-cell communication (Mathieu et al., 2019; Cai et al., 2021; Kameli et al., 2021).
EVs are small, lipid bilayer-enclosed vesicles containing various protein and RNA cargoes, and are released by cells of both eukaryotic and prokaryotic organisms into the extracellular space (Colombo et al., 2014; van Niel et al., 2018). EVs are a heterogeneous group of vesicles with different sizes and intracellular origins. For example, exosomes, microvesicles and apoptotic bodies each represent a major class of EVs, and originate from multivesicular bodies (MVBs), the plasma membrane, or apoptotic cells during apoptosis, respectively (Akers et al., 2013; Colombo et al., 2014; van Niel et al., 2018). In plants, EVs were initially observed in carrot cell cultures by transmission electron microscopy (TEM) in 1967 (Halperin and Jensen, 1967). Since then, plant EVs have been observed in plant-fungal interaction sites by TEM, such as in Blumeria graminis f. sp.hordei infected barley epidermal cells (An et al., 2006a; An et al., 2006b), Botrytis cinerea infected Arabidopsis leaf cells (Cai et al., 2018), and Rhizophagus irregularis arbuscules in rice root (Roth et al., 2019). TEM and confocal microscopy have both identified plant MVBs fused with the plasma membrane at fungal or oomycete invasion sites, suggesting that plant exosomes are released by MVB mediated secretion (An et al., 2006a; An et al., 2006b; An et al., 2007; Nielsen et al., 2012; Bozkurt et al., 2014; Cai et al., 2018).
It is worth noting that plant EVs are nanovesicles primarily from the apoplastic washing fluid (AWF). However, nanovesicles isolated from disrupted whole leaf tissue are not pure EVs, as they contain cytoplasmic intracellular membrane contaminants (Liu et al., 2020). Currently, plant EVs have been isolated from the AWF of several plant tissues, including Arabidopsis leaves (Rutter and Innes, 2017; Cai et al., 2018; He et al., 2021), sunflower seeds and seedlings (Regente et al., 2009; Regente et al., 2017), and Nicotiana benthamiana leaves (Movahed et al., 2019). In Arabidopsis leaves, to our knowledge, at least three known EV subtypes exist: Tetraspanin (TET) 8-positive EVs derived from MVBs, which can be considered plant exosomes (Cai et al., 2018; Cai et al., 2021), Penetration 1 (PEN1)-positive EVs (Rutter and Innes, 2017), and EVs produced by exocyst-positive organelle (EXPO)’s fusion with the plasma membrane (Wang et al., 2010; Ding et al., 2014). Recent work has shown that endogenous sRNAs are secreted by plant EVs as a defense mechanism against fungal pathogens (Cai et al., 2018). Further efforts revealed that TET8-positive exosomes are the major class of plant EVs responsible for sRNA transport, along with several RNA binding proteins which contribute to sRNA selective loading and stabilization in EVs (He et al., 2021).
Numerous methods for the isolation of EVs in mammalian systems have been developed in the last decade. Of these methods, separation by differential centrifugation is considered the standard, specifically for the isolation of small EVs or exosomes (Thery et al., 2006; Mathivanan et al., 2012). This method has several substeps, including centrifugation at 300 ×g to sediment cells, at 2000 ×g to remove dead cells and apoptotic bodies (large vesicles), at 10,000–15,000 ×g to remove cell debris and microvesicles (medium vesicles), and a final centrifugation step at ≥100,000 ×g (100,000 to 200,000 ×g) to pellet small EVs, especially exosomes (Thery et al., 2006; Crescitelli et al., 2013; Konoshenko et al., 2018; Willms et al., 2018; Jeppesen et al., 2019). The EV pellet is then washed once to remove non-EV proteins by resuspension and the final centrifugation step is repeated (Thery et al., 2006; Konoshenko et al., 2018). Specific subtypes of EVs can be further purified from this pellet through high-speed density gradient ultracentrifugation or bead-based immunoaffinity capture (Thery et al., 2006; Jeppesen et al., 2019).
While animal EVs have been well studied over the past decades, plant EVs remain poorly investigated. This is mainly due to the lack of accepted EV isolation protocols. Because plant EVs are present primarily in the apoplastic space, the most critical step of EV isolation is isolation of clean AWF, which is obtained by a simple, well-established infiltration-centrifugation method (Wang et al., 2005; Sanmartin et al., 2007; Hatsugai et al., 2009; O’Leary et al., 2014). Based on established animal EV purification protocols, plant EV separation involves differential centrifugation of AWF, with two consecutive steps of low speed centrifugation at 2,000 ×g and 10,000 ×g, to remove dead cells, cell debris, and large vesicles, followed by high speed centrifugation at 100,000 × g to pellet small EVs (Prado et al., 2014; Cai et al., 2018; Movahed et al., 2019; Liu et al., 2020; He et al., 2021). In some studies, a lower centrifugal force, 40,000 ×g, was used to isolate EVs. For example, this speed was used to isolate EVs derived from sunflower seeds and seedlings (Regente et al., 2009; Regente et al., 2017). Furthermore, EVs pelleted at this speed in Arabidopsis contain PEN1-positive EVs (Rutter and Innes, 2017). Note that in distinct protocols for plant EV isolation, the differences lie not only in the speed of centrifugation for the final EV sedimentation, but also in AWF collection. Thus far, there is no standard protocol for plant EV isolation from AWF across different plant species. Therefore, in this study, we compared the results from different EV isolation methods and propose a standardized method for plant EV isolation and purification from Arabidopsis. We utilized high-speed density gradient centrifugation to separate and purify EV subtypes based on their different densities. Furthermore, we also describe a recently developed immunoaffinity capture method, using bead-based antibodies that recognize the plant EV enriched TET8 protein, allowing the precise capture of the specific TET8-positive EV subtype.
RESULTS
Isolation of plant EVs by differential centrifugation
Differential centrifugation is the most commonly used method for EV isolation from cell culture supernatants and biological fluids (Thery et al., 2006; Willms et al., 2018). The methods used to isolate plant EVs share similarities with those used to isolate mammalian EVs, with the additional first step of AWF collection (Figure 1). Based on the commonly used infiltration-centrifugation method for plant AWF collection, we developed a protocol for the extraction of AWF from Arabidopsis leaves with an optimized vacuum infiltration and centrifugation method (O’Leary et al., 2014; Cai et al., 2018; He et al., 2021). Initially, fully expanded rosette leaves were detached from plants at the base of the leaf using a razor blade to limit contamination by the vasculature stream, which contains mobile RNAs and ribonucleoprotein complexes (Zhang et al., 2009; Liu and Chen, 2018). Cytoplasmic contaminants from damaged cells were removed by washing the cut leaves in distilled water (Figure 1A). The detached leaves were gently infiltrated with the infiltration buffer by negative pressure within a needleless syringe, then the leaves were carefully arranged to have the cut side of the leaf bases towards the bottom of the tube to avoid cell damage during centrifugation (Figure 1A). AWF was then collected by centrifugation at 900 ×g (Figure 1A). EVs were subsequently isolated from AWF by the following centrifugation steps (Figure 1B): (i) The AWF was centrifuged for 30 min at 4 °C at 2,000 × g to remove large cell debris (Figure S1). (ii) The supernatants were filtered through a 0.45 μm filter to remove the largest vesicles. (iii) The supernatant was moved into new ultracentrifuge tubes, and large vesicles were removed with another centrifugation step at 10,000 × g for 30 minutes at 4°C (Figure S1). (iv) The plant EV fraction was then pelleted using high speed (100,000 × g) centrifugation for 1 hour. (v) The pellet was washed to remove potential protein aggregates via a second round of ultracentrifugation at 100,000 × g. The EV pellet obtained through this step is called the P100 fraction (Figure S1). P100 fraction can be further used for the analysis of small RNA (sRNA) content in EVs. To confirm that sRNAs were located inside EVs, not out of EVs, the P100 fraction can be treated with nuclease or proteinase plus nuclease before RNA extraction (Figure 1C). If sRNAs were largely resistant to degradation by nuclease or proteinase plus nuclease treatment but became susceptible in the presence of Triton-X-100 (a detergent that can rupture EVs), these sRNAs can be determined to located inside EVs. For example, EV-enriched sRNAs, TAS1c-siR483, TAS2-siR453 and miRNA166 can be detected in nuclease-treated EVs or proteinase plus nuclease-treated EVs, indicating they located inside EVs (Cai et al., 2018) (Figure 1D).
Figure 1. Schematic of the plant EV isolation and small RNA (sRNA) detection.
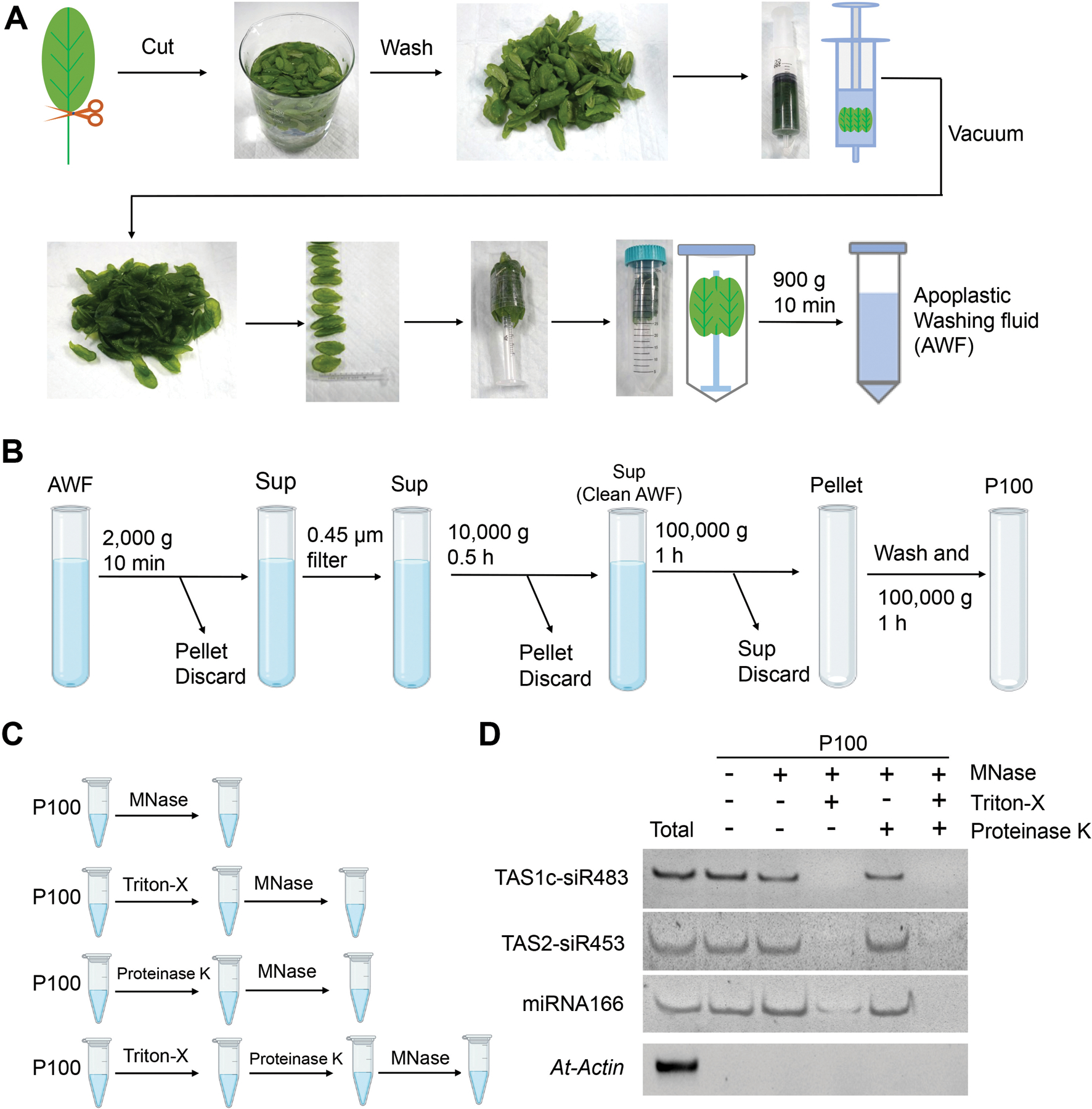
(A) Images show the various steps in apoplastic washing fluid (AWF) isolation from Arabidopsis (detached leaves protocol, Method 1 in Figure 2). The distinct proximal (petiole) part of leaves was removed using scissors, and the distal (blade) zones of leaves were collected. The leaves were placed in a syringe and gently vacuumed with infiltration buffer. The syringe with taped leaves was placed into a 50 ml conical tube, and then centrifuged at 900 × g to collect the apoplastic washing fluid. (B) Schematic of EVs isolated by differential centrifugation of AWF from Arabidopsis. The clean AWF was centrifuged at 100,000 × g to obtain the P100 EV fraction. Sup, Supernatant. (C) Schematic of EVs treated with micrococcal (MNase) and proteinase K. (D) EV-enriched sRNAs (TAS1c-siR483, TAS2-siR453 and miRNA166) were detected in nuclease-treated EVs or proteinase plus nuclease-treated EVs. Actin gene was used as the control. The “total” lane indicates total RNAs from leaves.
Technical evaluation of AWF collection from Arabidopsis leaves
Extraction of AWF is a crucial step to obtain high quality plant EVs with few contaminants. In parallel with the detached leaves method described above (Figure 1A, Method 1), we performed a second method (Figure S2, Method 2) in order to compare the purity and quality of EVs obtained. In Method 2, the entire aerial part of plants was harvested by cutting the stem directly above the roots. Plant tissues were then vacuumed and centrifuged to collect the AWF (Figure S2) (Rutter and Innes, 2017; Baldrich et al., 2019). Previously we showed that fungal infection increases EV secretion (Cai et al., 2018; He et al., 2021). Here, we used B. cinerea-infected Arabidopsis to increase the yield of isolated EVs in both methods. Ideally, AWF should be free of contamination from cell debris and cytoplasmic molecules, such as chlorophyll, the major pigment (green) in chloroplasts (O’Leary et al., 2014). However, AWF extracted by Method 2 was green in color indicating obvious contamination of cytoplasmic molecules, whereas AWF extracted by Method 1 was clear with no visible chlorophyll contamination (Figure 2A). Additionally, Western blot analysis demonstrated that both the AWF and P100 fraction obtained using Method 2 were enriched in Rubisco proteins as compared to the AWF and P100 fraction extracted by Method 1 (Figure 2B). To be more precise, we directly visualized the vesicles in the P100 fractions prepared from AWF extracted by Method 1 and Method 2 using TEM. The P100 fraction obtained via Method 2 contains large amounts of non-vesicle structures/materials, while very little non-vesicle material was observed in the P100 fraction obtained via Method 1 (Figure 2C). These results indicate that the detached leaves method (Method 1) is a superior choice for AWF collection to reduce contamination in EV preparations.
Figure 2. The detached leaves protocol (Method 1) for AWF isolation is better than the whole plant protocol (Method 2) in Arabidopsis.
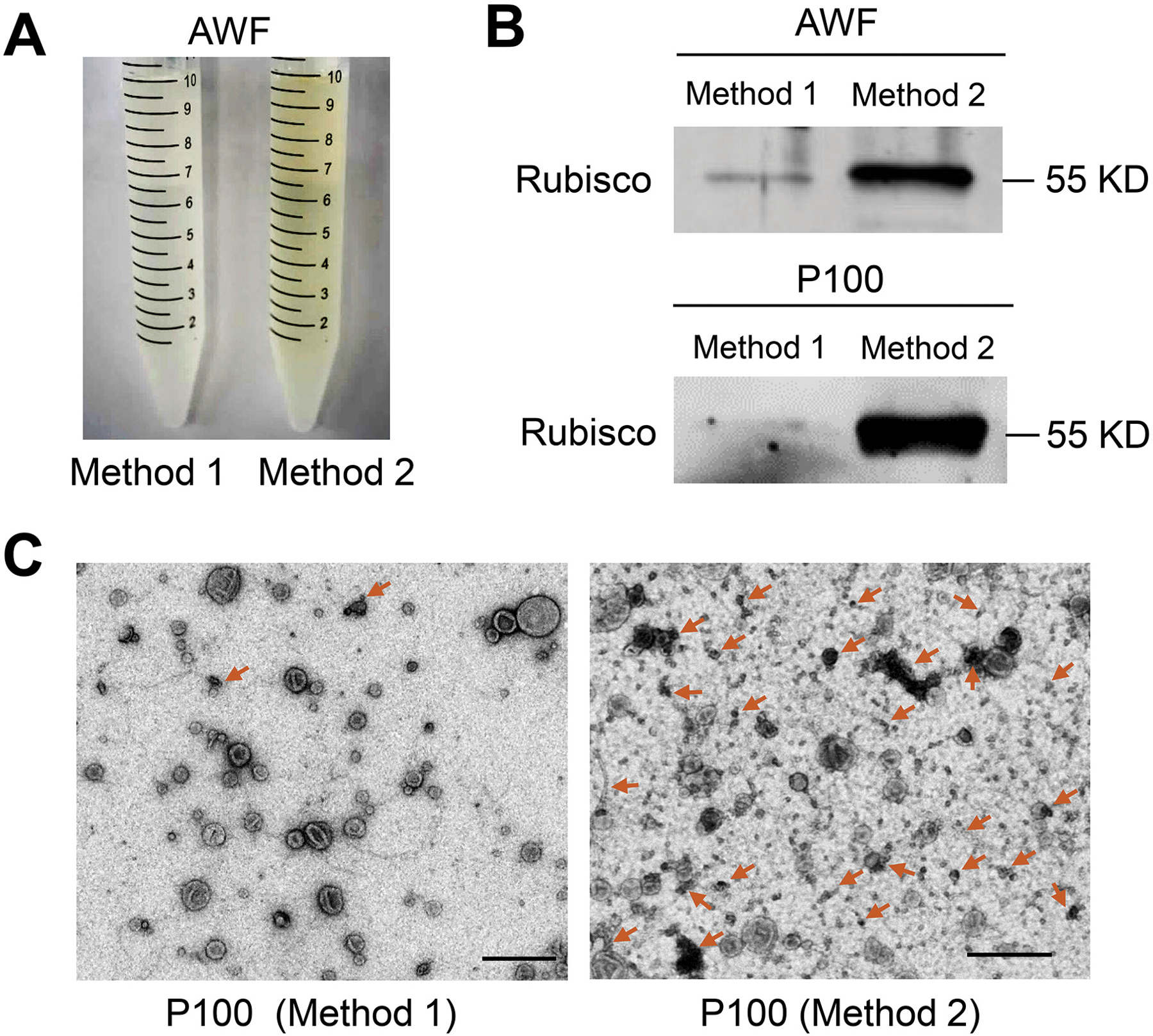
(A) Comparison of the color of AWF isolated by Method 1 and Method 2. The same amount of plants (50 plants) was used for both methods. (B) Detection of Rubisco protein in both AWF and their P100 EV fraction by Western blot using Rubisco antibody, protein size is indicated by KD. To perform the Western blot of AWF samples, equivalent amounts of AWF (10 μl) collected by both methods in (A) were used. To perform the Western blot of EV samples, all AWF collected in (A) was centrifuged at 100, 000 × g to get P100 fractions. Both P100 pellets were resuspended in 100 μl infiltration buffer, and 10 μl of this suspension was used for the Western blot. (C) Representative transmission electron microscopy (TEM) images of P100 fraction isolated from AWF collected by Method 1 and Method 2. Non-vesicle structures marked by arrows. Scale bars, 500 nm.
Technical evaluation of final ultracentrifugation speed for plant EV isolation
In differential centrifugation steps, the final supernatant is ultracentrifuged to pellet the EVs. In animal systems, genuine exosomes (or small EVs in general) are usually sedimented at speeds of 100,000–200,000 × g (Thery et al., 2006; Kowal et al., 2016; Konoshenko et al., 2018; Jeppesen et al., 2019). For plant EV isolation, two final ultracentrifuge speeds, 100,000 × g (Prado et al., 2014; Prado et al., 2015; Cai et al., 2018; Movahed et al., 2019; Liu et al., 2020; He et al., 2021) and 40,000 × g (Regente et al., 2009; Rutter and Innes, 2017; Baldrich et al., 2019), have been used in different studies. Here, we compared the EV fractions, P100 and P40, obtained by a final ultracentrifugation step at 100,000 × g, or 40,000 × g, respectively (Figure 3A). The pellet obtained from further centrifugation of the supernatant of the P40 fraction at 100,000 × g was named the P100 minus P40 (P100–40) fraction (Figure 3A). Subsequently, the morphology of EVs were examined by TEM (Jung and Mun, 2018). Plant EVs in the P100 fraction showed similar morphology to animal EVs isolated by centrifugation at 100,000 × g (Figure 3B) (Thery et al., 2006; Jung and Mun, 2018), and were unlikely to be deformed or broken during 100,000 × g centrifugation (Figure 3B). In comparison to the P100 fraction, there were fewer EVs present in the P40 fraction, while a substantial amount of EVs were isolated after centrifugation of the supernatant of the P40 fraction at 100,000×g (P100–40) (Figure 3B). Therefore, it is not appropriate to consider the supernatant of P40 a non-vesicle fraction (Baldrich et al., 2019). Corroborating this conclusion, isolation of EV fractions from transgenic plants co-expressing TET8-GFP and mCherry-PEN1 proteins contained a large amount of TET8-positive EVs in the P100–40 fraction (Figure 3C). In addition, TET8 positive EVs represent a majority of the EVs in both the P100–40 fraction (85%) and the P100 fraction (65%), while PEN1 positive EVs represent a majority of the EVs (72%) in the P40 fraction (Figure 4A). These results demonstrate that centrifugation at 100,000 × g collects much more EVs than centrifugation at 40,000 × g for plant EV isolation. Further, centrifugation at 40,000 × g results in a loss of large amounts of TET-positive EVs, the exosomes.
Figure 3. Centrifugation at 100,000 × g enriches plant EVs much more efficiently than at 40,000 × g.
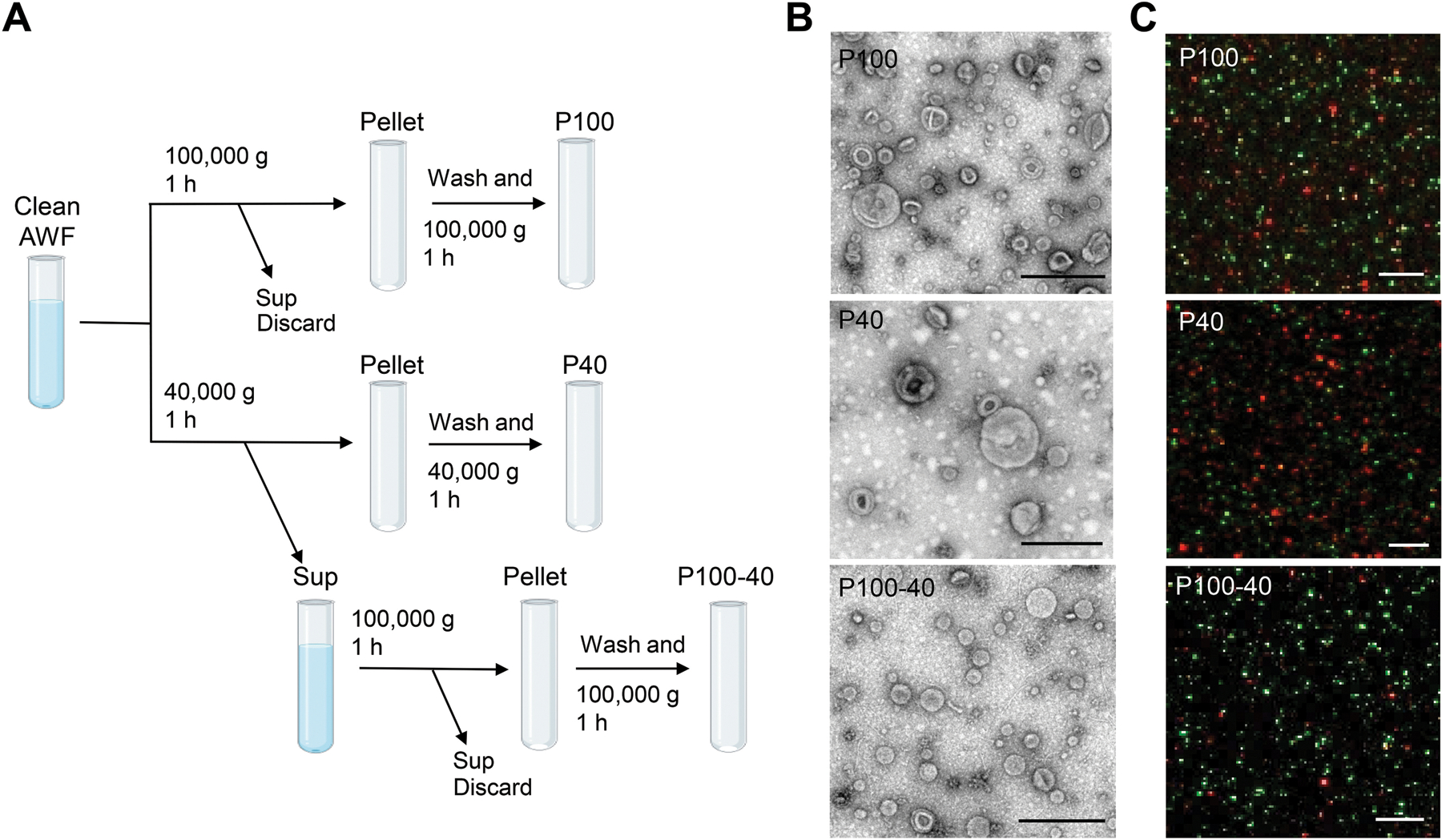
(A) Schematic of EV isolation by ultracentrifugation of AWF from Arabidopsis. EVs were isolated from clean AWF (isolated by Method 1) via ultracentrifugation at 40,000 × g (P40 fraction) and 100,000 × g (P100 fraction) for 1 hour. For the P100–40 fraction, the supernatant (Sup) of the P40 fraction was centrifuged a second time at 100,000 ×g for 1 hour. (B) Representative TEM images of P100 fraction, P40 fraction and P100–40 fraction isolated from B. cinerea-infected wild-type Arabidopsis. Scale bars, 500 nm. (C) Confocal microscopy of EV fractions (P100, P40 and P100–40) isolated from B. cinerea-infected TET8-GFP/mCherry-PEN1 double-fluorescence transgenic plants. Scale bars, 5 μm.
Figure 4. Quantification analysis of the distribution of EV subtypes and sizes in different centrifuge fractions.
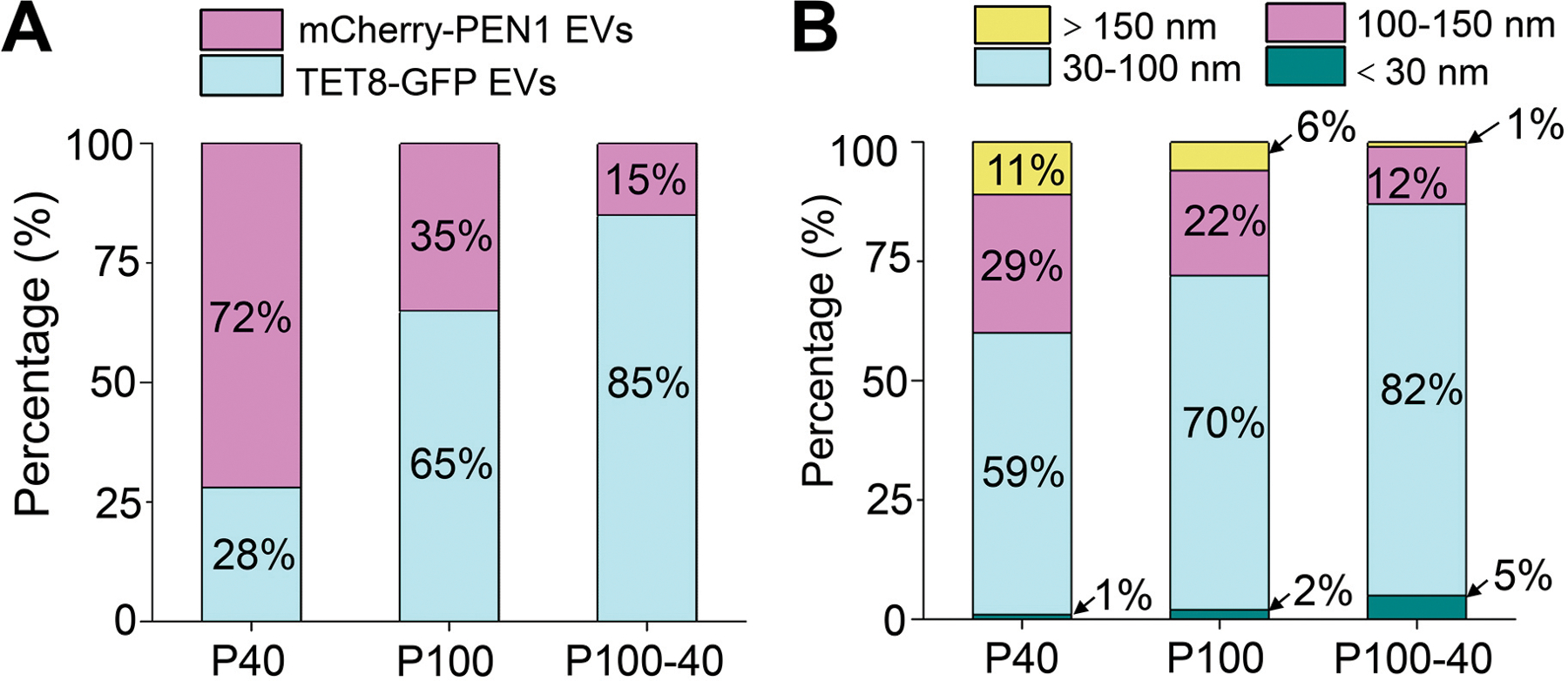
(A) Histograms for the distribution of TET8-GFP EVs and mCherry-PEN1 EVs isolated from TET8-GFP/mCherry-PEN1 double-fluorescence transgenic plants in P40 (1937 vesicles analyzed), P100 (2114 vesicles analyzed) and P100–40 (1245 vesicles analyzed) fractions from confocal images. (B) Histograms for the size distribution of EVs in P40 (226 vesicles analyzed), P100 (222 vesicles analyzed) and P100–40 (232 vesicles analyzed) fractions from TEM images.
We also analyzed plant EV size using TEM imaging. TEM micrographs of the P100 fraction showed that a majority of the vesicles (92%) had diameters ranging between 30 nm and 150 nm (Figure 4B). This result demonstrates that plant EVs in the P100 fraction are similar in size to an EV subtype termed exosomes (30 nm-150 nm in diameter) (Colombo et al., 2014; Kowal et al., 2016; Mathieu et al., 2019). In the P40 fraction, 40% of the vesicles observed had diameters larger than 100 nm, while only 13% of the vesicles in the P100–40 fraction were in this size range (Figure 4B), suggesting that centrifugation at 40,000 × g pellets larger vesicles. In addition, the majority of vesicles (82%) observed in the P100–40 fraction had diameters ranging between 30 nm and 100 nm (Figure 4B). These results suggest that centrifugation at 100,000 × g isolates plant EVs, especially small EVs like exosomes, at a much greater efficiency than centrifugation at 40,000 × g.
Density gradient fractionation separates plant EVs
Although Method 1 provides reasonably pure plant EVs (Figure 2), some applications may require extra purification steps. The overlapping sedimentation of exosomes, microvesicles and other large vesicles produces a mixture of vesicles in the ultracentrifugation fraction (Konoshenko et al., 2018). Density gradient fractionation separation is a classical method used to separate vesicles according to their floatation speed and equilibrium density (Colombo et al., 2014; Kowal et al., 2016; Jeppesen et al., 2019). This strategy separates EVs using sucrose or iodixanol gradient centrifugation of EV pellets prepared by differential centrifugation. Previous work has utilized sucrose gradients on EVs isolated from P100 fractions (He et al., 2021) and iodixanol (OptiPrep) gradients on EVs isolated from P40 fractions (Rutter and Innes, 2017) to facilitate the separation of subtypes of EVs. Because centrifugation at 100,000 × g enriches plant EVs much more efficiently than 40,000 × g, we used an iodixanol density gradient to further separate EVs from the P100 fraction and estimate their density using top-loading methods (Figure 5A). Using the TET8 antibody and TEM imaging, we identified that most of the TET8 positive EVs accumulated in the third fraction (F3) at an average density of 1.08 g/ml of iodixanol (Figure 5B, C). This is similar to the density of exosomes in animal systems (1.08–1.12 g/ml) (Wubbolts et al., 2003; Iliev et al., 2018; Jeppesen et al., 2019). We further analyzed the size of vesicles in the F3 fraction. The majority of vesicles (83%) in the F3 fraction had diameters ranging between 30 nm and 100 nm, which is similar to EV sizes obtained in the P100–40 fraction (Figure 4B, 5C, D).
Figure 5. Characterization of P100 EVs using iodixanol gradients.
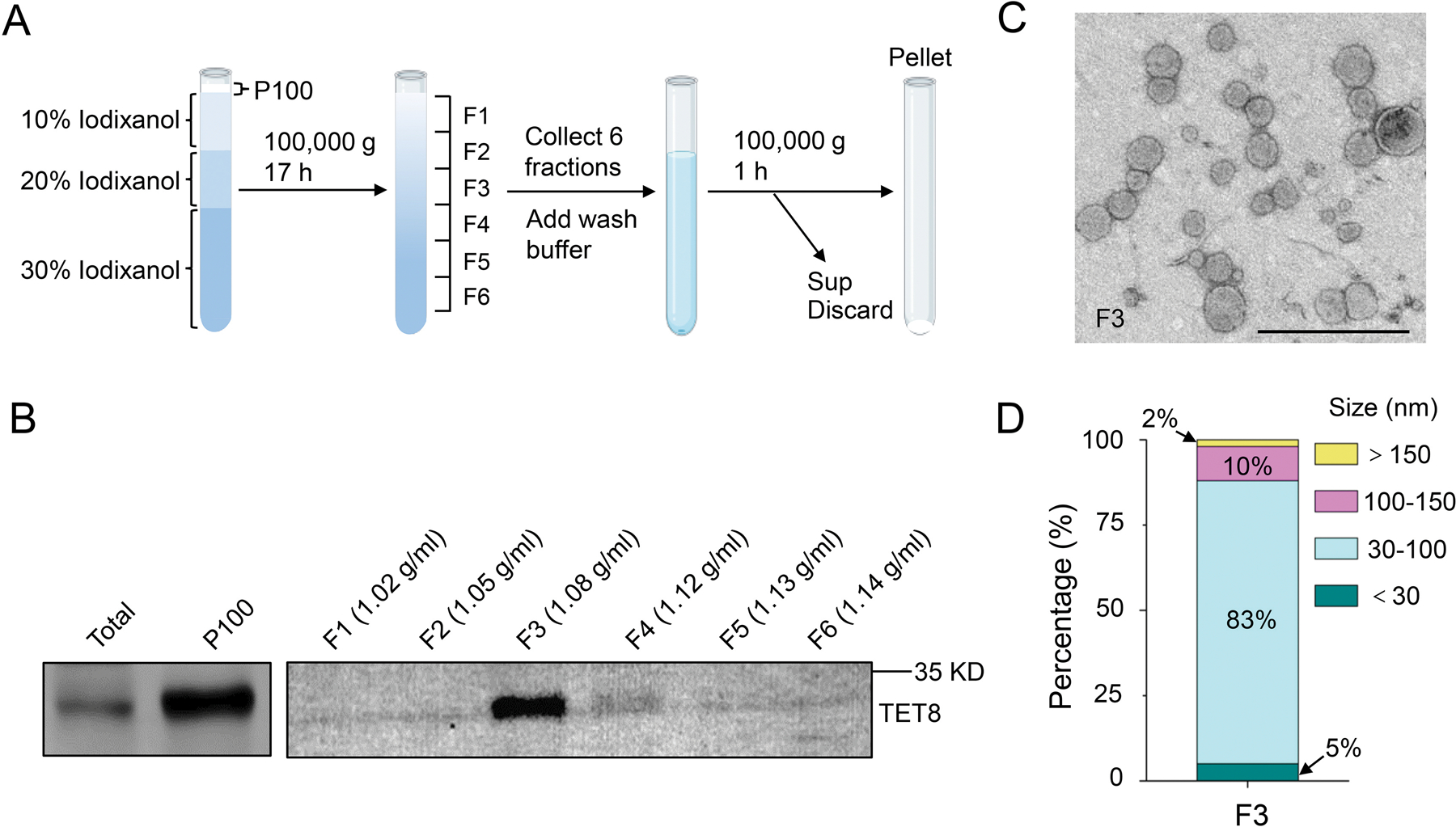
(A) P100 fraction obtained after 100,000×g centrifugation was allowed to float into an overlayed iodixanol gradient by top loading. Sup, Supernatant. (B) Six fractions were collected and analyzed by Western blot, showing the TET8-positive EVs enriched in a single fraction (F3) with TET8 native antibody. (C) Representative TEM images of F3 fraction in (B). Scale bars, 500 nm. (D) Histogram for the size distribution of EVs in F3 fraction in (C) (284 vesicles analyzed).
Immunoaffinity capture-based technique purifies specific classes of EVs
Immunoaffinity capture-based EV isolation is considered to be the most advanced method to purify specific classes of EVs (Thery et al., 2006; Kowal et al., 2016; Jeppesen et al., 2019; He et al., 2021). This technique relies on the use of an antibody to capture EVs with a specific protein marker on the surface of the EVs (Thery et al., 2006). Tetraspanins, such as CD81 or CD63, are ideal immuno-capture proteins since they are enriched on exosome membranes (Andreu and Yanez-Mo, 2014). He et al. developed an immunocapture purification method using beads coated with an antibody targeting the plant exosome marker TET8 (Figure 6A) (He et al., 2021). It is worth noting that antibody-recognized regions of the protein marker must be on the outer surface of the EVs. Thus, the antibody that specifically recognizes the large exposed extravesicular loop, EC2 domain of TET8, has been well designed to pull-down TET8 positive EVs from the P100 fraction (He et al., 2021). Using this method, TET8-positive EVs can be successfully purified from the P100 fraction (Figure 6B). Specificity of the immunoaffinity capture was examined using beads coated with an irrelevant antibody (IgG) (Figure 6B). Thus, this approach can be easily used for isolating/purifying a specific subtype of EVs in plants. By using this method, EV-enriched sRNAs and RNA binding proteins, such as Argonaute 1 (AGO1), RNA helicase (RH) 11, RH37, annexin (ANN) 1 and ANN2 were clearly detected in the TET8-positive EVs (He et al., 2021). Thus, immunoaffinity isolation is the ideal method for the precise analysis of the cargo contents of specific EV subtypes.
Figure 6. Immunoaffinity isolation is the most advanced method for the purification of specific subclasses of EVs in plants.
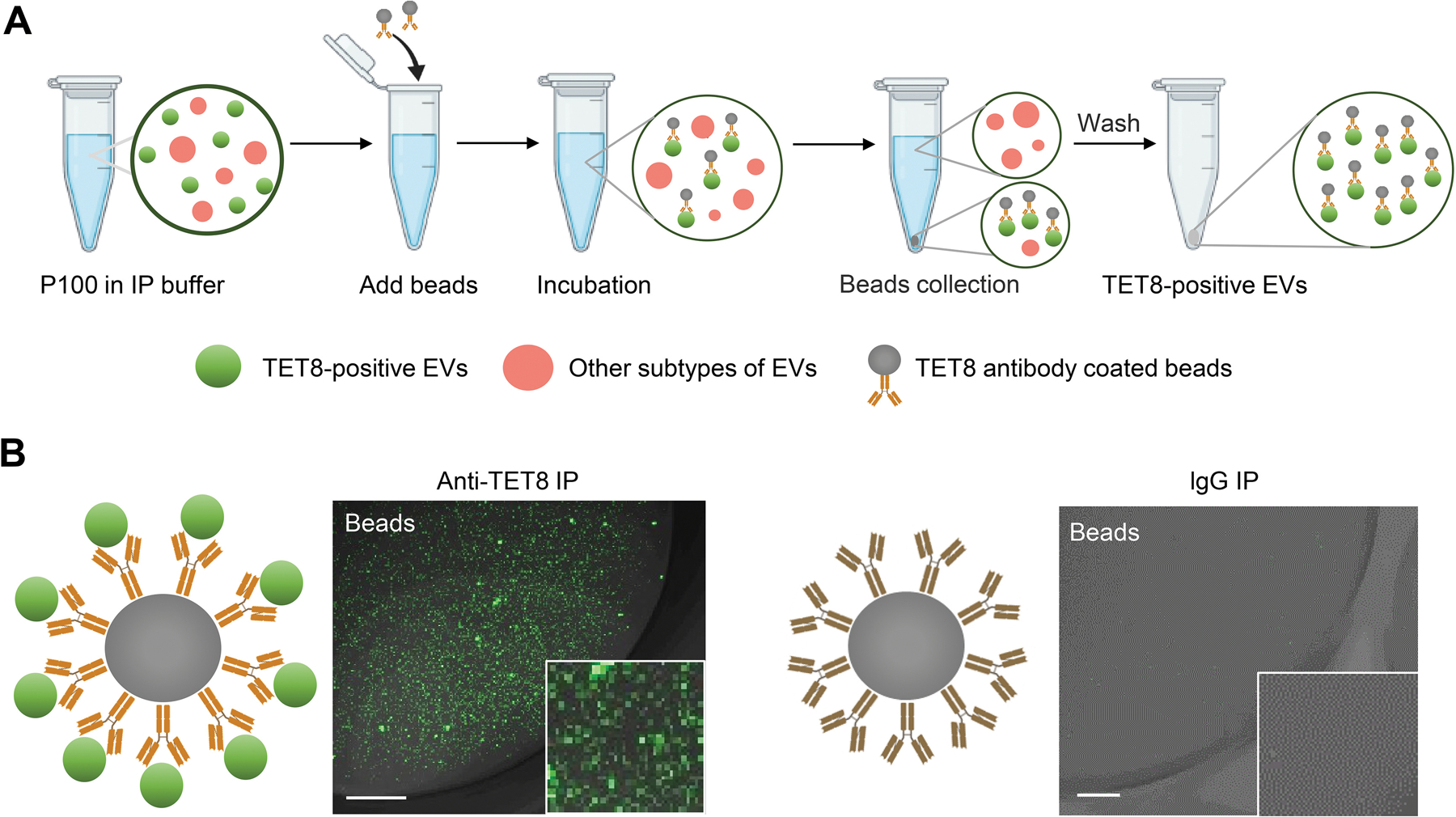
(A) Schematic of P100 fraction subjected to immuno-isolation with beads coupled to antibodies against TET8, or irrelevant Rabbit IgG as a negative control. (B) Confocal microscopy demonstrated that the TET8-positive EVs were pulled-down by TET8-specific antibody-linked beads. Scale bars, 10 μm.
DISCUSSION
In recent years, plant-derived EVs have garnered increased interest because of their essential role in cross-kingdom or cross-organism communication, and research in this field has exponentially increased (Cai et al., 2019a; Cai et al., 2021). Plant EVs have been isolated from the AWF of different plant tissues by differential centrifugation. So far, this method has been used for EVs isolated from Arabidopsis leaves (Rutter and Innes, 2017; Cai et al., 2018; He et al., 2021), sunflower imbibing seeds and seedlings (Regente et al., 2009; Regente et al., 2017), and Nicotiana benthamiana leaves (Movahed et al., 2019). Further, it has been reported that olive (Olea europaea) pollen grains released nanovesicles (28 to 60 nm in diameter) in media during pollen germination and pollen tube growth in vitro (Prado et al., 2014). These nanovesicles were isolated from the media by differential centrifugation (Prado et al., 2014). In addition to natural EVs, artificial plant-derived vesicles have been isolated from disrupted leaves or plant tissues via differential centrifugation (Zhang et al., 2016a; Kameli et al., 2021). Interestingly, these artificial vesicles have shown promising utility in drug delivery in human health and disease applications (Wang et al., 2013; Mu et al., 2014; Teng et al., 2018; Liu et al., 2020). Due to the existence of diverse EV separation methods developed in plants, in this study, we compared the quality of EVs isolated from two major published methods step-by-step and optimized a protocol for AWF collection and differential centrifugation with minimum contamination and high EV yield. Extra subsequent steps, such as immunoaffinity capture, were also introduced in detail to purify specific subclasses of EVs.
In animal systems, EVs have been isolated from diverse bodily fluids, including blood, urine, saliva, breast milk, semen and cell culture media (Colombo et al., 2014). Unlike in animals, isolation of plant EVs first requires the collection of clean apoplastic fluids. Due to the fragility of plant leaves, collection of clean apoplastic fluids has been a challenge. Here, we found that using only the detached Arabidopsis rosette leaves with no petioles for AWF collection can minimize the contamination of cytosolic contents and vasculature fluid (Method 1 in Figure 2). Because phloem stream contains large amounts of mobile RNAs and ribonucleoprotein complexes (Zhang et al., 2009; Liu and Chen, 2018), this detached leaf protocol minimizes contamination by the vasculature fluids, making subsequent analyses on the RNA content in the apoplastic EVs more accurate. On the contrary, the alternative AWF collection method which used the entire aerial part of a plant (Method 2 in Figure 2), can lead to cell breakage and contamination of intracellular contents and vasculature sap. Therefore, isolating AWF from detached leaves with no petioles is the preferable method for clean AWF collection before EV isolation.
Ultracentrifugation is the most commonly used technique for EV isolation from biofluids and cell culture supernatants. Similar to animal EV isolation, which overwhelmingly relies on a final ultracentrifugation step of 100,000 × g, plant EVs were also highly enriched in the fraction collected by ultracentrifugation at 100,000 × g from AWF (Cai et al., 2018; He et al., 2021). In this study, we have characterized and compared the quality of EVs isolated from both intermediate speed (40,000 × g) and high speed (100,000 × g) fractions. Consistent with our previous study (He et al., 2021), we found that a large amount of EVs, specifically TET-positive EVs, remains in the P100–40 fraction. Thus, centrifugation at 100,000 × g has greater separation efficiency resulting in higher EV yield and small EVs, whereas centrifugation at 40,000 × g largely reduces the yield, and favors the collection of large EVs. Because PEN1 positive EVs represent a majority (72%) of EVs in the P40 fraction, centrifugation at 40,000 × g is suitable for the isolation of PEN1-associated EVs (Rutter and Innes, 2017; He et al., 2021). The majority of EVs (70%) recovered from P100 centrifugation and 82% of the EVs recovered from the supernatant of P40 fraction (P100–40) had sizes ranging between 30 nm and 100 nm in diameter (Figure 4), similar to the animal exosomes (Colombo et al., 2014; Kowal et al., 2016; Mathieu et al., 2019). It is worth noting that a small fraction of other large EV types, possibly microvesicles and other large vesicles, was also co-sedimented into the pellet by centrifugation at 100,000 × g (Figure 4B).
Utilizing density gradient centrifugation techniques post-ultracentrifugation allows for the isolation of EVs with higher purity than those isolated with ultracentrifugation alone, and can separate distinct EV subtypes based on their densities (Konoshenko et al., 2018; Jeppesen et al., 2019). Previously, we found that in P100 fractions floated in a sucrose density gradient, TET8-positive EVs and EV-associated sRNAs were enriched in the EV fractions at densities of 1.12–1.19 g/ml (He et al., 2021). In this study, P100 fractions were floated in an iodixanol density gradient, and TET8-positive vesicles were enriched in the gradient fraction of 1.08 g/ml, on average. The different densities of TET8-positive EVs in sucrose versus iodixanol could be a result of differences in the osmotic pressure of these two gradients. This result was similar to a previous study which also found that vesicles in P100 pellets derived from human dendritic cells exhibited slightly different densities in sucrose versus iodixanol gradients (Kowal et al., 2016). Note that PEN1-positive EVs collected at 40,000 × g were enriched in the iodixanol gradient fraction at densities ranging from 1.029 g/ml to 1.056 g/ml (Rutter and Innes, 2017), demonstrating that TET8-positive EVs and PEN1-positive EVs represent two distinct sub-populations of EVs with different densities. Further study is required to determine the density of other EV subtypes, such as Exo70E2-positive EVs, by its marker lines or the specific antibodies.
Density gradient centrifugation still has some disadvantages, as it is complex, laborious, and time-consuming (up to 2 days). Additionally, it is difficult to separate different subtypes of EVs with similar densities. Therefore, immunoaffinity isolation is the most precise method for the purification of a specific subtype of EVs (Thery et al., 2006; Kowal et al., 2016; Jeppesen et al., 2019; He et al., 2021). Co-isolation of non-vesicular contaminants from the cytoplasm and other unwanted vesicles can be prevented by the highly specific affinity interactions that occur between an antigen and an antibody. Ideal antigens are EV biomarkers which are highly concentrated on the EV membrane, for example, the major histocompatibility complex (MHC) antigens and tetraspanins proteins (Kowal et al., 2016; Jeppesen et al., 2019). In plants, we showed that TET8-positive EVs can be successfully isolated from P100 fractions by an antibody that specifically recognizes the EC2 domain of TET8. For future research, it would be ideal to purify other subclasses of plant EVs using the immunoaffinity isolation method. In summary, these findings should serve as a guide to choose and further optimize EV isolation methods in the plant field for their desired downstream applications.
MATERIALS AND METHODS
Plant materials
Arabidopsis thaliana ecotype Columbia-0 (Col‐0) was used in this study. Arabidopsis marker lines TET8pro::TET8-GFP and TET8-GFP/mCherry-PEN1 double-fluorescence lines (Cai et al., 2018; He et al., 2021), were used as described previously. Arabidopsis seeds were grown in soil side-by-side at 22°C for 4 weeks under short-day periods (12 h of light followed by 12 h of darkness).
Apoplastic washing fluid collection from Arabidopsis leaves
Apoplastic washing fluid (AFW) collection from Arabidopsis leaves was modified from previous studies (O’Leary et al., 2014; Madsen et al., 2016). A typical experiment for EV isolation requires ~50 plants for each genotype/treatment. The distinct proximal (petiole) part of leaves was removed using scissors, and the distal (blade) zones of leaves were collected. After recording the biomass, leaves were washed 3 times with water. The leaves were carefully placed in a 200 ml syringe and gently vacuumed with an infiltration buffer (20 mM MES hydrate, 2 mM CaCl2, 0.1 M NaCl, pH 6.0) for 20 seconds. Excess infiltration buffer on the leaf surface was removed by a clean paper towel and the leaves were then fixed onto a small plastic stick. The small plastic sticks with leaves were then placed into a 50 ml conical tube, keeping the apex of the leaf facing upwards, and then centrifuged for 10 min at 4°C at 900 × g to collect the AWF.
Isolation of plant EVs by differential centrifugation
Plant EVs were isolated from Arabidopsis AWF. The AWF was centrifuged for 30 min at 4 °C at 2,000 × g to remove large cell debris and then filtered through a 0.45 μm filter. Next, the supernatants were transferred into new ultracentrifuge tubes and centrifuged for 30 min at 4 °C at 10,000 × g. After the pellet was discarded, the supernatants (the clean apoplastic washing fluid) were centrifuged for 1 hour at 4 °C at 100,000 × g or 40,000 × g to obtain the P100 EV fraction or P40 EV fraction. To obtain the P100-P40 EV fraction, the supernatants of P40 were centrifuged again for 1 hour at 4 °C at 100,000 × g. All pellets were washed in 10 ml of infiltration buffer and finally re-centrifuged at the same speed before being resuspended in infiltration buffer for further study.
MNase and proteinase K treatment of plant EVs
To clarify sRNAs inside or outside EVs, P100 vesicles were treated with micrococcal (MNase) and proteinase K. For MNase treatment, P100 vesicles was treated with 10 U of MNase (Thermo Fisher) for 15 minutes at 37°C with or without Triton X-100. For proteinase K plus MNase treatment, P100 vesicles was treated with 20 μg/ml proteinase K (Invitrogen) for 1 hour at 37°C with or without Triton X-100. The proteinase activity was inhibited by adding 5 mM phenylmethylsulfonyl fluoride for 10 minutes at room temperature. The sample was then treated with 10 U of MNase for 15 minutes at 37°C. For Triton-X-100 treatment, P100 vesicles were incubated with 1% (v/v) Triton-X-100 on ice for 30 minutes before proteinase and nuclease treatments. Immediately after proteinase and nuclease treatments, RNA was extracted for further detection of specific sRNAs. sRNA RT-PCR was performed as previously described (Cai et al., 2018).
Iodixanol gradient separation of plant EVs
Discontinuous iodixanol gradients (OptiPrep, STEMCELL) were prepared as described previously with slight modifications (Kowal et al., 2016). Working solutions of 10% (v/v), 20% (v/v) and 30% (v/v) idoixanol were made by diluting an aqueous 60% OptiPrep stock solution in infiltration buffer (20 mM MES hydrate, 2 mM CaCl2, 0.1 M NaCl, pH 6.0). The gradient was formed by successively layering 4.8 ml of 30% solution, 2.1 ml of 20% solution, and 2 ml of 10% solution in a 13PA tube (himac) from bottom to top. About 0.4 ml of EVs resuspended in infiltration buffer were layered on top of the gradient. The tube was centrifuged for 100,000 × g for 17 h at 4°C (P40ST, CP80NX, himac). After stopping the centrifuge without breaks, 0.4 ml was removed from the top of the tube, and then six fractions of 1.4 ml each were collected from the top of the tube. These fractions were each diluted to 12 ml with infiltration buffer and centrifuged at 100,000 × g for 1 h at 4°C to obtain pellets from each fraction.
Transmission electron microscope analysis of plant EVs
Sample preparation of EVs for TEM analysis followed the protocol by Maroto et al.(Maroto et al., 2017). 10 μl of EVs suspended in infiltration buffer were deposited onto 3.0 mm copper Formvar-carbon-coated electron microscopy grids (TED PELLA), and the sample was then wicked off using filter paper. The grids were negatively stained with 10 μl of 1% uranyl acetate and allowed to air dry before being imaged at 100 KV using a Transmission Electron Microscope (JEM-1400plus, JEOL). EV size was assessed with Image J software.
Immunoaffinity capture of plant EVs
Immunoaffinity capture of plant EVs was performed as described previously (He et al. 2021). Briefly, antibodies for immunoaffinity capture, Rabbit polyclonal anti-AtTET8 (Homemade) and normal rabbit immunoglobulin G (Thermo Fisher), were coated with protein A beads in IP buffer (20 mM MES hydrate, 2 mM CaCl2, 0.1 M NaCl, pH 7.5). Beads were then washed three times with IP buffer (containing 0.3% BSA), and resuspended in the same buffer, to which P100 fraction was added, followed by overnight incubation at 4°C with rotation. Bead-bound EVs were collected and washed by IP buffer for further analysis.
Confocal microscopy analyses of plant EVs
For visualization of EV-associated GFP-fluorescence and mCherry-fluorescence, EV pellets or EV coated beads suspended in infiltration buffer were examined using a 40× water immersion or dip-in lens mounted on a Confocal Laser Scanning Microscope equipped with an argon/krypton laser (Leica TCS SP5).
Supplementary Material
Figure S1. Representative transmission electron microscopy (TEM) images of fractions isolated by differential centrifugation of AWF from Arabidopsis
(A) Pellet of AWF centrifugation at 2,000 × g. Non-vesicle structures or cell debris marked by arrows. (B) Pellet of AWF centrifugation at 10,000 × g. (C) Pellet of AWF centrifugation at 100,000 × g (P100). Scale bars in (A–C), 500 nm.
Figure S2. Work-flow of isolation of AWF from Arabidopsis (Whole rosettes protocol, Method 2 in Figure 2)
Whole rosettes were harvested at root by using scissors. The rosettes were placed in a syringe and gently vacuumed with infiltration buffer, and then placed root down into a 30 ml tube, which was then put into 50 ml conical tube, and then centrifuged at 900 × g to collect the AWF.
ACKNOWLEDGEMENTS
We thank Rachael Hamby and Angela Chen for editing the paper. Work in the Q.C. laboratory was supported by grants from the National Natural Science Foundation of China (32070288). Work in the H.J. laboratory was supported by grants from the National Institute of Health (R35 GM136379), the National Science Foundation (IOS2017314), the United States Department of Agriculture National Institute of Food and Agriculture (2021-67013-34258 and. 2019-70016-29067), the Australian Research Council Industrial Transformation Research Hub (IH190100022), as well as the CIFAR Fungal Kingdom fellowship to H.J.
Biographies



Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Akers JC, Gonda D, Kim R, Carter BS, and Chen CC (2013). Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol 113: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Q, Ehlers K, Kogel KH, van Bel AJ, and Huckelhoven R (2006a). Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 172: 563–576. [DOI] [PubMed] [Google Scholar]
- An Q, Huckelhoven R, Kogel KH, and van Bel AJ (2006b). Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol. 8: 1009–1019. [DOI] [PubMed] [Google Scholar]
- An Q, van Bel AJ, and Huckelhoven R (2007). Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal Behav. 2: 4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu Z, and Yanez-Mo M (2014). Tetraspanins in extracellular vesicle formation and function. Front. Immunol 5: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrich P, Rutter BD, Zand KH, Podicheti R, Meyers BC, and Innes RW (2019). Plant extracellular vesicles contain diverse small RNA species and are enriched in 10 to 17 nucleotide “Tiny” RNAs. Plant Cell 31: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt TO, Belhaj K, Dagdas YF, Chaparro-Garcia A, Wu C, Cano LM, and Kamoun S (2014). Rerouting of plant late endocytic trafficking toward a pathogen interface. Traffic 16: 204–26 [DOI] [PubMed] [Google Scholar]
- Cai Q, He B, and Jin H (2019a). A safe ride in extracellular vesicles - small RNA trafficking between plant hosts and pathogens. Curr. Opin. Plant Biol 52: 140–148. [DOI] [PubMed] [Google Scholar]
- Cai Q, He B, Wang S, Fletcher S, Niu D, Mitter N, Birch PRJ, and Jin H (2021). Message in a bubble: Shuttling small RNAs and proteins between cells and interacting organisms using extracellular vesicles. Annu. Rev. Plant Biol 72: 497–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, He B, Weiberg A, Buck AH, and Jin H (2019b). Small RNAs and extracellular vesicles: New mechanisms of cross-species communication and innovative tools for disease control. PLoS Pathog. 15: e1008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Qiao L, Wang M, He B, Lin FM, Palmquist J, Huang SD, and Jin H (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360: 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Raposo G, and Thery C (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol 30: 255–289. [DOI] [PubMed] [Google Scholar]
- Crescitelli R, Lasser C, Szabo TG, Kittel A, Eldh M, Dianzani I, Buzas EI, and Lotvall J (2013). Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2: 20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang J, Lai JHC, Chan VHL, Wang X, Cai Y, Tan X, Bao Y, Xia J, Robinson DG, and Jiang L (2014). Exo70E2 is essential for exocyst subunit recruitment and EXPO formation in both plants and animals. Mol. Biol. Cell 25: 412–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin W, and Jensen WA (1967). Ultrastructural changes during growth and embryogenesis in carrot cell cultures. J. Ultrastruct. Res 18: 428–43. [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, and Hara-Nishimura I (2009). A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 23: 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Cai Q, Qiao L, Huang CY, Wang S, Miao W, Ha T, Wang Y, and Jin H (2021). RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 7: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Wang H, Hu P, Hamby R, and Jin H (2019). Small RNAs - Big players in plant-microbe interactions. Cell Host Microbe. 26: 173–182. [DOI] [PubMed] [Google Scholar]
- Iliev D, Strandskog G, Nepal A, Aspar A, Olsen R, Jorgensen J, Wolfson D, Ahluwalia BS, Handzhiyski J, and Mironova R (2018). Stimulation of exosome release by extracellular DNA is conserved across multiple cell types. FEBS J. 285: 3114–3133. [DOI] [PubMed] [Google Scholar]
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, and Coffey RJ (2019). Reassessment of Exosome Composition. Cell 177: 428–445 e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung MK, and Mun JY (2018). Sample preparation and imaging of exosomes by transmission electron microscopy. J. Vis. Exp 131: 56482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameli N, Dragojlovic-Kerkache A, Savelkoul P, and Stassen FR (2021). Plant-derived extracellular vesicles: Current findings, challenges, and future applications. Membranes 11: 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Anzai H, and Yamaguchi I (2001). Microbial toxins in plant-pathogen interactions: Biosynthesis, resistance mechanisms, and significance. J. Gen. Appl. Microbiol 47: 149–160. [DOI] [PubMed] [Google Scholar]
- Knip M, Constantin ME, and Thordal-Christensen H (2014). Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet. 10: e1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konoshenko MY, Lekchnov EA, Vlassov AV, and Laktionov PP (2018). Isolation of extracellular vesicles: General methodologies and latest trends. Biomed Res. Int 2018: 8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, and Thery C (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U. S. A 113: E968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, and Chen X (2018). Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 4: 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wu S, Koo Y, Yang A, Dai Y, Khant H, Osman SR, Chowdhury M, Wei H, Li Y, Court K, Hwang E, Wen Y, Dasari SK, Nguyen M, Tang EC, Chehab EW, de Val N, Braam J, and Sood AK (2020). Characterization of and isolation methods for plant leaf nanovesicles and small extracellular vesicles. Nanomedicine 29: 102271. [DOI] [PubMed] [Google Scholar]
- Madsen SR, Nour-Eldin HH, and Halkier BA (2016). Collection of apoplastic fluids from Arabidopsis thaliana leaves. Methods Mol. Biol 1405: 35–42. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Hakansson J, Ringstad L, and Bjorn C (2016). Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell Infect. Microbiol 6: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto R, Zhao Y, Jamaluddin M, Popov VL, Wang H, Kalubowilage M, Zhang Y, Luisi J, Sun H, Culbertson CT, Bossmann SH, Motamedi M, and Brasier AR (2017). Effects of storage temperature on airway exosome integrity for diagnostic and functional analyses. J. Extracell. Vesicles 6: 1359478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Martin-Jaular L, Lavieu G, and Thery C (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol 21: 9–17. [DOI] [PubMed] [Google Scholar]
- Mathivanan S, Fahner CJ, Reid GE, and Simpson RJ (2012). ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic. Acids Res 40: D1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahed N, Cabanillas DG, Wan J, Vali H, Laliberte JF, and Zheng H (2019). Turnip mosaic virus components are released into the extracellular space by vesicles in infected leaves. Plant Physiol. 180: 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Zhuang X, Wang Q, Jiang H, Deng ZB, Wang B, Zhang L, Kakar S, Jun Y, Miller D, and Zhang HG (2014). Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res 58: 1561–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen ME, Feechan A, Bohlenius H, Ueda T, and Thordal-Christensen H (2012). Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc. Natl. Acad. Sci. U. S. A 109: 11443–11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary BM, Rico A, McCraw S, Fones HN, and Preston GM (2014). The infiltration-centrifugation technique for extraction of apoplastic fluid from plant leaves using Phaseolus vulgaris as an example. J. Vis. Exp 94: 52113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado N, Alche JD, Casado-Vela J, Mas S, Villalba M, Rodriguez R, and Batanero E (2014). Nanovesicles are secreted during pollen germination and pollen tube growth: a possible role in fertilization. Mol. Plant 7: 573–577. [DOI] [PubMed] [Google Scholar]
- Prado N, De Linares C, Sanz ML, Gamboa P, Villalba M, Rodriguez R, and Batanero E (2015). Pollensomes as natural vehicles for pollen allergens. J. Immunol 195: 445–449. [DOI] [PubMed] [Google Scholar]
- Regente M, Corti-Monzon G, Maldonado AM, Pinedo M, Jorrin J, and de la Canal L (2009). Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett. 583: 3363–3366. [DOI] [PubMed] [Google Scholar]
- Regente M, Pinedo M, San Clemente H, Balliau T, Jamet E, and de la Canal L (2017). Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J. Exp. Bot 68: 5485–5495. [DOI] [PubMed] [Google Scholar]
- Robinson D, Ding Y, and Jiang L (2016). Unconventional protein secretion in plants: a critical assessment. Protoplasma 253: 31–43. [DOI] [PubMed] [Google Scholar]
- Roth R, Hillmer S, Funaya C, Chiapello M, Schumacher K, Lo Presti L, Kahmann R, and Paszkowski U (2019). Arbuscular cell invasion coincides with extracellular vesicles and membrane tubules. Nat. Plants 5: 204–211. [DOI] [PubMed] [Google Scholar]
- Rutte BD, and Innes RW (2017). Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173: 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartin M, Ordonez A, Sohn EJ, Robert S, Sanchez-Serrano JJ, Surpin MA, Raikhel NV, and Rojo E (2007). Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A 104: 3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, Hutchins E, Mu J, Deng Z, Luo C, Sundaram K, Sriwastva MK, Zhang L, Hsieh M, Reiman R, Haribabu B, Yan J, Jala VR, Miller DM, Van Keuren-Jensen K, Merchant ML, McClain CJ, Park JW, Egilmez NK, and Zhang HG (2018). Plant-derived exosomal micrornas shape the gut microbiota. Cell Host Microbe 24: 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G, and Clayton A (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3: Unit 3. 22. [DOI] [PubMed] [Google Scholar]
- Toruno TY, Stergiopoulos I, and Coaker G (2016). Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol 54: 419–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, D’Angelo G, and Raposo G (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Bio 19: 213–228. [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, and Dong X (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040. [DOI] [PubMed] [Google Scholar]
- Wang J, Ding Y, Wang J, Hillmer S, Miao Y, Lo SW, Wang X, Robinson DG, and Jiang L (2010). EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 22: 4009–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhuang X, Mu J, Deng ZB, Jiang H, Xiang X, Wang B, Yan J, Miller D, and Zhang HG (2013). Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat. Commu 4: 1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms E, Cabanas C, Mager I, Wood MJA, and Vader P (2018). Extracellular vesicle heterogeneity: Subpopulations, isolation techniques, and diverse functions in cancer progression. Front. Immunol 9: 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot JW, Geuze HJ, and Stoorvogel W (2003). Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem 278: 10963–10972. [DOI] [PubMed] [Google Scholar]
- Zhang HG, Cao P, Teng Y, Hu X, Wang Q, Yeri AS, Zhuang X, Samykutty A, Mu J, Deng ZB, Zhang L, Mobley JA, Yan J, Van Keuren-Jensen K, and Miller D (2016a). Isolation, identification, and characterization of novel nanovesicles. Oncotarget 7: 41346–41362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sun L, and Kragler F (2009). The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 150: 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhao YL, Zhao J, Wang S, Jin Y, Chen Z, Fang Y, Hua C, Ding S, and Guo H (2016b). Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2: 16153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Representative transmission electron microscopy (TEM) images of fractions isolated by differential centrifugation of AWF from Arabidopsis
(A) Pellet of AWF centrifugation at 2,000 × g. Non-vesicle structures or cell debris marked by arrows. (B) Pellet of AWF centrifugation at 10,000 × g. (C) Pellet of AWF centrifugation at 100,000 × g (P100). Scale bars in (A–C), 500 nm.
Figure S2. Work-flow of isolation of AWF from Arabidopsis (Whole rosettes protocol, Method 2 in Figure 2)
Whole rosettes were harvested at root by using scissors. The rosettes were placed in a syringe and gently vacuumed with infiltration buffer, and then placed root down into a 30 ml tube, which was then put into 50 ml conical tube, and then centrifuged at 900 × g to collect the AWF.


