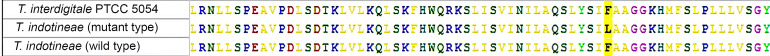Abstract
Drug resistance is one of the major challenges to skin fungal infections, especially in tropical and subtropical infections caused by dermatophytes. This study aimed to determine the antifungal susceptibility of clinically dermatophytes and evaluate point mutations in terbinafine-resistant isolates. A total number of 123 clinical dermatophyte isolates in eight species were evaluated in terms of sensitivity to seven major antifungals. Furthermore, the point mutation in squalene epoxidase (SQLE) gene responsible for terbinafine resistance was studied. The dermatophytes species were identified by morphological characteristics and confirmed by the ITS sequencing. Also, the phylogenetic tree was drawn using the RAxML analyses for 123 dermatophytes isolates. A new XXIX genotype was also found in 4 Trichophyton mentagrophytes isolates. Based on the results obtained, terbinafine was the most effective antifungal drug followed by itraconazole and voriconazole. Trichophyton rubrum and Trichophyton tonsurans were the most susceptible species (MIC50 = 0.01, 0.09 μg/ml), and T. mentagrophytes was the most resistant species (MIC50 = 0.125 μg/ml) to terbinafine. Of the 123 dermatophytes isolates, six isolates showed reduced susceptibility to terbinafine, and only Trichophyton indotineae had a mutation in SQLE gene as a Phe397Leu substitution. Overall, the antifungal susceptibility test is necessary for managing dermatophytosis. These results help physicians to control the course of the disease and provide further insights to select effective drugs for patients with dermatophytosis, especially in tropical and subtropical regions of the world, where dermatophytosis is still a public health problem.
Keywords: dermatophytes, antifungal susceptibility testing, point mutation, terbinafine resistance, Phe397Leu substitution, T. indotineae, dermatophytosis, squalene epoxidase
1 Introduction
Dermatophytes are a group of keratinophilic fungi with considerable morphological and genetic similarities (Dabas et al., 2017; Salehi et al., 2021). They can attack keratinized tissue of humans and animals and create dermatophytosis (Zareshahrabadi et al., 2020). According to the new classification, dermatophytes are classified into Trichophyton, Epidermophyton, Nannizzia, Paraphyton, Lophophyton, Microsporum, and Arthroderma (de Hoog et al., 2017).
According to the WHO’s evaluation, dermatophytosis affects 20%–25% of the world population (Ebert et al., 2020). In addition, studies have shown that dermatophyte species responses to antifungal drugs are not the same (Bhatia and Sharma, 2015). The confirmed therapeutic strategies for dermatophytosis infection include griseofulvin (GRI) drugs and systemic or topical triazole and allylamine drugs, which mostly include itraconazole (ITZ) and terbinafine (TRB). Currently, TRB is the first choice for the treatment of dermatophytosis given its stable clinical effect and fewer recurrence (Haugh et al., 2000; Niimi et al., 2010). On the other hand, therapeutic failure has become an alarming trend, and health systems around the world pay heavy costs to treat the disease every year (Siopi et al., 2021). In addition, there are an increased number of studies about resistance to antifungal drugs and TRB in particular in dermatophyte species (Haugh et al., 2000; Mukherjee et al., 2003; Osborne et al., 2005; Niimi et al., 2010; Singh et al., 2018; Nenoff et al., 2020; Siopi et al., 2021).
The TRB is a synthetic allylamine derivative that, by the activity of squalene epoxidase (SQLE), causes the accumulation of squalene and decreases ergosterol of the cell membrane, which leads to cellular death (Ghannoum et al., 2004; Osborne et al., 2005; Afshari et al., 2016).
The genetic basis of TRB resistance in fungi including Aspergillus fumigatus, Aspergillus nidulans, and Trichophyton rubrum has shown that the resistance may arise from the overexpression of SQLE gene, or it could result from mutations in other genes that indirectly affect antimycotic susceptibility (Rocha et al., 2002; Liu et al., 2004; Santos et al., 2018). It has been shown that resistance to TRB in dermatophyte species is related to the mutation of salicylate 1-monooxygenase (sa1A) and SQLE (Mukherjee et al., 2003; Yamada et al., 2017). Studies have shown that the resistance is more related to the substitutions at one of the amino acid positions of Leu393, Phe397, Phe415, and His440 in SQLE protein (Mukherjee et al., 2003; Osborne et al., 2005; Osborne et al., 2006). Furthermore, a recent study by Nenoff et al. (2020) identified two substitutions of amino acids at Ser395Pro and Ser443Pro positions of SQLE in Trichophyton strains resistant to TRB. It has been shown that replacement in the F397L position of SQLE is the most common type of substitution. There are reports of mutation in different positions of SQLE in T. tonsurans, T. interdigitale, T. mentagrophytes,, and T. rubrum isolates resistant to TRB (Rocha et al., 2002; Singh et al., 2018). In addition, Taghipour et al. showed that there was a relationship between SQLE mutation in the species resistant to TRB and ITS genotype. So the resistant strains of T. mentagrophytes are categorized only in VIII genotype, and resistant species of T. interdigitale are categorized in II genotype. On the other hand, according to the new classification of T. mentagrophytes, subtype VIII is considered as a separate species named Trichophyton indotineae (Kano et al., 2020).
However, the emergence of TRB-resistant dermatophyte strains in the south of Asia and an increase in resistance of the strains to antifungal drugs are survival mechanisms of dermatophyte fungi, which lead to failure of treatment and recurrence of disease (Ebert et al., 2020). Therefore, in this study, to accurately identify dermatophyte species, ITS region sequence was utilized, and ITS genotypes of T. mentagrophytes and T. interdigitale were determined. In addition, antifungal activity assessment of TRB drugs, ITZ, ketoconazole (KTZ), fluconazole (FLZ), posaconazole (PCZ), voriconazole (VCZ), and amphotericin B (AMB) was done through Clinical and Laboratory Standards Institute (CLSI) broth microdilution M38-A2 method against 123 clinical isolates and five standard isolates of dermatophyte. In addition, the point mutation in SQLE gene was investigated in TRB-resistant strains.
2 Material and Methods
2.1 Clinical Fungal Isolation
This experimental study included 123 dermatophyte isolates obtained from the patients visiting the Mycology Department, Pasteur Institute of Iran, between 2018 and 2019. The isolates were identified using microscope and culture, and for final confirmation, ITS sequence was used. In addition, five standard strains were provided from Persian Type Culture Collection (PTCC), Iranian Research Organization for Science and Technology (Karaj-Iran), including T. mentagrophytes PTCC 5054, Microsporum canis PTCC 5069, Nannizzia gypsea PTCC 130396, Trichophyton verrucosum PTCC 10694, and T. rubrum PTCC 5808, which were used as quality control. The species under study were T. mentagrophytes/T. interdigitale complex (include T. indotineae), T. rubrum, T. tonsurans, Epidermophyton floccosum, T. verrucosum, N. gypsea, Nannizzia fulva, and M. canis (Table 1).
Table 1.
Clinical features of the 123 clinical dermatophyte strains based on the infection body area.
| Clinical manifestation | Etiologic agents, no. (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Trichophyton mentagrophytes | Trichophyton interdigitale | Trichophyton indotineae | Trichophyton rubrum | Trichophyton tonsurans | Trichophyton verrucosum | Epidermophyton floccosum | Microsporum canis | Nannizzia gypsea | Nannizzia fulva | Total no. (%) | |
| Tinea cruris | 2 (1.62) | 11 (8.84) | 5 (4.06) | 5 (4.06) | 3 (2.43) | – | 14 (11.38) | 1 (0.81) | – | – | 41 (33.33) |
| Tinea pedis | 2 (1.62) | 9 (7.31) | 3 (2.43) | 7 (5.69) | – | – | 3 (2.43) | – | – | – | 24 (19.51) |
| Tinea corporis | 1 (0.81) | 1 (0.81) | 2 (1.62) | – | 2 (1.62) | – | 1 (0.81) | 5 (4.06) | – | – | 12 (9.75) |
| Tinea capitis | – | 2 (1.62) | – | – | 8 (6.50) | – | – | 4 (3.25) | 2 (1.62) | – | 16 (13.00) |
| Tinea unguium | – | – | – | – | – | – | – | – | 4 (3.25) | 2 (1.62) | 6 (4.87) |
| Tinea faciei | – | – | – | – | 3 (2.43) | 5 (4.06) | – | – | – | – | 8 (6.50) |
| Tinea manuum | 1 (0.81) | 5 (4.06) | – | 3 (2.43) | – | 2 (1.62) | 1 (0.81) | 4 (3.25) | – | – | 16 (13.00) |
| Total | 6 | 28 | 10 | 15 | 16 | 7 | 19 | 14 | 6 | 2 | 123 |
2.2 Molecular Identification by ITS Region
All fungal strains were cultured on Mycobiotic agar (Merck, Darmstadt, Germany) and incubated at 27°C for 7 days (Osborne et al., 2005). In summary, fungal cell fragmentation was performed by liquid nitrogen and added to the extracted cells of DNA extraction buffer containing 200 M of Tris-HCl, pH 8, 25 mM of EDTA, SDS 0.5% W/V, and NaCl 250 mM. The DNA extraction method was based on phenol/chloroform/isoamyl alcohol (25:24:1) and proteinase K. After extraction, the resulting DNA was re-dissolved in 50 μl of Tris-EDTA (TE) buffer and stored at −20°C (Salehi et al., 2018a). The ITS region was PCR amplified using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990). The final volume of PCRs was 25 μl, containing 12.5 μl of premix (Ampliqon, Odense, Denmark), 1 μl of DNA template, 0.5 μM of forward and reverse primers, and distilled water. The PCR cycling conditions were as follows: 5 min initial pre-incubation at 95°C, followed by 35 cycles consisting of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 45 s, with a final extension at 72°C for 5 min (Salehi et al., 2020). Five microliters of the PCR products was electrophoresed on the 1% agarose gel in Tris/Borate/EDTA (TBE) buffer (Yurkov et al., 2012). The sequences of isolates were edited manually and subjected to ClustalW pairwise alignment using the MEGA10 software. The sequences deposited in GenBank are shown in Table 2. ITS genotyping determined T. interdigitale/T. mentagrophytes species complex according to the studies by Heidemann et al. (2010) and Taghipour et al. (2019). In addition to examining the relationship between the genotype and resistance to TRB, the ITS genotype of complex isolates T. mentagrophytes/T. interdigitale/T. indotineae was determined.
Table 2.
Age and sex distribution of dermatophyte species isolated from clinical specimens.
| Dermatophytes species | Sex (no.) | Age (no.) | Accession no. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| F | M | 0–10 | 11–20 | 21–30 | 31–40 | 41–50 | 51–60 | 61–70 | 71–80 | ||
| Trichophyton mentagrophytes (n = 6) | 4 | 2 | 1 | – | 2 | 1 | 1 | 1 | – | – | MZ983790- MZ983485- MZ994488- MZ994650- MZ994652- MZ994491 |
| Trichophyton interdigitale (n = 28) | 12 | 16 | 1 | – | 3 | 11 | 5 | 6 | 2 | – | OK 35221- OK 35220- OK 110565- OK 110585- OK 110591- OK 110574- OK 110568- OK 110566- OK 110586- OK 110567- OK 110580- OK 110572- OK 110578- OK 110576- OK 110577- OK 35231- OK 110583- OK 110570- OK 110569- OK 110589- OK 110571- OK 35266-o OK 110575- OK 110581- OK 110573- OK 110579- OK 110587-OK110582 |
| Trichophyton indotineae (n = 10) | 4 | 6 | – | 1 | 1 | 5 | 2 | 1 | – | – | OM366332 to OM366341 |
| Trichophyton rubrum (n = 15) | 3 | 12 | – | – | 2 | 2 | 7 | 1 | 2 | 1 | MT188699- MZ427314- MZ434885- MT188700- MT150739- MZ434887- MZ434886- MZ427316- MT191357- MT152325 |
| Trichophyton tonsurans (n = 16) | 3 | 13 | 4 | 6 | 3 | 1 | 2 | – | – | – | MT041242- MT041041- MT041256- MT066197- MT051844 |
| Trichophyton verrucosum (n = 7) | – | 7 | – | – | 2 | 2 | 2 | 1 | – | – | MT318679- MT318720 |
| Epidermophyton floccosum (n = 19) | 5 | 14 | 1 | – | 4 | 6 | 3 | 2 | 3 | – | MT040969- MT040750- MT150728- MT040755- MZ363671- MT040763- MZ363673- MZ363722- MZ363721- MT040762- MZ363674 |
| Microsporum canis (n = 14) | 9 | 5 | 4 | 2 | 3 | 3 | – | 2 | – | – | MT129526- MT067649- MT183698- Mz363857- MT136105- MT129500 |
| Nannizzia gypsea (n = 6) | 3 | 3 | 2 | 2 | 1 | – | – | 1 | – | – | MT318651- MZ434959- MZ435310- MT394865 |
| Nannizzia fulva (n = 2) | 2 | – | 1 | 1 | – | – | – | – | – | – | – |
Then, the sequences were analyzed by RAxML version 8.2 (Stamatakis, 2014) running on CIPRES Science Gateway (Miller et al., 2010). Optimization in RAxML was carried out using the GTRCAT option. Bootstrap values for maximum likelihood were 1,000 replicates with one search replicate per bootstrap replicate and Fusarium solani as the outgroup.
2.3 Antifungal Susceptibility Testing
2.3.1 Chemical Antifungal Drugs
The drug susceptibility test was performed through minimum inhibitory concentration (MIC) microdilution broth. The drugs related to the MIC test were prepared according to the M38-A2 protocol for filamentous fungi (Wayne, 2008).
2.3.2 Drug Susceptibility Testing Using Microdilution Broth
The broth microdilution was used following M38-A2 CLSI protocol to examine and assess MIC in all strains (Wayne, 2008). According to the CLSI standard, drug stocks were prepared in dimethyl sulfoxide (DMSO). Different concentrations (100 µl) were poured into 96-well round-bottom microplates from the lowest concentration to the highest concentration. According to the CLSI standard, the range of antifungals was as follows: 0.001–32 μg/ml for TRB; 0.01–16 μg/ml for ITZ, KTZ, VCZ, PCZ, and AMB; and 0.06–64 μg/ml for FLZ. Then the prepared suspensions (100 µl) of each strain containing 1-3 × 103 ml/CFU spore were added to the wells. The plates were incubated at 35°C and visually assessed for fungal growth after 96 h. The MIC range, geometric mean, MIC50, and MIC90 were calculated for all the isolates tested.
2.4 PCR Assay Targeting the SQLE Region
To investigate mutations in SQLE gene, the strains with less susceptibility to TRB were evaluated with the primers Drsq1 (5′-TTGCCAACGGGGTGTAAAG-3′) and Drsq2 (5′-GGGGCCATCTATAATTCAGACTC-3′) (Osborne et al., 2006). The primer for replacing amino acids in Leu393Phe, Leu393Ser, Phe397Leu, and Gln408Leu in SQLE was used. The length of the fragments for Trichophyton was 500 bp, while it was 520 bp for Epidermophyton and Nannizzia. According to CLSI, T. rubrum strains with MIC > 0.5 µg/ml and other strains with MIC > 0.25 µg/ml were selected as the strains with less sensitivity to TRB.
The PCRs were prepared with the final volume of 50 μl containing 25 μl of premix (Ampliqon, Denmark), 3 μl of DNA template, 0.6 μM of forward and reverse primers, and distilled water. The PCR cycling conditions were as follows: 5 min initial pre-incubation at 95°C, followed by 35 cycles consisting of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 45 s, with a final extension at 72°C for 5 min (Salehi et al., 2018b). The PCR products of the SQLE region were sequenced by the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit. The forward and reverse sequences of isolates showing reduced susceptibility to TRB were subjected to ClustalW pairwise alignment using the MEGA10 software and edited manually to improve the alignment accuracy.
2.5 Statistical Analysis
The quantitative data from MIC, the geometric mean, MIC50, and MIC90 were calculated using the SPSS statistical package.
3 Results
3.1 Characteristics of the Studied Dermatophyte Isolates
Clinical data of 123 patients including 82 men (66.66%) and 41 women (33.33%) are listed in Table 1. In addition, the age range of the patients was 2–80 years with a mean age of 41. The most common clinical manifestations were tinea cruris (34.95%) followed by tinea pedis (17.88%), tinea manuum (19.51%), and tinea capitis (16.26%). The dominant species in tinea cruris, tinea pedis, and tinea capitis were T. tonsurans, T. rubrum, and E. floccosum, respectively. In addition, the isolated species from tinea faciei only included T. tonsurans and T. verrucosum. It is significant that N. fulva was only isolated from tinea unguium and that T. verrucosum was only isolated from the male cases.
3.2 Molecular Identification by ITS Region
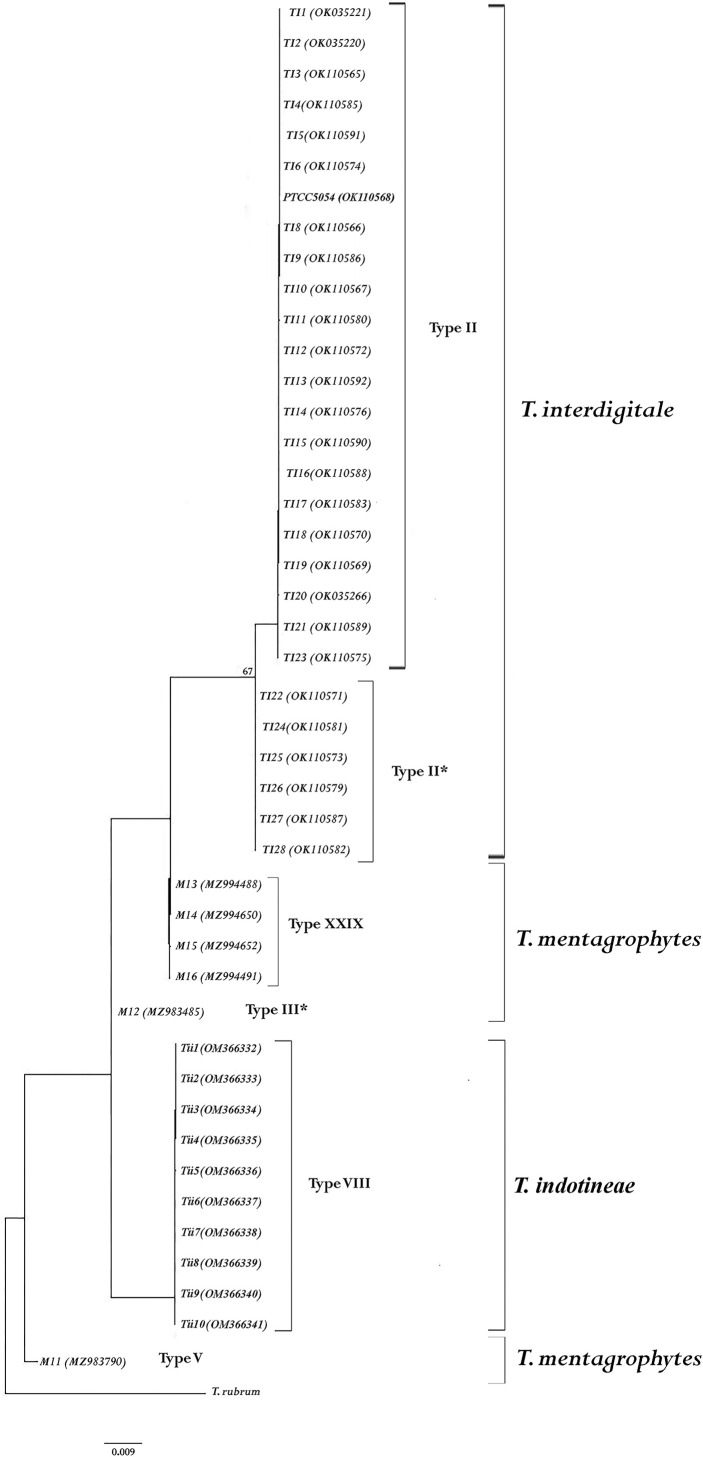
All the dermatophyte species were identified by determining the sequence of ITS-rDNA regions. The results of the molecular method indicated species T. mentagrophytes (6), T. interdigitale (28), T. indotineae (10), E. floccosum (19), T. tonsurans (16), T. rubrum (15), M. canis (14), T. verrucosum (7), N. gypsea (6), and N. fulva (2). After manually editing and blast analysis, the sequences were deposited in GenBank (Table 2). In addition, ITS genotype examination results showed that out of 6 T. mentagrophytes isolates, there was one isolate in each of the V and III* genotypes. T. indotineae was located between genotypes V and III* of T. mentagrophytes. Also, a new genotype was found in four isolates of T. mentagrophytes (XXIX). T. interdigitale isolates were only seen in two genotypes II (n = 22) and II* (n = 6) (Figure 1).
Figure 1.
The genotyping tree was constructed based on ITS region sequences by RAxML analysis from Trichophyton mentagrophytes/Trichophyton interdigitale complex, and the accession numbers are given in parentheses. Trichophyton rubrum was used as an outgroup to root the dendrogram. The bootstrap value greater than 65% is shown above the branches.
The phylogenetic tree was drawn through RAxML analysis for all species (Figure S1). The tree illustrates the phylogenetic relationships between all isolates under study. Based on phylogenetic analysis, all isolates were placed in six clades (T. mentagrophytes, T. interdigitale, and T. indotineae were in separate clusters). In addition, N. gypsea and N. fulva were observed in one clade with high support. In addition, the M. canis clade was in the base position compared to other species (Figure S1). Supports of bootstrap higher than 85% were represented in RAxML analysis. In addition, the phylogenetic tree did not demonstrate a relationship between the isolates that showed a decrease in sensitivity to TRB.
3.3 Antifungal Susceptibility Testing
MIC range, geometric mean MIC, MIC50, and MIC90 were computed for all dermatophyte strains (Table 2). In general, among all strains, the highest antifungal effects were observed with TRB followed by ITZ, VCZ, KTZ, FLZ, AMB, and PCZ. The most sensitive strains to TRB were T. rubrum, T. tonsurans, T. interdigitale, M. canis, T. verrucosum, N. gypsea, N. fulva, T. mentagrophytes, E. floccosum, and T. indotineae in a descending order. In addition, the results showed that M. canis (MIC50 = 0.12 μg/ml) had the highest MIC and T. indotineae (MIC50 = 1 μg/ml) had the lowest MIC against ITZ. In addition, KTZ had a low suppressive effect only against E. floccosum (G mean = 1.07 μg/ml). Also, among T. mentagrophytes, T. interdigitale, and T. indotineae species, the lowest and highest sensitivity to seven antifungal drugs tested was observed in T. indotineae and T. interdigitale, respectively. Furthermore, TRB had the highest antifungal effects against T. indotineae.
Moreover, VCZ had a high suppressive effect on N. gypsea species. The FLZ had a lower suppressive effect on T. rubrum and T. mentagrophytes species compared to other species. In general, the lowest suppressive effect on all dermatophyte species was observed with PCZ (Table 3).
Table 3.
Antifungal susceptibility profile of 123 dermatophyte strains to seven antifungal agents by broth microdilution method.
| Dermatophyte species | Antifungal drugs | MIC range (µg/ml) | MIC50/MIC90 (µg/ml) | G mean (µg/ml) | MIC (µg/ml) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 64 | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.125 | 0.06 | 0.03 | 0.015 | 0.007 | 0.003 | 0.001 | |||||
| Trichophyton mentagrophytes (n = 6) | TRB | 0.015–16 | 0.09/– | 0.13 | 1 | 2 | 1 | 1 | 1 | |||||||||||
| ITZ | 0.125–8 | 0.75/– | 0.70 | 1 | 2 | 1 | 1 | 1 | ||||||||||||
| KTZ | 0.06–8 | 1.25/– | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||
| FLZ | 0.125–16 | 1.25/– | 1.2 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||
| VCZ | 0.125–8 | 0.3/– | 0.5 | 1 | 1 | 1 | 1 | 2 | ||||||||||||
| PCZ | 0.125–16 | 1.25/– | 1.09 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||
| AMB | 0.125–8 | 0.75/– | 0.79 | 1 | 2 | 1 | 1 | 1 | ||||||||||||
| Trichophyton interdigitale (n = 28) | TRB | 0.003–0.025 | 0.06/0.125 | 0.04 | 7 | 8 | 6 | 4 | 2 | 1 | ||||||||||
| ITZ | 0.03–16 | 0.25/8.8 | 0.38 | 2 | 1 | 2 | 2 | 1 | 4 | 4 | 9 | 2 | 1 | |||||||
| KTZ | 0.03–16 | 0.5/8.8 | 0.37 | 2 | 1 | 2 | 1 | 3 | 6 | 2 | 7 | 2 | 2 | |||||||
| FLZ | 0.125–64 | 0.25/35.2 | 0.78 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 3 | 4 | 10 | |||||||
| VCZ | 0.125–16 | 0.37/16 | 0.70 | 4 | 3 | 1 | 2 | 4 | 6 | 8 | ||||||||||
| PCZ | 1–16 | 1.5/16 | 1.55 | 6 | 3 | 3 | 2 | 4 | 3 | 2 | 4 | 1 | ||||||||
| AMB | 0.125–8 | 0.25/8 | 0.56 | 5 | 2 | 3 | 3 | 6 | 9 | |||||||||||
| Trichophyton indotineae (n = 10) | TRB | 0.015–32 | 0.125/30.4 | 0.24 | 1 | 1 | 5 | 1 | 1 | 1 | ||||||||||
| ITZ | 0.06–16 | 1/15.2 | 0.9 | 1 | 1 | 1 | 2 | 1 | 3 | 1 | ||||||||||
| KTZ | 0.125–16 | 0.5/16 | 0.79 | 2 | 2 | 2 | 2 | 2 | ||||||||||||
| FLZ | 0.125–16 | 0.75/14.8 | 0.93 | 1 | 3 | 1 | 1 | 2 | 2 | |||||||||||
| VCZ | 0.125–8 | 0.37/7.4 | 0.53 | 1 | 1 | 2 | 1 | 3 | 2 | |||||||||||
| PCZ | 0.125–16 | 0.75/16 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 2 | ||||||||||
| AMB | 0.125–16 | 0.5/15.2 | 0.81 | 1 | 1 | 1 | 1 | 2 | 3 | 1 | ||||||||||
| Trichophyton rubrum (n = 15) | TRB | 0.003–0.006 | 0.015/0.06 | 0.01 | 3 | 2 | 3 | 2 | 5 | |||||||||||
| ITZ | 0.06–16 | 0.25/14 | 0.30 | 1 | 1 | 1 | 2 | 3 | 2 | 5 | ||||||||||
| KTZ | 0.125–16 | 0.25/16 | 0.75 | 2 | 2 | 1 | 2 | 4 | 4 | |||||||||||
| FLZ | 0.125–16 | 0.5/16 | 1.04 | 3 | 1 | 1 | – | 2 | 2 | 4 | 2 | |||||||||
| VCZ | 0.06–8 | 0.25/5.6 | 0.39 | 1 | 1 | 1 | 1 | 3 | 2 | 5 | 1 | |||||||||
| PCZ | 0.25–16 | 1/16 | 1.58 | 3 | 1 | 1 | 1 | 3 | 4 | 2 | ||||||||||
| AMB | 0.125–8 | 0.5/8 | 0.79 | 3 | 1 | 1 | 2 | 2 | 3 | 3 | ||||||||||
| Trichophyton tonsurans (n = 16) | TRB | 0.003–0.125 | 0.09/0.125 | 0.07 | 8 | 5 | 3 | |||||||||||||
| ITZ | 0.06–8 | 0.37/8 | 0.51 | 2 | 2 | 1 | 1 | 2 | 2 | 4 | 2 | |||||||||
| KTZ | 0.125–16 | 0.5/10.4 | 0.64 | 1 | 2 | 2 | 4 | 5 | 2 | |||||||||||
| FLZ | 0.125–32 | 0.5/20.8 | 0.67 | 1 | 1 | 1 | 2 | 5 | 4 | 2 | ||||||||||
| VCZ | 0.06–16 | 0.25/16 | 0.49 | 3 | 1 | 2 | 3 | 5 | 2 | |||||||||||
| PCZ | 0.125–16 | 0.75/16 | 1.24 | 4 | 2 | 2 | 2 | 3 | 3 | |||||||||||
| AMB | 0.06–16 | 0.25/16 | 0.56 | 3 | 2 | 2 | 3 | 2 | 3 | 1 | ||||||||||
| Trichophyton verrucosum (n = 7) | TRB | 0.06–4 | 0.125/– | 0.16 | 1 | 4 | 2 | |||||||||||||
| ITZ | 0.06–1 | 0.25/– | 0.30 | 2 | 1 | 2 | 1 | 1 | ||||||||||||
| KTZ | 0.125–1 | 0.5/– | 0.37 | 2 | 2 | 1 | 2 | |||||||||||||
| FLZ | 0.125–16 | 0.5/– | 0.90 | 1 | 1 | 1 | 1 | 1 | 2 | |||||||||||
| VCZ | 0.03–16 | 0.5/– | 0.40 | 1 | 1 | 2 | 1 | 1 | 1 | |||||||||||
| PCZ | 0.125–16 | 0.5/– | 1.21 | 2 | 1 | 1 | 2 | 1 | ||||||||||||
| AMB | 0.06–2 | 0.25/– | 0.30 | 1 | 1 | 1 | 1 | 2 | 1 | |||||||||||
| Epidermophyton floccosum (n = 19) | TRB | 0.06–16 | 0.125/0.25 | 0.17 | 1 | 6 | 9 | 3 | ||||||||||||
| ITZ | 0.06–16 | 0.5/16 | 0.80 | 3 | 2 | 1 | 2 | 2 | 5 | 3 | 1 | |||||||||
| KTZ | 0.125–16 | 0.5/16 | 1.07 | 4 | 2 | 1 | 1 | 1 | 3 | 3 | 5 | |||||||||
| FLZ | 0.125–32 | 0.37/32 | 0.77 | 2 | 2 | 1 | 1 | 3 | 7 | 3 | ||||||||||
| VCZ | 0.03–8 | 0.5/8 | 0.80 | 5 | 1 | 2 | 3 | 3 | 4 | 1 | ||||||||||
| PCZ | 0.125–16 | 0.5/16 | 1 | 3 | 2 | 1 | 1 | 2 | 3 | 4 | 3 | |||||||||
| AMB | 0.06–16 | 0.25/16 | 0.57 | 3 | 2 | 4 | 3 | 5 | 2 | |||||||||||
| Microsporum canis (n = 14) | TRB | 0.03–0.125 | 0.09/0.125 | 0.07 | 7 | 5 | 2 | |||||||||||||
| ITZ | 0.03–16 | 0.125/8.5 | 0.19 | 1 | 1 | 1 | 3 | 3 | 4 | 1 | ||||||||||
| KTZ | 0.125–16 | 0.5/20.5 | 0.93 | 2 | 1 | 1 | 3 | 3 | 4 | |||||||||||
| FLZ | 0.125–32 | 0.37/20 | 0.67 | 1 | 2 | 1 | 1 | 2 | 3 | 4 | ||||||||||
| VCZ | 0.125–16 | 0.5/16 | 0.86 | 2 | 1 | 1 | 1 | 3 | 5 | 1 | ||||||||||
| PCZ | 0.125–16 | 1.5/16 | 1.72 | 4 | 1 | 1 | 1 | 1 | 2 | 3 | 1 | |||||||||
| AMB | 0.25–16 | 1/16 | 1.28 | 3 | 1 | 1 | 3 | 2 | 4 | |||||||||||
| Nannizzia gypsea (n = 6) | TRB | 0.015–16 | 0.09/– | 0.13 | 1 | 2 | 1 | 1 | 1 | |||||||||||
| ITZ | 0.125–8 | 0.37/– | 0.70 | 2 | 1 | 1 | 2 | |||||||||||||
| KTZ | 0.125–16 | 0.37/– | 0.89 | 1 | 1 | 1 | 2 | 1 | ||||||||||||
| FLZ | 0.125–16 | 0.62/– | 0.62 | 1 | 1 | 1 | 2 | 1 | ||||||||||||
| VCZ | 0.125–16 | 0.18/– | 0.35 | 1 | 2 | 3 | ||||||||||||||
| PCZ | 0.25–16 | 1.25/– | 1.58 | 3 | 1 | 2 | ||||||||||||||
| AMB | 0.125–8 | 0.37/– | 0.70 | 1 | 1 | 1 | 2 | 1 | ||||||||||||
| Nannizzia fulva (n = 2) | TRB | 0.125–0.25 | –/– | – | 1 | 1 | ||||||||||||||
| ITZ | 0.25–0.5 | –/– | – | 1 | 1 | |||||||||||||||
| KTZ | 0.125–2 | –/– | – | 1 | 1 | |||||||||||||||
| FLZ | 0.25–1 | –/– | – | 1 | 1 | |||||||||||||||
| VCZ | 0.25–0.5 | –/– | – | 1 | 1 | |||||||||||||||
| PCZ | 0.5–1 | –/– | – | 1 | 1 | |||||||||||||||
| AMB | 0.5–1 | –/– | – | 1 | 1 | |||||||||||||||
MIC, minimum inhibitory concentration; TRB, terbinafine; ITZ, itraconazole; KTZ, ketoconazole; FLZ, fluconazole; VCZ, voriconazole; PCZ, posaconazole; AMB, amphotericin B.
A comparison of MIC50 in II and II* genotypes of T. interdigitale showed that genotype II had a higher MIC50 with all drugs. In addition, isolates of XXIX genotype of T. mentagrophytes had a higher MIC50 as compared to T. indotineae.
In general, a comparison of Trichophyton, Epidermophyton, Microsporum, and Nannizzia genera showed that TRB and ITZ had the highest effect as compared to other drugs. Trichophyton genus had the lowest G mean against all drugs followed by Nannizzia. On the other hand, for all the drugs under study, Epidermophyton had the highest MIC50.
In Nannizzia genus, TRB and VCZ had a strong effect, KTZ had a weak effect, and AMB was inefficient (Table 3).
Among anthropophilic, geophilic, and zoophilic species, anthropophilic species had a high sensitivity to all drugs. In addition, the lowest and highest G mean in the anthropophilic group was observed in T. rubrum and E. floccosum isolates, respectively. On the other hand, AMB and PCZ had a low inhibitory effect against E. floccosum in the anthropophilic group. In general, geophilic and zoophilic species compared to anthropophilic species had a lower sensitivity to TRB and ITZ, respectively. Moreover, among zoophilic and geophilic species, M. canis and N. gypsea had a high level of sensitivity to TRB, respectively. Overall, azoles had the highest inhibitory effect against T. tonsurans species and the lowest inhibitory effect against T. indotineae.
Among all isolates, T. indotineae (n = 2), T. mentagrophytes (n = 1), E. floccosum (n = 1), Trichophyton verrucosum (n = 1), and N. gypsea (n = 1) demonstrated a decrease in sensitivity to TRB (MIC > 32 µg/ml). In addition, among all strains, two isolates (1.62%) were resistant to all drugs under study. The highest cross-resistance was observed between FLZ and ITZ (16.26%), and cross-resistance among azole antifungals was observed in 11 isolates (8.94%).
3.4 Point Mutation in SQLE Gene (Terbinafine-Resistant Strains)
To examine mutation in SQLE gene, all the six strains with a lower sensitivity to TRB were reproduced using Drsq primers and sequenced. The band length of the PCR product for Trichophyton was 500 bp, while it was 520 bp for Epidermophyton and Nannizzia. Forward and reverse sequences were edited using MEGA10 for each isolate, and then, the sequences were compared with sensitive strain sequences in GenBank using BLASTn. Afterward, the nucleic acid sequence was converted into an amino acid sequence. Among the six strains with a lower sensitivity to TRB, a point mutation was seen only in one strain of T. indotineae with MIC > 32, so Phe397Leu replacement of SQLE protein was observed. Replacement of C with A in SQLE gene leads to the replacement of Phe with Leu (Figure 2). The DNA sequence of the TRB-resistant isolate was recorded with accession no. OM373652 of T. indotineae in GenBank.
Figure 2.
Comparison of the amino acid sequence of squalene epoxidase in terbinafine-sensitive strain and terbinafine-resistant strain.
4 Discussion
In the light of continuing emergence of resistant dermatophytes to antifungal drugs around the world, monitoring drug resistance is essential (Siopi et al., 2021). In general, the identification of dermatophytes at the species level is a major issue for treating patients (Falahati et al., 2018). In this study, 123 clinical isolates of dermatophyte and five standard strains were examined. All the strains were identified by determining the ITS region sequence, and then the sensitivity pattern for seven antifungal drugs was determined using antifungal susceptibility testing (AFST). As the results showed, the number of men was twice the number of women, and tinea cruris was the dominant form. Other studies have reported tinea pedis and tinea capitis as the prevalent forms of dermatophytosis (Zareshahrabadi et al., 2020).
The current new taxonomy of dermatophytes separates T. mentagrophytes from its clonal offshoot T. interdigitale (Lipner and Scher, 2019). In fact, this classification is highly important from a clinical viewpoint, as T. interdigitale is only anthropophilic, and it is usually isolated from non-inflammatory cases such as tinea unguium and tinea pedis. In contrast, T. indotineae is mostly zoophilic and a cause of inflammatory symptoms, which mostly cause tinea corporis and tinea cruris. This is a key factor in selecting the right treatment (Taghipour et al., 2019; Siopi et al., 2021). In this study, T. interdigitale was the main causative agent of tinea pedis, while E. floccosum was responsible for most cases of tinea cruris. Siopi et al. reported that T. interdigitale species was mostly isolated from tinea pedis, which is similar to our results (Siopi et al., 2021).
In this study, the highest frequency of ITS genotypes of T. interdigitale and T. mentagrophytes belonged to II and VIII genotypes, respectively. Taghipour et al. showed that the VIII genotype was the most common in T. mentagrophytes species (Taghipour et al., 2019). In addition, a new XXIX genotype was found in T. mentagrophytes in this study. Dabas et al. showed that out of 123 dermatophyte isolates, 56% were T. interdigitale, which had been first mistakenly identified as T. mentagrophytes. Through determining the sequence of ITS regions, these two species were differentiated (Dabas et al., 2017), which is in accordance with our findings.
The phylogenetic tree using RAxML analysis showed that the sequence of ITS region can effectively differentiate dermatophyte species. Despite that Baert et al. failed to differentiate Nannizzia and Epidermophyton genera using the sequence of the ITS and BT2 regions, the results showed that the sequence of ITS regions can differentiate these two genera (Baert et al., 2020). Our results are consistent with Gräser et al. (1999).
According to the therapeutic protocols, dermatophytosis TRB is the first choice as a systemic treatment. Also, the latest studies showed the number of resistant cases to TRB is increasing (Dogra et al., 2017). SQLE gene mutation changes the protein structure and interrupts the drug attachment to the target enzyme (Shankarnarayan et al., 2020; Shaw et al., 2020; Kong et al., 2021). Here, six strains showed a low sensitivity to TRB, and among them, only one isolate (T. mentagrophytes) had a mutation on the Phe397Leu amino acid position. The cause of resistance in other strains might be replacement in other areas of SQLE gene or other intervening mechanisms of resistance.
According to Hiruma et al., the highest TRB resistance was observed in T. rubrum with L393F replacement in SQLE gene (Hiruma et al., 2021). Salehi et al. examined mutation in SQLE gene and reported replacement in Phe397Leu amino acid in T. tonsurans and T. rubrum species (Salehi et al., 2018b). In addition, Rezaei-Matehkolaei et al. showed SQLE gene mutation in five strains of TRB-resistant T. interdigitale and T. mentagrophytes (Rezaei-Matehkolaei et al., 2013). Singh et al. reported a replacement in Phe397Leu and Leu393Phe positions in TRB-resistant T. interdigitale strains (Singh et al., 2018). On the other hand, Yamada et al. found the replacement of amino acid in Phe397Leu position of SQLE gene in TRB-resistant T. rubrum species (Yamada et al., 2017). A similar examination was conducted by Lagowski et al. on resistant strains of T. mentagrophytes, and four strains with a mutation at the Leu393Phe region were reported (Lagowski et al., 2020).
In a study by Taghipour et al. of 45 strains of T. mentagrophytes, 5 strains (11.11%) were TRB resistant and had substitutions in Ala448Thr, Leu393Ser, and Phe397Leu positions in SQLE protein (Dabas et al., 2017). All the TRB-resistant isolates of T. mentagrophytes in other studies belonged to the VIII type of ITS genotype. In our study, out of 16 T. mentagrophytes, 3 strains were less sensitive to TRB, of which only 1 strain had a mutation in SQLE gene. This finding can be explained by the low number of T. mentagrophytes isolates or the low number of the VIII type isolates (n = 10). Also, in this study, contrary to the report by Heidemann et al. (2010), no significant correlation was seen between ITS genotype and clinical data, which is in line with the results of Salehi et al. It seems that conclusions about these data require further studies and samples.
In addition to TRB, AFST was carried out on six antifungal drugs. In general, AFST results of all isolates showed that, despite the resistant strains, TRB still is the most efficient drug against dermatophyte species. In addition, the results indicated that T. indotineae and E. floccosum had the lowest sensitivity to TRB. This finding is consistent with Lagowski et al. (2020), Rezaei-Matehkolaei et al. (2013), Rudramurthy et al. (2018), and Taghipour et al. (2019). Furthermore, in studies by Pourpak et al. (Pourpak and Firooz, 2021) and Kano et al. (2020), low sensitivity against TRB was reported in T. indotineae. Moreover, Taghipour et al. (2019) and Salehi et al. (2018b) showed that T. interdigitale species were more sensitive to TRB than T. mentagrophytes and T. indotineae, which is also consistent with our study.
In addition to TRB, ITZ is also prescribed for systemic treatment of dermatophytosis (Siopi et al., 2021). Here, ITZ had the highest and lowest effects on M. canis (G mean = 0.19 μg/ml) and T. indotineae (G mean = 0.9 μg/ml), respectively. On the other hand, FLZ demonstrated high levels of MIC, which is also consistent with Curatolo et al. (2020) and Barros et al. (2006). Curatolo et al. (2020) showed that TRB and ITZ had a good suppressive effect, which is consistent with the findings of the present study.
Aneke et al. (2018) reported that TRB and VCZ had the highest resistance effect on dermatophyte species and M. canis species in particular. This is not consistent with our finding of VCZ. It has been shown that in T. mentagrophytes/T. interdigitale complex, genotype XXIX of T. mentagrophytes and genotype II of T. interdigitale had a higher MIC50 with all drugs under study. This finding indicates that specific genotypes of this complex species had a higher MIC than others. Further examination of this topic can lead to the identification of more important species.
Among all tested isolates in our study, three strains were resistant to all drugs. In five strains (4.06%) out of all species, cross-resistance between TRB and other azoles drugs was observed, which is expected given that both groups of drugs suppress SQLE and cytochrome on the ergosterol biosynthesis path. It is suggested that in addition to major point mutations in SQLE gene (L393S, L393F, F397L, and Q408L), which were examined in the present study, other less frequent mutations including F415S, H440Y, and A448T are further investigated. Likewise, selecting more isolates from less frequent dermatophyte species of different geographic regions will help to provide more accurate data about antifungal susceptibility and genotype variations within the population.
5 Conclusion
With respect to the increasing prevalence of dermatophytosis and the growing numbers of antifungal drug resistance in dermatophyte species, precise identification of the etiologic species by molecular methods is believed to be crucial to achieve more effective treatments. In addition, epidemiological changes and lack of drug susceptibility testing have led to the failure of treatment and subsequent recurrence of the infection. Determining genotypes can improve our epidemiological information because specific genotypes have higher and different AFST and MIC results, which can help us to have a better diagnosis and treatment. Apparently, determining choice-effective drugs and drug-resistant strains through AFST can bring us closer to more efficient therapeutic goals. Given that TRB is the frontline defense against dermatophytosis, the growing resistance to TRB is a considerable challenge. Altogether, our results showed that precise identification of etiologic dermatophyte species and prescribing antifungal drugs with more caution can prevent resistance in strains, effectively reducing frequently recurrent infections, and prevent the distribution of the infection within the population.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by IR.PII.REC.1397.021. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
NP performed the experiments, contributed to the analysis of the data, and drafted the manuscript. MS-G, ACN, MA and ZS assisted in the interpretation of the molecular data, data analysis and editing the manuscript. MR-A supervised the study, participated in the study design and data analysis, and approved the final draft. All authors have read and approved the final version of the manuscript.
Funding
This work was supported financially by the Pasteur Institute of Iran.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors gratefully thank the personnel of the Mycology Department of the Pasteur Institute of Iran for helpful assistance in isolate preparation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.851769/full#supplementary-material
References
- Afshari M., Shams-Ghahfarokhi M., Razzaghi-Abyaneh M. (2016). Antifungal Susceptibility and Virulence Factors of Clinically Isolated Dermatophytes in Tehran, Iran. Iran J. Microbiol. 8 (1), 36–46. [PMC free article] [PubMed] [Google Scholar]
- Aneke C. I., Otranto D., Cafarchia C. (2018). Therapy and Antifungal Susceptibility Profile of Microsporum canis . J. Fungi 4 (3). doi: 10.3390/jof4030107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baert F., De Hoog G. S., Packeu A., Hendrickx M. (2020). Updating the Taxonomy of Dermatophytes of the BCCM/IHEM Collection According to the New Standard: A Phylogenetic Approach. Mycopathologia 185 (1), 161–168. doi: 10.1007/s11046-019-00338-7 [DOI] [PubMed] [Google Scholar]
- Barros M. E. S., Santos D. D. A., Hamdan J. S. (2006). In Vitro Methods for Antifungal Susceptibility Testing of Trichophyton spp. Mycol Res. 110 (11), 1355–1360. doi: 10.1016/j.mycres.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Bhatia V. K., Sharma P. C. (2015). Determination of Minimum Inhibitory Concentrations of Itraconazole, Terbinafine and Ketoconazole Against Dermatophyte Species by Broth Microdilution Method. Indian J. Med. Microbiol. 33, 533–537. doi: 10.4103/0255-0857.167341 [DOI] [PubMed] [Google Scholar]
- Curatolo R., Juricevic N., Leong C. H., Bosshard P. P. (2020). Antifungal Susceptibility Testing of Dermatophytes: Development and Evaluation of an Optimized Broth Microdilution Method. Mycoses 64 (3), 282–291. doi: 10.1111/myc.13202 [DOI] [PubMed] [Google Scholar]
- Dabas Y., Xess I., Singh G., Pandey M., Meena S. (2017). Molecular Identification and Antifungal Susceptibility Patterns of Clinical Dermatophytes Following CLSI and EUCAST Guidelines. J. Fungi 3 (2), 17. doi: 10.3390/jof3020017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoog G. S., Dukik K., Monod M., Packeu A., Stubbe D., Hendrickx M., et al. (2017). Toward a Novel Multilocus Phylogenetic Taxonomy for the Dermatophytes. Mycopathologia 182, 5–31. doi: 10.1007/s11046-016-0073-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra S., Kaul S., Yadav S. (2017). Treatment of Dermatophytosis in Elderly, Children, and Pregnant Women. Indian Dermatol. Online J. 8 (5), 310–318. doi: 10.4103/idoj.IDOJ-169-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A., Monod M., Salamin K., Burmester A., Uhrlaß S., Wiegand C., et al. (2020). Alarming India-Wide Phenomenon of Antifungal Resistance in Dermatophytes: A Multicentre Study. Mycoses 63 (7), 717–728. doi: 10.1111/myc.13091 [DOI] [PubMed] [Google Scholar]
- Falahati M., Fateh R., Nasiri A., Zaini F., Fattahi A., Farahyar S. H. (2018). Specific Identification and Antifungal Susceptibility Pattern of Clinically Important Dermatophyte Species Isolated From Patients With Dermatophytosis in Tehran, Iran. Arch. Clin. Infect. Dis. 13 (3), e63104. doi: 10.5812/archcid.63104 [DOI] [Google Scholar]
- Ghannoum M., Chaturvedi V., Espinel-Ingroff A., Pfaller M., Rinaldi M., Lee-Yang W., et al. (2004). Intra and Inter Laboratory Study of a Method for Testing the Antifungal Susceptibilities of Dermatophytes. J. Clin. Microbiol. 42, 2977–2979. doi: 10.1128/JCM.42.7.2977-2979.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräser Y., El Fari M., Vilgalys R., Kuijpers A. F. A., De Hoog G. S., Presber W., et al. (1999). Phylogeny and Taxonomy of the Family Arthrodermataceae (Dermatophytes) Using Sequence Analysis of the Ribosomal ITS Region. Med. Mycol. 37 (2), 105–114. doi: 10.1080/02681219980000171 [DOI] [PubMed] [Google Scholar]
- Haugh M., Helou S., Boissel J. P., Cribier B. J. (2000). Terbinafine in Fungal Infections of the Nails: A Meta-Analysis of Randomized Clinical Trials. Br. J. Dermatol. 147 (1), 118–121. doi: 10.1046/j.1365-2133.2002.04825.x [DOI] [PubMed] [Google Scholar]
- Heidemann S., Monod M., Graser Y. (2010). Signature Polymorphisms in the Internal Transcribed Spacer Region Relevant for the Differentiation of Zoophilic and Anthropophilic Strains of Trichophyton nterdigitale and Other Species of T. mentagrophytes Sensu Lato. Br. J. Dermatol. 162 (2), 282–295. doi: 10.1111/j.1365-2133.2009.09494.x [DOI] [PubMed] [Google Scholar]
- Hiruma J., Noguchi H., Hase M., Tokuhisa Y., Shimizu T., Ogawa T., et al. (2021). Epidemiological Study of Terbinafine-Resistant Dermatophytes Isolated From Japanese Patients. J. Dermatol. 48 (4), 564–567. doi: 10.1111/1346-8138.15745 [DOI] [PubMed] [Google Scholar]
- Kano R., Kimura U., Kakurai M., Hiruma J., Kamata H., Suga Y., et al. (2020). Trichophyton indotineae sp. Nov.: A New Highly Terbinafine-Resistant Anthropophilic Dermatophyte Species. Mycopathologia 185, 947–958. doi: 10.1007/s11046-020-00455-8 [DOI] [PubMed] [Google Scholar]
- Kong X., Tang C. H., Singh A., Ahmed S. A., Al-Hatmi A. M. S., Chowdhary A., et al. (2021). Antifungal Susceptibility and Mutations in the Squalene Epoxidase Gene in Dermatophytes of the Trichophyton mentagrophytes Species Complex. Antimicrob. Agents Chemother. 16 65 (8), e0005621. doi: 10.1128/AAC.00056-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagowski D., Gnat S., Nowakiewicz A., Osinska M., Dyląg M. (2020). Intrinsic Resistance to Terbinafine Among Human and Animal Isolates of Trichophyton m entagrophytes Related to Amino Acid Substitution in the Squalene Epoxidase. Infection 48 (6), 889–897. doi: 10.1007/s15010-020-01498-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipner S. R., Scher R. K. (2019). Onychomycosis: Treatment and Prevention of Recurrence. J. Am. Acad. Dermatol. 80 (4), 853–867. doi: 10.1016/j.jaad.2018.05.1260 [DOI] [PubMed] [Google Scholar]
- Liu W., May G. S., Lionakis M. S., Levis R. E., Kontoyiannis D. P. (2004). Extra Copies of the Aspergillus fumigatus Squalene Epoxidase Gene Confer Resistance to Terbinafine: Genetic Approach to Studying Gene Dose-Dependent Resistance to Antifungals in A. fumigatus . Antimicrob. Agent Chemother. 48 (7), 2490–2496. doi: 10.1128/AAC.48.7.2490-2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2010). Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. Gateway Computing Environments Workshop (GCE), 1–8. doi: 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Mukherjee P., Leidich S., Isham N., Leitner I., Ryder N., Ghannoum M. (2003). Clinical Trichophyton Rubrum Strain Exhibiting Primary Resistance to Terbinafine. Antimicrob. Agents Chemother. 47 (1), 82–86. doi: 10.1128/AAC.47.1.82-86.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenoff P., Verma S. B., Ebert A., Süß A., Fischer E., Auerswald E., et al. (2020). Spread of Terbinafine-Resistant Trichophyton mentagrophytes Type VIII (India) in Germany–”The Tip of the Iceberg. J. Fungi 6 (4), 207. doi: 10.3390/jof6040207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi M., Firth N. A., Cannon R. (2010). Antifungal Drug Resistance of Oral Fungi. Odontology 98 (1), 15–25. doi: 10.1007/s10266-009-0118-3 [DOI] [PubMed] [Google Scholar]
- Osborne C., Leitner I., Favre B., Ryder N. (2005). Amino Acid Substitution in Trichophyton rubrum Squalene Epoxidase Associated With Resistance to Terbinafine. Antimicrob. Agents Chemother. 49 (7), 2840–2844. doi: 10.1128/AAC.49.7.2840-2844.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne C., Leitner I., Hofbauer B., Fielding C., Favre B., Ryder N. (2006). Biological, Biochemical, and Molecular Characterization of a New Clinical Trichophyton rubrum Isolate Resistant to Terbinafine. Antimicrob. Agents Chemother. 50, 2234–2236. doi: 10.1128/AAC.01600-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourpak Z., Firooz A. (2021). Multidrug-Resistant Trichophyton mentagrophytes Genotype VIII in an Iranian Family With Generalized Dermatophytosis: Report of Four Cases and Review of Literature. Int. J. Dermatol. 60 (6), 686–692. doi: 10.1111/ijd.15226 [DOI] [PubMed] [Google Scholar]
- Rezaei-Matehkolaei A., Makimura K., De Hoog S., Shidfar M. R., Zaini F., Eshraghian M., et al. (2013). Molecular Epidemiology of Dermatophytosis in Tehran, Iran, a Clinical and Microbial Survey. Med. Mycol. 51 (2), 203–207. doi: 10.3109/13693786.2012.686124 [DOI] [PubMed] [Google Scholar]
- Rocha E. M. F., Almeida C. B., Martinez-Rossi N. M. (2002). Identification of Genes Involved in Terbinafine Resistance in Aspergillus nidulans . Lett. Appl. Microbiol. 35 (3), 228–232. doi: 10.1046/j.1472-765x.2002.01174.x [DOI] [PubMed] [Google Scholar]
- Rudramurthy S. H. M., Shankarnarayan S. H. A., Dogra S., Shaw D., Mushtaq K. H., Paul R. A., et al. (2018). Mutation in the Squalene Epoxidase Gene of Trichophyton Interdigitale and Trichophyton rubrum Associated With Allylamine Resistance. Antimicrob. Agents Chemother. 62 (5), e02522–e02517. doi: 10.1128/AAC.02522-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi Z., Shams-Ghahfarokhi M., Fattahi A., Ghazanfari M., Yazdanparast S. A. (2018. a). A Head-to-Head Comparison of Four Cryopreservation Protocols of Dermatophyte Species. Infect. Epidemiol 4 (3), 109–114. [Google Scholar]
- Salehi Z., Shams-Ghahfarokhi M., Razzaghi-Abyaneh M. (2021) Molecular Epidemiology, Genetic Diversity, and Antifungal Susceptibility of Major Pathogenic Dermatophytes Isolated From Human Dermatophytosis. Front. Microbiol. 4. doi: 10.3389/fmicb.2021.643509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi Z., Shams-Ghahfarokhi M., Razzaghi-Abyaneh M. (2018. b). Antifungal Drug Susceptibility Profile of Clinically Important Dermatophytes and Determination of Point Mutations in Terbinafine-Resistant Isolates. Eur. J. Clin. Microbiol. Infect. Dis. 37 (10), 1841–1846. doi: 10.1007/s10096-018-3317-4 [DOI] [PubMed] [Google Scholar]
- Salehi Z., Shams-Ghahfarokhi M., Razzaghi-Abyaneh M. (2020). Internal Transcribed Spacer rDNA and TEF-1α Gene Sequencing of Pathogenic Dermatophyte Species and Differentiation of Closely Related Species Using PCR-RFLP of the Topoisomerase II . Cell J. 22 (1), 85–91. doi: 10.22074/cellj.2020.6372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos H. L., Lang E. A. S., Fernando Segato F., Rossi A., Martinez-Rossi N. M. (2018). Terbinafine Resistance Conferred by Multiple Copies of the Salicylate 1-Monooxygenase Gene in Trichophyton rubrum . Med. Mycol. 56 (3), 378–381. doi: 10.1093/mmy/myx044 [DOI] [PubMed] [Google Scholar]
- Shankarnarayan S. H. A., Shaw D., Sharma A., Chakrabarti A., Dogra S., Kumaran M. S., et al. (2020). Rapid Detection of Terbinafine Resistance in Trichophyton Species by Amplified Refractory Mutation System Polymerase Chain Reaction. Sci. Rep. 10, 1297. doi: 10.1038/s41598-020-58187-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw D., Singh S., Dogra S., Jayaraman J., Bhat R., Panda S., et al. (2020). MIC and Upper Limit of Wild-Type Distribution for 13 Antifungal Agents Against a Trichophyton mentagrophytes- Trichophyton interdigitale Complex of Indian Origin. Antimicrob. Agents Chemother. 64 (4), e01964–e01919. doi: 10.1128/AAC.01964-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Masih A., Khurana A., Singh P., Gupta M., Hagen F., et al. (2018). High Terbinafine Resistance in Trichophyton interdigitale Isolates in Delhi, India Harbouring Mutations in the Squalene Epoxidase (SQLE) Gene. Mycoses 61 (7), 477–484. doi: 10.1111/myc.12772 [DOI] [PubMed] [Google Scholar]
- Siopi M., Efstathiou I., Theodoropoulos K., Pournaras S., Meletiadis J. (2021). Molecular Epidemiology and Antifungal Susceptibility of Trichophyton Isolates in Greece: Emergence of Terbinafine-Resistant Trichophyton mentagrophytes Type VIII Locally and Globally. J. Fungi 7 (6), 419. doi: 10.3390/jof7060419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 3, 1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghipour S., Pchelin I. M., Mahmoudabadi A. Z., Ansari S., Katiraee F., Rafiei A., et al. (2019). Trichophyton mentagrophytes and T. interdigitale Genotypes Are Associated With Particular Geographic Areas and Clinical Manifestations. Mycoses 62 (11), 1084–1091. doi: 10.1111/myc.12993 [DOI] [PubMed] [Google Scholar]
- Wayne P. A. (2008). “CLSI Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi. Approved Standard”, in CLSI Document M38–A2, 2nd ed (Wayne PA 19087 USA:Clinical and Laboratory Standards Institute; ). [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR protocols: a guide to methods and applications. In: PCR Protocols, pp. 315–22. New York: Academic Press, Inc., Harcourt Brace Jovanovich Publishers. doi: 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Yamada T., Maeda M., Alshahni M., Tanaka R., Yaguchi T., Bontems O., et al. (2017). Terbinafine Resistance of Trichophyton Clinical Isolates Caused by Specific Point Mutations in the Squalene Epoxidase Gene. Antimicrob. Agents Chemother. 61 (7), e00115–e00117. doi: 10.1128/AAC.00115-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkov A., Krüger D., Begerow D., et al. (2012). Basidiomycetous Yeasts From Boletales Fruiting Bodies and Their Interactions With the Mycoparasite Sepedonium chrysospermum and the Host Fungus Paxillus . Microb. Ecol. 63 (2), 295–303. doi: 10.1007/s00248-011-9923-7 [DOI] [PubMed] [Google Scholar]
- Zareshahrabadi Z., Totonchi A., Rezaei-Matehkolaei A., Ilkit M., Ghahartars M., Arastehfar A., et al. (2020). Molecular Identification and Antifungal Susceptibility Among Clinical Isolates of Dermatophytes in Shiraz, Iran (2017-2019). Mycoses 64 (4), 385–393. doi: 10.1111/myc.13226 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.