Abstract
Single-cell RNA sequencing (scRNA-seq) has greatly advanced our understanding of cellular heterogeneity by profiling individual cell transcriptomes. However, cell dissociation from the tissue structure causes a loss of spatial information, which hinders the identification of intercellular communication networks and global transcriptional patterns present in the tissue architecture. To overcome this limitation, novel transcriptomic platforms that preserve spatial information have been actively developed. Significant achievements in imaging technologies have enabled in situ targeted transcriptomic profiling in single cells at single-molecule resolution. In addition, technologies based on mRNA capture followed by sequencing have made possible profiling of the genome-wide transcriptome at the 55-100 μm resolution. Unfortunately, neither imaging-based technology nor capture-based method elucidates a complete picture of the spatial transcriptome in a tissue. Therefore, addressing specific biological questions requires balancing experimental throughput and spatial resolution, mandating the efforts to develop computational algorithms that are pivotal to circumvent technology-specific limitations. In this review, we focus on the current state-of-the-art spatially resolved transcriptomic technologies, describe their applications in a variety of biological domains, and explore recent discoveries demonstrating their enormous potential in biomedical research. We further highlight novel integrative computational methodologies with other data modalities that provide a framework to derive biological insight into heterogeneous and complex tissue organization.
Keywords: Spatially resolved transcriptomics, Single-cell RNA sequencing, Integrative computational algorithm, Multimodal data analysis
INTRODUCTION
Single-cell RNA sequencing (scRNA-seq) technique has enabled us to investigate deeper into the functions and characteristics of each cell, looking into the details of its genetic compositions of mRNA that are transiently active at the time of the data acquisition (1). Compared to bulk-RNA sequencing, this is a tremendous step forward that has allowed scientists to gain information about the functional differences between individual cell types (2). However, scRNA-seq loses spatial information of individual cells, interactions among adjacent cells, and local signaling networks. For example, the tumor microenvironment has remained elusive (3, 4). In order to overcome this limitation, transcriptomic techniques that capture the spatial information for tissues of interest have been actively developed (5, 6).
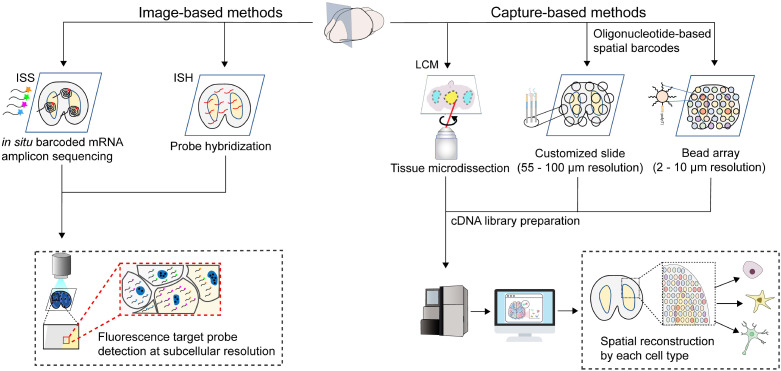
Spatially resolved transcriptomics has achieved significant progress in the biomedical research field with advances in imaging and next-generation sequencing (NGS) technology. Spatially resolved transcriptomics has two main categories: 1) image-based in situ hybridization (ISH) and in situ sequencing (ISS) methods; 2) technologies based on mRNA capture, including laser capture microdissection (LCM) that are spatially barcoded on slides or beads. The different spatially resolved transcriptomic methods have their key features, such as the number of transcripts detected, cellular resolution, and size of the region captured. While image-based methods require prior knowledge of the genes of interest, NGS-based methods capture the whole transcriptome from tissue sections unbiasedly. Also, the spatial resolution of image-based methods for tissue imaging is limited only by the optical diffraction limit and, therefore, suitable for analyzing a subcellular organization. In contrast, the spatial resolution of NGS-based methods is limited by the physical size of a spot that captures cells. Although both methods have improved considerably in the past few years, efforts to refine the sensitivity and increase the cellular resolution of these methods are still ongoing. The overview of two main spatially resolved transcriptomic techniques is described in Fig. 1.
Fig. 1.
Overview of two main categories of spatially resolved transcriptomic techniques: image-based (Left) and capture-based (Right) methods. Imaging-based methods detect target genes by sequencing directly into a fixed tissue section (ISS) or hybridizing a complementary fluorescent probe (ISH). Both image-based methods are suitable for analyzing subcellular transcript patterns of a cell. Capture-based methods are divided into three sub-categories: directly obtaining a specific region of interest from a tissue section using laser capture microdissection (LCM), customized slides, or bead arrays to capture mRNAs by oligonucleotide-based spatial barcodes followed by NGS. Combining computational strategies enables the comprehensive mapping of cell types in spatially resolved transcriptomic data with a specific spatial resolution by each method.
In this review, we describe how different spatially resolved transcriptomic techniques have evolved based on the needs resulting from distinct tissue environments (Table 1) and how those methods are combined with the high-throughput scRNA-seq data to complement each method’s technical and functional constraints (Table 2).
Table 1.
Spatially resolved transcriptomic techniques
| Techniques | Features | Target genes | Application | Programming language | Reference |
|---|---|---|---|---|---|
| Image-based spatially resolved transcriptomics: ISH | |||||
| smFISH smHCR |
Short DNA probes complementary to mRNA targets trigger chain reactions | 39 probes (smHCR) | Zebra fish, mouse brain | MATLAB | Shah et al., 2016 Development (7) |
| osmFISH | Binding of 20 nucleotide-long fluorescently labeled DNA probes | 33 marker genes | Mouse brain | Python | Codeluppi et al., 2018 Nat Methods (8) |
| MERFISH | Using chemical cleavage instead of photobleaching to remove fluorescent signals | 130 genes in up to 100,000 cells | Cultured U-2 OS cells | MATLAB | Moffitt et al., 2016 PNAS (9) |
| MERFISH-based analysis platform | In situ cell-type identification and mapping in combination with scRNA-seq | Targeting a set of 155 genes | Mouse hypothalamic preoptic region | MATLAB | Moffitt et al., 2018 Science (10) |
| seqFISH+ | Enables visualization of the subcellular localization | 10,000 genes in single cells | NIH/3T3, mouse brain | MATLAB | Eng et al., 2019 Nature (11) |
| SABER | Additional signal amplification or applying serial imaging with DNA-Exchange | 18,000 probes targeting a 3.9-Mb region | Mouse retinal tissue | MATLAB & Python | Kishi et al., 2019 Nat Methods (12) |
| Split-FISH | Alternative approach to reduce off-target background fluorescence by integrating split-probe strategy with multiplexed FISH | 317 genes in single cell | Mouse brain, liver, kidney, ovary | Python | Goh et al., 2020 Nat Methods (13) |
| Image-based spatially resolved transcriptomics: ISS | |||||
| STARmap | Integrated with hydrogel-tissue chemistry and targeted signal amplification | 160 to 1,020 genes simultaneously | Mouse brain | Python | Wang et al., 2018 Science (15) |
| INSTA-seq | Sequences two bases simultaneously from both ends of the cDNA fragments | Up to 443,304 UMIs in total | Drosophila retina | R | Fürth et al., 2019 BioRxiv (16) |
| HybISS | New barcoding system via sequence-by-hybridization chemistry | 119 genes for PLP design | Mouse visual cortex, human brain | MATLAB | Gyllborg et al., 2020 Nucleic Acids Res (17) |
| pciSeq | Bayesian algorithm derived from scRNA-seq clusters data | Designed 755 probes for 99 genes | Mouse CA1 interneuron | Python | Qian et al., 2020 Nat Methods (18) |
| ExSeq | cDNA amplicons are eluted from the sample and re-sequenced | Up to 3,039 genes with untargeted approach | Mouse brain, mouse visual cortex, human breast cancer | MATLAB | Alon et al., 2021 Science (19) |
| Capture-based spatially resolved transcriptomics: LCM | |||||
| exome- capture RNA- sequencing | Optimized standard protocol for hematoxylin and eosin (H&E) staining | Whole exome | 7 tumor samples of TNBC | MATLAB | Romanens et al., 2020 BioRxiv (20) |
| immuno-LCM- RNAseq | RNA quality was significantly improved using modified protocol | Up to 60 cells were demonstrated to be sufficient quality | Mouse small intestine | Python | Zhang et al., 2021 BioRxiv (21) |
| PIC | Photo-irradiated cells were suppressed cDNA amplification | 8,000 genes were detected with 7 × 104 unique read counts | Mouse embryo | R | Honda et al., 2021 Nat Commun (22) |
| Capture-based spatially resolvedtranscriptomics: Oligonucleotide-based spatial barcode on slide | |||||
| ST | Arrayed reverse transcription primers with unique positional barcodes | Up to 200 million oligonucleotides in each of 1007 features | Mouse brain, human breast cancer | R | Ståhl et al., 2016 Science (24) |
| Salmén et al., 2018 Nat Protoc (23) | |||||
| Multimodal analysis | Combined single-cell RNA sequencing with ST | Median depth of 1,629 UMIs/spot and 967 genes/spot | Human cSCC | MATLAB & R | Ji et al., 2020 Cell (29) |
| ST | Combined ST and ISS | Mean 31,283 UMIs and 6,578 unique genes per TD | AD mouse model, mouse and human brain | Python | Chen et al., 2020 Cell (28) |
| Capture-based spatially resolvedtranscriptomics: Oligonucleotide-based spatial barcode on bead array | |||||
| Slide-seq | DNA-barcoded beads with known positions (in situ indexing) | 1.5 million beads, of which 770,000 can be analyzed | Mouse cerebellum and hippocampus | MATLAB & R | Rodriques et al., 2019 Science (30) |
| HDST | Barcoded poly(d)T oligonucleotides into 2-μm wells with a randomly ordered bead array-based | 2,893,865 individual barcoded beads | Mouse brain, primary breast cancer | Python | Vickovic et al., 2019 Nat Methods (31) |
| Slide-seq V2 | Improvements in library generation, bead synthesis and array indexing | Mean 45,772 UMIs in 110 μm diameter area | Mouse hippocampus | MATLAB & R & Python | Stickels et al., 2021 Nat Biotechnol (32) |
| Seq-Scope | Based on a solid-phase amplification using an Illumina sequencing platform | Up to 5.88-19.7 genes were identified per HDMI pixel | Human liver and colon | Python | Cho et al., 2021 Cell (33) |
| Stereo-seq | Combined DNA nanoball pattern arrays and tissue RNA capture | Up to 133,776 UMIs per 100 μm diameter | Mouse brain | Not identified yet | Chen et al., 2021 BioRxiv (34) |
Table 2.
Integration of spatially resolved transcriptomic data with other methods
| Techniques | Features | Input data | Application | Programming language | Reference |
|---|---|---|---|---|---|
| Combination with scRNA-seq | |||||
| pciSeq | Bayesian algorithm derived from scRNA-seq clusters data with ISS | Designed 755 probes for 99 genes | Mouse CA1 interneuron | Python | Qian et al., 2020 Nat Methods (18) |
| seqFISH | Computing the ratio of the performance and prediction scores with scRNA-seq data | Each cell contained avg 196 mRNA from 93.2 genes | Embryo development in brain and gut | R | Lohoff et al., 2021 Nat Biotechnol (35) |
| Multiple spatial transcriptomics | Unbiased approach with additional in situ hybridization using RNAscope and multi-molecule ISS | Mean 31,283 UMIs and 6,578 unique genes per tissue domain | AD mouse model, mouse and human brain | Python | Chen et al., 2020 Cell (25) |
| Slide-seq | NMFreg that reconstructs expression of each cell type signatures defined by scRNA-seq | 1.5 million beads, of which 770,000 could be analyzed | Mouse cerebellum and hippocampus | MATLAB & R | Rodriques et al., 2019 Science (30) |
| Integrating microarray-base d ST and MIA | Enrichment analysis that two-tailed Student’s t-test were used to compare expression of those marker genes | 2,500-3,300 UMIs and 1,400-1,700 unique expressed genes per single cell | Pancreatic ductal adeno-carcinoma | R | Moncada et al., 2020 Nat Biotechnol (36) |
| Deep learning-based spatial information | |||||
| DEEPsc | Deep-learning network was trained with spatial position feature vectors as simulated scRNA-seq data | Started with top 3,000 highly variable genes | Drosophila embryo, Zebrafish embryo, murine frontal cortex | MATLAB | Maseda et al., 2021 Front Genet (37) |
| BayesSpace | Bayesian statistical method that uses the information from spatial neighborhoods to achieve super-resolution images | 10X Genomics Visium data, does not require independent single-cell data or marker gene preselection | Brain, melanoma, invasive ductal carcinoma, ovarian adeno-carcinoma | R | Zhao et al., 2021 Nat Biotechnol (38) |
| SPICEMIX | Enhances the NMF of gene expression with a graphical representation of the spatial relationship of cells | 2,470 genes in 523 cells (seqFISH+), 930 cells and 1,020 genes (STARmap) | Mouse primary visual cortex | Python | Chidester et al., 2020 BioRxiv (39) |
| SpaOTsc | Infer the spatial distance between every pair of cells by computing the optimal transport distance | 851-15,413 cells and 10,495-45,789 genes (scRNA-seq), 64-1,549 spatial positions and 47-1,020 genes | Zebrafish embryo, Drosophila embryo, mouse visual cortex | Python | Cang et al., 2020 Nat Commun (4) |
| Deconvolution of spatially resolved transcriptomics | |||||
| RCTD | Statistical model assumed to be Poisson distributed and maximum-likelihood estimation (MLE) used to infer the cell types | Slide-seq and 10X Genomics Visium data | Mouse brain | R | Cable et al., 2021 Nat Biotechnol (40) |
| SPOTlight | NMF along with non-negative least squares (NNLS) model with both the basis and coefficient matrices with cell type marker genes | 41,986 cells were merged to identify a total of 10,623 immune cells | Mouse brain, pancreatic adeno-carcinoma | R | Elosua-Bayes et al., 2021 Nucleic Acids Res (41) |
| SpatialDWLS | Dampened weighted least squares (DWLS) model with cell-type specific gene signatures from a public scRNA-seq dataset as a reference | 10,000 genes in 523 cells (seqFISH+) | Mouse brain, human heart | R | Dong et al., 2021 Genome Biol (42) |
| DestVI | Bayesian model for multi-resolution deconvolution of cell types using Variational Inference | Pair of ST and scRNA-seq from same tissue | Murine lymph node, mouse tumor model | Python | Lopez et al., 2021 BioRxiv (43) |
| Cell type inference via image-based machine learning | |||||
| ST-Net | Deep learning algorithm that combines ST and histology images to predict the target gene expression of each spot | 30,612 spots in 68 breast tissue sections | Breast cancer | Python | He et al., 2020 Nat Biomed Eng (44) |
| HisToGene | Employs a modified Vision Transformer model for gene expression prediction from histology images | 9,612 spots and 785 genes in breast cancer tissue | Breast cancer | PyTorch | Pang et al., 2021 BioRxiv (45) |
| stLearn | Deep neural network model to predict hotspots where cell-cell interactions are more likely to occur | Feature vectors from H&E images of the tissue section | Mouse brain, human brain, breast cancer | Python | Pham et al., 2020 BioRxiv (46) |
| SpaCell | Normalized count data and H&E staining images were trained with convolutional neural network | Tissue morphology and spatial gene expression data | Prostate cancer, amyotrophic lateral sclerosis | Python | Tan et al., 2020 Bioinformatics (47) |
| CoSTA | Clustering by Gaussian mixture model (GMM) and weight updating as commonly performed in training neural networks | Image-type matrix of MERFISH and Slide-seq data | Mouse brain, brain Injury | Python | Xu et al., 2021 BMC Bioinformatics (48) |
| STUtility | NMF to decompose ST data and identification and extraction of neighbouring capture-spots | 10x Genomics Visium data | Mouse brain, breast cancer tissue, lymph node, rheumatoid arthritis | R | Bergenstråhle et al., 2020 BMC Genomics (49) |
| ISST | Image-based in silico spatial transcriptomics without hybridization or sequencing | 12 sections from the mouse olfactory bulb | Mouse olfactory bulb, human breast cancer | Python | Bergenstråhle et al., 2020 BioRxiv (50) |
| Cellular protein information | |||||
| CITE-seq | Oligonucleotide-labeled antibodies are used to integrate cellular protein and transcriptome measurements | Common immune subpopulation markers (CD8a, CD3e, CD19, CD56, CD16, CD11c and CD14) | Human HeLa, mouse 4T1 cell, immune subpopulation | R | Stoeckius et al., 2017 Nat Methods (52) |
| IMC | Epitope-based imaging methods that employ a mass spectrometer for readout to infer RNA-to-protein correlations | Detected three mRNA simultaneously (HER2, CK19 and CXCL10) | Breast cancer | MATLAB & R & Python | Schulz et al., 2018 Cell Syst (53) |
| nanoPOTS | Unique proteins were identified via combination of LCM and ultrasensitive nanoLC-MS/MS | > 2,000 proteins with 100 μm spatial resolution | Mouse luminal epithelial cell, stromal cell, glandular epithelial cell | R | Piehowski et al., 2020 Nat Commun (54) |
| Spatial ATAC-seq | |||||
| sciMAP-ATAC | Spatially resolved, single-cell profiling of chromatin states from a single tissue punch | Mean 12,052 - 30,212 passing reads per cell | Mouse and human brain, cerebral ischemia model system | R | Thornton et al., 2021 Nat Commun (55) |
| Spatial-ATAC-seq | DNA barcode solutions were introduced to the tissue surface using an array of microchannels | 36,303-100,786 unique fragments per pixel | Mouse embryos, human tonsil tissue | R | Deng et al., 2021 BioRxiv (56) |
RESULTS
Development of spatially resolved transcriptomic techniques
Image-based spatially resolved transcriptomics: There are two major approaches that visualize targeted genes of interest by fluorescent labels. First, in situ hybridization (ISH) is a technique that detects mRNA molecules in thick tissue samples to find gene expression patterns in single cells within the native tissue environment. In detail, it detects target sequences using fluorescent images generated by hybridization of a complementary probe. This approach has advantages in visualizing RNA molecules directly in the original locations of the biological environment. However, the high autofluorescence background of tissue samples present limitations in detecting a large set of different transcripts simultaneously.
In 2016, Shah et al. developed single-molecule fluorescence in situ hybridization (smFISH) methods with multi-color and multi-RNA imaging in deep tissues using single-molecule hybridization chain reaction (smHCR) (7). The central innovation of smHCR was the fluorescent amplification of probes complementary to the target mRNA to increase the sensitivity for gene detection. This technique was essential to ensure that puncta generated by the fluorescence were bright enough for high sensitivity. Using this approach, deeper tissue was successfully visualized with higher sensitivity as a single mRNA was detected in situ, even in thick tissues in the intact vertebrate embryo of zebrafish. However, they were only small enough for diffraction-limited resolution. Similarly, unamplified cyclic-ouroboros smFISH (osmFISH) visualizes each RNA molecule by binding 20 nucleotide-long fluorescently labeled DNA probes (8). Subsequently, the expression of 33 marker genes was successfully analyzed in mouse somatosensory cortex and hippo-campus by conventional epifluorescence microscope.
To understand the spatial context of cells, direct imaging of individual RNA molecules within intact cells and tissues is vital. The multiplexed fluorescence in situ hybridization (FISH) technique has been widely used for this purpose. MERFISH (multiplexed error-robust fluorescence in situ hybridization) identifies RNA through a combinational labeling approach that consists of RNA molecules with error-robust barcodes followed by sequential rounds of smFISH to read out these barcodes (9). With this approach, an imaging process with an ensemble of fluorescent spots that define binary barcodes (on or off fluorescent signal) allowed the characterization of 130 genes in 100,000 cultured U-2 OS cells. The same group improved MERFISH-based imaging where scRNA-seq was combined with the MERFISH-based images for in situ cell-type identification and mapping (10). This upgraded analysis platform successfully identified functionally important 155 marker genes from ∼70 neuronal populations and spatial organizations in the hypothalamic preoptic region of mice. Another strategy to increase the number of distinguished mRNA molecules was achieved by applying four colors with eight barcoding rounds into pseudocolors. Using seqFISH+, images of mRNA for 10,000 genes in a single cell were acquired with high accuracy and sub-diffraction-limit resolution using a conventional confocal microscope (11). This method identified a subcellular localization pattern of 10,000 mRNAs in the brain, and ligand-receptor pairs from potential cell-cell interactions were suggested by the observed mRNA expression patterns in the cells. This proves that seqFISH+ can overcome not only optical crowding and provide a tenfold or greater improvement over existing methods, but also enables super-resolved imaging with conventional confocal microscopes.
Higher intensity of FISH signals is important to improve the gene detection performance. Signal amplification by exchange reaction (SABER) technique was designed to amplify the intensity of quantitative FISH signals (12). In brief, DNA and RNA FISH probes were first chemically synthesized with a primer sequence of their 3’ end extended into primer-exchange reaction (PER). In SABER, a multitude of PER-concatemerized probes sets can be hybridized to their targets simultaneously and read out in sequential rounds of imaging. With this approach, 18,000 probes in total targeting a 3.9-Mb region of human metaphase spreads and interphase cells were mapped to three colors, which all colocalized as expected. However, a key challenge of multiplexed FISH is that it is difficult to detect individual target probes in complex tissue environments due to the noisy background of tissue. To address this issue, Goh et al. adopted an alternative approach to reduce off-target background fluorescence by integrating split-probe strategy with multiplexed FISH (13). By improving the bridge probe design to hybridize with sufficient complementary base pairing, split-FISH is capable of accurate transcriptomic profiling on four mouse tissue types, even in opaque tissue. However, taking sequential images with multiple hybridization rounds for several target genes can sometimes be misaligned because of possible technical or experimental errors. Therefore, correcting these misalignments is an essential step; thus, tools for precise alignments of multiple images have been developed (14).
Another imaging-based approach for spatially resolved transcriptomics is in situ sequencing (ISS). The main advantage of ISS over ISH is that ISS can detect more genes (∼10,000) than ISH in a non-targeted manner. In ISS-based methods, transcripts were sequenced directly in fixed tissues. Specifically, mRNA molecules are reverse transcribed and amplified by rolling circle amplification (RCA). The micrometer- or nanometer-sized RCA products are then subjected to sequencing-by-ligation (SBL), and the barcode within the probe is simultaneously decoded.
Wang et al. devised the spatially resolved transcript amplicon readout mapping (STARmap) methods to detect and quantify 160 gene sets simultaneously in the mouse primary visual cortex. STARmap incorporates hydrogel chemistry, improved padlock-probe technology, and error-robust SBL methods (15). Also, the extended STARmap can image a larger number of genes successfully with a better spatial resolution (1,020 genes detected simultaneously at millimeter-scale volumes encompassing ∼30,000 cells). To overcome the limitation of short reads in ISS, a new method utilizing bidirectional sequencing chemistry and an imaging transcript-specific barcode was developed. With this bidirectional sequencing method, the generated reads were efficiently extracted and assembled into longer reads using NGS technology (16). One of the most important aspects of sequencing methodology is maximizing throughput for reliable gene detection. Hybridization-based in situ sequencing (HybISS) achieved this goal by replacing the SBL technique of ISS with sequencing-by-hybridization (SBH) to detect hundreds of target genes and successfully applied in mouse and human brains. The SBH enabled detecting genes with increased signal-to-noise using autofluorescence quenching (17). When classifying cell types, the low throughput of ISS technology might not be sufficient to have reliable outcomes. To address this issue, probabilistic cell typing by in situ sequencing (pciSeq) introduced an approach that uses ample scRNA-seq classification to identify cell types using multiplexed in situ RNA detection (18). Specifically, the probability of assigning each read to a given cell type was calculated using a Bayesian algorithm derived from scRNA-seq cluster data.
Another attempt has been made with ExSeq where cDNA amplicons are eluted from the sample and re-sequenced ex situ with NGS for long-read untargeted and targeted in situ RNA sequencing (19). ExSeq yielded the readout of thousands of genes, including splice variants, and enabled highly multiplexed mapping of RNAs when applied in mouse hippocampal neuron cells and tissues.
Capture-based spatially resolved transcriptomics: Tissues preserved in FFPE blocks can be stored from 3-10 years. Using laser capture microdissection (LCM), Romanens et al. extracted tissues related to breast cancer that were preserved in FFPE blocks using an optimized protocol for hematoxylin and eosin (H&E) staining with the best parameters for dyes, incubation time, the thickness of tissue, and surface area for microdissection (20). In the study, Romanens and colleagues spatially characterized tumor and immune cells in triple-negative breast cancer. Using the immunofluorescence-guided laser capture microdissection (immuno-LCM-RNAseq) technique, it became easier to dissect tissues and study particular genes of cells that have functions that are highly dependent on the spatial organization in the tissue (21). In their study, the quality of RNA was immensely improved by a modified high-salt protocol and RNase inhibitor, which enabled the investigation of full-length transcripts and isoforms.
Compared with the LCM-seq method, Photo-isolation chemistry has the advantage in spatial resolution because its photochemical isolation technique uses photo-caged oligo-deoxynucleotides for in situ reverse transcription (22). This approach has allowed to analyze cells in complicated and fine structures, detecting 8,000 genes with 7 × 104 unique read counts per single cell in mouse embryos. These advanced LCM can profile more than 10,000 genes from dozens of cells with spatial resolution up to subcellular or subnuclear levels. While the sequencing depth is high enough, the analysis is restricted to a limited number of regions.
Another strategy to obtain spatially resolved transcriptomic data is to use pre-arranged set of barcoded reverse transcription (RT)-primers on glass slides. Spatial Transcriptomics (ST) analyzes the transcriptome in individual tissue sections while maintaining two-dimensional positional information of tissues. Specifically, slide sections are delivered to a glass slide that bears RT-primers and sets of DNA barcodes in situ. After generating cDNA libraries, NGS was carried out with the sequencing libraries. Then, the spatially resolved whole-transcriptome information is provided by aligning tissue images with the sorted RNA-seq data that include spatial information carried by the barcodes. However, the spatial resolution of ST is limited to 100 μm with a center-to-center distance of 200 μm which yields around 50 to 100 cells per single spot (23, 24). The higher resolution of ST was achieved with the 10X Genomics Visium platform that features a physical spot size of 55 μm diameter and a center-to-center distance of 100 μm. As a result, the 10X Genomics Visium platform has been one of the most popular choices for mapping spatial profiles of tissues in interest in various fields, including neuroscience, cancer biology, and developmental study (25-27). Spatial cellular resolution and the quality of analysis can be further improved by combining two or more different spatially resolved transcriptomic techniques. Chen et al. combined ISS and capture-based ST methods to identify a plaque-induced gene network in micro- and astroglia cells and oligodendrocytes in the Alzheimer’s disease model of mouse and human. Then, the oligodendrocyte gene responses were confirmed at the single-cell level using ISS with target-specific probes with known cell type markers. Combining those two techniques elucidated how amyloid plaques are involved in the neurodegenerative process (28). In another study, Ji et al. carried out three techniques of single-cell, spatially resolved transcriptomics, and multiplexed ion beam imaging (MIBI) to investigate the tumor microenvironment of cutaneous squamous cell carcinoma (29). Spatially resolved transcriptomes from 17,064 spots were obtained utilizing both conventional ST and 10X Genomics Visium kits. The combination of three methods revealed ligand-receptor networks of specific cell types, suggesting tumor-specific keratinocytes as a signaling hub for intercellular communication.
The techniques for spatial transcriptomics with barcoded oligonucleotide capture array described, however, have limitation in the spatial resolution of up to 55-100 μm due to the physical size of capturing spots. To resolve the issue of low spatial resolution in capture-based sequencing methodology, bead-based capturing sequencing was developed. In 2019, Rodriques et al. developed Slide-seq for higher cellular resolution genome-wide analysis using DNA-barcoded 10 μm beads onto a rubber-coated glass slide (30). In Slide-seq, each bead’s distinct barcoded sequence is determined via SBL methods. Using the beads, the resolution comparable to the size of an individual cell was achieved by NGS of barcoded RNA-seq libraries and successfully applied to the hippocampus area in mice. Slide-seq defined finer cellular subpopulations of cerebellar cell types, which had not been identified in previous spatially resolved single-cell sequencing studies. Additionally, high-definition spatial transcriptomics (HDST) was developed as another effort to accomplish higher resolution for barcoded transcripts (31). HDST randomly deposits barcoded poly(d)T oligonucleotides into a 2 μm well with bead array-based fabrication. Mouse brain and primary breast cancers were profiled with 25× higher resolution than that of Slide-seq using this method, resulting in a reconstruction of the spatial architecture of mouse brain and primary breast cancer tissues. Although Slide-seq has a superior spatial resolution compared to other techniques, it has a lower transcript detection sensitivity that limits biological applications. To circumvent these restrictive factors of the initial version, Slide-seqV2 was developed to offer higher capturing efficiency for transcripts by implementing a readily available monobase-sequencing strategy instead of dibase-sequencing, leading to a dramatic increase the capture efficiency (32). Another advancement from Slide-seq was the application of an error robust computational approach using NGS. With the higher number of unique molecular identifier (UMI) per bead (494 UMIs per 10 μm), which is higher than HDST, Slide-seqV2 demonstrated that it can detect a rare transcript, such as dendritic mRNA in CA1 neurons. Cho et al. developed a high-resolution spatial barcoding technology, Seq-Scope, with a 0.5-0.8 μm center-to-center resolution (33). This technique was developed based on the NGS technology that utilizes randomly barcoded single-molecule oligonucleotides with an outstanding transcriptome capture capacity (∼4,700 UMIs/cell on average), comparable to the conventional scRNA-seq methods. This technical improvement enabled visualizing the histological organization of the transcriptome architecture in liver tissues at subcellular level. More recently, another high- resolution spatial barcoding technology, SpaTial Enhanced Re-solution Omics-sequencing (Stereo-seq) was introduced with a center-to-center resolution of 500-715 nm (34). This method applies microfluidic channels perpendicularly for unique pair-wise barcoding on each spot of the tissue section. Stereo-seq can capture up to 133,775 UMI per 100 μm diameter bin, superior to other available technologies, including Seq-Scope compared to the same bin resolution. Stereo-seq was capable of profiling whole mouse embryo, demonstrating that it can be applied to a much larger tissue such as whole human brain sections.
Integration of spatially resolved transcriptomic data with other methods
While scRNA-seq and spatially resolved transcriptomic data have limitations, a computational integration of two or more data modalities can better characterize spatial cell type compositions and local cell states in the tissues. Here, we describe the improvements benefiting from the novel in silico computational algorithms, which integrate spatially resolved transcriptomic data with additional data modalities, such as scRNA-seq, and several other Omics approaches to better understand spatial architecture of tissues and to derive biological insights.
Combination with scRNA-seq: probabilistic cell typing by in situ sequencing (pciSeq) was used to integrate scRNA-seq data with ISS for cluster analysis (18). To design a gene panel, genes were grouped leveraging prior scRNA-seq cell type classification. Using pciSeq, Qian et al. were able to map the inhibitory neurons of hippocampal area CA1, making it feasible to confirm results for ISS. Before this study, the ISS RNA detection had not been proven to distinguish fine cortical cell types identified from previous reports. Defining how cell states correlate with spatial cell distribution and investigating how the local signaling environment impacts molecular signatures and, ultimately, the fate of cells is inevitably important to understand mouse embryo changes during early development. Lohoff and colleagues delineated the precise location of distinct cell types within a single reference scaffold by combining a high-resolution seqFISH map with scRNA-seq (35). From this study, integration of single-cell transcriptome with spatial context enabled not only identification of 387 target genes from seqFISH but also the imputation of the spatially resolved map at cellular resolution in a mouse organogenesis model.
To improve the low spatial resolution of capture-based spatial methods, the two data modalities from each scRNA-seq and spatially resolved transcriptomic methods were integrated into other studies. Cell type signature defined by scRNA-seq was integrated with Slide-seq data, facilitating the discovery of spatially defined gene expression patterns from mouse hippocampus. For reconstructing expression of each Slide-seq data, non-negative matrix factorization regression (NMFreg) was utilized as a combination of cell-type signatures defined in the reference scRNA-seq data (30). In addition, several computational techniques for combining single-cell and MIBI or multiple spatial transcriptomics were reported to achieve a higher cellular resolution (28, 29). Moncada et al. integrated scRNA-seq with the ST method to study the microenvironment of primary pancreatic tumors by introducing multimodal intersection analysis (MIA) and reported that fibroblast-specific genes in the tissue of pancreatic ductal adenocarcinoma overlap significantly with the set of genes specific to the cancer region from the ST data (36). The study also identified the colocalization of inflammatory fibroblasts and cancer cells expressing a stress-response gene module.
Deep learning-based spatial information: Machine learning algorithms have been extensively used as an imputation method for predicting cell types based on the context of relevant datasets. For example, DEEPsc, an artificial neural network model for categorizing cell types, was trained to predict a specific cell type with sets of genes from the scRNA-seq reference atlas dataset (37) and achieved an accuracy comparable to several existing methods (2-norm, infinity norm, mean percent difference, and large margin nearest neighbor) for applications utilizing 3,000 highly variable genes. Additionally, BayesSpace applies a Bayesian statistical method that takes the spatial gene expression profile from neighborhoods as a prior and achieves a super-resolution image (38). It is noteworthy that BayesSpace does not require separate scRNA-seq gene expression signature or preselected marker genes. SPICEMIX, another approach using non-negative matrix factorization (NMF) based on probabilistic latent variable modeling, calculates spatial affinity between the metagenes and their proportions of neighboring cells (39). Data acquired from seqFISH+ and STARmap were used to demonstrate its ability to refine the identification of cell types in mouse primary visual cortex. SpaOTsc was developed to better understand cell-cell communications in the spatially resolved transcriptomic datasets and inferred the spatial distance between every pair of cell types by computing the optimal transport distance measured by utilizing spatial measurements of a relatively small number of genes without the scRNA-seq dataset (4). The results from SpaOTsc indicate that signal sender cells exhibit more spatial localization patterns, while the locations of the signal receivers are more scattered over throughout the tissues of zebrafish and the mouse visual cortex. SpaOTsc can be used both to integrate non-spatial single-cell measurements with the spatial data and to reconstruct spatial cellular dynamics in tissues.
Deconvolution of spatially resolved transcriptomics: Owing to the lower cellular resolution of spatial barcoding capture-based methods, the proportions of discrete cell types for a given spot have been inferred by various deconvolution algorithms. Robust cell type decomposition (RCTD) is based on statistical model maximum-likelihood estimation to approximate the proportions of spatially localized cellular subtypes in spatially resolved transcriptomic data such as Slide-seq or 10X Genomics Visium datasets (40). RCTD accurately recapitulated known cell type spatial distribution in both Slide-seq and 10X Genomics Visium in the mouse brain tissues. Alternatively, SPOTlight uses NMF alongside non-negative least squares for accurate and sensitive cell-type detection, seamlessly applied in mouse brain and 22 immune subpopulations from pancreatic adenocarcinoma samples with a curated annotation (41). Recently, SpatialDWLS was shown to have a higher degree of sensitivity and accuracy than RCTD and SPOTlight using the dampened weighted least squares (DWLS) algorithm in which the weight is selected to minimize the overall relative error rate to infer cell-type composition (42). By applying SpatialDWLS to spatial transcripto-mics dataset of mouse brain and human embryonic heart, increased abundance of different cell types was observed during development. Finally, DestVI was developed to alleviate the problem of the complicated deconvolution, especially when there is a continuum of cell states that cannot be clearly distinguished (43). To address this problem, deconvolution of spatially resolved transcriptomic profiles used Variational Inference (DestVI), a Bayesian model-based multi-resolution cell- type deconvolution algorithm, was developed. DestVI learns cell type-specific profiles and sub-cell type variations using a conditional deep generative model. By combining scRNA-seq and spatially resolved transcriptomic data from the same tissue, DestVI was used to study the immune interplay within lymph nodes and explore the spatial tissue architecture of the mouse tumor microenvironment. Notably, DestVI outperforms existing discrete deconvolution approaches such as RCTD, SPOTlight, and Seurat.
Cell type inference via image-based machine learning: H&E-stained histology images are easy and cheap to obtain and routinely generated in clinics. Several studies integrated spat-ially resolved transcriptomics and histopathology images to extract feature information and make predictions by various machine learning algorithms. ST-Net implements a convolutional neural network model and was trained on 68 breast tissue sections from 23 patients with breast cancer to predict the gene expression based on histopathology images (44). In addition, HisToGene adopts Vision Transformer for image recognition and predicts super-resolution gene expression. Using the same dataset from ST-Net as a training set, HisToGene outperformed ST-Net in both gene expression prediction and clustering tissue regions accuracy (45). stLearn also integrates three types of datasets, including spatial dimensionality, tissue morphology, and genome-wide transcriptional profile using a deep learning network model (46) and predict cell type clustering, intercellular interaction, and reconstruction of spatial transition gradients, which were all successfully conducted with brain and breast cancer datasets. SpaCell and CoSTA also utilize a convolutional neural network model to predict malignant cells in prostate cancer and quantify the level of spatial expression relationships between each pair of genes from mouse brain samples, respectively (47, 48). Alternatively, STUtility uses raw 10X Genomics Visium RNA-seq and image data as input and processes the images, aligns consecutive stacked tissue images, and finally visualize them all together in 3D. Another useful functionality of STUtility includes NMF to decompose 10X Genomics Visium data into a lower dimension and a method to identify neighboring capture spots in a spatial network (49). This approach was applied in human breast cancer tissues to define the leading edge of the tumor region, eventually leading to the delineation of tumor heterogeneity between the tumor core and the tumor front. Bergenstråhle et al. also constructed a deep generative model for spatial data fusion by combining low-sensitivity, low-resolution in situ RNA capturing expression data with high-resolution histological image data (50). This method inferred de-noised full-transcriptome spatial gene expression at the same resolution as the image data in mouse olfactory bulb by jointly training a recognition neural network that maps the image data to the variational parameters of the latent state to optimize model parameters.
Cellular protein information: Single-cell profiling via proteomics approach has become progressively comprehensive, and unbiased profiling of protein expression has had a broad impact in biomedical research (51). Recent advances in techniques such as epitope-based imaging, mass cytometry, and mass spectrometry enable protein expression to be mapped across tissue with high resolution. Stoeckius et al. introduced a method named cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) that integrates cellular protein and transcriptome measurements in an effort to provide phenotypic information such as cell-surface protein expression levels (52). This multimodal analysis on cellular protein, using oligonucleotide-labeled antibodies and high-throughput scRNA-seq, revealed phenotypes including a detailed multimodal characterization of immune subpopulations that had not been ascertained by using scRNA-seq alone. The study carried out by Schulz et al. used extended imaging mass cytometry (IMC), based on the simultaneous staining followed by multiplexed detection of mRNA and proteins in tissue (53). This approach that couples the RNAscope-based ISH with simultaneous antibody staining was used in 70 breast cancer samples and demonstrated a moderate correlation between HER2 and CK19 mRNA and protein expression levels. However, IMC has technical challenges for probing spatial distribution due to the limited number of labels and/or antibodies for labeling proteins. To get around this, Piehowski et al. developed an automated imaging approach, Nanodroplet Processing in One plot for Trace Samples (nanoPOTS), utilizing label-free nanoproteomics to analyze tissue voxels (54). nanoPOTS increased the number of target proteins to more than 2,000 with spatial resolution of 100 μm. Combined with LCM and ultrasensitive nanoLC-MS/MS, large sample sets were collected and processed with high sensitivity capturing the unique proteome mapping in a mouse uterus model.
Spatial ATAC-seq: To capture spatial epigenetic information in tissue at the single-cell level and genome scale, single-cell combinatorial indexing on microbiopsies that is assigned to positions for the assay for transposase accessible chromatin (sciMAP-ATAC) was introduced (55). sciMAP-ATAC produced data of similar quality to non-spatial sci-ATAC of cells within a 214-micron cubic area and was submitted to the adult mouse primary somatosensory cortex and human primary visual cortex to successfully characterize the spatial progressive nature of cerebral ischemic infarction. Integration of sciMAP-ATAC with single-nucleus RNA-seq and single-cell chromatin accessibility datasets demonstrated high concordance for most cell types. Deng et al. also reported spatially resolved chromatin accessibility profiling method (spatial-ATAC-seq) (56). The basic concept of ATAC-seq utilizing NGS with Tn5 transposition chemistry was improved with a microfluidic barcoding system, introducing two barcode solutions to the tissue surface using an array of microchannels for in situ ligation perpendicularly. By integrating the spatial-ATAC-seq data with the scRNA-seq data, each cell type and organ-specific cell type was assigned in mouse embryos. Furthermore, differences between the epigenetic state and protein expression of specific marker genes enabled distinguishing cell types in mouse brain and human tonsil.
DISCUSSION
While spatially resolved transcriptomic technology offers tremendous opportunity to discover spatial heterogeneity in the disease state, characterize spatial expression blueprints during development, and elucidate spatial architecture at the molecular level, its potential lies beyond that as it is still in the early days of development. Unfortunately, none of the currently available spatially resolved transcriptomic technologies are perfect, and the choice of methods depends on study design, the biological question, and often a balance between the cell and/or transcript throughput and spatial resolution. A few overlooked caveats include the requirement for a large number of pseudocolors and barcodes in image-based techniques and a low spatial resolution in capture-based methods. This has brought new technological challenges. The integrative computational algorithms that combine the spatially resolved transcriptomic data and other data modalities have significantly contributed to not only overcoming the key challenges faced by current spatially resolved transcriptomic technologies but gaining fundamental biological insights. The development of novel computational tools will continue to play a significant role in ex-ploring large-scale spatially resolved transcriptomic datasets, translating the consequences of newly acquired spatial patterns, and elucidating principles of the underlying biology. The advent of such integrative approaches will ultimately shed light on the mechanisms that explain the essential differences in spatial architectures of healthy and diseased tissues.
ACKNOWLEDGEMENTS
The authors are grateful to Junho Song for critical reading of the manuscript. This research was supported by the National Research Foundation of Korea (NRF) grants funded by the South Korean government (2020R1F1A1076705).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
References
- 1.Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50:1–14. doi: 10.1038/s12276-018-0071-8.f40e2b868673466fbfe21e5f399da102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20:631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 3.Roth R, Kim S, Kim J, Rhee S. Single-cell and spatial transcriptomics approaches of cardiovascular development and disease. BMB Rep. 2020;53:393–399. doi: 10.5483/BMBRep.2020.53.8.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cang Z, Nie Q. Inferring spatial and signaling relationships between cells from single cell transcriptomic data. Nat Commun. 2020;11:2084. doi: 10.1038/s41467-020-15968-5.f4a6e22defa144b7ba4de8cfceb89169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asp M, Bergenstråhle J, Lundeberg J. Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays. 2020;42:e1900221. doi: 10.1002/bies.201900221. [DOI] [PubMed] [Google Scholar]
- 6.Dries R, Chen J, Del Rossi N, Khan MM, Sistig A, Yuan GC. Advances in spatial transcriptomic data analysis. Genome Res. 2021;31:1706–1718. doi: 10.1101/gr.275224.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah S, Lubeck E, Schwarzkopf M, et al. Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development. 2016;143:2862–2867. doi: 10.1242/dev.138560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Codeluppi S, Borm LE, Zeisel A, et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat Methods. 2018;15:932–935. doi: 10.1038/s41592-018-0175-z. [DOI] [PubMed] [Google Scholar]
- 9.Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci U S A. 2016;113:11046–11051. doi: 10.1073/pnas.1612826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moffitt JR, Bambah-Mukku D, Eichhorn SW, et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018;362:6416. doi: 10.1126/science.aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eng CL, Lawson M, Zhu Q, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568:235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kishi JY, Lapan SW, Beliveau BJ, et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat Methods. 2019;16:533–544. doi: 10.1038/s41592-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goh JJL, Chou N, Seow WY, et al. Highly specific multiplexed RNA imaging in tissues with split-FISH. Nat Methods. 2020;17:689–693. doi: 10.1038/s41592-020-0858-0. [DOI] [PubMed] [Google Scholar]
- 14.Borovec J, Kybic J, Arganda-Carreras I, et al. ANHIR: automatic non-rigid histological image registration challenge. IEEE Trans Med Imaging. 2020;39:3042–3052. doi: 10.1109/TMI.2020.2986331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Allen WE, Wright MA, et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018;361:6400. doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fürth D, Hatini V, Lee JH. In situ transcriptome accessibility sequencing (INSTA-seq) bioRxiv. 2019:722819. doi: 10.1101/722819. [DOI] [Google Scholar]
- 17.Gyllborg D, Langseth CM, Qian X, et al. Hybridization-based in situ sequencing (HybISS) for spatially resolved transcriptomics in human and mouse brain tissue. Nucleic Acids Res. 2020;48:e112. doi: 10.1093/nar/gkaa792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian X, Harris KD, Hauling T, et al. Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat Methods. 2020;17:101–106. doi: 10.1038/s41592-019-0631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alon S, Goodwin DR, Sinha A, et al. Expansion sequencing: spatially precise in situ transcriptomics in intact biological systems. Science. 2021;371:6528. doi: 10.1126/science.aax2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romanens L, Chaskar P, Tille JC, et al. Spatial transcriptomics of tumor microenvironment in formalin-fixed paraffin-embedded breast cancer. bioRxiv. 2020:2020.2001.2031.928143. doi: 10.1101/2020.01.31.928143. [DOI] [Google Scholar]
- 21.Zhang X, Hu C, Huang C, et al. Robust acquisition of high resolution spatial transcriptomes from preserved tissues with immunofluorescence based laser capture microdissection. bioRxiv. 2021:2021.2007.2013.452123. doi: 10.1101/2021.07.13.452123. [DOI] [Google Scholar]
- 22.Honda M, Oki S, Kimura R, et al. High-depth spatial transcriptome analysis by photo-isolation chemistry. Nat Commun. 2021;12:4416. doi: 10.1038/s41467-021-24691-8.bb1fc3bbebd6485c9ec92dde46e117b8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salmén F, Ståhl PL, Mollbrink A, et al. Barcoded solid-phase RNA capture for spatial transcriptomics profiling in mammalian tissue sections. Nat Protoc. 2018;13:2501–2534. doi: 10.1038/s41596-018-0045-2. [DOI] [PubMed] [Google Scholar]
- 24.Ståhl PL, Salmén F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 25.Yao Z, van Velthoven CTJ, Nguyen TN, et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell. 2021;184:3222–3241.e3226. doi: 10.1016/j.cell.2021.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu SZ, Al-Eryani G, Roden DL, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53:1334–1347. doi: 10.1038/s41588-021-00911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fawkner-Corbett D, Antanaviciute A, Parikh K, et al. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell. 2021;184:810–826.e823. doi: 10.1016/j.cell.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen WT, Lu A, Craessaerts K, et al. Spatial transcriptomics and in situ sequencing to study Alzheimer's Disease. Cell. 2020;182:976–991.e919. doi: 10.1016/j.cell.2020.06.038. [DOI] [PubMed] [Google Scholar]
- 29.Ji AL, Rubin AJ, Thrane K, et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. 2020;182:497–514.e422. doi: 10.1016/j.cell.2020.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriques SG, Stickels RR, Goeva A, et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vickovic S, Eraslan G, Salmén F, et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stickels RR, Murray E, Kumar P, et al. Highly sen-sitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat Biotechnol. 2021;39:313–319. doi: 10.1038/s41587-020-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho CS, Xi J, Si Y, et al. Microscopic examination of spatial transcriptome using Seq-Scope. Cell. 2021;184:3559–3572. doi: 10.1016/j.cell.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen A, Liao S, Cheng M, et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball patterned arrays. bioRxiv. 2021:2021.2001.2017.427004. doi: 10.1101/2021.01.17.427004. [DOI] [PubMed] [Google Scholar]
- 35.Lohoff T, Ghazanfar S, Missarova A, et al. Integration of spatial and single-cell transcriptomic data elucidates mouse organogenesis. Nat Biotechnol. 2022;40:74–85. doi: 10.1038/s41587-021-01006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moncada R, Barkley D, Wagner F, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38:333–342. doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 37.Maseda F, Cang Z, Nie Q. DEEPsc: a deep learning-based map connecting single-cell transcriptomics and spatial imaging data. Front Genet. 2021;12:636743. doi: 10.3389/fgene.2021.636743.edd320083dc144ed84fed6fad7e47248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao E, Stone MR, Ren X, et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat Biotechnol. 2021;39:1375–1384. doi: 10.1038/s41587-021-00935-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chidester B, Zhou T, Ma J. SPICEMIX: integrative single-cell spatial modeling for inferring cell identity. bioRxiv. 2021:2020.2011.2029.383067. doi: 10.1101/2020.11.29.383067. [DOI] [Google Scholar]
- 40.Cable DM, Murray E, Zou LS, et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat Biotechnol. 2021 doi: 10.1038/s41587-021-00830-w. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elosua-Bayes M, Nieto P, Mereu E, Gut I, Heyn H. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 2021;49:e50. doi: 10.1093/nar/gkab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong R, Yuan GC. SpatialDWLS: accurate deconvolution of spatial transcriptomic data. Genome Biol. 2021;22:145. doi: 10.1186/s13059-021-02362-7.938a4d0a4a9c40ffa6224e9a224b678a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez R, Li B, Keren-Shaul H, et al. Multi-resolution deconvolution of spatial transcriptomics data reveals cont-inuous patterns of inflammation. bioRxiv. 2021:2021.2005.2010.443517. doi: 10.1101/2021.05.10.443517. [DOI] [Google Scholar]
- 44.He B, Bergenstråhle L, Stenbeck L, et al. Integrating spatial gene expression and breast tumour morphology via deep learning. Nat Biomed Eng. 2020;4:827–834. doi: 10.1038/s41551-020-0578-x. [DOI] [PubMed] [Google Scholar]
- 45.Pang M, Su K, Li M. Leveraging information in spatial transcriptomics to predict super-resolution gene expression from histology images in tumors. bioRxiv. 2021:2021.2011.2028.470212. doi: 10.1101/2021.11.28.470212. [DOI] [Google Scholar]
- 46.Pham D, Tan X, Xu J, et al. stLearn: integrating spatial location, tissue morphology and gene expression to find cell types, cell-cell interactions and spatial trajectories within undissociated tissues. bioRxiv. 2020:2020.2005.2031.125658. doi: 10.1101/2020.05.31.125658. [DOI] [Google Scholar]
- 47.Tan X, Su A, Tran M, Nguyen Q. SpaCell: integrating tissue morphology and spatial gene expression to predict disease cells. Bioinformatics. 2020;36:2293–2294. doi: 10.1093/bioinformatics/btz914. [DOI] [PubMed] [Google Scholar]
- 48.Xu W, Gao Y, Wang Y, Guan J. Protein-protein interaction prediction based on ordinal regression and recurrent convolutional neural networks. BMC Bioinformatics. 2021;22:485. doi: 10.1186/s12859-021-04369-0.56123d2a5f71479c88645bcce4b9a51f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergenstråhle J, Larsson L, Lundeberg J. Seam-less integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genomics. 2020;21:482. doi: 10.1186/s12864-020-06832-3.51b162de24e2441ca4bcaffdb981c4b7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergenstråhle L, He B, Bergenstråhle J, et al. Super-resolved spatial transcriptomics by deep data fusion. bioRxiv. 2020:2020.2002.2028.963413. doi: 10.1101/2020.02.28.963413. [DOI] [PubMed] [Google Scholar]
- 51.Kelly RT. Single-cell proteomics: progress and prospects. Mol Cell Proteomics. 2020;19:1739–1748. doi: 10.1074/mcp.R120.002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz D, Zanotelli VRT, Fischer JR, et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 2018;6:25–36.e25. doi: 10.1016/j.cels.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piehowski PD, Zhu Y, Bramer LM, et al. Automated mass spectrometry imaging of over 2000 proteins from tissue sections at 100-μm spatial resolution. Nat Commun. 2020;11:8. doi: 10.1038/s41467-019-13858-z.e3b5a3a1d7b147dd83097e22370e56dc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thornton CA, Mulqueen RM, Torkenczy KA, et al. Spatially mapped single-cell chromatin accessibility. Nat Commun. 2021;12:1274. doi: 10.1038/s41467-021-21515-7.1ad12e0e2a2a4ba0b41d734d7e7f4e32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng Y, Bartosovic M, Ma S, et al. Spatial-ATAC-seq: spatially resolved chromatin accessibility profiling of tissues at genome scale and cellular level. bioRxiv. 2021:2021.2006.2006.447244. doi: 10.1101/2021.06.06.447244. [DOI] [Google Scholar]