Using laser trapping with recombinant Ndc80 and Dam1 kinetochore complexes, Flores et al. show that interactions between three distinct regions of the complexes support strong, stable attachments to dynamic microtubule tips. Phosphorylating two of the regions reduces both attachment strength and stability. Phosphorylating the third region specifically reduces long-term stability.
Abstract
Accurate mitosis requires kinetochores to make persistent, load-bearing attachments to dynamic microtubule tips, thereby coupling chromosome movements to tip growth and shortening. This tip-coupling behavior depends on the conserved Ndc80 complex and, in budding yeast, on the Dam1 complex, which bind each other directly via three distinct interacting regions. The functional relevance of these multiple interactions was mysterious. Here we show that interactions between two of these regions support the high rupture strengths that occur when applied force is rapidly increased and also support the stability of tip-coupling when force is held constant over longer durations. The contribution of either of these two regions to tip-coupling is reduced by phosphorylation by Aurora B kinase. The third interaction region makes no apparent contribution to rupture strength, but its phosphorylation by Aurora B kinase specifically decreases the long-term stability of tip-coupling. The specific reduction of long-term stability relative to short-term strength might have important implications for mitotic error correction.
Introduction
A central role during mitosis is played by the kinetochores, multiprotein machines that assemble onto specialized sites on each chromosome. Kinetochores couple chromosomes to the dynamic tips of spindle microtubules, thereby enabling the filaments as they grow and shorten to exert forces and move the chromosomes. Microtubules grow and shorten by addition and loss of tubulin subunits from their tips. Kinetochores must form persistent attachments to these tips under diverse and sometimes rapidly changing loading conditions, a fundamental behavior that we refer to as “tip-coupling.” How they do so, and how these attachments are selectively inhibited during mitotic error correction, is not well understood.
The Ndc80 complex, an essential microtubule-binding element of the outer kinetochore, is highly conserved across eukaryotes. In yeast, all four subunits of the Ndc80 complex are essential for viability, as loss-of-function mutations in any of one of them causes chromosomes to detach from spindle microtubules and prevents activation of the spindle assembly checkpoint (Gillett et al., 2004; He et al., 2001; Janke et al., 2001; Wigge et al., 1998; Wigge and Kilmartin, 2001; Zheng et al., 1999), resulting in chromosome missegregation. Inactivation of the Ndc80 complex in many other organisms leads to similar defects in chromosome segregation and checkpoint activation (Appelgren et al., 2003; Bharadwaj et al., 2004; Cheeseman et al., 2004; DeLuca et al., 2005; DeLuca et al., 2002; Desai et al., 2003; Hori et al., 2003; Howe et al., 2001; Martin-Lluesma et al., 2002; McCleland et al., 2003; McCleland et al., 2004; Meraldi et al., 2004; Nabetani et al., 2001; Zheng et al., 1999). The Ndc80 complex links the centromere-binding elements of the inner kinetochore to the microtubule. Previous work shows that individual Ndc80 complexes bind loosely and diffuse along the microtubule lattice and that multiple Ndc80 complexes can track with disassembling microtubule tips (McIntosh et al., 2008; Powers et al., 2009; Zaytsev et al., 2015).
The Ndc80 complex interacts directly with the Dam1 complex, another essential outer kinetochore component in budding yeast, on microtubules (Kim et al., 2017). This interaction with the Dam1 complex increases the ability of couplers based on the Ndc80 complex to bear load (Powers et al., 2009; Tien et al., 2010) and allows them to track more stably with growing and shortening microtubule tips (Lampert et al., 2010; Tien et al., 2010). The Dam1 complex is a heterodecamer containing at least three subunits that interact with the Ndc80 complex (Kim et al., 2017). Although the Dam1 complex has been found only in fungi, the Ska complex likely serves an analogous function in human and other organisms that lack the Dam1 complex (Chan et al., 2012; Daum et al., 2009; Gaitanos et al., 2009; Hanisch et al., 2006; Helgeson et al., 2018; Schmidt et al., 2012; van Hooff et al., 2017). The Dam1 complex contributes to microtubule tip-coupling and force production, and it is a target for regulation of kinetochore–microtubule attachments (Asbury et al., 2006; Cheeseman et al., 2002; Franck et al., 2007; Tanaka et al., 2007; Westermann et al., 2006). In the presence of microtubules, it forms oligomeric rings around the filaments (Miranda et al., 2005; Westermann et al., 2005). Although ring formation is not strictly required for microtubule-driven movement of the Dam1 complex (Gestaut et al., 2008), its oligomerization is essential for proper chromosome segregation (Umbreit et al., 2014) and for strong coupling to microtubule tips in vitro (Umbreit et al., 2014; Volkov et al., 2013).
High-resolution molecular structures for key portions of the fungal Ndc80 and Dam1 complexes are now available (Jenni and Harrison, 2018; Valverde et al., 2016), but their precise arrangement at kinetochores in cells remains unclear. A recent study using cryo-electron tomography found many partial and full Dam1 complex rings near the tips of spindle microtubules and, occasionally, two rings per microtubule (Ng et al., 2019). Efforts to quantify the average number of Dam1 complexes per kinetochore have reported values of 16–32 (Joglekar et al., 2006; Lawrimore et al., 2011). The latter would be sufficient to form two complete rings.
The accuracy of chromosome segregation is ensured in many organisms by Aurora B kinase (Biggins and Murray, 2001; Biggins et al., 1999; Cheeseman et al., 2006; DeLuca et al., 2006; DeLuca et al., 2011; Haase et al., 2017; Hauf et al., 2003; Janke et al., 2002; Musacchio, 2011; Musacchio and Desai, 2017; Pinsky et al., 2006; Shang et al., 2003), which selectively phosphorylates aberrant kinetochore–microtubule attachments, causing their release, while leaving correctly bioriented attachments unaffected. Tension probably provides the signal on which this discrimination is based (Biggins et al., 1999; Li and Nicklas, 1997; Pinsky and Biggins, 2005; Tanaka et al., 2002). Correctly bioriented kinetochores come under tension, which is thought to protect them from Aurora B phosphorylation, allowing opposing phosphatases to stabilize their attachments. Conversely, erroneously attached kinetochores are relaxed and therefore detached by Aurora B phosphorylation. However, this view of error correction leads to a conundrum: how can a detached, phosphorylated kinetochore sustain enough force upon reattachment to allow suppression of Aurora B kinase activity and dephosphorylation, as needed for stabilization, rather than releasing prematurely?
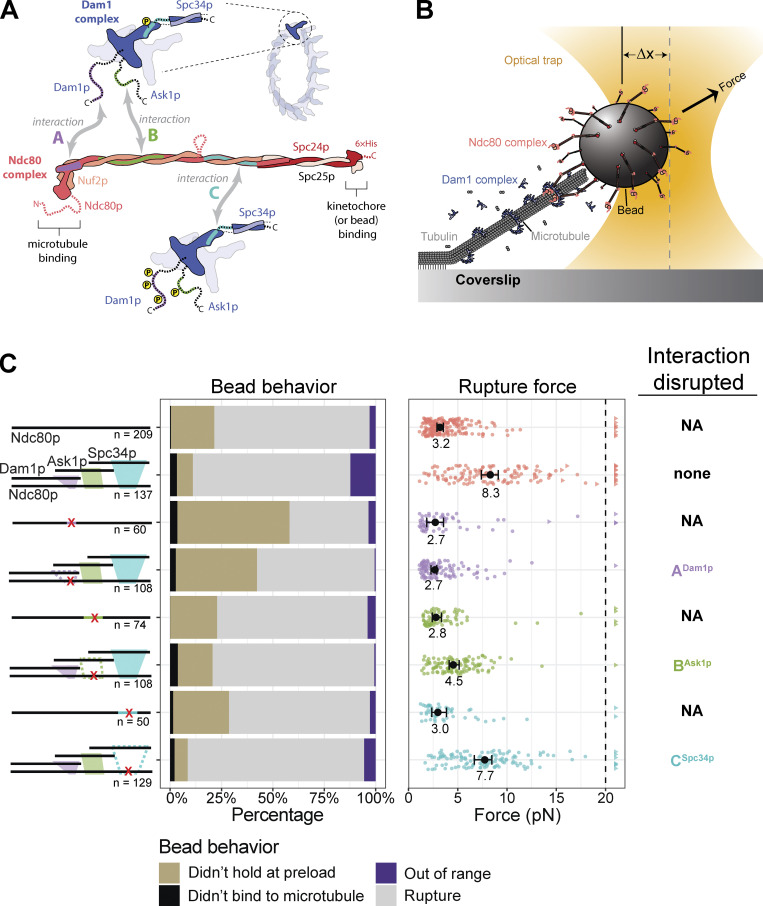
Our previous work identified three distinct interactions between different regions of the Dam1 and Ndc80 complexes (Fig. 1 A; Kim et al., 2017). All three interacting regions can be independently disrupted, either by adding insertion mutations into the participating regions of Ndc80p or by systematically phosphorylating the corresponding regions of the Dam1 complex with the yeast homologue of Aurora B, Ipl1p kinase (Kim et al., 2017; Tien et al., 2013). Insertion mutations in the Ndc80p interaction regions are lethal: they disrupt localization of the Dam1 complex to the kinetochore in vivo, and they cause errors in chromosome segregation (Kim et al., 2017). The importance of the three interacting regions for supporting load-bearing attachments to dynamic microtubule tips has not previously been tested.
Figure 1.
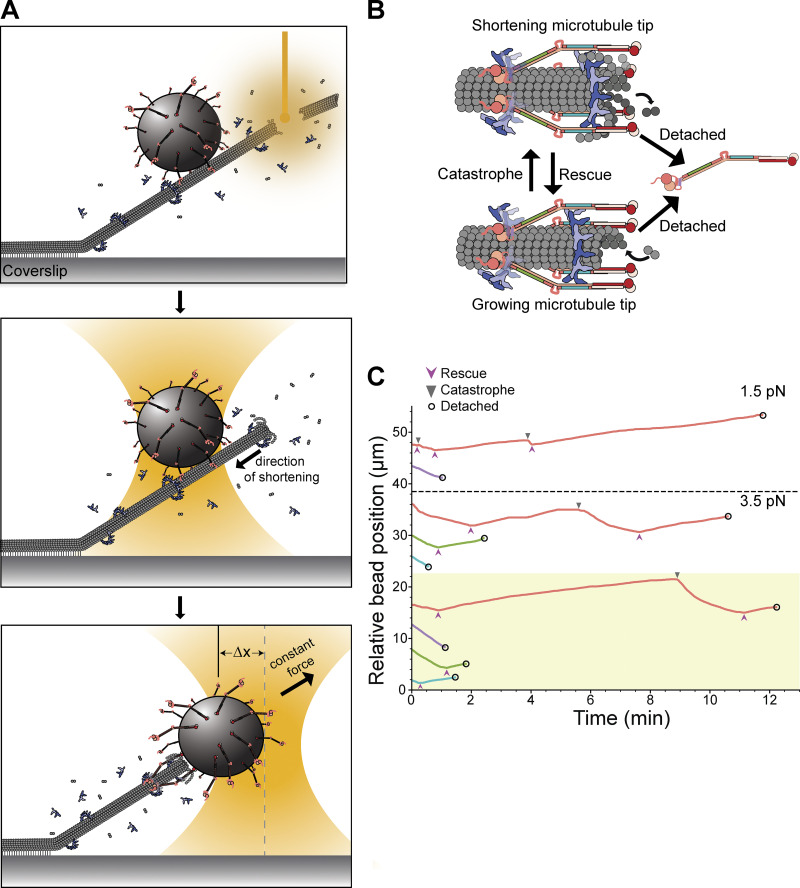
Regions ANdc80p and BNdc80p support load-bearing interactions with the Dam1 complex on growing microtubule tips. (A) The Ndc80 and Dam1 complexes interact via three distinct regions on each complex. Region ANdc80p near the microtubule-binding head of Ndc80p interacts with a corresponding region ADam1p within the disordered C-terminal extension of Dam1p (highlighted in purple). Region BNdc80p interacts with BAsk1p within the C-terminal extension of Ask1p (green). Region CNdc80p interacts with CSpc34p on the C-terminal portion of Spc34p (turquoise). The Ndc80 and Dam1 complexes and their three interacting regions are shown approximately to scale, based on their high-resolution structures (Jenni and Harrison, 2018; Valverde et al., 2016). (B) Schematic of a rupture force experiment. Beads were coated either with wild-type Ndc80 complexes or with mutant Ndc80 complexes carrying insertions that disrupted regions ANdc80p, BNdc80p, or CNdc80p. Individual beads were attached using a laser trap to the tips of single growing microtubules. In the presence or absence of 30 nM Dam1 complex in solution, their attachment strength was measured by ramping the force (at 0.25 pN · s−1) until rupture occurred. (C) Rupture strengths for Ndc80-decorated beads measured in the presence or absence of Dam1 complex. Schematics on the left depict the complexes used in each experiment, including either wild-type Ndc80 complexes or mutant Ndc80 complexes with disruptions in ANdc80p, BNdc80p, or CNdc80p. The Ndc80 mutations are indicated by red X symbols, and the disrupted interactions with the Dam1 complex are shown as dashed outlines. The stacked bar graph in the middle shows the fraction of tested beads that exhibited each of the following four behaviors: (i) did not bind to microtubule (black), (ii) attached to the microtubule but did not hold the 1-pN preload force (gold), (iii) ruptured at a force >1 pN (gray), or (iv) right-censored (purple), when the bead reached the maximum trap force before rupturing. The graph on the right shows the measured rupture forces. Each colored circle represents a single rupture event. Each colored triangle represents right-censored data, when a bead reached the maximum trap force before rupturing. The total number of measurements for each condition, including ruptures and right-censored events, are indicated by n values below the schematics. The black circles represent the median rupture forces, with 95% CIs represented as black bars. Numbers below the black circles indicate median values.
To study the contributions of the three regions to load-bearing interactions between the Ndc80 and Dam1 complexes, we engineered and purified a series of recombinant complexes carrying mutations or phosphorylations that specifically disrupt each region, individually and in combination. We then directly measured the strength and stability of attachments based on the Ndc80 and Dam1 complexes to the tips of individual dynamic microtubules, using feedback-controlled laser trapping. Our results indicate that two regions (A and B) support interactions required for tip-coupling, contributing to the rupture strength measured when applied force is rapidly increased and also supporting the stability of attachment when force is held constant over longer durations. Both activities are inhibited by Ipl1p phosphorylation of regions A or B. One region (C) is required for tip-coupling only when force is held constant over longer durations. Phosphorylating this region specifically reduces the long-term stability of attachment without reducing short-term rupture strength, an effect that might help explain how newly bioriented attachments can sustain enough force to allow dephosphorylation and stabilization.
Results
Two regions of the Ndc80 complex support strong load-bearing interactions with the Dam1 complex on growing microtubule tips
Our prior study showed that mutations inserted into the protein Ndc80p at each of three distinct regions, which we refer to as ANdc80p, BNdc80p, and CNdc80p (Fig. 1 A), can disrupt the interaction of the Ndc80 complex with the Dam1 complex (Kim et al., 2017). Fluorescence-based assays in that study allowed measurement of the affinity of the Ndc80 and Dam1 complexes for one another on microtubules but did not include external force, which kinetochores must sustain almost continuously during mitosis.
To begin assessing the contributions of each interaction region to the formation of load-bearing tip attachments, we performed rupture force assays (Fig. 1 B). Polystyrene microbeads were coated with wild-type Ndc80 complex, or with one of three mutant complexes carrying insertion mutations that disrupt region ANdc80p, BNdc80p, or CNdc80p. The beads were then introduced into a chamber containing dynamic microtubules growing actively from coverslip-anchored seeds, with purified tubulin and GTP in solution, and in the presence or absence of wild-type Dam1 complex in solution. Individual beads were captured using a laser trap, placed onto a growing microtubule tip, and then pulled with a steadily increasing force ramp (increasing the force at a rate of 0.25 pN · s−1) until they ruptured from the tip. Many ruptures were recorded for each condition, to allow statistically rigorous comparisons of median strengths. The microbeads were decorated with a relatively high density of Ndc80 complexes, such that many bead-bound Ndc80 complexes, up to a maximum of ∼90, could probably interact simultaneously with the individual dynamic microtubule tip (see Materials and methods). Free Dam1 complexes were included at concentrations sufficient to make a clear contribution to rupture strength, as detailed below, but they were not visible in the trapping microscope, so their precise structural arrangement at the Ndc80 complex–microtubule interface was unknown.
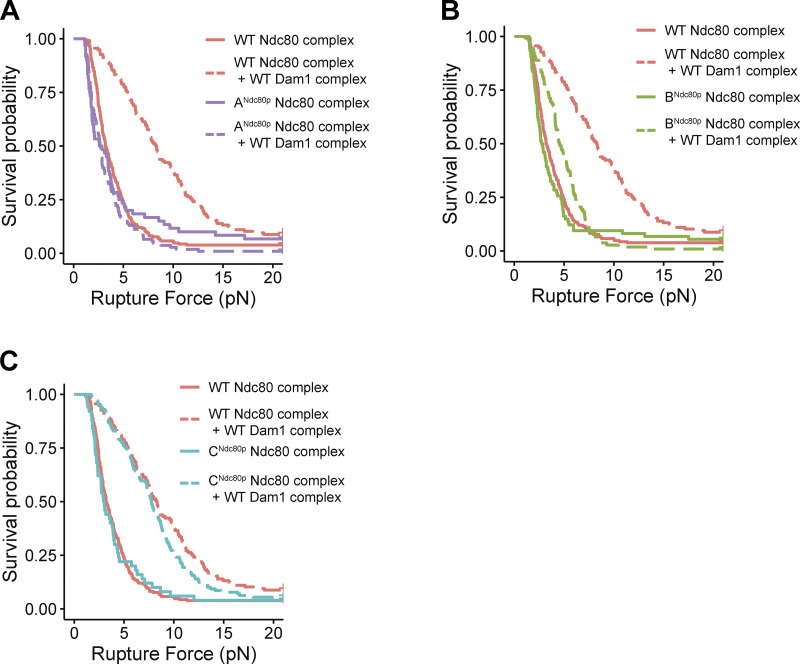
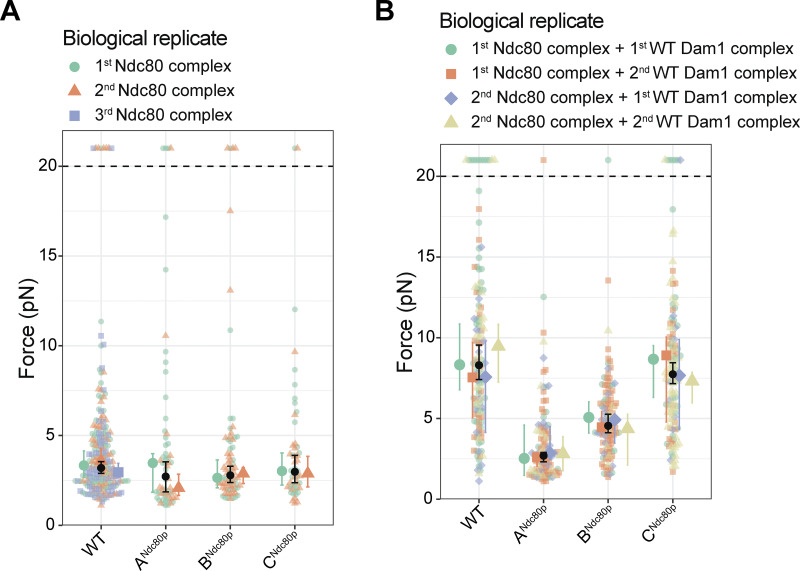
Disrupting region BNdc80p or CNdc80p had no effect on the ability of the Ndc80 complex alone to form attachments or to bear load during tip growth (Fig. 1 C, Fig. S1, B and C; and Table S1). However, beads coated with the mutant ANdc80p Ndc80 complex failed more frequently to support the minimum preload force, ∼1 pN (Fig. 1 C and Table S1). Nevertheless, a significant fraction supported loads >1 pN and gave measurable rupture strengths, slightly weaker than beads coated with wild-type Ndc80 complex (Figs. 1 C and S1 A and Table S1). Adding free Dam1 complex to tip-couplers based on the wild-type Ndc80 complex increased their median rupture strength from 3.2 to 8.3 pN, consistent with previous findings (Figs. 1 C and S1; Tien et al., 2010). In contrast, addition of the Dam1 complex completely failed to strengthen tip-couplers based on the mutant ANdc80p Ndc80 complex and only partially strengthened couplers based on mutant BNdc80p, raising their median rupture strength to only 4.5 pN (Fig. 1 C, Fig. S1, A and B; and Table S1). Couplers based on the mutant CNdc80p Ndc80 complex were strengthened upon addition of Dam1 complex by an amount that was indistinguishable from the strengthening of wild-type Ndc80–based couplers (Figs. 1 C and S1 C and Table S1). Biological replicates were in close agreement (Fig. S2, A and B; and Table S2). These observations indicate that regions ANdc80p and BNdc80p support load-bearing interactions with the Dam1 complex that are important for strong coupling to growing microtubule tips, whereas region CNdc80p did not appear to contribute under the conditions of our rupture force assay. Our observations also indicate that region ANdc80p is important for tip-coupling by the Ndc80 complex alone.
Figure S1.
Distributions of rupture force for couplers based on wild-type and mutant Ndc80 complexes, displayed as Kaplan-Meier survival curves. (A–C) Kaplan–Meier survival curves comparing wild-type Ndc80 complex versus Ndc80 mutants that disrupt region ANdc80p (A), region BNdc80p (B), and region CNdc80p (C). Solid curves were measured with Ndc80 complex–decorated beads alone, in the absence of free Dam1 complex. Dashed curves were measured in the presence of 30 nM wild-type Dam1 complex in solution. A single dataset recorded with wild-type Ndc80 complex is shown on all three graphs for comparison.
Figure S2.
Agreement of rupture force distributions across biological replicates. (A) Superplot showing the rupture forces measured using beads coated with Ndc80 complex alone (in the absence of free Dam1 complex), colored separately for the first biological replicate (aquamarine), the second biological replicate (orange), and the third biological replicate (only for wild type; blue). Data points above the horizontal dashed line were right-censored, when a bead reached the maximum trap force before rupturing. Larger colored symbols represent medians and 95% CIs for each biological replicate. Black symbols represent medians and 95% CIs for all the data combined and are identical to the medians shown in Fig. 1 C. (B) Superplot showing rupture force measured using beads coated with Ndc80 complex in the presence of 30 nM free Dam complex in solution. Aquamarine data were collected using the first biological replicate of both the Ndc80 and Dam1 complexes. Orange data were collected using the first biological replicate of the Ndc80 complex and the second replicate of the Dam1 complex. Blue data were collected using the second biological replicate of Ndc80 complex and the first biological replicate of Dam1 complex. Tan data were collected using the second biological replicate of both Ndc80 and Dam1 complexes. Data points above the horizontal dashed line were right-censored, when a bead reached the maximum trap force before rupturing. Larger colored symbols represent medians and 95% CIs for each biological replicate. Black symbols represent medians and 95% CIs for all the data combined and are identical to the medians shown in Fig. 1 C.
The two corresponding regions of the Dam1 complex support strong load-bearing interactions with the Ndc80 complex on growing microtubule tips
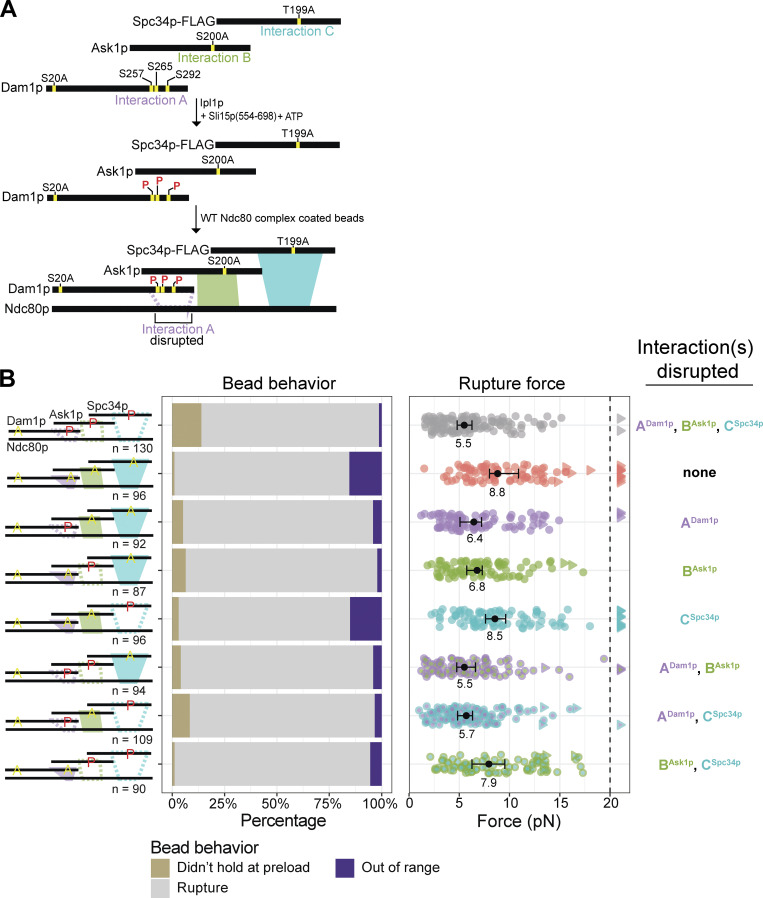
The three previously identified regions of the Ndc80 complex, ANdc80p, BNdc80p, and CNdc80p, interact with three corresponding regions of the Dam1 complex, which we refer to as ADam1p, BAsk1p, and CSpc34p (with superscripts denoting the protein subunits involved; Kim et al., 2017). Each interaction can be inhibited by Ipl1p phosphorylation specifically at Ser/Thr residues within the corresponding regions of the Dam1 complex (Kim et al., 2017). Prior work shows that concurrent phosphorylation of all three of these regions has little to no effect on oligomerization of the Dam1 complex, nor on its intrinsic affinity for microtubules (Gestaut et al., 2008; Lampert et al., 2010; Tien et al., 2010; Zelter et al., 2015). However, oligomerization can be inhibited by phosphorylation specifically at residue S20 on Dam1p (Zelter et al., 2015). For this reason, we included a phospho-blocking S20A mutation in all our phosphorylated Dam1 constructs, so that we could specifically inhibit their interactions with the Ndc80 complex, without affecting their oligomerization.
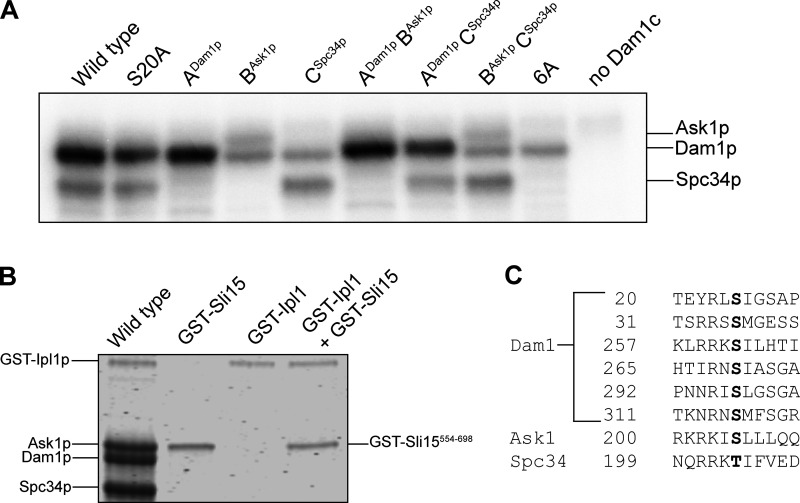
To test how phosphorylating regions ADam1p, BAsk1p, or CSpc34p affects the strength of tip-coupling, we purified a series of recombinant Dam1 complexes carrying Ala substitutions at selected Ipl1p target residues. Treating the mutant Dam1 complexes with purified Ipl1p kinase led to phosphorylation specifically of regions ADam1p, BAsk1p, or CSpc34p, individually and in different combinations (Fig. 2 A). Assaying the incorporation of [γ-32P]ATP confirmed high levels of phosphorylation, with 2–3 phosphoryl groups incorporated per molecule of Dam1p, ∼0.7 phosphoryl groups per molecule of Ask1p, and ∼1 phosphoryl group per molecule of Spc34p (Table 1 and Fig. S3 A). As expected, Ala substitutions at the Ipl1p target residues in regions BAsk1p (at Ser 200) or CSpc34p (at Thr 199) blocked all detectable phosphorylation of Ask1p or Spc34p, respectively. However, blocking all four known phosphorylation sites in Dam1p (i.e., Ser 20, Ser 257, Ser 265, and Ser 292) only partially blocked its phosphorylation, suggesting the presence of previously unidentified Ipl1p target residues (Table 1 and Fig. S3 A). Indeed, mass spectrometry revealed additional phosphorylation of Dam1p at Ser 31 and Ser 311, residues that fall within sequences similar to the Ipl1p consensus motif (Cheeseman et al., 2002; Fig. S3 C). Phosphorylation at these two new target residues did not affect the strength of tip-couplers based on the Ndc80 and Dam1 complexes (Fig. S4 and Table S3).
Figure 2.
Regions ADam1p and BAsk1p support load-bearing interactions with the Ndc80 complex on growing microtubule tips. (A) Method for phosphorylating specific interaction regions within the Dam1 complex. In the ADam1p complex, alanine substitutions at Ask1pS200 and Spc34pT199 block regions BAsk1p and CSpc34p, so treatment of ADam1p complex with Ipl1p kinase and ATP led to phosphorylation specifically of the region ADam1p sites. Analogous approaches were also used to specifically phosphorylate regions BAsk1p or CSpc34p or various combinations of regions ADam1p, BAsk1p, and CSpc34p. (B) Rupture strengths for Ndc80-decorated beads measured in the presence of Dam1 complex, phosphorylated at indicated sites (red P’s) and carrying phospho-blocking alanine substitutions (yellow A’s), as diagrammed at left. Interaction regions that were disrupted by phosphorylation are shown in the diagram as dashed outlines. The stacked bar graph in the middle shows the fraction of tested beads that exhibited each of the following three behaviors: (i) attached to the microtubule but did not hold the 1-pN preload force (gold), (ii) ruptured at a force >1 pN (gray), or (iii) right-censored, when the bead reached the maximum trap force before rupturing (purple). The graph on the right shows the measured rupture forces. Each colored circle represents a single rupture event. Each colored triangle represents right-censored data, when a bead reached the maximum trap force before rupturing. The total number of measurements for each condition, including ruptures and right-censored events, for each condition are indicated by n values below the schematics. Black circles represent median rupture forces, with bars showing 95% CIs. Numbers below the black circles indicate median values.
Table 1.
Alanine substitutions and phosphorylation levels measured for the engineered Dam1 complexes used in this study.
| Complex | Ndc80 interaction regions | Other phospho-sites | Incorporation of [γ-32P]ATP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Interactions | A | B | C | (NA) | Moles phosphoryl group per mole Dam1p | Moles phosphoryl group per mole Ask1p | Moles phosphoryl group per mole Spc34p | ||||
| Dam1c subunit | Dam1p | Ask1p | Spc34p | Dam1p | |||||||
| Residues | S257 | S265 | S292 | S200 | T199 | S20 | S31 | S311 | |||
| Wild-type | S | S | S | S | T | S | S | S | 2.6 ± 0.41 | ND | 1.2 ± 0.04 |
| S20A | S | S | S | S | T | A | S | S | 2.1 ± 0.10 | ND | 1.0 ± 0.20 |
| ADam1p | S | S | S | A | A | A | S | S | 2.3 ± 0.51 | None | None |
| BAsk1p | A | A | A | S | A | A | S | S | 0.90 ± 0.05 | 0.7 ± 0.09 | None |
| CSpc34p | A | A | A | A | T | A | S | S | 0.77 ± 0.14 | None | 0.93 ± 0.15 |
| ADam1pBAsk1p | S | S | S | S | A | A | S | S | 2.6 ± 0.87 | ND | None |
| ADam1pCSpc34p | S | S | S | A | T | A | S | S | 1.9 ± 0.56 | None | 0.75 ± 0.03 |
| BAsk1pCSpc34p | A | A | A | S | T | A | S | S | 0.80 ± 0.11 | 0.61 ± 0.10 | 0.92 ± 0.14 |
| 6A | A | A | A | A | A | A | S | S | 0.72 | None | None |
Alanine substitutions blocked phosphorylation at indicated sites (bold A's), so treatment with Ipl1 kinase and ATP led to phosphorylation specifically at sites that remained unmodified (S or T). Phosphorylation levels on Dam1p, Spc34p, and when possible, Ask1p were quantified as described in Materials and methods. NA, not applicable; ND, not determined.
Figure S3.
Analysis of phosphorylation levels on wild-type and mutant Dam1 complexes. (A) Autoradiograph of an SDS-PAGE gel (8–14%) showing incorporation of [γ-32P]ATP into different interaction regions of the Dam1 complex. Wild-type Dam1 complex contains no alanine mutations. The S20A complex has that single mutation in Dam1p. The ADam1p complex has mutations: Dam1p S20A, Ask1p S200A, and Spc34p T199A, so only region ADam1p can be phosphorylated. The BAsk1p complex has mutations: Dam1p S20A, S257A, S265A, and S292A and Spc34p T199A, so only region BAsk1p can be phosphorylated. The CAsk1p complex has mutations: Dam1p S20A, S257A, S265A, and S292A and Ask1p S200A, so only region CSpc34p can be phosphorylated. The ADam1pBAsk1p complex has mutations: Dam1p S20A and Spc34p T199A, so only regions ADam1p and BAsk1p can be phosphorylated. The ADam1pCSpc34p complex has mutations: Dam1p S20A and Ask1p S200A, so only regions ADam1p and CAsk1p can be phosphorylated. The BAsk1pCSpc34p complex has mutations: Dam1p S20A, S257A, S265A, and S292A, so only regions BAsk1p and CSpc34p can be phosphorylated. The 6A complex has mutations: Dam1p S20A, S257A, S265A, and S292A; Ask1p S200A; and Spc34p T199A, so none of the interaction regions can be phosphorylated. The rightmost lane shows a control reaction performed in the absence of the Dam1 complex, with only Ipl1p and Sli15p. (B) Coomassie blue–stained SDS-PAGE gel (8–14%) of wild-type Dam1 complex with GST-Sli15554–698 and GST-Ipl1p (wild-type), GST-Sli15554–698 alone, GST-Ipl1p alone, and GST-Sli15554–698 together with GST-Ipl1p. (C) Highlighted in black are residues that are phosphorylated under the conditions of our phosphorylation assay. Source data are available for this figure: SourceData FS3.
Figure S4.
Phosphorylation of two new Ipl1p target residues in Dam1p, S31 and S311, does not affect rupture strength. Rupture strengths for Ndc80-decorated beads measured in the presence of Dam1 complex phosphorylated at indicated sites (red P’s) and carrying phospho-blocking alanine substitutions (yellow A’s), as diagrammed at left. Interaction regions that were disrupted by phosphorylation are shown in the diagram as dashed outlines. The stacked bar graph in the middle shows the fraction of tested beads that exhibited each of the following three behaviors: (i) attached to the microtubule but did not hold the 1-pN preload force (gold), (ii) ruptured at a force >1 pN (gray), or (iii) right-censored, when the bead reached the maximum trap force before rupturing (purple). The graph on the right shows the measured rupture forces. Each colored circle represents a single rupture event. Each colored triangle represents right-censored data, when a bead reached the maximum trap force before rupturing. The total number of measurements for each condition, including ruptures and right-censored events, for each condition are indicated by n values below the schematics. Black circles represent median rupture forces, with bars showing 95% CIs. Numbers below the black circles indicate median values. To avoid oligomerization deficiency, S20 on Dam1p was mutated to alanine in all experiments shown here. Yellow shaded regions (rows 1 and 3) indicate experiments that used Dam1 complexes phosphorylated by treatment with Ipl1p, Sli15p, and ATP. Unshaded regions (rows 2 and 4) indicate experiments that used mock-treated Dam1 complexes (i.e., treated with Ipl1p and Sli15p, but in the absence of ATP).
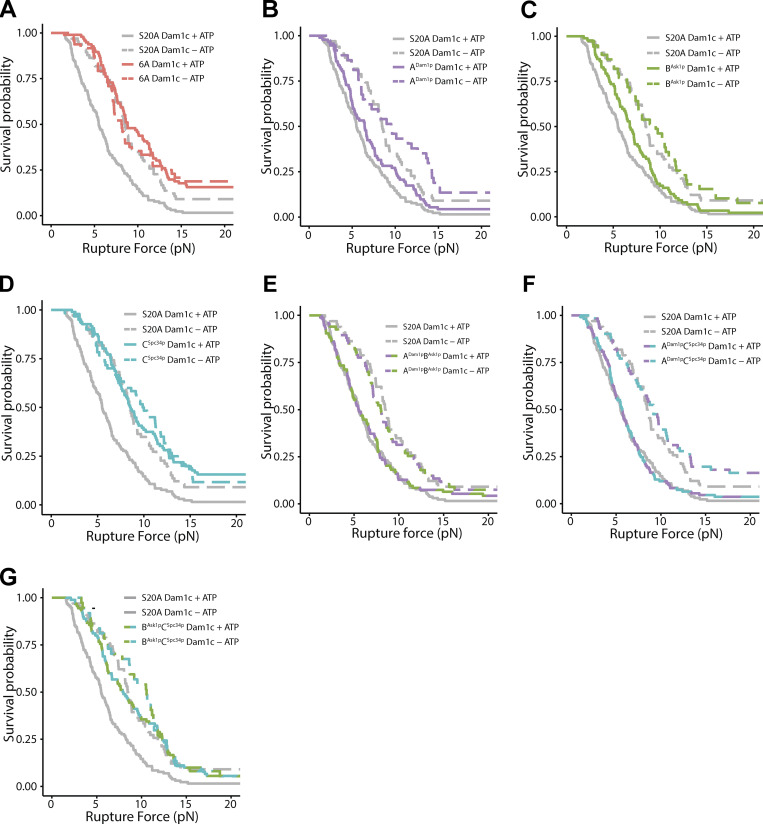
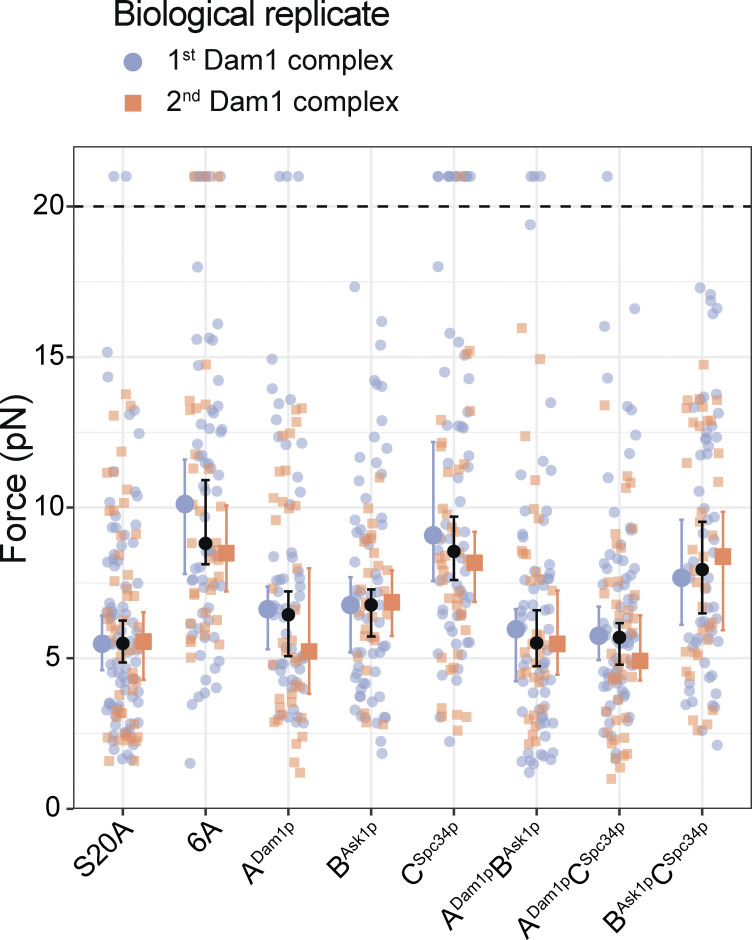
Individually phosphorylating region ADam1p of the Dam1 complex reduced the median rupture strength of Ndc80 complex–based couplers to 6.4 pN, a value that is weaker than the strength measured using the unphosphorylated Dam1 complex (8.3 pN) or using a phospho-deficient 6A Dam1 complex, carrying six Ala substitutions that blocked Ipl1p phosphorylation at all three regions (8.8 pN; Figs. 2 B and S5 B and Table S4). Phosphorylating only region BAsk1p reduced the median rupture strength to 6.8 pN, a value that is also lower than the phospho-deficient 6A Dam1 complex (Figs. 2 B and S5 C and Table S4). Phosphorylating region CSpc34p caused no significant weakening, despite nearly stoichiometric phosphorylation of Spc34p (Figs. 2 B and S5 D and Table S4). Different biological replicates of the phosphorylated Dam1 complexes gave similar results (Fig. S6 and Table S5). Together, these data confirm that regions ADam1p and BAsk1p of the Dam1 complex support load-bearing interactions with the Ndc80 complex that are important for coupling to growing microtubule tips, and that region CSpc34p does not appear to contribute, mirroring the rupture strengths recorded when corresponding regions of the Ndc80 complex were disrupted. Moreover, they show that interactions A and B are both weakened by Ipl1p phosphorylation of the participating regions of the Dam1 complex, ADam1p and BAsk1p.
Figure S5.
Rupture force distributions for Ndc80-decorated beads measured in the presence of phosphorylated and mock-phosphorylated Dam1 complex, displayed as Kaplan-Meier survival curves. (A–G) Kaplan–Meier survival curves comparing the S20A Dam1 complex with the 6A Dam1 complex (A), the ADam1p complex (B), the BAsk1p complex (C), the CSpc34p complex (D), the ADam1pBAsk1p complex (E), the ADam1pCSpc34p complex (F), and the BAsk1pCSpc34p complex (G). Solid curves were measured with ATP-treated Dam1 complex. Dashed curves were measured with mock-treated Dam1 complex (i.e., without the addition of ATP).
Figure S6.
Agreement of rupture force distributions across biological replicate Dam1 complexes. Superplot showing the rupture forces measured using beads coated with wild-type Ndc80 complex in the presence of the first biological replicates (blue) and the second biological replicates (orange) of the indicated Dam1 complexes, which were prephosphorylated in vitro (the same wild-type Ndc80 complex was used in all cases). Data points above the horizontal dashed line were right-censored when the bead reached the maximum trap force before rupturing. Larger colored symbols represent medians and 95% CIs for each biological replicate. Black symbols represent medians and 95% CIs for all the data combined and are identical to the medians shown in Fig. 2 B.
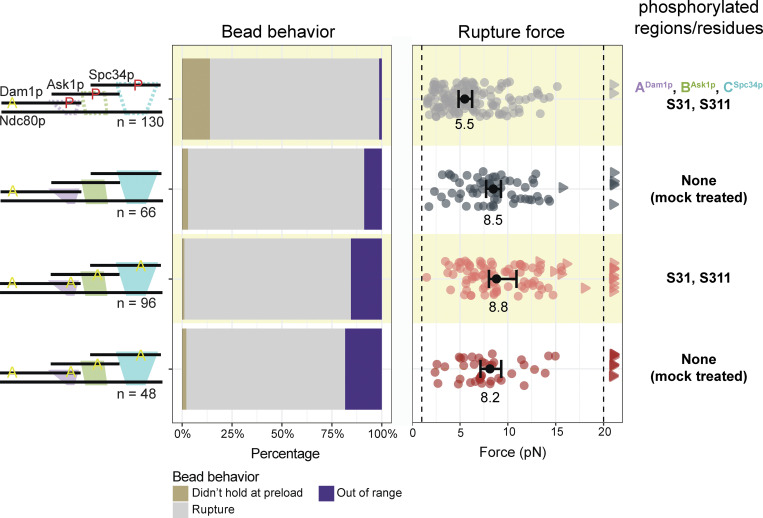
When measured in the presence of Dam1 complex phosphorylated at all three regions, ADam1p, BAsk1p, and CSpc34p, the median strength of Ndc80 complex-based couplers was weak (5.5 pN; Fig. 2 B), as expected, but higher than the strength of couplers based on wild-type Ndc80 complex alone (3.2 pN; Fig. 1 C). This observation indicates that our phosphorylation of all three regions of the Dam1 complex did not completely disrupt its load-bearing interactions with the Ndc80 complex, perhaps because we were unable to fully saturate all the phosphorylation sites on Dam1p and Ask1p (Table 1 and Fig. S3). Additionally, there was a higher percentage of beads that could not withstand 1 pN of load as compared with the phospho-deficient 6A Dam1 complex (Fig. 2 B and Tables S3 and S4). Phosphorylating regions ADam1p and BAsk1p together caused no further weakening compared with phosphorylation of ADam1p alone, suggesting redundancy in the load-bearing function of regions ADam1p and BAsk1p (Fig. 2 B, Fig. S5, E–G; and Table S4).
All three interacting regions of the Ndc80 and Dam1 complexes contribute to attachment stability under constant load
During mitosis, kinetochores maintain persistent, load-bearing attachments to spindle microtubules that oscillate between phases of growth and shortening. To test the importance of the three distinct interactions between the Ndc80 and Dam1 complexes over longer durations, and across different microtubule tip states, we performed force clamp assays. As in the earlier rupture force assays, polystyrene microbeads were coated with either wild-type or mutant Ndc80 complex and introduced into a chamber containing dynamic microtubules growing from coverslip-anchored seeds, and with tubulin and Dam1 complex free in solution. In this case, however, individual beads were attached initially to the sides of the microtubules using the laser trap, such that the growing filament tips extended past the beads (Fig. 3 A). Laser scissors were then used to sever the growing tips, causing the microtubules to rapidly shorten (Franck et al., 2010; Walker et al., 1989). When a shortening tip encountered a side-bound bead, the bead often began tracking with the tip as the microtubule continued shortening. Initiating the experiment in this manner facilitated collection of many episodes of tip-tracking during shortening. Sometimes the tip subsequently underwent one or more switch events, resuming growth, switching back into shortening, etc., before bead detachment (Fig. 3 B). The laser trap was programmed to maintain a constant tensile force as the bead tracked with the dynamic tip until a detachment occurred (Fig. 3 C). For measurements using the mutant BNdc80p or CNdc80p Ndc80 complexes, we applied 3.5 pN of tension, a level chosen to give modest detachment rates in the presence of wild-type Dam1 complex. Likewise, 3.5 pN of tension was applied to beads coated with wild-type Ndc80 complexes in the presence of either phospho-deficient 6A Dam1 complex or Dam1 complexes phosphorylated at regions ADam1p, BAsk1p, or CSpc34p. A lower tension of 1.5 pN was applied when using the mutant ANdc80p Ndc80 complex due to its relative weakness. Control experiments using the wild-type Ndc80 and Dam1 complexes were performed at both levels of force for comparison. To measure detachment rates specifically during microtubule growth or shortening, we collected many traces for each type of coupler, counted the numbers of detachments observed during growth or shortening, and then divided these counts by the total observation times spent in growth or shortening. The same approach was used to measure the rates of switching from growth into shortening (i.e., catastrophe rates) and from shortening into growth (rescue rates).
Figure 3.
Together the Ndc80 and Dam1 complexes maintain persistent, load-bearing attachments to growing and shortening microtubule tips. (A) Schematic of force clamp assay. A bead coated with Ndc80 complex and in the presence of free Dam1 complex is initially placed onto the side (lattice) of a microtubule. Laser scissors are then used to sever the microtubule tip (top panel), which induces tip shortening (middle panel). Eventually, the shortening microtubule tip reaches the bead, whereupon a constant force is applied to the bead as it tracks with the dynamic tip (bottom panel). (B) A coupler can detach from a microtubule tip either during growth (assembly) or during shortening (disassembly), with distinct rates for each type of detachment event. The tip can interconvert between growth and shortening via transitions called catastrophe and rescue. (C) Examples of tip-coupled movement recorded while constant tensile force was applied using a feedback-controlled laser trap. The upper five traces, plotted in the unshaded (white) region, were recorded using beads decorated with either wild-type Ndc80 complex (red), or with mutant Ndc80 complexes carrying insertions that disrupted regions ANdc80p (purple), BNdc80p (green), or CNdc80p (turquoise), all in the presence of free wild-type Dam1 complex. The lower four traces, plotted in the yellow shaded region, were recorded using beads decorated with wild-type Ndc80 complex in the presence of free phosphodeficient 6A Dam1 complex (red), as a control, or in the presence of Dam1 complex phosphorylated at region ADam1p (purple), BAsk1p (green), or CSpc34p (turquoise). Traces above the dashed line were recorded with 1.5 pN of continuously applied tension, and those below the dashed line were recorded with 3.5 pN. Arrowheads mark catastrophe (▼) and rescue (▲) events. Open circles indicate detachments.
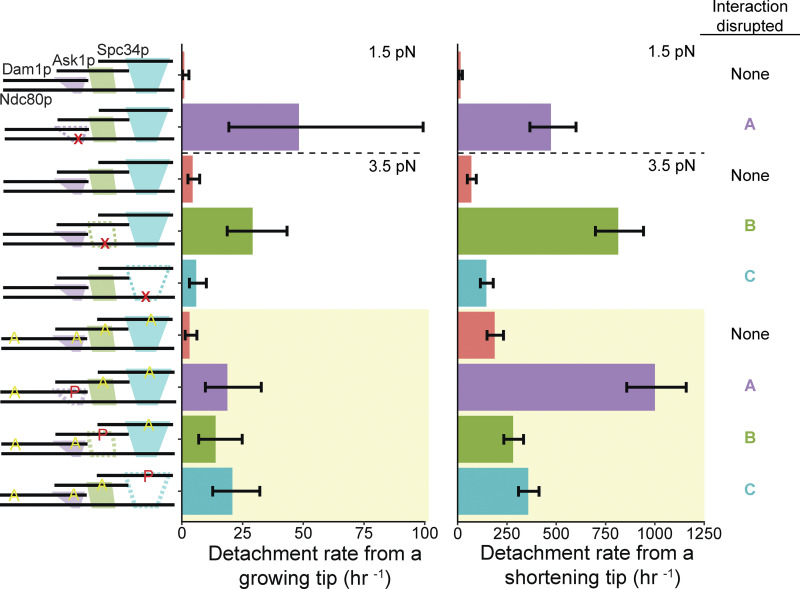
In the absence of the Dam1 complex, very few Ndc80 complex–coated beads could track persistently with shortening microtubule tips under the applied force of 1.5 pN (only 3 of 41 beads tested, or 7%) and none could track at 3.5 pN (0 of 33 beads tested). However, the addition of free wild-type Dam1 complex enabled more robust tracking (8 of 16 at 1.5 pN, 50%; 19 of 56 at 3.5 pN, 34%), confirming that the Dam1 complex is crucial for Ndc80 complex–based couplers to sustain piconewton loads while attached to a dynamic microtubule tip, as previously reported (Tien et al., 2010).
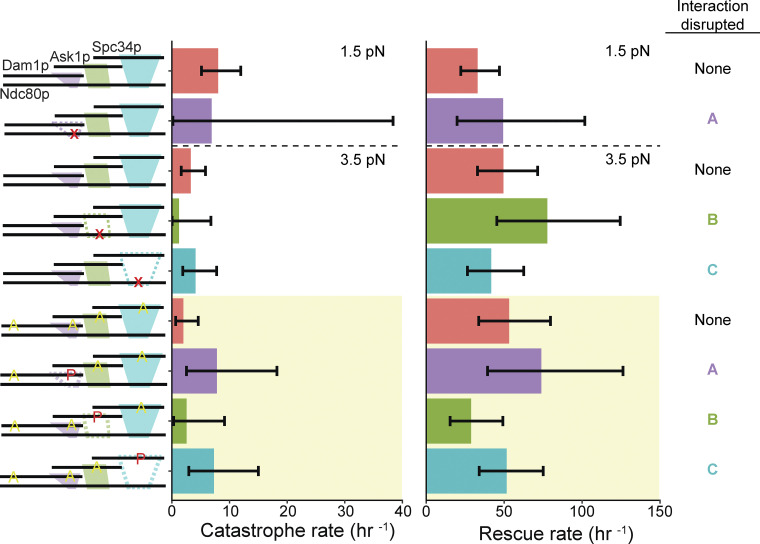
In the presence of Dam1 complex, detachment rates measured during force clamp experiments usually followed a trend similar to that seen earlier in rupture force measurements. Mutating regions ANdc80p or BNdc80p of the Ndc80 complex or phosphorylating regions ADam1p or BAsk1p of the Dam1 complex caused significantly higher detachment rates from growing and shortening microtubule tips compared with controls, in which none of the interactions were disrupted (Fig. 4 and Tables S6 and S7). These observations provide further evidence that interactions A and B are both important for sustaining load-bearing attachments to dynamic microtubule tips. The detachment rates measured after perturbing interaction C followed a different trend compared with the earlier rupture force measurements. Whereas mutating region CNdc80p or phosphorylating the corresponding region CSpc34p had no impact on rupture strengths (Figs. 1 and 2), these same perturbations significantly reduced the stability of attachments during force clamp experiments. Mutating region CNdc80p of the Ndc80 complex, for example, increased the detachment rate during microtubule shortening (Fig. 4 and Tables S6 and S7). Phosphorylating region CSpc34p of the Dam1 complex accelerated detachments during both microtubule growth and shortening (Fig. 4 and Tables S6 and S7). These observations indicate that interaction C can contribute to the stability of tip-coupling under load, depending on the conditions. Phosphorylating either region ADam1p or CSpc34p also resulted in a threefold higher catastrophe rate compared with the controls using phospho-deficient 6A Dam1 complex (Fig. S7 and Tables S6 and S7).
Figure 4.
All three interactions between the Ndc80 and Dam1 complexes contribute to attachment stability under constant load. Detachment rates from either growing or shortening microtubule tips, measured using beads coated with 5 nM Ndc80 complex and 5 nM Dam1 complex in solution. Schematics on the left depict the complexes used in each experiment, including either wild-type or mutant Ndc80 complexes with disruptions (indicated by red X symbols) in ANdc80p, BNdc80p, or CNdc80p and with unphosphorylated Dam1 complexes, or including wild-type Ndc80 complexes with Dam1 complexes that were either unphosphorylated or were phosphorylated at indicated sites (red P’s) and carried phospho-blocking alanine substitutions (yellow A’s), as diagrammed. Interactions that were disrupted are shown as dashed outlines in the schematics on the left and are also listed on the far right. Bars above the dashed horizontal lines on the graphs represent rates measured at 1.5 pN of constant tension. Those below the dashed horizontal lines represent rates measured at 3.5 pN of constant tension. The yellow shaded region indicates rates measured using Dam1 complexes that were phosphorylated by treatment with Ipl1p, Sli15p, and ATP. Error bars represent 95% CIs, which were estimated using the exact method.
Figure S7.
Catastrophe and rescue rates measured using beads coated with 5 nM Ndc80 complex and 5 nM Dam1 complex in solution. Schematics on the left depict the complexes used in each experiment, including either wild-type or mutant Ndc80 complexes with disruptions (indicated by red X symbols) in ANdc80p, BNdc80p, or CNdc80p and with unphosphorylated Dam1 complexes, or including wild-type Ndc80 complexes with Dam1 complexes that were either unphosphorylated or were phosphorylated at indicated sites (red P’s) and carried phospho-blocking alanine substitutions (yellow A’s), as diagrammed. Interactions that were disrupted are shown as dashed outlines in the schematic on the left and are also listed on the far right. Bars above the dashed horizontal lines on the graphs represent rates measured at 1.5 pN of constant tension. Those below the dashed horizontal lines represent rates measured at 3.5 pN of constant tension. The yellow shaded region indicates rates measured using Dam1 complexes that were phosphorylated by treatment with Ipl1p, Sli15p, and ATP. Error bars represent 95% CIs, which were estimated using the exact method.
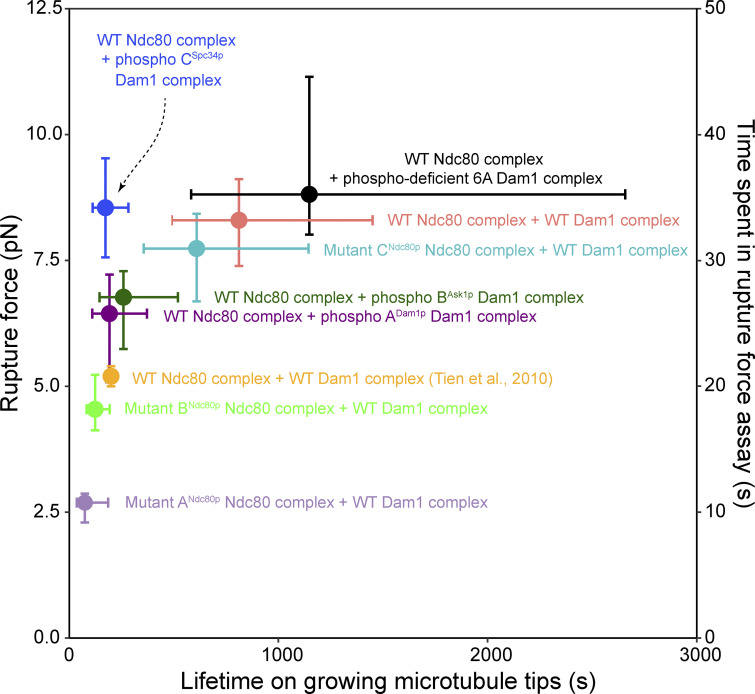
In prior laser trap studies using purified recombinant kinetochore complexes (Helgeson et al., 2018; Tien et al., 2010) or isolated native kinetochore particles (Akiyoshi et al., 2010; Miller et al., 2016; Sarangapani et al., 2013), higher rupture strengths have correlated with longer-lived attachments. All the couplers we studied here also follow this trend, except our couplers using Dam1 complex phosphorylated at region CSpc34p, which exhibit relatively high rupture strengths but relatively low stability under constant force (Fig. 5). Notably, the rupture strength measurements are quick, requiring only ∼30 s to ramp up to the median rupture force measured with wild-type Ndc80 and Dam1 complexes (8.3 pN), whereas the force clamp measurements often last many minutes. This difference in duration seems likely to underlie the differential importance of interaction C in the two experiments and suggests that phosphorylation at region CSpc34 specifically lowers the long-term stability of the attachments while preserving their short-term strength.
Figure 5.
Phosphorylation at region CSpc34p uniquely reduces stability under constant force while preserving rupture strength. Rupture strengths from Figs. 1 and 2 are replotted against attachment lifetimes measured for the same couplers on growing microtubule tips under constant force (1.5–3.5 pN). Data were obtained using beads coated with either wild-type Ndc80 complexes (salmon) or mutant Ndc80 complexes with disruptions in region ANdc80p (light purple), BNdc80p (lime green), or CNdc80p (turquoise) in the presence of unphosphorylated Dam1 complexes, or using beads coated with wild-type Ndc80 complexes in the presence of either phospho-deficient 6A Dam1 complexes (black) or Dam1 complexes phosphorylated at region ADam1p (dark purple), BAsk1p (dark green), or CSpc34p (blue).
Discussion
Electron micrographs of Dam1 complex rings on microtubules showed that Ndc80 complexes can bridge across two Dam1 complex rings in vitro and that all three of the interacting regions are required for this bridging to occur (Kim et al., 2017). A different arrangement for the two complexes, with all three interacting regions of the Ndc80 complex contacting a single Dam1 complex ring, is also possible (Jenni and Harrison, 2018), and multiple different arrangements are seen at kinetochores in vivo (Ng et al., 2019). The results we present here demonstrate clearly that all three interacting regions are important for supporting strong and persistent load-bearing attachments to dynamic microtubule tips. We found that two of the three interactions, A and B, are important both for short-term rupture strength, measured when applied force is rapidly increased, and for the longer-term stability of tip-coupling when force is held constant. The third interaction, C, apparently contributes only to longer-term stability.
Individually mutating any of the three regions of the Ndc80 complex, ANdc80p, BNdc80p, or CNdc80p, partially reduces its affinity for the Dam1 complex in single-molecule fluorescence assays (Kim et al., 2017). Here we found that when the two complexes were coupled to dynamic microtubule tips, mutating region ANdc80p completely abolished their load-bearing interaction, such that tip attachments based on the mutant ANdc80p Ndc80 complex were indistinguishably weak whether or not the Dam1 complex was present. Additionally, about half of the beads coated with the mutant ANdc80p Ndc80 complex failed to withstand even 1 pN of load, suggesting that mutant ANdc80p has a major, intrinsic defect in load-bearing ability. By comparison, mutant BNdc80p was less impaired in its ability to form load-bearing interactions with the Dam1 complex, and mutant CNdc80p was unimpaired when tested in the rupture strength assay. These findings demonstrate that affinity does not always correlate precisely with rupture strength, and they also explain the more severe phenotype of mutant ANdc80p, which disrupts the arrangement of kinetochores and the localization of the Dam1 complex more dramatically in vivo than the B or C mutants (Kim et al., 2017).
Region ANdc80p is located on the “helical hairpin” of Ndc80p (Valverde et al., 2016), a key structural feature previously implicated in Ndc80–Dam1 complex interactions. Mutations in the N-terminal helix of this hairpin are lethal in vivo and, curiously, lead to a higher affinity for the microtubule lattice that is not further enhanced by the addition of Dam1 complex in fluorescence-based in vitro assays (Tien et al., 2013). Similarly, deletion of the nearby small disordered region connecting the CH domain “head” of Ndc80p to the hairpin confers a slow-growth phenotype and prevents the Dam1 complex from enhancing the ability of the Ndc80 complex to bind microtubules in sedimentation assays (Lampert et al., 2013). Our data further confirm the importance of the hairpin for interaction with the Dam1 complex and establish its role specifically in the transmission of force to a dynamic microtubule tip, which represents the physiologically relevant substrate.
Interactions A, B, and C are regulated by the yeast Aurora B homologue, Ipl1p kinase, which phosphorylates the participating regions of the Dam1 complex (Cheeseman et al., 2002; Kim et al., 2017). Phosphorylation decreases the affinity of the Dam1 complex for the Ndc80 complex in both fluorescence (Tien et al., 2010) and sedimentation (Lampert et al., 2010) assays in vitro, and it is important for accurate chromosome segregation in vivo (Cheeseman et al., 2002; Jin et al., 2017). Together, these observations have led to the view that Ipl1p phosphorylation triggers release of kinetochores from microtubules in part by disrupting the Dam1–Ndc80 complex interface (Cheeseman et al., 2002; Doodhi et al., 2021; Kim et al., 2017; Tien et al., 2010). We demonstrate here that Ipl1p phosphorylation goes beyond regulating the affinity of the Dam1 and Ndc80 complexes for each other, also modulating the strength and stability of their interaction during tip coupling.
To our knowledge, the possibility that regulatory mechanisms might differentially affect long-term stability versus short-term strength has not previously been considered. Our work reveals, unexpectedly, that these two aspects of tip-coupling can be differentially affected in reconstitutions containing only two microtubule-binding kinetochore complexes, by phosphorylating different intermolecular interfaces between the two complexes. Ipl1p phosphorylation at region CSpc34p causes no defect in rupture strength but a large defect in attachment lifetimes measured in the force clamp assay. This specific reduction in long-term stability without loss of short-term strength could have interesting and important consequences for error correction in vivo. An initially unattached kinetochore is relaxed and therefore expected to be readily phosphorylated by Ipl1p/Aurora B. Nevertheless, when it establishes a new, correctly bioriented attachment, it must withstand the forces of biorientation long enough for the tension to halt Aurora B phosphorylation while dephosphorylation occurs and stabilizes the attachment (Khodjakov and Pines, 2010). The ability of attachments in which CSpc34p is phosphorylated to survive high force might help to preserve bioriented attachments long enough for their dephosphorylation to occur. We speculate that the phosphorylation of other kinetochore substrates regulated by Aurora B, such as the human Ndc80 and Ska complexes (Chan et al., 2012; DeLuca et al., 2006; Zaytsev et al., 2015), will also be found to differentially reduce long-term stability while maintaining short-term strength.
Materials and methods
Plasmids and constructs
All expression vectors are listed in Table S8.
Protein expression, purification, and phosphorylation
Ndc80 complex
Saccharomyces cerevisiae Ndc80 and Dam1 complexes were expressed in Escherichia coli using polycistronic vectors, as previously described (Kim et al., 2017; Powers et al., 2009; Tien et al., 2010; Wei et al., 2005). All Ndc80 complex constructs were expressed from two bicistronic vectors encoding Ndc80/Nuf2 and Spc24/Spc25 (pEM033). Spc24p of the Ndc80 complex contained a C-terminal 6X His tag. BL21 Rosetta 2 DE3 cells were transformed with wild-type (pETDuet, pEM033), mutant BNdc80p (pJOK004, pEM033), or mutant CNdc80p (pJOK005, pEM033). Cultures were grown to a density of 40 Klett units in NZ medium, protein expression was induced by the addition of 2 mM IPTG, and the culture was incubated at 30°C for 8 h while shaking at 240 rpm. Arctic Express DE3 cells were transformed with pJOK003 (ANdc80p/Nuf2) and pEM033 (Spc24-6X His/Spc25). Cultures were grown to a density of 40 Klett units and cooled by incubating at 8°C for 30 min. Protein expression was induced by the addition of 2 mM IPTG, and the culture was incubated at 8°C for 16 h while shaking at 240 rpm.
For purification of mutant and wild-type Ndc80 complexes, cells were collected by centrifugation, resuspended in 50 mM HEPES buffer (pH 7.6) containing 300 mM NaCl, 1 protease tablet (Roche), 1 mM PMSF, and 5 mM imidazole, and lysed using a French press. Lysates were cleared twice by centrifugation (20 min, 25,000 g, 4°C). All Ndc80 constructs were first purified via nickel affinity column chromatography (Bio-Rad) and eluted in 50 mM HEPES buffer (pH 7.6) containing 200 mM NaCl and 400 mM imidazole. Samples were then subjected to gel filtration column chromatography (Superdex 200; Bio-Rad) in 50 mM HEPES buffer (pH 7.6) containing 200 mM NaCl. Wild-type, mutant BNdc80p, and mutant CNdc80p were concentrated to 1–3 μM. Mutant ANdc80p was not concentrated.
Dam1 complex
All Dam1 complex constructs were expressed from a single polycistronic vector (pJT44; Tien et al., 2010) and purified as previously described (Gestaut et al., 2008; Kim et al., 2017; Miranda et al., 2005; Tien et al., 2010). The Spc34p component of the Dam1 complex contained a C-terminal FLAG tag. BL21 Rosetta 2 DE3 cells were transformed with wild-type Dam1 complex (pJT44), S20A Dam1 complex (pRLF002), 6A Dam1 complex (pRLF009), ADam1p Dam1 complex (pRLF008), BAsk1p Dam1 complex (pRLF007), CSpc34p Dam1 complex (pRLF006), ADam1pBAsk1p Dam1 complex (pRLF005), ADam1pCSpc34p (pRLF003), or BAsk1pCSpc34p Dam1 complex (pRLF004). Cells were grown up to an OD600 of 0.6, and then protein expression was induced by the addition of 2 mM IPTG and the culture was incubated at 18C for 16 h with shaking at 240 rpm. Cells were collected by centrifugation, resuspended in 50 mM sodium phosphate buffer (pH 6.9) containing 500 mM NaCl, 1 mM PMSF, and 1 protease tablet and lysed with a French press. Lysates were cleared as stated above. All Dam1 constructs were first purified by affinity chromatography using FLAG resin (GE Healthcare Biosciences) under gravity flow and eluted with 50 mM sodium phosphate buffer (pH 6.9) containing 500 mM NaCl and 100μg/ml FLAG peptide before being purified by gel filtration chromatography in 50 mM sodium phosphate buffer (pH 6.9) containing 500 mM NaCl.
Ipl1p and Sli15p
GST-Ipl1p (pSB196; Sue Biggins, Fred Hutchinson Cancer Research Center) and GST-Sli15p (residues 554–698, pSB503; Sue Biggins) were purified as previously described (Gestaut et al., 2008; Kim et al., 2017; Tien et al., 2010; Zelter et al., 2015). Briefly, GST-Sli15p and GST-Ipl1p were expressed at 37°C and 23°C, respectively, for 2 h. GST-Ipl1p was purified using GSTrap HP (GE Healthcare Biosciences) following the manufacturer’s instructions, except that the elution buffer was 50 mM Tris buffer (pH 8.0) containing 250 mM KCl and 10 mM glutathione. A HiTrap 26/10 desalting column (GE Healthcare) was used to exchange the buffer to 50 mM HEPES buffer (pH 7.4) containing 100 mM NaCl. GST-Sli15p was purified with glutathione-Sepharose 4B resin (GE Healthcare) following the manufacturer’s instructions. Elution buffer was 20 mM Tris buffer (pH 8.0) containing 200 mM NaCl, 1 mM β-mercaptoethanol, 1 mM EDTA, and 10 mM glutathione.
Dam1 complex phosphorylation
The Dam1 complex phosphorylation assay was performed as previously described (Kim et al., 2017). Briefly, 4 μM recombinant Dam1 complex was incubated with 0.5 μM GST-Ipl1p and 0.5 μM GST-Sli15p in 50 mM HEPES buffer (pH 7.4) containing 200 mM NaCl, 10 mM ATP, and 25 mM MgCl2. Reactions were incubated for 90 min at 30°C while rotating. Mock-treated (nonphosphorylated) controls of the Dam1 complex were the same as the phosphorylated samples except that dH2O replaced ATP.
Another phosphorylation assay was performed to measure the number of phosphoryl groups incorporated per molecule of Dam1p, Ask1p, and Spc34p. The assay was performed as above but with the addition of 25 μCi of [γ-32P]ATP. Sample buffer was added after the incubation period, and the samples were subjected to SDS-PAGE on an 8–14% polyacrylamide gel. Gels were dried between two membranes and exposed to a phosphor imaging screen (Azure Biosystems) for 24 h. The screen was imaged using an Azure Biosystems imager and Sapphire software v1.1.0315.0. Standards were imaged with the gel, and the standards and gel were quantified using ImageJ.
Mass spectrometry
Protein samples (15 μl of each sample at 0.87 µg/μl) were diluted to 0.3 µg/μl by the addition of 28.5 μl of 50 mM ammonium bicarbonate. 2.2 μl of 2% PPS silent surfactant (21011; Expedeon) plus 1.21 μl of 200 mM tris(2-carboxyethyl)phosphine was added, and samples were reduced at 60°C for 1 h, cooled to room temperature, and alkylated by the addition of 1.19 μl of 0.25 M iodoacetamide and incubation for 20 min at room temperature in the dark. 2.2 μl trypsin in water (0.4 μl/μl) was added, and the samples were digested at 37°C for 4 h in an Eppendorf Thermomixer with shaking (1,000 rpm). After digestion, 5 M HCl was added to a final concentration of 250 mM, and PPS was allowed to cleave for 1 h at room temperature. Samples were spun at maximum speed in a benchtop microfuge for 10 min, and supernatant was transferred to autosampler vials and stored at −80°C until MS analysis.
For mass spectrometry data acquisition, 3 μl of digested protein was loaded by autosampler onto a 150-μm Kasil fritted trap packed with 2 cm of ReprosilPur C18AQ (3-μm bead diameter; Dr. Maisch) at a flow rate of 2 μl per min. After desalting with 8 μl of 0.1% formic acid plus 2% acetonitrile, the trap was brought online with a fused silica capillary tip column (75 μm inner diameter) packed with 30 cm of ReprosilPur C18AQ (3-μm bead diameter; Dr. Maisch). Peptides were eluted from the column at 0.25 μl/min using an acetonitrile gradient. A Thermo Fisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer was used to perform mass spectrometry in data-dependent acquisition mode with both MS (centroid, 60K resolution) and MS/MS (centroid, 30K resolution) spectra acquired in the orbitrap. Acquired spectra were converted into mzML using msconvert from ProteoWizard (Chambers et al., 2012). MS data were searched to identify peptides, proteins, and phosphorylation using Comet 2018.01 rev 2 (Eng et al., 2013). Oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were defined as allowed variable modifications. A statistically meaningful q value was assigned to each peptide spectrum match through analysis of the target and decoy peptide spectrum match distributions using Percolator v3.02.1 (Käll et al., 2007). Target databases consisted of the complete E. coli proteome and the expressed protein sequences and common contaminant proteins. Decoy databases consisted of the corresponding set of reversed protein sequences. All raw MS data, configuration files and results have been made available using Limelight at https://limelight.yeastrc.org/limelight/d/pg/project/57.
Optical trap rupture force assay
Design, calibration, and use of the optical trap was essentially as described previously (Franck et al., 2010). Response of the bead-position sensor was mapped by using the piezo specimen stage to raster scan coverslip-anchored beads through the trap laser beam. Trap stiffness was then calibrated using the drag force, equipartition, and power spectrum methods (Lang et al., 2002). Custom LabView software was used for implementing stage-based feedback control and for recording trap data, with bead-trap separation sampled at 40 kHz and stage position updated at 50 Hz to control the level of tension on the bead.
Recombinant His6-tagged Ndc80 complex was linked to 0.56-μm-diameter streptavidin-coated polystyrene microbeads (Spherotech) using biotinylated His5 antibody (Qiagen), as previously described (Asbury et al., 2006; Franck et al., 2007; Powers et al., 2009). We obtained the same median rupture force using beads incubated with 30 nM Ndc80 complex as with beads incubated with 10 nM Ndc80 complex. This observation, together with simple geometric considerations (Hamilton et al., 2020), implies that a maximum of ∼90 Ndc80 complexes could interact simultaneously with the microtubule.
For all rupture force assays, a final concentration of 30 nM Ndc80 complex was incubated with 3.5 pM beads for 1 h, and the beads were washed twice with AB solution (1× BRB80 [80 mM PIPES buffer, pH 6.9, containing 1 mM MgCl2, and 1 mM EGTA], 2 mg/ml BSA, and 1 mM DTT). Glass slides and functionalized coverslips were used to construct flow channels. A coverslip was adhered to a glass slide with double-sided tape, leaving a channel between two adjacent strips of tape. Channels were functionalized by adding 5 mg/ml biotinylated BSA (Vector Labs) and incubating for 15 min inside a humidity chamber, washing with 1× BRB80, and then incubating with 0.3 mg/ml avidin DN (Vector Labs) for 5 min inside the humidity chamber. The channel was washed again with 1× BRB80, and biotinylated microtubule seeds stabilized with GMPCPP were added and allowed to incubate for 5 min inside the humidity chamber. Growth buffer (1× BRB80, 8 mg/ml BSA, 1 mM GTP, and 1 mg/ml κ-casein) was added to the channel and incubated for 5 min in the humidity chamber. Ndc80-coated beads were added to a reaction mix (1× BRB80, 8 mg/ml BSA, 1 mM GTP, 40 mM glucose, and 1 mM DTT) and sonicated for 10 s. After sonication, an oxygen scavenging system (250 μg/ml glucose oxidase, 30 μg/ml catalase, and 4.5 mg/ml glucose) was added. Dam1 complex was also added to the reaction mix to achieve a final concentration of 30 nM. Lastly, ∼1.5–2 mg/ml of purified bovine brain tubulin (Castoldi and Popov, 2003) was added to the reaction mixture. The reaction mixture was subsequently added to the channel, which was sealed with nail polish.
Data were collected for a total of 1 h after addition of tubulin. To determine whether an Ndc80-coated bead was able to bind to microtubules in the absence of force, it was placed onto a microtubule and the trap was shuttered. Microtubule-attached beads were pulled at 1 pN of force to determine whether they were able to withstand 1 pN of force and to slide them to the microtubule plus ends. Gradually increasing force was applied (at 0.25 pN · s−1) until the bead ruptured from the microtubule tip or the maximum trapping force was reached (∼20 pN under the conditions used here). Rupture forces were analyzed using Igor Pro (WaveMetrics). We also noted whenever a bead exceeded the maximum force before rupturing, yielding a right-censored value.
95% confidence intervals (CIs) were generated for the median rupture forces by performing bootstrapping on each dataset using the boot package in R. Statistical significance for the differences in medians between each sample were determined using a Mann–Whitney U test in R. Statistical significance for the differences in bead behavior between different samples was determined using Fisher’s exact test in R.
Optical trap force clamp assay
Recombinant His6-tagged Ndc80 complex was linked to 0.56-μm-diameter streptavidin-coated polystyrene microbeads using biotinylated His5 antibody, essentially as described above for the rupture force assay, except that a final concentration of 5 nM Ndc80 complex was mixed with 3.5 pM beads. Slide preparation and bead washing were performed as described above.
Each of the rates shown in Figs. 4 and S7 was calculated from a set of >17 individual events lasting a total of 0.14–3 h (as indicated in Table S6). We define detachment rate during assembly as
where Nassembly represents the total number of detachments observed during assembly, and Tassembly represents the total time recorded in assembly. A catastrophe occurs when a microtubule switches from a growing (assembling) state to a shortening (disassembling) state. Catastrophe rate was defined as
where Ncatastrophe represents the total number of catastrophes observed, which is divided by Tassembly because catastrophe events can occur only when a microtubule tip is assembling. Similarly, the detachment rate during disassembly was defined as
where Ndisassembly represents the total number of detachments observed during disassembly, and Tdisassembly represents the total time recorded in disassembly.
A rescue occurs when a microtubule switches from a shortening (disassembling) state to a growing (assembling) state. Sometimes during force clamp experiments, a bead was carried far enough while tracking with a shortening microtubule tip to approach or reach the stabilized seed, from which the microtubule was nucleated, before the tip resumed growth. To distinguish true spontaneous rescues from cases where disassembly was stopped by the seed, we scored events as rescues only when the microtubule length, measured immediately after the end of disassembly, was greater than the length of 95% of microtubule seeds. To measure the distribution of microtubule seed lengths, a channel slide was created as above, with coverslip-anchored seeds but without the addition of free tubulin, Dam1 complex, or Ndc80 complex–coated beads. Images of the seeds were recorded, and their lengths were measured using ImageJ. Likewise, videos were recorded during all force clamp experiments and analyzed to determine the lengths of the microtubules immediately after a bead stopped exhibiting disassembly-driven motion and the microtubule potentially had rescued. If the microtubule length at the moment of the putative rescue was greater than the length of 95% of the microtubule seeds, then the event was considered a true rescue. The rescue rate was then defined as
where Nrescue is the total number of true rescues observed, which was divided by Tdisassembly because rescue events can occur only when a microtubule tip is disassembling. Each of the four rates was assumed to describe a Poisson process, and their 95% CIs were calculated using the exact method (Garwood, 1936; Ulm, 1990).
Data analysis and figure preparation
Data from the optical trap assays were analyzed in Igor Pro, R (Kabacoff, 2011; Kassambra and Kosiniski, 2018; R Core Team, 2013; Wickham, 2016), Matlab, ImageJ, and Python. Figures were produced using Python, R, Adobe Illustrator, and Adobe InDesign.
Online supplemental material
Fig. S1 (related to Fig. 1) shows Kaplan–Meier survival curves comparing the rupture force distributions for wild-type Ndc80 complex versus Ndc80 mutants. Fig. S2 (related to Fig. 1) is a superplot showing rupture forces measured across three biological replicates of the wild-type Ndc80 complex and two biological replicates of the wild-type Dam1 complex. Fig. S3 (related to Fig. 2) shows incorporation of [γ-32P]ATP into different interaction regions of the Dam1 complex. Fig. S4 (related to Fig. 2) shows that phosphorylation at residues S31 and S311 of Dam1p did not affect the strength of tip-couplers based on the Ndc80 and Dam1 complexes. Fig. S5 (related to Fig. 2) shows Kaplan–Meier survival curves comparing the rupture force distributions measured with phosphorylated and mock-treated Dam1 complexes. Fig. S6 (related to Fig. 2) is a superplot showing rupture forces measured across two biological replicates of prephosphorylated Dam1 complexes. Fig. S7 (related to Fig. 4) shows catastrophe and rescue rates measured in force clamp experiments. Table S1 provides statistical comparisons for Fig. 1 C. Table S2 provides statistical comparisons for Fig. S2, A and B. Table S3 provides statistical comparisons for Fig. S4. Table S4 provides statistical comparisons for Fig. 2 B. Table S5 provides statistical comparisons for Fig. S5. Table S6 provides a summary of force clamp data for Figs. 4 and S7. Table S7 provides statistical comparisons for Figs. 4 and S7. Table S8 details all the plasmids used in this study.
Supplementary Material
lists statistical comparisons for Fig. 1 C.
lists statistical comparisons for Fig. S2, A and B.
lists statistical comparisons for Fig. S4.
lists statistical comparisons for Fig. 2 B.
lists statistical comparisons for Fig. S5.
lists summary of force clamp data, Figs. 4 and S7.
lists statistical comparisons for force clamp experiments, Figs. 4 and S7.
lists plasmids used in this study.
is the source file for Fig. S3.
Acknowledgments
We thank the entire Davis lab, Asbury lab, and Seattle Mitosis Club for helpful feedback and discussions.
This work was supported by the National Science Foundation Graduate Research Fellowship (awarded to R.L. Flores), the University of Washington’s Proteomics Resource (UWPR95794), and the National Institutes of Health (R01GM040506 and R35GM130293 to T.N. Davis and R01GM079373 and R35GM134842 to C.L. Asbury).
The authors declare no competing financial interests.
Author contributions: R.L. Flores was responsible for conceptualization, data curation, formal analysis, investigation, methodology, funding acquisition, figure visualization, creating code for data analysis, validation, writing the original draft, and review and editing of the manuscript. Z.E. Peterson was responsible for investigation. A. Zelter was responsible for data curation, formal analysis, investigation, methodology, resources, and software. M. Riffle contributed formal analysis and software and assisted in reviewing and editing the manuscript. C.L. Asbury contributed conceptualization, formal analysis, funding acquisition, investigation, resources, software, supervision, and review and editing of the manuscript. T.N. Davis contributed conceptualization, formal analysis, funding acquisition, investigation, resources, supervision, and review and editing of the manuscript.
References
- Akiyoshi, B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., and Biggins S.. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579. 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelgren, H., Kniola B., and Ekwall K.. 2003. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 116:4035–4042. 10.1242/jcs.00707 [DOI] [PubMed] [Google Scholar]
- Asbury, C.L., Gestaut D.R., Powers A.F., Franck A.D., and Davis T.N.. 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA. 103:9873–9878. 10.1073/pnas.0602249103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj, R., Qi W., and Yu H.. 2004. Identification of two novel components of the human NDC80 kinetochore complex*[boxs]. J. Biol. Chem. 279:13076–13085. 10.1074/jbc.m310224200 [DOI] [PubMed] [Google Scholar]
- Biggins, S., and Murray A.W.. 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15:3118–3129. 10.1101/gad.934801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., and Murray A.W.. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544. 10.1101/gad.13.5.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi, M., and Popov A.V.. 2003. Purification of brain tubulin through two cycles of polymerization–depolymerization in a high-molarity buffer. Protein Expr. Purif. 32:83–88. 10.1016/s1046-5928(03)00218-3 [DOI] [PubMed] [Google Scholar]
- Chambers, M.C., Maclean B., Burke R., Amodei D., Ruderman D.L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., et al. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30:918–920. 10.1038/nbt.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, Y.W., Jeyaprakash A.A., Nigg E.A., and Santamaria A.. 2012. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J. Cell Biol. 196:563–571. 10.1083/jcb.201109001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Anderson S., Jwa M., Green E.M., Kang J.s., Yates J.R., Chan C.S.M., Drubin D.G., and Barnes G.. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. 10.1016/s0092-8674(02)00973-x [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., Niessen S., Anderson S., Hyndman F., Yates J.R. 3rd, Oegema K., and Desai A.. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. 10.1101/gad.1234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum, J.R., Wren J.D., Daniel J.J., Sivakumar S., McAvoy J.N., Potapova T.A., and Gorbsky G.J.. 2009. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr. Biol. 19:1467–1472. 10.1016/j.cub.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, J.G., Dong Y., Hergert P., Strauss J., Hickey J.M., Salmon E.D., and McEwen B.F.. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531. 10.1091/mbc.e04-09-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- DeLuca, J.G., Moree B., Hickey J.M., Kilmartin J.V., and Salmon E.D.. 2002. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159:549–555. 10.1083/jcb.200208159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, K.F., Lens S.M.A., and DeLuca J.G.. 2011. Temporal changes in Hec1 phosphorylation control kinetochore–microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634. 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., and Oegema K.. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doodhi, H., Kasciukovic T., Clayton L., and Tanaka T.U.. 2021. Aurora B switches relative strength of kinetochore-microtubule attachment modes for error correction. J. Cell Biol. 220:e202011117. 10.1083/jcb.202011117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, J.K., Jahan T.A., and Hoopmann M.R.. 2013. Comet: an open-source MS/MS sequence database search tool. Proteomics. 13:22–24. 10.1002/pmic.201200439 [DOI] [PubMed] [Google Scholar]
- Franck, A.D., Powers A.F., Gestaut D.R., Davis T.N., and Asbury C.L.. 2010. Direct physical study of kinetochore-microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods. 51:242–250. 10.1016/j.ymeth.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck, A.D., Powers A.F., Gestaut D.R., Gonen T., Davis T.N., and Asbury C.L.. 2007. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat. Cell Biol. 9:832–837. 10.1038/ncb1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos, T., Santamaria A., Arulanandam D.A.J., Wang B., Conti E., and Nigg E.. 2009. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 28:1442–1452. 10.1038/emboj.2009.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood, F.. 1936. Fiducial limits for the Poisson distribution. Biometrika. 28:437–442. 10.2307/2333958 [DOI] [Google Scholar]
- Gestaut, D.R., Graczyk B., Cooper J., Widlund P.O., Zelter A., Wordeman L., Asbury C.L., and Davis T.N.. 2008. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10:407–414. 10.1038/ncb1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett, E.S., Espelin C.W., and Sorger P.K.. 2004. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164:535–546. 10.1083/jcb.200308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase, J., Bonner M.K., Halas H., and Kelly A.E.. 2017. Distinct roles of the chromosomal passenger complex in the detection of and response to errors in kinetochore-microtubule attachment. Dev. Cell. 42:640–654.e5. 10.1016/j.devcel.2017.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, G.E., Helgeson L.A., Noland C.L., Asbury C.L., Dimitrova Y.N., and Davis T.N.. 2020. Reconstitution reveals two paths of force transmission through the kinetochore. Elife. 9:e56582. 10.7554/eLife.56582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch, A., Silljé H.H.W., and Nigg E.A.. 2006. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 25:5504–5515. 10.1038/sj.emboj.7601426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf, S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., and Peters J.M.. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281–294. 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Rines D.R., Espelin C.W., and Sorger P.K.. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 106:195–206. 10.1016/s0092-8674(01)00438-x [DOI] [PubMed] [Google Scholar]
- Helgeson, L.A., Zelter A., Riffle M., MacCoss M.J., Asbury C.L., and Davis T.N.. 2018. Human Ska complex and Ndc80 complex interact to form a load-bearing assembly that strengthens kinetochore-microtubule attachments. Proc. Natl. Acad. Sci. USA. 115:2740–2745. 10.1073/pnas.1718553115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, T., Haraguchi T., Hiraoka Y., Kimura H., and Fukagawa T.. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347–3362. 10.1242/jcs.00645 [DOI] [PubMed] [Google Scholar]
- Howe, M., McDonald K.L., Albertson D.G., and Meyer B.J.. 2001. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 153:1227–1238. 10.1083/jcb.153.6.1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Ortiz J., Lechner J., Shevchenko A., Shevchenko A., Magiera M.M., Schramm C., and Schiebel E.. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20:777–791. 10.1093/emboj/20.4.777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Ortíz J., Tanaka T.U., Lechner J., and Schiebel E.. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181–193. 10.1093/emboj/21.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenni, S., and Harrison S.C.. 2018. Structure of the DASH/Dam1 complex shows its role at the yeast kinetochore-microtubule interface. Science. 360:552–558. 10.1126/science.aar6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, F., Bokros M., and Wang Y.. 2017. The phosphorylation of a kinetochore protein Dam1 by Aurora B/Ipl1 kinase promotes chromosome bipolar attachment in yeast. Sci. Rep. 7:11880. 10.1038/s41598-017-12329-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar, A.P., Bouck D.C., Molk J.N., Bloom K.S., and Salmon E.D.. 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8:581–585. 10.1038/ncb1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabacoff, R.. 2011. R in Action: Data Analysis and Graphics with R. Manning Publications Co., Shelter Island, NY. 447 pp. [Google Scholar]
- Käll, L., Canterbury J.D., Weston J., Noble W.S., and MacCoss M.J.. 2007. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 4:923–925. 10.1038/nmeth1113 [DOI] [PubMed] [Google Scholar]
- Kassambra, A., and Kosiniski M.. 2018. survminer: drawing survival curves using ‘ggplot2’. [Google Scholar]
- Khodjakov, A., and Pines J.. 2010. Centromere tension: a divisive issue. Nat. Cell Biol. 12:919–923. 10.1038/ncb1010-919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.O., Zelter A., Umbreit N.T., Bollozos A., Riffle M., Johnson R., MacCoss M.J., Asbury C.L., and Davis T.N.. 2017. The Ndc80 complex bridges two Dam1 complex rings. Elife. 6:e21069. 10.7554/eLife.21069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert, F., Hornung P., and Westermann S.. 2010. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189:641–649. 10.1083/jcb.200912021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert, F., Mieck C., Alushin G.M., Nogales E., and Westermann S.. 2013. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J. Cell Biol. 200:21–30. 10.1083/jcb.201210091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, M.J., Asbury C.L., Shaevitz J.W., and Block S.M.. 2002. An automated two-dimensional optical force clamp for single molecule studies. Biophys. J. 83:491–501. 10.1016/S0006-3495(02)75185-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore, J., Bloom K.S., and Salmon E.D.. 2011. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 195:573–582. 10.1083/jcb.201106036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., and Nicklas R.B.. 1997. Tension-sensitive kinetochore phosphorylation and the chromosome distribution checkpoint in praying mantid spermatocytes. J. Cell Sci. 110:537–545. 10.1242/jcs.110.5.537 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma, S., Stucke V.M., and Nigg E.A.. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 297:2267–2270. 10.1126/science.1075596 [DOI] [PubMed] [Google Scholar]
- McCleland, M.L., Gardner R.D., Kallio M.J., Daum J.R., Gorbsky G.J., Burke D.J., and Stukenberg P.T.. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17:101–114. 10.1101/gad.1040903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland, M.L., Kallio M.J., Barrett-Wilt G.A., Kestner C.A., Shabanowitz J., Hunt D.F., Gorbsky G.J., and Stukenberg P.T.. 2004. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr. Biol. 14:131–137. 10.1016/j.cub.2003.12.058 [DOI] [PubMed] [Google Scholar]
- McIntosh, J.R., Grishchuk E.L., Morphew M.K., Efremov A.K., Zhudenkov K., Volkov V.A., Cheeseman I.M., Desai A., Mastronarde D.N., and Ataullakhanov F.I.. 2008. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 135:322–333. 10.1016/j.cell.2008.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi, P., Draviam V.M., and Sorger P.K.. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60. 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Miller, M.P., Asbury C.L., and Biggins S.. 2016. A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell. 165:1428–1439. 10.1016/j.cell.2016.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, J.J.L., De Wulf P., Sorger P.K., and Harrison S.C.. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138–143. 10.1038/nsmb896 [DOI] [PubMed] [Google Scholar]
- Musacchio, A.. 2011. Spindle assembly checkpoint: the third decade. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3595–3604. 10.1098/rstb.2011.0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio, A., and Desai A.. 2017. A molecular view of kinetochore assembly and function. Biology (Basel). 6:5. 10.3390/biology6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani, A., Koujin T., Tsutsumi C., Haraguchi T., and Hiraoka Y.. 2001. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 110:322–334. 10.1007/s004120100153 [DOI] [PubMed] [Google Scholar]
- Ng, C.T., Deng L., Chen C., Lim H.H., Shi J., Surana U., and Gan L.. 2019. Electron cryotomography analysis of Dam1C/DASH at the kinetochore-spindle interface in situ. J. Cell Biol. 218:455–473. 10.1083/jcb.201809088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky, B.A., and Biggins S.. 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15:486–493. 10.1016/j.tcb.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Pinsky, B.A., Kung C., Shokat K.M., and Biggins S.. 2006. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 8:78–83. 10.1038/ncb1341 [DOI] [PubMed] [Google Scholar]
- Powers, A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., and Asbury C.L.. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875. 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing, R foundation for statistical computing, Vienna, Austria. [Google Scholar]
- Sarangapani, K.K., Akiyoshi B., Duggan N.M., Biggins S., and Asbury C.L.. 2013. Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proc. Natl. Acad. Sci. USA. 110:7282–7287. 10.1073/pnas.1220700110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, J.C., Arthanari H., Boeszoermenyi A., Dashkevich N.M., Wilson-Kubalek E.M., Monnier N., Markus M., Oberer M., Milligan R.A., Bathe M., Wagner G., et al. 2012. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 23:968–980. 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, C., Hazbun T.R., Cheeseman I.M., Aranda J., Fields S., Drubin D.G., and Barnes G.. 2003. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell. 14:3342–3355. 10.1091/mbc.e02-11-0765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Kitamura E., Kitamura Y., and Tanaka T.U.. 2007. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 178:269–281. 10.1083/jcb.200702141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J.R., and Nasmyth K.. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329. 10.1016/s0092-8674(02)00633-5 [DOI] [PubMed] [Google Scholar]
- Tien, J.F., Fong K.K., Umbreit N.T., Payen C., Zelter A., Asbury C.L., Dunham M.J., and Davis T.N.. 2013. Coupling unbiased mutagenesis to high-throughput DNA sequencing uncovers functional domains in the Ndc80 kinetochore protein of Saccharomyces cerevisiae. Genetics. 195:159–170. 10.1534/genetics.113.152728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien, J.F., Umbreit N.T., Gestaut D.R., Franck A.D., Cooper J., Wordeman L., Gonen T., Asbury C.L., and Davis T.N.. 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189:713–723. 10.1083/jcb.200910142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, K.. 1990. Simple method to calculate the confidence interval of a standardized mortality ratio (SMR). Am. J. Epi. 131:373–375. 10.1093/oxfordjournals.aje.a115507 [DOI] [PubMed] [Google Scholar]
- Umbreit, N.T., Miller M.P., Tien J.F., Ortolá J.C., Gui L., Lee K.K., Biggins S., Asbury C.L., and Davis T.N.. 2014. Kinetochores require oligomerization of Dam1 complex to maintain microtubule attachments against tension and promote biorientation. Nat. Commun. 5:4951. 10.1038/ncomms5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, R., Ingram J., and Harrison S.C.. 2016. Conserved tetramer junction in the kinetochore Ndc80 complex. Cell Rep. 17:1915–1922. 10.1016/j.celrep.2016.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooff, J.J.E., Snel B., and Kops G.J.P.L.. 2017. Unique phylogenetic distributions of the Ska and Dam1 complexes support functional analogy and suggest multiple parallel displacements of Ska by Dam1. Genome Biol. Evol. 9:1295–1303. 10.1093/gbe/evx088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov, V.A., Zaytsev A.V., Gudimchuk N., Grissom P.M., Gintsburg A.L., Ataullakhanov F.I., McIntosh J.R., and Grishchuk E.L.. 2013. Long tethers provide high-force coupling of the Dam1 ring to shortening microtubules. Proc. Natl. Acad. Sci. USA. 110:7708–7713. 10.1073/pnas.1305821110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, R.A., Inoue S., and Salmon E.D.. 1989. Asymmetric behavior of severed microtubule ends after ultraviolet-microbeam irradiation of individual microtubules in vitro. J. Cell Biol. 108:931–937. 10.1083/jcb.108.3.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, R.R., Sorger P.K., and Harrison S.C.. 2005. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA. 102:5363–5367. 10.1073/pnas.0501168102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, S., Avila-Sakar A., Wang H.W., Niederstrasser H., Wong J., Drubin D.G., Nogales E., and Barnes G.. 2005. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 17:277–290. 10.1016/j.molcel.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Westermann, S., Wang H.W., Avila-Sakar A., Drubin D.G., Nogales E., and Barnes G.. 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 440:565–569. 10.1038/nature04409 [DOI] [PubMed] [Google Scholar]
- Wickham, H.. 2016. Ggplot2: elegant graphics for data analysis. In Use R! Springer, Switzerland. 1 online resource. [Google Scholar]
- Wigge, P.A., Jensen O.N., Holmes S., Soues S., Mann M., and Kilmartin J.V.. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141:967–977. 10.1083/jcb.141.4.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P.A., and Kilmartin J.V.. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349–360. 10.1083/jcb.152.2.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev, A.V., Mick J.E., Maslennikov E., Nikashin B., DeLuca J.G., and Grishchuk E.L.. 2015. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol. Biol. Cell. 26:1829–1844. 10.1091/mbc.E14-11-1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelter, A., Bonomi M., Kim J.O., Umbreit N.T., Hoopmann M.R., Johnson R., Riffle M., Jaschob D., MacCoss M.J., Moritz R.L., and Davis T.N.. 2015. The molecular architecture of the Dam1 kinetochore complex is defined by cross-linking based structural modelling. Nat. Commun. 6:8673. 10.1038/ncomms9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., Chen Y., and Lee W.H.. 1999. Hec1p, an evolutionarily conserved coiled-coil protein, modulates chromosome segregation through interaction with SMC proteins. Mol. Cell Biol. 19:5417–5428. 10.1128/MCB.19.8.5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists statistical comparisons for Fig. 1 C.
lists statistical comparisons for Fig. S2, A and B.
lists statistical comparisons for Fig. S4.
lists statistical comparisons for Fig. 2 B.
lists statistical comparisons for Fig. S5.
lists summary of force clamp data, Figs. 4 and S7.
lists statistical comparisons for force clamp experiments, Figs. 4 and S7.
lists plasmids used in this study.
is the source file for Fig. S3.