Abstract
Ethylene response factors (ERFs) are downstream components of ethylene-signaling pathways known to play critical roles in ethylene-controlled climacteric fruit ripening, yet little is known about the molecular mechanism underlying their mode of action. Here, we demonstrate that SlERF.F12, a member of the ERF.F subfamily containing Ethylene-responsive element-binding factor-associated Amphiphilic Repression (EAR) motifs, negatively regulates the onset of tomato (Solanum lycopersicum) fruit ripening by recruiting the co-repressor TOPLESS 2 (TPL2) and the histone deacetylases (HDAs) HDA1/HDA3 to repress the transcription of ripening-related genes. The SlERF.F12-mediated transcriptional repression of key ripening-related genes 1-AMINO-CYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2 (ACS2), ACS4, POLYGALACTURONASE 2a, and PECTATE LYASE is dependent on the presence of its C-terminal EAR motif. We show that SlERF.F12 interacts with the co-repressor TPL2 via the C-terminal EAR motif and recruits HDAs SlHDA1 and SlHDA3 to form a tripartite complex in vivo that actively represses transcription of ripening genes by decreasing the level of the permissive histone acetylation marks H3K9Ac and H3K27Ac at their promoter regions. These findings provide new insights into the ripening regulatory network and uncover a direct link between repressor ERFs and histone modifiers in modulating the transition to ripening of climacteric fruit.
The SlERF.F12–TPL2–HDA1/3 complex represses fruit ripening initiation by promoting histone deacetylation in the promoter regions of ripening-related genes.
IN A NUTSHELL.
Background: The ripening of fleshy fruits is a complex, genetically programmed process. Tomato (Solanum lycopersicum) has been widely used as a model system for studying fleshy fruit ripening. Although ripening is orchestrated by a complex multi-phytohormonal control, the plant hormone ethylene has long been accepted as the main trigger of ripening in climacteric fruits, and its downstream transcriptional regulators ethylene response factors (ERFs) are responsible for the ethylene signal. Among these ERFs, several members have an Ethylene-responsive element-binding factor-associated Amphiphilic Repression (EAR) motif, the most common transcriptional repressor motif identified in plants to date. However, the functional significance of EAR motif-containing ERF proteins has yet to be determined in the context of fruit ripening and associated regulatory mechanisms.
Question: We identified the ERF gene, named SlERF.F12, which encodes a protein with an EAR motif and whose expression levels dramatically decrease at the transition to ripening, therefore being an ideal candidate to play an important role in controlling this process. However, the role of SlERF.F12 in fruit ripening and its regulatory mechanism in fruit ripening remains unclear.
Findings: We demonstrate that SlERF.F12 negatively regulates the onset of tomato fruit ripening by recruiting the co-repressor TOPLESS protein 2 (TPL2) and the histone deacetylases (HDAs) HDA1/HDA3 to repress the transcription of ripening-related genes. We show that SlERF.F12 interacts with the co-repressor TPL2 via its C-terminal EAR motif and recruits HDAs to form a tripartite complex. This complex actively represses transcription of ripening genes by decreasing the level of acetylation at their promoter regions.
Next steps: We would like to know whether this regulatory module is conversed across other fruit species such as kiwifruit, apple, pear, and banana. Based on our findings, we wish to develop strategies for the application of our results to control the ripening time and shelf life of fruits.
Introduction
The ripening of fleshy fruits is a complex, genetically programmed process involving a series of physiological and biochemical changes leading to profound alterations in fruit color, texture, and flavor (Klee and Giovannoni, 2011; Seymour et al., 2013). Fleshy fruits are traditionally classified as climacteric and nonclimacteric types depending on whether they experience an increase in respiration and ethylene production at the onset of ripening (McMurchie et al., 1972; Lelièvre et al., 1997). Climacteric fruits such as tomatoes (Solanum lycopersicum), apples (Malus domestica), and bananas (Musa sp.) exhibit a rapid rise in respiration and a burst of ethylene production during ripening initiation, whereas nonclimacteric fruits including citrus, strawberry (Fragaria × ananassa), and grape (Vitis vinifera) lack these characteristic bursts. Tremendous progress has been achieved in uncovering the regulatory mechanism underlying climacteric fruit ripening using tomato as a model system (Liu et al., 2015a; Giovannoni et al., 2017; Li et al., 2020a, 2020b, 2021).
Although fruit ripening is most likely orchestrated by complex multi-phytohormonal control (Hao et al., 2015; Shin et al., 2019), the plant hormone ethylene has long been accepted as the main trigger of climacteric fruit ripening (Burg and Burg, 1962; Alexander and Grierson, 2002; Grierson, 2013), and blocking ethylene production or signal transduction via mutation or downregulation of key genes of ethylene biosynthesis or signaling pathways efficiently blocks the ripening process (Lin et al., 2009; Liu et al., 2015a). In addition to ethylene, transcription factors such as RIPENING INHIBITOR (RIN), NONRIPENING (NOR), COLORLESS NONRIPENING (CNR), APETALA 2a (AP2a), and TOMATO AGAMOUS-LIKE 1 regulate fruit ripening (Klee and Giovannoni, 2011; Karlova et al., 2014; Liu et al., 2015a; Li et al., 2020a, 2020b). Moreover, epigenetic modifications such as DNA methylation, RNA methylation, and histone modifications also play important roles in climacteric fruit ripening (Zhong et al., 2013; Liu et al., 2015b; Lang et al., 2017; Lü et al., 2018; Zhou et al., 2019; Liang et al., 2020). Importantly, the roles of both transcription factors and epigenetic modifications in regulating fruit ripening are mostly ethylene-dependent, further emphasizing the central role of ethylene in regulating climacteric ripening.
Much progress has been made toward deciphering the mechanisms by which plants perceive and respond to ethylene. The currently accepted model states that a linear signal transduction pathway leads to the activation of downstream transcriptional regulators known as ERFs(Benavente and Alonso, 2006; Ju and Chang, 2015). Despite many studies showing that ethylene signaling is instrumental in climacteric fruit ripening, how ethylene targets and modulates the expression of specific ripening-related genes remains largely unknown. ERFs form one of the largest plant transcription factor families and are thought to directly regulate ethylene-responsive gene expression. In this regard, these transcription factors can potentially mediate the diversity of ethylene responses such as those seen in various aspects of climacteric fruit ripening (Pirrello et al., 2012; Liu et al., 2016). ERFs belong to the large AP2/ERF multi-gene family defined by the presence of the AP2/ERF domain, which consists of approximately 60–70 amino acids (Riechmann et al., 2000). ERF proteins bind to the GCC box or dehydration-responsive element/C-repeat (DRE/CRT) cis-acting elements in the promoter regions of ethylene-responsive genes (Ohme-Takagi and Shinshi, 1995; Hao et al., 2002; Pirrello et al., 2012) and play important roles in biotic and abiotic stress responses in various plant species (Hao et al., 2002; Müller and Munné-Bosch, 2015; Gu et al., 2017). While the involvement of ERFs in phytohormone signaling and fruit ripening is widely accepted (Li et al., 2007; Lee et al., 2012; Liu et al., 2014, 2016), the specific roles and modes of action of most ERF remain rather elusive.
Using tomato as a model plant, we previously identified 77 ERFs, which were divided into nine subfamilies (A–J) based on their structural features (Pirrello et al., 2012; Liu et al., 2016). Among these nine subfamilies, ERF.F subfamily members are characterized by the presence of an Ethylene-responsive element-binding factor-associated Amphiphilic Repression (EAR) motif, the most common transcriptional repression motif identified in plants to date (Kagale and Rozwadowski, 2011). Proteins with an EAR motif can repress gene expression via the recruitment and action of co-repressors, such as SWITCH INDEPENDENT 3 (SIN3), SIN3-ASSOCIATED POLYPEPTIDE 18 (SAP18), and TPL/TPL-RELATED, as well as histone deacetylases (HDAs) (Song et al., 2005; Song and Galbraith, 2006; Hill et al., 2008; Kagale and Rozwadowski, 2011; Causier et al., 2012; Wang et al., 2013; Ryu et al., 2014; Kim et al., 2019). EAR motif-containing ERFs have been shown to be involved in abiotic or biotic stress responses such as salt, wounding, cold, drought, or pathogen attack in several plant species (Ohta et al., 2001; Song et al., 2005; Cao et al., 2006; Trujillo et al., 2008; Dong and Liu, 2010; Pan et al., 2010; Zhang et al., 2010; Lu et al., 2011; Dong et al., 2012, 2015). However, the functional significance of EAR motif-containing ERF proteins in fruit ripening and in their associated regulatory mechanisms are yet to be elucidated.
In this study, we demonstrate that the ERF.F subfamily gene SlERF.F12, encoding a protein containing two typical EAR motifs (LxLxL and DLNxxP), acts as a transcriptional repressor of ripening-related genes. Our findings show that SlERF.F12 represses fruit ripening by recruiting the co-repressor TPL2 and the chromatin modifier proteins HDA1/HDA3 to epigenetically repress the expression of ripening-related genes. This study reveals the role and mode of action of an EAR motif-containing ERF in fruit ripening and sheds new light on the regulatory mechanism of climacteric fruit ripening.
Results
SlERF.F12 acts as a transcriptional repressor and its expression decreases during fruit ripening initiation
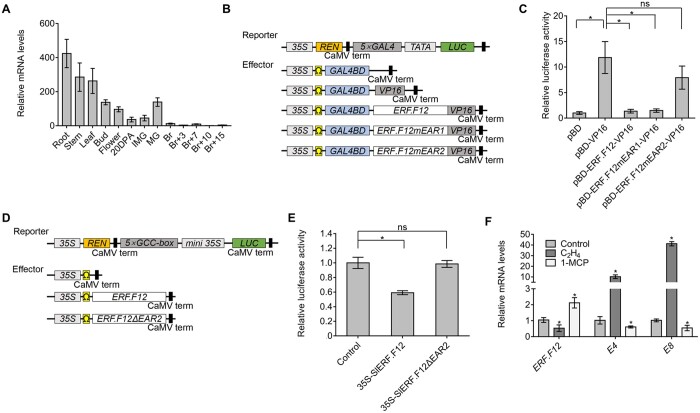
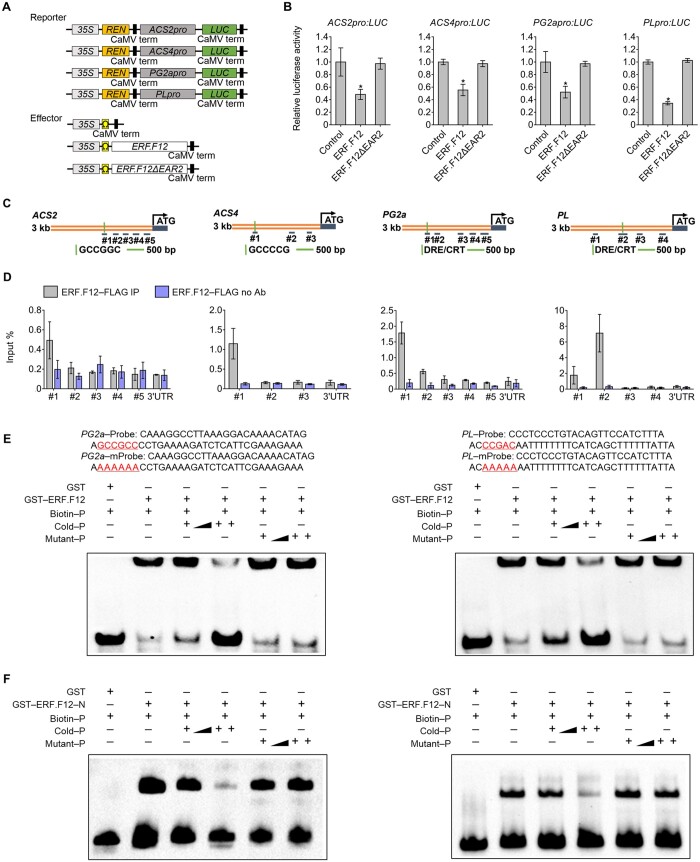
Members of the ERF.F subfamily are potential transcriptional repressors (Pirrello et al., 2012; Liu et al., 2016), but to date, their role and mode of action in fruit ripening remain largely unknown. Mining the latest tomato reference genome (SL4.0) to explore the ERF.F subfamily identified four novel members (SlERF.F10, SlERF.F11, SlERF.F12, and SlERF.F13) (Supplemental Figure S1A; Supplemental Table S1). Notably, of these, SlERF.F12 (Solyc02g077840) contained two typical EAR motifs, EAR1 and EAR2, located in the middle and the C-terminal regions, respectively (Supplemental Figure S1, A, and B). Remarkably, SlERF.F12 exhibited a dramatic decrease in expression in tomato fruits at the transition to ripening. SlERF.F12 showed relatively high transcript levels in vegetative tissues and in immature green (IMG) fruits, followed by a sharp decrease at the onset of ripening, then remaining low at the post-breaker (Br) stages (Figure 1A). The decrease in SlERF.F12 expression occurred concomitantly with the initiation of fruit ripening, suggesting that downregulation of this ERF might be required for a normal ripening progression. To investigate whether SlERF.F12 acts as a transcriptional repressor, we generated constructs to examine the ability of SlERF.F12, and variants with defective EAR motifs (named SlERF.F12-mEAR1 and SlERF.F12-mEAR2), to inhibit transactivation mediated by the strong VP16 activator from Herpes simplex virus in transient expression assays (Figure 1B). Both the intact (SlERF.F12) and SlERF.F12-mEAR1 variant, mutated in the first EAR motif, repressed VP16-promoted firefly luciferase (LUC) activity (Figure 1C). In contrast, mutations in the C-terminal EAR motif (EAR2) led to a loss of SlERF.F12 repression potential (Figure 1C), suggesting that the transcriptional inhibition of SlERF.F12 is mostly dependent on this C-terminal EAR2 motif. We also generated a reporter construct with the ethylene-inducible GCC box upstream of LUC in a plasmid that overexpresses Renilla LUC (REN) as an internal control (Figure 1D). Transient expression assays revealed that SlERF.F12 represses the transcriptional activity of the LUC reporter, in contrast to the mutated version SlERF.F12-ΔEAR2 lacking the C-terminal EAR motif (Figure 1E). These data indicated that SlERF.F12 represses the transcription of GCC box-containing promoters in an EAR2-dependent manner.
Figure 1.
ERF.F12 displays a ripening-related expression pattern and encodes a transcriptional repressor. A, Relative ERF.F12 transcript levels in different tissues, as assessed by RT-qPCR. 20 DPA, 20 DPA; Br+3–15, 3–15 days after the Br stage. B, Schematic diagram of the double-reporter and effector plasmids in the dual LUC assay for transcriptional inhibition assays. C, Transcriptional repression assays of ERF.F12. The dual LUC/REN reporter was co-transfected with individual effector plasmids into N. benthamiana leaf protoplasts. ERF.F12mEAR1, ERF.F12 with the core Leu residues of EAR1 changed to Ser. ERF.F12mEAR2, ERF.F12 with the core DLN motif of EAR2 changed to SSS. pBD, empty vector, negative control. pBD-VP16, VP16 transcriptional activator domain, positive control. Asterisk indicates statistical significance using Student’s t test, P < 0.05. D, Schematic diagram of the double-reporter and effector plasmids in the dual LUC assay for measuring transcriptional repressor ability of ERF.F12 on a promoter containing the GCC box. E, ERF.F12 represses transcription from a promoter containing a synthetic GCC box. The dual LUC/REN reporter was co-transfected with individual effector plasmids into N. benthamiana leaf protoplasts. ERF.F12ΔEAR2, ERF.F12 with a deletion of EAR2. Asterisk indicates statistical significance using Student’s t test, P < 0.05. F, ERF.F12 transcript levels in WT fruits at the MG stage treated with ethylene (50 μL L−1) for 8 h or 1-MCP (1.0 μL L−1) for 12 h, as determined by RT-qPCR. E4 and E8 are ethylene-responsive marker genes. Asterisks indicate statistical significance using Student’s t test, P < 0.05.
The ripening-associated expression pattern of SlERF.F12 prompted us to investigate whether its expression is under the control of ethylene. To this end, performed RT-quantitative PCR (RT-qPCR) to assess SlERF.F12 transcript levels in mature green (MG) fruits treated with exogenous ethylene or with 1-methylcyclopropene (1-MCP), an inhibitor of ethylene perception. We used two known ethylene-responsive genes, E4 and E8, as controls to validate the efficacy of the treatments. Relative SlERF.F12 transcript levels were lower in response to exogenous ethylene treatment but increased in response to 1-MCP (Figure 1F), in line with the decreased expression of this gene during ripening when tomato fruits undergo elevated ethylene production. We also examined the ethylene response of SlERF.F12 in the vegetative tissues roots, stems, and leaves. SlERF.F12 transcript abundance decreased upon ethylene treatment in both roots and leaves but not in stems, while it increased in response to treatment with 1-MCP (Supplemental Figure S2). These data motivated an exploration of the physiological significance of SlERF.F12 as a putative repressor of tomato fruit ripening.
SlERF.F12 represses the transition from the unripe to ripe fruit stage
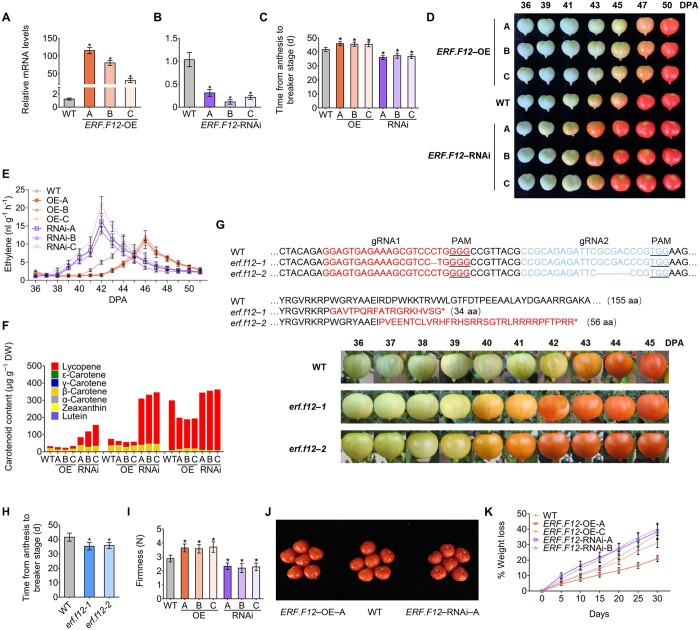
The downregulation of SlERF.F12 transcript levels at the transition from MG to Br stage, together with its negative regulation by ethylene at the MG stage, suggested that this gene may be a negative regulator of fruit ripening. To test this hypothesis, we generated tomato lines with lower (by RNA interference [RNAi]) or higher (by overexpression driven by the cauliflower mosaic virus [CaMV] 35S promoter) SlERF.F12 expression. We obtained 10 independent homozygous 35S:ERF.F12-OE lines, from which we selected three lines (ERF.F12-OE-A, ERF.F12-OE-B, and ERF.F12-OE-C) with representative phenotypes and different expression levels for further phenotypic and molecular analyses (Figure 2A). Likewise, we generated eight ERF.F12-RNAi lines, from which we selected three representative lines (ERF.F12-RNAi-A, ERF.F12-RNAi-B, and ERF.F12-RNAi-C) for thorough characterization (Figure 2B). We validated the specificity of SlERF.F12 downregulation in the three RNAi lines by examining the relative expression of all other members of the ERF.F subfamily by RT-qPCR (Supplemental Figure S3).
Figure 2.
ERF.F12 represses the transition to fruit ripening and affects shelf life in tomato. A and B, Relative ERF.F12 transcript levels, in fruits at the Br stage in WT, ERF.F12-OE (A), and ERF.F12-RNAi lines (B), as assessed by RT-qPCR. ERF.F12-OE-A, -B, and -C are three independent ERF.F12-overexpressing lines. ERF.F12-RNAi-A, -B, and -C are three independent ERF.F12-RNAi lines. Asterisks indicate statistical significance using Student’s t test, P < 0.05. C, Time from anthesis to the Br stage in WT, ERF.F12-OE, and ERF.F12-RNAi lines. Asterisks indicate statistical significance using Student’s t test, P < 0.05. D, Different ripening stages in WT, ERF.F12-OE, and ERF.F12-RNAi lines. Fruits from overexpression lines show a delayed ripening phenotype while RNAi lines ripen earlier. E, Ethylene production in WT, ERF.F12-OE, and ERF.F12-RNAi fruits at different ripening stages. Values represent means of at least 15 individual fruits. F, Accumulation of carotenoids in WT, ERF.F12-OE, and ERF.F12-RNAi lines at different ripening stages. G, Genotype and different ripening stages in WT and mutant lines (erf.f12-1 and erf.f12-2) generated by CRISPR/Cas9 genome editing. The protospacer adjacent motif is underlined. Fruits from two independent lines show an earlier ripening phenotype. H, Time period from anthesis to Br in WT, erf.f12-1, and erf.f12-2 mutants. Asterisks indicate statistical significance using Student’s t test, P < 0.05. I, Fruit firmness in WT, ERF.F12-OE, and ERF.F12-RNAi lines at the Br + 7 stage. Average values were calculated for 20 individual fruits. Asterisks indicate statistical significance using Student’s t test: *P < 0.05. J, WT, ERF.F12-OE, and ERF.F12-RNAi fruits were harvested at the Br + 7 stage and photographs were taken after storing the fruits at room temperature for 20 days. K, PLW in WT and ERF.F12 transgenic fruits. The weight loss per fruit was measured every 5 days over 30 days of storage. Data are shown as means ± standard deviation (sd) (n = 20).
Notably, all three SlERF.F12-OE tomato lines displayed a significantly delayed onset of fruit ripening (P、 < 0.05; Figure 2, C and D), consistent with the hypothesis that SlERF.F12 negatively regulates the transition to ripening. In contrast, the downregulation of SlERF.F12 led to an advanced onset of ripening by 3–4 days (Figure 2, C and D). Indeed, wild-type (WT) fruits reached the Br stage at 42-day postanthesis (DPA) (Figure 2C), whereas the average time from anthesis to Br stage extended to 45 DPA in SlERF.F12-OE lines and shortened to 39 DPA in SlERF.F12-RNAi lines. We next investigated climacteric ethylene production in SlERF.F12-OE and SlERF.F12-RNAi fruits by monitoring ethylene production from 36 to 51 DPA. The peak of climacteric ethylene production shifted from 44 DPA in the WT to 46 DPA in SlERF.F12-OE fruits (Figure 2E) and occurred 2 days earlier in SlERF.F12-RNAi fruits than in WT. The amount of ethylene produced did not differ substantially between WT and OE lines but increased in RNAi lines. Because the accumulation of carotenoids is a hallmark of tomato fruit ripening, we performed ultra-performance liquid chromatography (UPLC) analysis of WT, SlERF.F12-OE, and SlERF.F12-RNAi fruits at 41, 44, and 47 `DPA, respectively. Total carotenoid contents were lower in SlERF.F12-OE fruits compared to the WT from 41 to 47 DPA (Figure 2F), consistent with the delayed ripening of SlERF.F12-OE fruits. In contrast, total carotenoid contents (primarily due to the accumulation of lycopene) were higher in SlERF.F12-RNAi fruits compared to the WT at all three stages examined (Figure 2F).
To further explore the role of SlERF.F12 in fruit ripening, we also generated knockout (ko) mutants using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system. Accordingly, we designed two specific single-guide RNAs and transformed the corresponding constructs into the tomato cultivar Micro-Tom (Figure 2G). We obtained two independent homozygous mutants in the T2 generation whose sequencing across the target sites identified a 1-bp deletion in the first target and a 5-bp deletion in the second target, giving rise to the erf.f12-1 and erf.f12-2 mutants, respectively (Figure 2G). Both mutations caused frameshifts predicted to result in truncated proteins, likely resulting in complete loss-of-function. Interestingly, consistent with the early ripening initiation seen in SlERF.F12-RNAi fruits, both erf.f12-1 and erf.f12-2 mutants reached the Br Stages 3–4 days earlier than the WT (Figure 2, G and H). Moreover, SlERF.F12 ko fruits exhibited earlier ethylene emission and softening (Supplemental Figure S4, A and B). In addition, the early accumulation of transcripts for key ripening-related genes was consistent with the advanced ripening initiation in SlERF.F12 ko mutants (Supplemental Figure S4C), supporting the idea of a repressor function for SlERF.F12 in fruit ripening. Given that both RNAi and ko lines displayed similar phenotypes with regards to fruit ripening, and because the RNAi lines were obtained first, we conducted subsequent physiological and transcriptomic characterization using the RNAi lines.
SlERF.F12 inhibits fruit softening and prolongs shelf life
To examine the effects of the altered expression of SlERF.F12 on fruit softening, a major ripening-associated phenomenon, we assessed fruit firmness in the WT, SlERF.F12-OE, and SlERF.F12-RNAi lines at the red-ripe (Br + 7) stage. Compared to WT, SlERF.F12-RNAi fruits displayed accelerated softening (Figure 2I), while SlERF.F12-OE fruits exhibited higher firmness than the WT (Figure 2I). To address the influence of SlERF.F12 overexpression and silencing on tomato fruit shelf life, we harvested WT, SlERF.F12-OE, and SlERF.F12-RNAi fruits at the Br + 7 stage and stored them at room temperature. Control WT fruits started to wrinkle after 10–15 days of storage at room temperature (Figure 2J), while SlERF.F12-RNAi fruits exhibited the first wrinkling symptoms as early as 7–10 days into storage (Figure 2J). In contrast, SlERF.F12-OE fruits displayed delayed senescence, occurring after 15–20 days of storage (Figure 2J). These data indicated that upregulation of SlERF.F12 in SlERF.F12-OE lines results in extended fruit shelf life and a delayed appearance of wrinkling symptoms. In addition, downregulation of SlERF.F12 resulted in higher physiological loss of water (PLW) values than in the WT, while SlERF.F12 overexpression led to lower PLW values (Figure 2K), further supporting the effects of disrupting SlERF.F12 expression on fruit shelf life.
The expression of ripening-related genes is altered in SlERF.F12 transgenic fruits
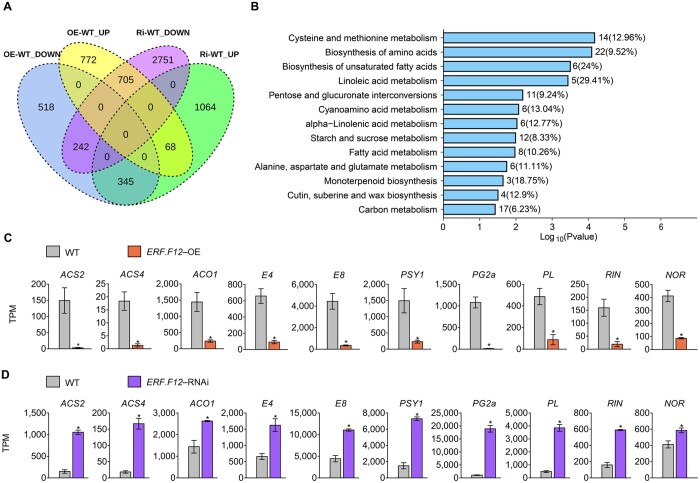
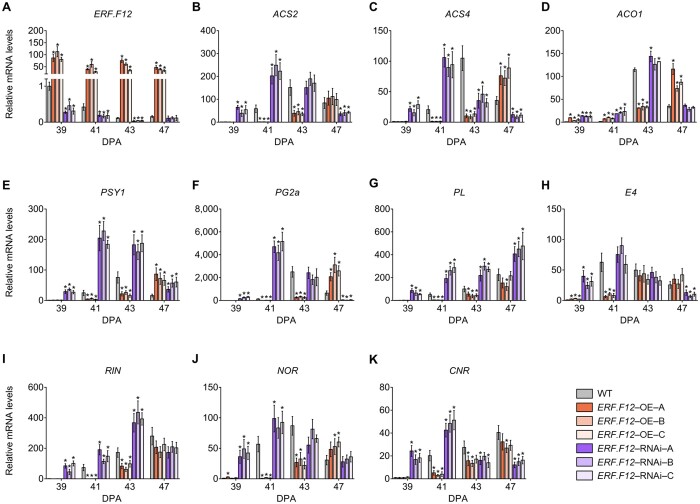
To gain molecular insight into the modified ripening processes in the SlERF.F12 transgenic lines, we performed transcriptome deep sequencing (RNA-seq) of SlERF.F12-OE, SlERF.F12-RNAi, and WT fruits at 41 DPA. We identified a total of 2,650 differentially expressed genes (DEGs) between the WT and SlERF.F12-OE lines, of which 1,105 were downregulated and 1,545 were upregulated in SlERF.F12-OE fruits (Figure 3A; Supplemental Data Set 1). In addition, we detected 3,698 downregulated genes and 1,477 upregulated genes in SlERF.F12-RNAi lines compared to the WT (Figure 3A; Supplemental Data Set 2). Because of the contrasting fruit ripening phenotype between SlERF.F12-OE and SlERF.F12-RNAi lines, we focused on the DEGs with contrasting expression in SlERF.F12-OE and RNAi fruits. Overall, 1,050 genes displayed an opposite expression pattern in SlERF.F12-OE and RNAi fruits, consisting of 345 genes upregulated in SlERF.F12-RNAi fruits but downregulated in OE lines, and 705 genes downregulated in SlERF.F12-RNAi fruits but upregulated in OE lines (Figure 3A; Supplemental Data Set 3). Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation pathway analysis revealed that multiple metabolic pathways are affected, such as carbon metabolism, starch and sucrose metabolism, fatty acid metabolism, linoleic acid metabolism, monoterpenoid biosynthesis, and biosynthesis of amino acids (Figure 3B), indicating that multiple processes are affected during ripening when the normal expression of SlERF.F12 is disrupted. Consistent with the accelerated ripening in SlERF.F12-RNAi lines and the delayed ripening of SlERF.F12-OE lines, key ripening-related genes were induced in SlERF.F12-RNAi fruits and repressed in OE fruits, such as ethylene biosynthetic and responsive genes (1-AMINO-CYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2 [ACS2], ACS4, 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE 1 [ACO1], E4, E8), lycopene biosynthesis gene (PHYTOENE SYNTHASE 1 [PSY1]), fruit softening genes (POLYGALACTURONASE 2a [PG2a], PECTATE LYASE [PL]), and key ripening regulators (RIN, NOR) (Figure 3, C and D; Supplemental Data Set 3). To confirm the regulatory role of SlERF.F12 in the expression of fruit ripening-related genes, we assessed their transcript levels in WT, SlERF.F12-OE, and SlERF.F12-RNAi fruits from 39 to 47 DPA by RT-qPCR. SlERF.F12 transcript levels were higher in SlERF.F12-OE lines compared to WT and lower in SlERF.F12-RNAi lines, as expected (Figure 4A). The peaks of transcript abundance for ACS2, ACS4, and ACO1, three key genes involved in climacteric ethylene production, occurred with a delay in SlERF.F12-OE fruits but shifted earlier in SlERF.F12-RNAi fruits compared to the WT (Figure 4, B–D). This shift in peak expression timing matched the timing of ethylene peaks in SlERF.F12-OE and SlERF.F12-RNAi fruits. In addition, transcript levels of other ripening-related genes, such as the carotenoid biosynthesis gene PSY1; the fruit softening-related genes PL and PG2a; the ethylene response gene E4; and the key ripening regulator genes RIN, NOR, and CNR were lower in SlERF.F12-OE fruits and higher in SlERF.F12-RNAi fruits than in WT during early ripening stages (from 39 to 41 DPA) (Figure 4, E–K). These results were in agreement with the delayed ripening initiation in SlERF.F12 overexpression lines and the advanced onset of ripening in RNAi lines.
Figure 3.
RNA-seq profiling of ERF.F12-OE and ERF12-RNAi fruits. A, Venn diagram showing the overlap between downregulated and upregulated genes that were differentially expressed in ERF.F12-OE and ERF.F12-RNAi fruits compared to WT at the 41 DPA stage. B, KEGG pathway analysis of 1,050 genes with an opposite expression pattern in ERF.F12-OE and ERF.F12-RNAi fruits. C and D, Comparison of ripening-related gene expression patterns obtained by RNA-seq in ERF.F12-OE (C) and ERF.F12-RNAi fruits (D). Data are shown as means ± standard deviation (sd) from three biological replicates. Asterisks indicate statistical significance using Student’s t test, P < 0.05.
Figure 4.
Relative expression of ripening-related genes in WT, ERF.F12-OE, and ERF.F12-RNAi lines during the ripening process. A, Relative ERF.F12 expression in WT, ERF.F12-OE, and ERF.F12-RNAi lines. B–K, Relative expression of ethylene biosynthesis genes ACS2 (B), ACS4 (C), ACO1 (D); carotenoid synthesis genes PSY1 (E); fruit softening-related genes PG2a (F), PL (G); ethylene-responsive genes E4 (H); and ripening regulators RIN (I), NOR (J), and CNR (K) in WT, ERF.F12-OE, and ERF.F12-RNAi lines. Total RNA was extracted from the indicated fruits at different ripening stages (39, 41, 43, and 47 DPA). Relative mRNA levels of each gene in WT at 39 DPA were normalized to 1, using SlActin gene as an internal control. Data are shown as means ± sd from six biological replicates. Asterisks indicate statistical significance using Student’s t test, P < 0.05.
SlERF.F12 affects plant growth and regulation of flowering time
Given the relatively high expression of SlERF.F12 in vegetative tissues, we examined its function in vegetative growth and development. Notably, the size of 6-week-old SlERF.F12-OE plants was not significantly different from that of WT, but SlERF.F12-RNAi plants were shorter (Supplemental Figure S5, A–C). In addition, SlERF.F12-RNAi plants exhibited an early flowering phenotype (Supplemental Figure S5, B and D), suggesting a potential role for SlERF.F12 in delaying the initiation of flowering. To investigate whether the altered plant growth and flowering time were related to ethylene, we assessed the transcript levels of genes involved in ethylene biosynthesis or response in 6-week-old leaves. We detected higher transcript levels for ethylene biosynthesis genes (ACS2 and ACS6) and ethylene responsive genes (E4 and E8) in SlERF.12-RNAi lines (Supplemental Figure S5E). We also investigated the effect of the altered expression of SlERF.F12 on leaf senescence, but we observed no obvious difference in leaf senescence between OE or RNAi lines and the WT after incubation in the dark for 2 weeks (Supplemental Figure S6, A and B). In contrast, etiolated SlERF.F12-RNAi seedlings were hypersensitive to ethylene, while sensitivity to the phytohormone was slightly lower in seedlings overexpressing SlERF.F12 (Supplemental Figure S7). In addition, triple response assays showed that etiolated erf.f12 seedlings are more sensitive to ethylene than WT (Supplemental Figure S8). Moreover, erf.f12 plants exhibited reduced size and early flowering compared to the WT, indicating altered vegetative growth and reproductive development in SlERF.F12 ko mutants (Supplemental Figure S9, A–C). In agreement, the expression of ACS2, ACS6, E4, and E8 was upregulated in erf.f12 plants (Supplemental Figure S9D). Leaf senescence assays showed no obvious differences in chlorophyll loss between SlERF.F12 ko mutants and the WT (Supplemental Figure S9, E and F). Taken together, these results indicate that, in addition to its role in the initiation of fruit ripening, SlERF.F12 also participates in plant growth and flowering time, at least partly through alteration of ethylene production or sensitivity.
SlERF.F12-mediated repression of ripening-related genes is dependent on the EAR motif
As shown above, the delayed ripening of SlERF.F12-OE fruits was associated with the downregulation of ripening-related genes, suggesting that SlERF.F12 may act as a negative regulator of the transcriptomic reprogramming associated with fruit ripening. To investigate this possibility, we examined the ability of SlERF.F12 to directly regulate the transcription of ripening-related genes using transient expression assays. We generated dual LUC reporter plasmids by individually fusing the ACS2, ACS4, PG2a, and PL promoter sequences to the firefly LUC reporter, using REN LUC driven by the 35S promoter as an internal control (Figure 5A). Co-transfection of the proACS2:LUC, proACS4:LUC, proPG2a:LUC, or proPL:LUC reporter constructs with the pro35S:SlERF.F12 effector in Nicotiana benthamiana leaf protoplasts resulted in significantly reduced luminescence intensity (Figure 5B), revealing the capacity of SlERF.F12 to repress the transcription of these ripening-related genes. Notably, this repressing activity was lost in the effector construct 35S:SlERF.F12ΔEAR2 lacking the C-terminal EAR motif (Figure 5B). These results suggested that SlERF.F12-mediated repression of the ripening-related genes is dependent on the presence of the C-terminal EAR motif (EAR2).
Figure 5.
ERF.F12 represses the transcription of ripening-related genes by binding to their promoters. A, Schematic diagram of the double-reporter and effector plasmids used in the transient expression assay. B, The repressive effect of ERF.F12 on the transcription of ACS2, ACS4, PG2a, and PL is dependent on the presence of the C-terminal EAR motif. The double reporter plasmid was co-transfected with individual effector plasmids into N. benthamiana protoplasts. ERF.F12ΔEAR2, ERF.F12 lacking the C-terminal EAR motif. Each value represents the mean of six biological replicates. Asterisks indicate statistical significance using Student’s t test, P < 0.05. C, Schematic diagram of promoters and primer positions used in ChIP-qPCR assays. D, Anti-FLAG ChIP-qPCR showing specific binding of ERF.F12 to the promoters of ACS2, ACS4, PG2a, and PL. Control ChIP was performed without antibody. E and F, EMSA showing the direct binding of intact ERF.F12 (E) and truncated ERF.F12 (F) to the promoters of PG2a and PL via the GCC box or DRE box. The sequences of the WT probes containing the GCC box or DRE box were labeled with biotin. Competition for ERF.F12 binding was performed with 50× and 500× cold probes containing the WT GCC box (GCCGCC), DRE box (CCGAC) or mutated controls (AAAAAA or AAAAA). The symbols − and + represent absence and presence, respectively, and ++ indicates increasing amounts.
To further examine the ability of SlERF.F12 to bind to the promoter of ripening-related genes, we conducted chromatin immunoprecipitation (ChIP) followed by qPCR (ChIP-qPCR) experiments using 39-DPA tomato fruit expressing the SlERF.F12-FLAG using anti-FLAG antibodies for immunoprecipitation. We designed specific primers across promoter regions of the four ripening-associated genes that include the putative ERF binding cis-elements, while primers across the 3′-untranslated region served as negative control (Figure 5C). We normalized the binding of SlERF.F12 to the input DNA fragments. SlERF.F12 bound to the promoters of the ripening-related genes ACS2 and ACS4 (ethylene biosynthetic genes), as well as PG2a and PL (cell wall-related genes), as determined by qPCR (Figure 5D). Moreover, electrophoretic mobility shift assays (EMSAs) showed that both SlERF.F12 and its truncated SlERF.F12 variant lacking the C-terminal EAR motif can bind directly to a DNA probe containing the GCC box or DRE/CRT box present in the PG2a and PL promoters (Figure 5, E and F). These results indicated that SlERF.F12 represses the transcription of ripening-related genes through direct binding to cis-elements in their promoters.
SlERF.F12 interacts with TPL2 through the C-terminal EAR motif
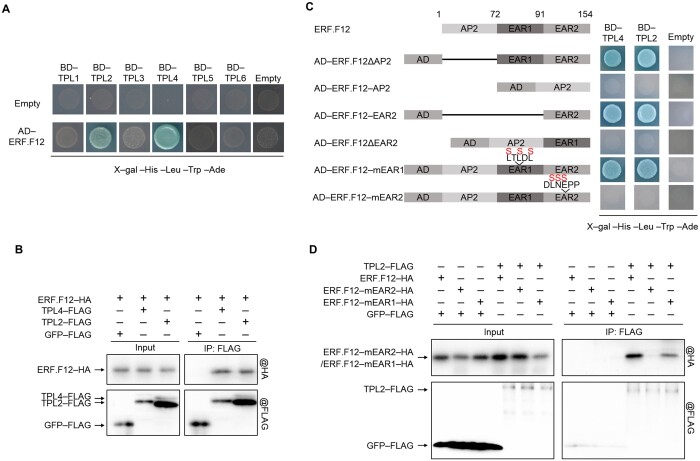
To elucidate the molecular mechanisms of SlERF.F12-mediated transcriptional repression, we performed a yeast two-hybrid (Y2H) screen to identify putative interactors, using SlERF.F12 as bait to screen a tomato fruit Y2H cDNA library. From a total of 201 positive clones, we identified 19 different genes, 2 of which were predicted to encode TOPLESS proteins (TPL2 and TPL4) (Supplemental Table S2). Because TOPLESS (TPL) proteins were previously reported to interact with EAR motif-containing transcription factors (Kagale and Rozwadowski, 2011), we performed a targeted Y2H assay to investigate the potential for interaction between SlERF.F12 and all known members of the tomato TOPLESS protein family (SlTPL). Only SlTPL2 and SlTPL4 exhibited a specific interaction with SlERF.F12 (Figure 6A). To validate these interactions in vivo, we performed co-immunoprecipitation (Co-IP) assays following transient co-infiltration of SlERF.F12-HA and SlTPL2/SlTPL4-FLAG constructs in N. benthamiana leaves. We observed clear Co-IP of SlERF.F12-HA with both SlTPL2-FLAG and SlTPL4-FLAG, but not with GFP-FLAG control (Figure 6B). Since SlERF.F12 contains two EAR motifs, we generated versions of SlERF.F12 lacking either `EAR1 or EAR2 to investigate whether both motifs are essential for the interaction with TPL2/TPL4. Y2H assays showed that deletion of the C-terminal EAR2 (SlERF.F12ΔEAR2) or its mutation (SlERF.F12mEAR2) abolishes the interaction between SlERF.F12 and TPLs (Figure 6C), whereas altering the other motif had no effect, indicating that EAR2, but not EAR1, is essential for the interaction between SlERF.F12 and SlTPL2/TPL4. In agreement, SlTPL2 and SlERF.F12 only co-immunoprecipitated when the EAR2 motif was present, confirming that EAR2 is the key motif required for the interaction between the two proteins in vivo (Figure 6D). The interaction of SlERF.F12 with TPL2/TPL4 suggested that these proteins might form a transcriptional repressor complex to negatively regulate the expression of ripening-related genes.
Figure 6.
ERF.F12 interacts with TPLs via the C-terminal EAR motif. A, Y2H assays between ERF.F12 and TPLs. TPL1, TPL2, TPL3, TPL4, TPL5, and TPL6 were used as bait, and ERF.F12 used as prey. Empty-BD and Empty-AD were co-transformed as negative controls. B, In vivo Co-IP assays of ERF.F12 with TPL2 and TPL4. ERF.F12-HA was transiently co-infiltrated with TPL2-FLAG or TPL4-FLAG in N. benthamiana leaves by Agrobacterium-mediated infiltration. GFP-FLAG was used as a negative control. Total proteins were extracted from infiltrated leaves and used for immunoprecipitation with anti-FLAG antibody. Immunoblots were probed with anti-FLAG antibody to detect TPLs and with anti-HA antibody to detect ERF.F12. C, The interaction between ERF.F12 and TPL2/4 is dependent on the C-terminal EAR motif. TPL2 and TPL4 were used as bait. ERF.F12 protein fragments generated by different deletions and mutated ERF.F12 in different EAR motifs were used as prey. ERF.F12ΔAP2 lacks the N-terminal AP2/ERF domain deleted; ERF.F12-AP2 harbors the AP2 domain but lacks both EAR motifs; ERF.F12-EAR2 is the C-terminal EAR motif; ERF.F12ΔEAR2 lacks the C-terminal EAR motif. ERF.F12-mEAR1 has a mutated EAR1 motif; ERF.F12-mEAR2 has a mutated EAR2 motif. D, ERF.F12 interacts with TPL2 and TPL4 through the C-terminal EAR motif in vitro. In an in vitro Co-IP assay, TPL2-FLAG and TPL4-FLAG bind to ERF.F12-HA and ERF.F12mEAR1-HA, but not ERF.F12mEAR2-HA. GFP-FLAG was used as a negative control.
SlERF.F12-TPL2 forms a protein complex that represses the transcription of ripening-related genes
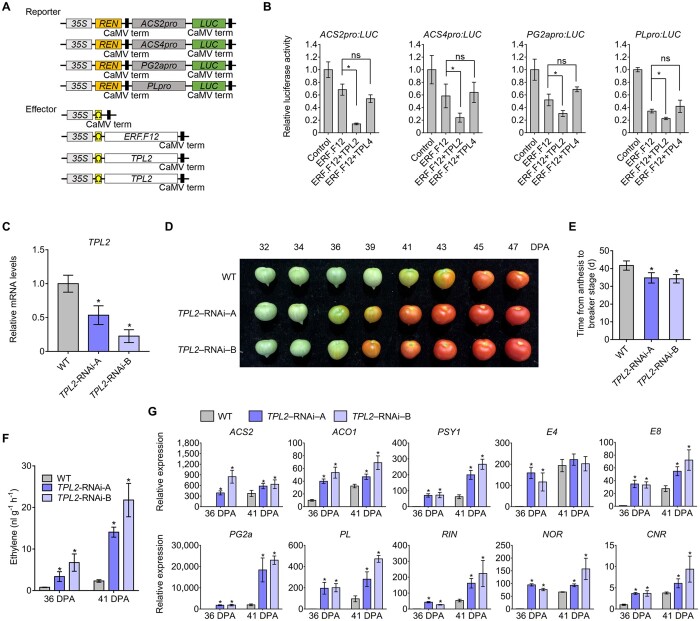
To investigate whether the interaction between SlERF.F12 and TPL2/TPL4 is involved in the transcriptional repression of ripening-related genes, we conducted transient expression assays using SlERF.F12 and SlTPL2/TPL4 as effectors with the promoters of ripening-related genes driving the transcription of firefly LUC as reporter constructs (Figure 7A). Co-transfection of the reporter constructs with SlERF.F12 and TPL2 effector constructs in N. benthamiana leaf protoplasts repressed transcription of the reporter gene (Figure 7B). In contrast, co-transfection of SlERF.F12 with TPL4 did not result in a significant repression of the transcription of the reporter genes. These results suggested that SlERF.F12 forms a repression complex in vivo specifically with TPL2, but not with TPL4, to negatively regulate the transcription of ripening-related genes.
Figure 7.
TPL2 enhances the repression of transcription of ripening-related genes by ERF.F12 and represses fruit ripening. A, Schematic diagram of the double-reporter and effector plasmids used in the transient expression assay. B, Repression by ERF.F12 co-transformed with TPL2 or TPL4 of the transcription of ripening-related genes ACS2, ACS4, PL, and PG2a. The double reporter construct was co-transfected with individual effector plasmids into N. benthamiana protoplasts. Each value represents the means of six biological replicates. Asterisks indicate statistical significance using Student’s t test, P < 0.05. C, Relative TPL2 transcript levels assessed by RT-qPCR in fruits at the Br stage in WT and TPL2-RNAi lines. Asterisks indicate statistical significance using Student’s t test, P < 0.05. D, Ripening phenotype of TPL2-RNAi lines. WT, TPL2-RNAi-A, and TPL2-RNAi-B fruits at 32, 34, 36, 39, 41, 43, 45, and 47 DPA are shown. E, Time from anthesis to the Br stage in WT, TPL2-RNAi-A, and TPL2-RNAi-B lines. Asterisks indicate statistical significance using Student’s t test, P < 0.05. F, Ethylene production in WT, TPL2-RNAi-A, and TPL2-RNAi-B fruits at 36 DPA and 41 DPA. Values represent means of measurements of at least 15 individual fruits. Asterisks indicate statistical significance using Student’s t test, P < 0.05. G, Relative expression of ethylene biosynthesis genes ACS2, ACO1; carotenoid biosynthesis genes PSY1; ethylene-responsive genes E4, E8; fruit softening-related genes PG2a, PL; and ripening regulators RIN, NOR, and CNR in WT, TPL2-RNAi-A, and TPL2-RNAi-B lines. Total RNA was extracted from the indicated fruits at 36 and 41 DPA. Relative transcript levels of each gene in WT at 36 DPA were normalized to 1, with SlActin as an internal control. Data are shown as means ± sd from six biological replicates. Asterisks indicate statistical significance using Student’s t test, P < 0.05.
These data raised the possibility that SlTPL2 might be involved in the regulation of fruit ripening. Accordingly, we generated stable tomato RNAi lines targeting SlTPL2. We obtained ten independent SlTPL2-RNAi lines showing significant downregulation of SlTPL2, from which we selected two representative lines (TPL2-RNAi-A and TPL2-RNAi-B) for phenotypic and molecular characterization (Figure 7C). SlTPL2-RNAi fruits exhibited an advanced transition to ripening compared to the WT, reaching the Br stage 4–5 days earlier than WT (Figure 7, D and E). In line with the early ripening phenotype, ethylene production was higher in SlTPL2-RNAi fruits at both 36 and 41 DPA (Figure 7F). Genes involved in ethylene biosynthesis (ACS2 and ACO1), carotenoid metabolism (PSY1), cell wall degradation (PG2a, PL), and ripening regulation (RIN, NOR, and CNR) as well as ethylene-responsive genes (E4 and E8) were all upregulated in SlTPL2-RNAi fruits relative to the WT, as shown by RT-qPCR (Figure 7G). These data favored the hypothesis that the early transition to ripening in SlTPL2-RNAi lines is due to the premature expression of key regulators of ripening, thus revealing that TPL2 also plays a negative role in fruit ripening, consistent with its interaction with SlERF.F12.
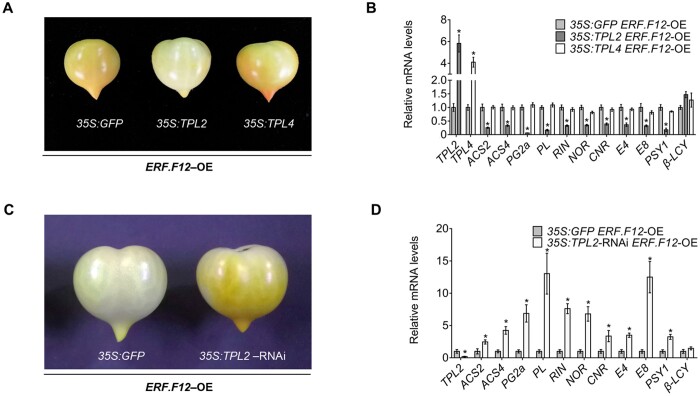
To gain more insight into the role of the SlERF.F12–TPL2 complex in regulating fruit ripening, we transiently expressed TPL2 or TPL4 in ERF.F12-OE-A fruits at the MG stage. Overexpression of SlTPL2 in ERF.F12-OE-A fruits resulted in a delayed initiation of fruit ripening compared to control ERF.F12-OE-A fruits expressing the mock GFP construct (Figure 8A). In contrast, similar experiments with SlTPL4 had no effect. Consistent with the observed delayed ripening, the relative transcript levels of the ripening-related genes ACS2, ACS4, PG2a, and PL were lower in TPL2 overexpressing fruits compared to those overexpressing TPL4 or GFP in the same ERF.F12-OE-A genetic background 5 days after infiltration (Figure 8B). In addition, we also transiently expressed a 35S:TPL2-RNAi construct in ERF.F12-OE-A fruits at the MG stage, which revealed that silencing of SlTPL2 in the SlERF.F12-OE genetic background advances the initiation of ripening compared to the control expressing 35S:GFP (Figure 8C). These data suggested that the repressive effect of SlERF.F12 on ripening initiation can be mitigated by the downregulation of SlTPL2. The increased transcript levels of ripening-related genes were in line with the advanced ripening initiation in the SlERF.F12-OE lines silenced for TPL2 (Figure 8D). Altogether, the data further supported the notion that the recruitment of TPL2 by SlERF.F12 plays an important role in negatively regulating the initiation of tomato fruit ripening through the transcriptional repression of a set of ripening-related genes.
Figure 8.
The repression of fruit ripening by ERF.F12 is dependent on TPL2. A, Ripening phenotype produced by transient overexpression of TPL2 or TPL4 in ERF.F12-OE-A fruits at 35 DPA. The photograph was taken 5 days after Agrobacterium-mediated infiltration. 35S:GFP was used as negative control. B, Relative transcript levels of TPL2, TPL4, ACS2, ACS4, PG2a, PL, RIN, NOR, CNR, E4, E8, PSY1, and β-LCY in fruits transiently expressing either GFP, TPL2, or TPL4 5 days after Agrobacterium-mediated infiltration. Each value represents the means of six biological replicates. Asterisks indicate statistical significance by Student’s t test, P < 0.05. C, Ripening phenotype produced by transient TPL2 silencing by RNAi in ERF.F12-OE-A fruits at 35 DPA. The photograph was taken 5 days after Agrobacterium-mediated infiltration. D, Relative expression levels of TPL2, ACS2, ACS4, PG2a, PL, RIN, NOR, CNR, E4, E8, PSY, and β-LCY in fruits transiently expressing either GFP or silencing TPL2 by RNAi 5 days after Agrobacterium-mediated infiltration. Each value represents the means of six biological replicates. Asterisks indicate statistical significance by Student’s t test, P < 0.05.
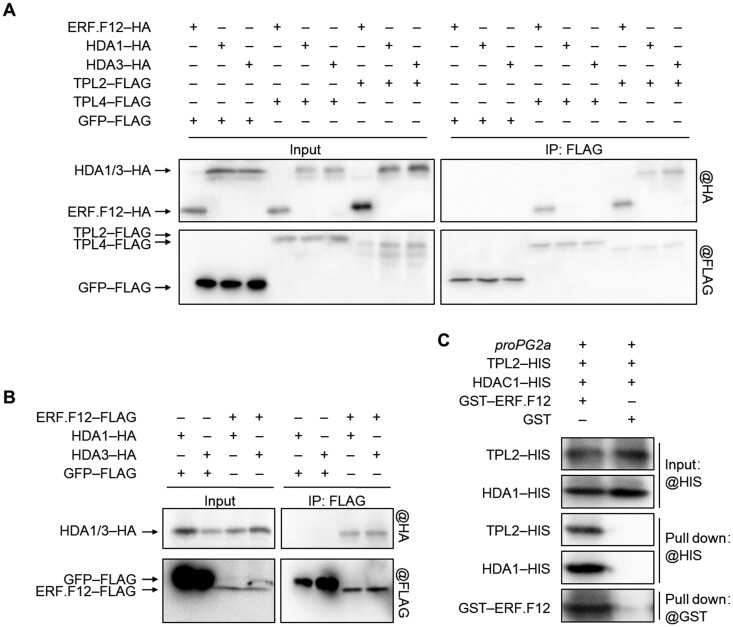
SlERF.F12 forms an in vivo tripartite complex with TPL2 and HDA1/3
HDAs such as HDA19 and HDA6 are additional components of the TPL-dependent transcriptional repression complex in Arabidopsis (Arabidopsis thaliana; Long et al., 2006; Wang et al., 2013). To explore whether the SlERF.F12–TPL2 complex recruits HDAs to repress the transcription of ripening-related genes in tomato, we performed Co-IP assays to test the in vivo interaction between the SlERF.F12–TPL2 complex and SlHDA1 and SlHDA3, two key HDAs previously reported to regulate tomato fruit ripening (Guo et al., 2017, 2018). Interestingly, SlERF.F12-HA co-immunoprecipitated with both SlTPL2-FLAG and SlTPL4-FLAG, but not with GFP-FLAG (Figure 9A), confirming the ability of SlERF.F12 to interact with these two TPLs. Moreover, SlHDA1-HA and SlHDA3-HA co-immunoprecipitated with SlTPL2-FLAG, but not with SlTPL4-FLAG (Figure 9A), revealing the specific interaction between SlTPL2 and SlDHA1/3 in vivo. In addition, our Co-IP assays showed that SlERF.F12 can interact with both SlDHA1 and SlHDA3 in vivo (Figure 9B). To examine whether SlERF.F12 recruits TPL2 and HDA1 to the promoters of SlERF.F12 target genes, we performed DNA pull-down assays with a biotin-labeled PG2a promoter. The PG2a promoter pulled down SlTPL2 and SlHDA1 only in the presence of SlERF.F12 (Figure 9C), suggesting that SlERF.F12, SlTPL2, and SlHDA1/3 may form a tripartite complex to repress the transcription of ripening-related genes in vivo and that SlTPL2 might be required as an adaptor protein between SlERF.F12 and SlHDA1/3.
Figure 9.
ERF.F12 interacts with TPL2 and HDA1/3 in vivo. A, In vivo Co-IP assays of ERF.F12 with TPL2/4 and TPL2/4 with HDA1/3. TPL2-FLAG or TPL4-FLAG was co-infiltrated with ERF.F12-HA, HDA1-HA, or HDA3-HA in N. benthamiana leaves by Agrobacterium-mediated infiltration. Total proteins were extracted and immunoprecipitated with anti-FLAG antibody. GFP-FLAG was used as negative control. Immunoblots were probed with anti-FLAG antibody to detect TPLs and anti-HA antibody to detect ERF.F12 and DHA1/3. B, In vivo Co-IP assays of ERF.F12 with HDA1 and HDA3. ERF.F12-FLAG was co-infiltrated with HDA1-HA or HDA3-HA in N. benthamiana leaves by Agrobacterium-mediated infiltration. GFP-FLAG was used as negative control. Total proteins were extracted and immunoprecipitated with anti-FLAG antibody. Immunoblots were probed with anti-FLAG antibody to detect ERF.F12 and with anti-HA antibody to detect HDA1 and HDA3. C, TPL2 and HDA1 bind to the PG2a promoter through ERF.F12. Recombinant TPL2-HIS and HDA1-HIS were incubated with a biotin-labeled PG2a promoter DNA fragment (500 bp) together with GST or GST-ERF.F12 and pulled down with streptavidin agarose beads. Immunoblots were probed with anti-GST or anti-HIS antibody.
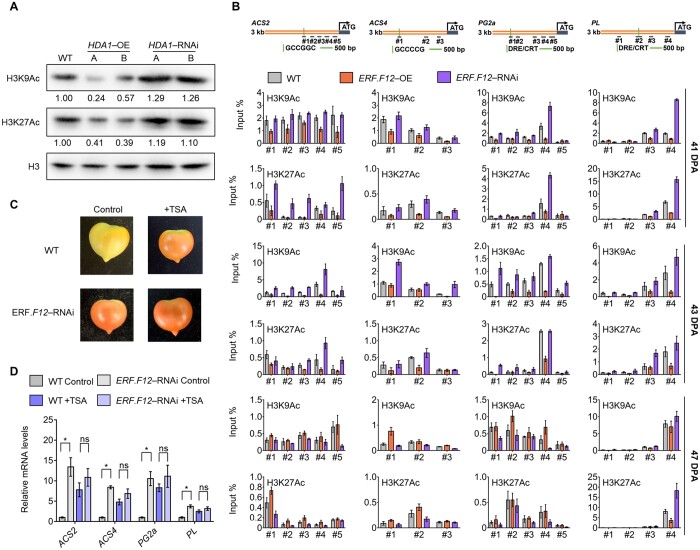
Since SlERF.F12 repressed the transcription of ripening-related genes in combination with SlTPL2 and SlHDA1/3, we postulated that SlERF.F12 transgenic fruits may exhibit altered histone acetylation levels at ripening-related genes. To test this hypothesis, we first examined whether the H3K9Ac and H3K27Ac histone marks were regulated by SlHDA1 by assessing global acetylation levels in SlDHA1-OE and SlDHA1-RNAi fruits at 41 DPA. The levels of both H3K9Ac and H3K27Ac were lower in SlHDA1-OE fruits but increased in SlHDA1-RNAi lines compared to control WT lines (Figure 10A). This result suggested that SlHDA1 can deacetylate the two histone H3 residues in tomato fruits. We also performed ChIP assays using antibodies against H3K9Ac and H3K27Ac to assess the acetylation levels at the promoter regions of ripening-related genes in SlERF.F12-OE, SlERF.F12-RNAi, and WT fruits at 41, 43, and 47 DPA ripening stages. Compared to the WT, H3K9Ac and H3K27Ac levels were lower in SlERF.F12-OE fruits at the promoter regions of ACS2, ACS4, PG2a, and PL, which are targets of SlERF.F12 (Figure 10B). In contrast, H3K9Ac and H3K27Ac levels at the promoter regions of these ripening-related genes were higher in SlERF.F12-RNAi fruits than in WT (Figure 10B), consistent with their higher transcript levels in these transgenic lines.
Figure 10.
ERF.F12 repression of fruit ripening requires HDA1 activity. A, Global H3K9Ac and H3K27Ac acetylation levels in WT, HDA1-OE, and HDA1-RNAi fruits at 41 DPA. Band intensities were normalized relative to total histone H3 loading controls. B, ChIP analysis of H3K9Ac and H3K27Ac levels at the ACS2, ACS4, PG2a, and PL promoters in WT, ERF.F12-OE-A, and ERF.F12-RNAi-B fruits at 41, 43, and 47 DPA. Data are shown as means ± sd with six biological replicates. C, TSA treatment inhibits promotion of fruit ripening in ERF.F12-RNAi lines. Fruits were infiltrated with 10-μM TSA at 36 DPA. The photographs were taken 7 days after infiltration. D, Relative transcript levels of ACS2, ACS4, PG2a, and PL 5 days after TSA treatment. Each value represents the mean of six biological replicates. Asterisks indicate statistical significance by Student’s t test, P < 0.05.
To validate the requirement for HDA activity in SlERF.F12-mediated ripening inhibition, we treated WT and SlERF.F12-RNAi fruits at 36 DPA with the HDA inhibitor trichostatin A (TSA). We observed an acceleration of ripening in TSA-treated WT fruits compared to untreated control fruits (Figure 10C). However, we saw no change in the ripening of SlERF.F12-RNAi lines treated with TSA relative to untreated controls (Figure 10C). Notably, RT-qPCR analysis showed that the relative expression levels of ripening-related genes ACS2, ACS4, PG2a, and PL are higher in TSA-treated WT fruits, whereas we observed no significant change in TSA-treated SlERF.F12-RNAi fruits compared to their controls (Figure 10D). This result indicated that HDA activity is required for the repression of ripening-related genes by SlERF.F12. Taken together, this study supports a model whereby SlERF.F12 recruits the TPL2-HDA1/3 repression complex to repress the transcription of key ripening-related genes by affecting histone acetylation levels at the promoter regions (Figure 11). Interestingly, the role of ERF.F12 in the transition to ripen appears to be conserved in several climacteric fruit species. This conclusion is supported by the mining of publicly available transcriptomic data, which indicated that SlERF12 homologs in kiwifruit (Actinidia chinensis), apple, banana (Musa acuminata), and pear (Pyrus bretschneideri) are downregulated at the onset of ripening, similar to our results in tomato (Supplemental Figure S10).
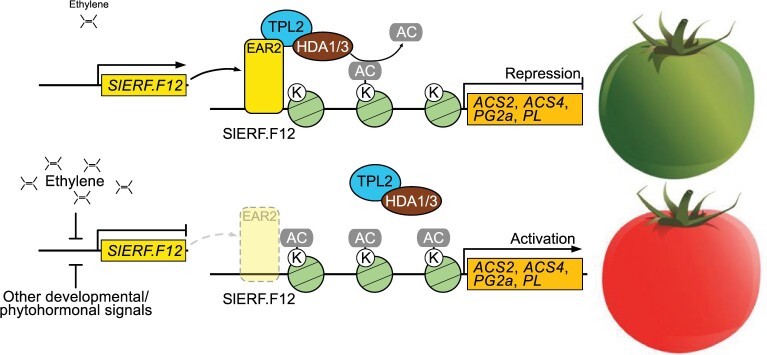
Figure 11.
Tentative model of the role of ERF.F12 in repression of ripening initiation. A, Before the onset of ripening initiation (i.e. MG stage), ERF.F12 is highly expressed and its encoded protein interacts with the co-repressors TPL2 and recruits HDA1 and HDA3 to form a repressor complex that epigenetically represses the transcription of ripening-related genes such as ACS2, ACS4, PG2a, and PL. B, During the onset of ripening initiation (i.e. Br stage), ERF.F12 transcript levels decrease upon climacteric ethylene production or other developmental/phytohormonal signals, which result in a reduction of ERF.F12 available to form an ERF.F12–TPL2–HDA1/3 complex, leading to histone modification and activation of the expression of ripening-related genes.
Discussion
ERFs are downstream components of ethylene signaling by directly regulating ethylene-responsive gene expression, suggesting that these transcription factors play important roles in ethylene-dependent developmental processes, including ripening of climacteric fruit (Pirrello et al., 2012; Liu et al., 2015a, 2016; Li et al., 2020a, 2020b). However, although several ERF genes have been shown to be involved in fruit ripening (Li et al., 2007; Lee et al., 2012; Liu et al., 2014; Sun et al., 2018), the roles and modes of action of most ERFs in regulating climacteric fruit ripening have remained elusive. Here, we demonstrate that SlERF.F12, an EAR motif-containing ERF.F subfamily member, acts as a repressor of fruit ripening by forming a repressor complex with SlTPL2 and SlHDA1/SlHDA3 to negatively regulate the transcription of key ripening-related genes in tomato. Our study uncovers a direct link between repressor ERFs and histone modifiers such as HDAs in modulating the transition to ripening. This new result broadens our knowledge of the regulatory network controlling climacteric fruit ripening and provides insights into the roles of EAR motif-containing ERFs in regulating the ultimate steps of fleshy fruit development.
ERFs are classified as transcriptional activators or repressors (Fujimoto et al., 2000; McGrath et al., 2005; Pirrello et al., 2012) and we show here that SlERF.F12 contains two typical EAR motifs known to confer a transcriptional repressor activity (Ohta et al., 2001; Song et al., 2005; Yang et al., 2005; Yin et al., 2010; Han et al., 2016; Li et al., 2019). SlERF.F12 repressed the expression of ripening-related genes ACS2, ACS4, PG2a, and PL by binding to their promoters (Figure 5). This repressor activity was alleviated at the onset ripening by the downregulation of SlERF.F12, which can also be induced by exogenous ethylene treatment, thus promoting the progression of ripening. Conversely, the overexpression of SlERF.F12 resulted in delayed fruit ripening, concomitantly with the downregulation of a set of key ripening-related genes (Figures 3 and 4). The transition to ripening in fleshy fruits like tomato is genetically programmed and involves an intricate interplay between multiple phytohormones and developmental factors. Although the precise underlying mechanisms remain unclear, it is largely accepted that both activators and repressors are at play (Liu et al., 2015a, 2015b; Li et al., 2021). Therefore, some activators may decrease the repressive effects of SlERF.F12 on ripening-related genes, which might explain why the delayed onset of ripening was not very strong in the overexpression (OE) lines. Combining reverse genetics approaches, physiological methods, Co-IP, and ChIP-qPCR assays, our study clearly showed that the transcriptional repressor SlERF.F12 plays a negative role in the transition to ripening in tomato fruits by recruiting the co-repressor TPL2 and the HDAs HDA1/HDA3 to repress the transcription of ripening-related genes. In this regard, our data uncovered a new layer of complexity of the mechanisms controlling the initiation of ripening.
Members of class II ERFs containing the EAR motif, initially identified in Arabidopsis (Ohta et al., 2001), have been shown to function as negative regulators by interacting with co-repressors like TPL and SAP18, as well as HDA19 (Kagale and Rozwadowski, 2011; Wang et al., 2013; Ryu et al., 2014; Li et al., 2019). In Arabidopsis, the class II repressor ERFs ERF3, ERF4, and ERF7 play important roles in regulating abscisic acid and abiotic stress responses by interacting with the co-repressors SAP18 or SIN3, which in turn interact and form a repressor complex with HDA19 (Song et al., 2005; Yang et al., 2005; Song and Galbraith, 2006). Arabidopsis ERF12, another EAR motif-containing ERF, was also recently shown to interact with TPL to regulate seed dormancy by repressing the expression of DELAY OF GERMINATION 1 (Li et al., 2019). MaERF11 and CpERF9, two EAR-containing transcriptional repressors, have been reported to repress the expression of ripening-related genes by directly recruiting MaHDA1 and CpHDA3 in banana and papaya (Carica papaya), respectively (Han et al., 2016; Fu et al., 2019). Here, we demonstrated that tomato SlERF.F12 interacts with the co-repressor SlTPL2 via its C-terminal EAR motif to recruit the ripening-associated HDASs SlHDA1 and SlHDA3, forming a complex that represses the transcription of ripening-related genes. Our findings suggest that the recruitment of chromatin-remodeling factors such as HDAs to epigenetically repress gene expression appears to be an important mechanism for the functions of these transcriptional repressors. Our study also demonstrates that an EAR motif-containing ERF interacts with TPL and recruits HDA proteins to regulate fruit ripening in tomato.
Altogether, the data support a model where SlERF.F12 acts as a negative regulator of tomato fruit ripening, prior to the onset of ripening through its interaction with the co-repressor TPL2 and the histone modifiers HDA1 and HDA3. These proteins form a complex to epigenetically repress the transcription of ripening-related genes. At the onset of ripening, SlERF.F12 expression is downregulated, likely due, at least in part, to the rise in climacteric ethylene production. Low SlERF.F12 abundance prevents the formation of the tripartite complex SlERF.F12-TPL2-HDA1/3 and therefore favors higher acetylation levels of permissive histone marks at the promoter regions of ripening-related genes, which promotes their active transcription (Figure 11). Overall, our study uncovers the molecular factors and underlying mechanisms that link ERFs and epigenetic control of transcription during the transition to ripening of tomato fruits. Interestingly, as a preliminary investigation, mining publicly available transcriptomic data corresponding to the ripening initiation of kiwifruit, apple, pear, and banana suggested a putatively conserved role for ERF.F12 homologs in the transition to ripen in several climacteric fruit species. It remains to be determined whether this regulatory network is ubiquitous to all climacteric fruits.
Materials and Methods
Identification of ERF.F subfamily members
The Pfam (Mistry et al., 2021) domain of the AP2/ERF domain (PF00847) was used to identify the tomato ERF family by the HMMER tool (P-value < 1e–5) referring to the classification of Nakano et al. (2006) and Pirrello et al. (2012). The phylogenetic tree of the ERF.F subfamily was generated by MEGA X software (Kumar et al., 2018). The aligned sequences were analyzed using DNAMAN version 8.0 software (Lynnon Biosoft, San Ramon, CA, USA; https://www.lynnon.com/).
Construction of plasmids and plant transformation
The full-length coding sequences of SlERF.F12 or SlERF.F12 with a 3xFlag sequence were cloned into the pBI121 vector to generate SlERF.F12-OE or SlERF.F12-FLAG constructs. A 424-bp fragment of SlERF.F12 and its reverse complement sequence was cloned into pBI121 on either side of an intron from ACTIN2 (At3g18780) for the RNAi construct. The pFASTCas9/ccdB binary vector for plant CRISPR/Cas9-mediated genome editing was kindly provided by Dr Li Zhengguo (College of Life Sciences, Chongqing University). Two target sequences of SlERF.F12 were designed with the CRISPR-P online tool (http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR). Double-stranded DNA of target sequences was generated by PCR and cloned into pFASTCas9/ccdB binary vector by Golden Gate Assembly. The final constructs pBI121-SlERF.F12-OE, pBI121-SLERF.F12-FLAG, pBI121-SlERF.F12-RNAi, and pFASTCas9/ccdB-SlERF.F12 were sequenced and transformed into the tomato cultivar Micro-Tom via Agrobacterium (Agrobacterium tumefaciens)-mediated transformation. Stable inheritance of transgenes was followed until theT2 generation in 10 independent overexpression lines and 8 independent RNAi lines. Six independent transgenic lines (ERF.F12-OE-A, ERF.F12-OE-B, ERF.F12-OE-C, ERF.F12-RNAi-A, ERF.F12-RNAi-B, and ERF.F12-RNAi-C) were used for subsequent experiments. Similarly, two distinct homozygous mutants (erf.f12-1 and erf.f12-2) derived from two different guide RNAs of the T2 generation were used for further analysis.
Plant materials and growth conditions
Tomato plants (S. lycopersicum L. cv Micro-Tom) and derived transgenic lines were grown under standard greenhouse conditions under a 14-h-day/10-h-night photoperiod, 25°C/20°C day/night temperature, 80% relative humidity, and 250-µmol m−2 s−1 light intensity supplemented with fluorescent lights (Foshan Electrical and Lighting Co., Ltd, Foshan, China; LED T8, 16W). Flowers were tagged on the day of anthesis to evaluate fruit developmental and ripening stages. MG refers to the stage when fruits start to develop a white color (around 35–38 DPA for WT) prior to color development, while Br indicates the stage when one part of the fruit starts to generate a slight yellow color (around 41 DPA for WT). The fruit samples were harvested at different developmental and ripening stages at 39, 41, 43, and 47 DPA. Upon harvesting, the pericarps were frozen immediately in liquid nitrogen, and stored at −80°C until use.
RNA isolation and RT-qPCR analysis
Total RNA was extracted from fruit pericarp at various stages of ripening using Plant RNA Purification Reagent (Invitrogen, Waltham, MA, USA; 12322-012) according to the manufacturer’s instructions. First-strand cDNAs were reverse transcribed from 2 μg of total RNA using an Omniscript Reverse Transcription kit (Takara, Shiga, Japan; RR047) following the manufacturer’s instructions. Gene-specific primers were designed with Primer Express software (PE-Applied Biosystems, Waltham, MA, USA) and verified by BLAST analysis against the tomato genome sequence (SL4.0). qPCR was performed as described by Pirrello et al. (2006) using 2×SYBR Green qPCR Mix (Vazyme, Nanjing, China; Q431-02) on a Bio-Rad CFX384 Real-Time PCR System (BIO-RAD, Hercules, CA, USA). The primer sequences used in this study are listed in Supplemental Data Set 4. A melting curve was generated for each sample at the end of each run to ensure the purity of the amplified products. SlActin (Solyc11g005330) was used as the internal control. The data were calculated using the internal control and the 2(−ΔΔCt) method (Pirrello et al., 2006). Six biological replicates were analyzed, each consisting of three technical replicates.
Transcriptome deep sequencing (RNA-seq)
Total RNA was extracted from 41-DPA fruits for the WT, SlERF.F12-OE, and SlERF.F12-RNAi lines with three independent biological replicates. The RNA was then sent for RNA-seq library construction and high-throughput sequencing at Novogene (Beijing, China). The libraries were sequenced on an Illumina NovaSeq 6000 instrument as 150-bp paired-end reads. For data analysis, paired reads were mapped to the tomato reference genome SL4.0 with the ITAG4.0 annotation using HISAT 2 (Kim et al., 2015) with default parameters. In total, more than 188 million clean reads were obtained, of which ∼96% mapped to the tomato reference genome (SL4.0). A P <0.05 and an absolute Log2 ratio >2 were used as the threshold to judge the significance of gene expression differences (Stéphane and Claverie, 1997). KEGG analysis of the DEGs was performed using the ClusterProfiler package in R (Yu et al., 2012).
Ethylene and 1-MCP treatments in different tissues
Four-week-old WT tomato plants were placed in sealed boxes and treated with 50-μL L−1 ethylene or 1.0-μL L−1 1-MCP for 24 h. The plants were separated into three tissues: roots, stems, and leaves. The separated tissues were frozen immediately in liquid nitrogen and stored at −80°C until RNA isolation. WT fruits were harvested at the Br stage and treated with 50-μL L−1 ethylene or 1.0-μL L−1 1-MCP for 24 h in sealed boxes. Fruit pericarps were immediately frozen in liquid nitrogen and stored at –80°C until use. Total RNA was extracted from roots, stems, leaves, and fruit pericarps; RT-qPCR was performed as described by Pirrello et al. (2006). The primer sequences used in this study are listed in Supplemental Data Set 4.
Ethylene production measurements
Ethylene production was measured as described by Liu et al. (2014) with minor modifications. Briefly, fruits at different ripening stages were harvested and placed in open 120-mL jars for 2 h to eliminate the effect of wounding stress. The jars were sealed and incubated for 2 h at room temperature, and 1 mL of headspace gas was injected into an Agilent 7890B gas chromatograph equipped with a flame ionization detector. Ethylene production from fruits was compared with ethylene standards of known concentrations and normalized by fruit weight.
Fruit firmness
At least 20 fruits per line were harvested at the Br + 7 stage and their firmness was assessed using Harpenden calipers (British Indicators Ltd., Burgess Hill, UK) as described by Ecarnot et al. (2013).
Determination of carotenoid contents
Tomato fruits of different lines were harvested at 41, 44, and 47 DPA and the pericarp tissue was freeze-dried and ground into a fine powder. Each sample (100 mg of fruit powder) was extracted in hexane: acetone: ethanol (2:1:1, v/v/v) containing 0.01% (v/v) butylated hydroxytoluene (BHT). After 30 s of agitation and 20 min of ultrasonic treatment, the mixture was centrifuged at 13,000g for 5 min. The supernatant was collected and the extraction steps were repeated twice as above. The extracts were dissolved in a 6:3:1 (v/v/v) solution of acetonitrile: methanol: methyl tert-butyl ether (MTBE) and subjected to UPLC analysis. For carotenoid analysis, a Dionex Ultimate 3000 Series UPLC (Thermo Scientific, Waltham, MA, USA) was used and the samples were separated on a 100 × 2.1 mm 1.9-μm Hypersil Gold C16 column (Thermo Scientific) using 75% (v/v) acetonitrile and 25% (v/v) methanol as mobile phase A and MTBE with 0.1% (v/v) BHT as mobile phase B. The run parameters were set as follows: 0.8 mL/min at 28°C: 0–2 min, 15% B; 2–2.5 min, 15%–25% B; 2.5–3 min, 25%–60% B; 3–4 min, 60%–95% B; 4–4.2 min, 95%–15% B; and 4.3–6 min, 15% B. Detection was performed at 448 nm for lutein, zeaxanthin, α-carotene, β-carotene, γ-carotene, and lycopene. Three independent biological replicates were used for the analysis. Compounds were quantified using standards purchased from Sigma-Aldrich (St Louis, MO, USA; https://www.sigmaaldrich.com/).
Triple response assay
The triple response assay was performed as described previously (Liu et al., 2013) with modifications. Surface sterilized seeds of SlERF.F12-OE and SlERF.F12-RNAi lines were allowed to germinate and grow on half-strength Murashige and Skoog (MS) medium in darkness at 25°C for 7 days. The seedlings were then treated with 20-μL L−1 ethylene or 1.0-μL L−1 1-MCP for 16–24 h in darkness. The seedling triple response was scored by assessing hypocotyl and root length. For each line, at least 50 seedlings were measured.
Dark-induced leaf senescence assay
Dark-induced leaf senescence experiments were performed based on the method described by Ma et al. (2018). Briefly, 10-week-old leaves from WT and transgenic plants were detached and placed on water-soaked filter papers in petri dishes with the adaxial side facing up. The petri dishes were then placed in darkness at room temperature for 14 days with the filter papers changed every 5 days. Chlorophyll was extracted from leaves with a mixture of ethanol and acetone (2:1, v/v) and the absorbance values at 663 and 645 nm were measured with a V-1000 spectrophotometer (AOE INSTRUMENTS Ltd, shanghai, China). Total chlorophyll contents were calculated following the formula:
Chlorophylla + b = (8.02 × OD645 + 20.21 × OD663) × solvent volume/leaf weight.
Transient expression assays
The transcriptional activity assay was performed based on the method described by Han et al. (2016) with modifications. The effector constructs were generated by amplifying the coding sequences of SlERF.F12, SlERF.F12mEAR1 (mutation of the first EAR motif), and SlERF.F12mEAR2 (mutation of the second, C-terminal, EAR motif) without stop codon and inserted into pBD-VP16 vector, while the GAL4:LUC reporter construct containing five copies of the GAL4-binding element driving expression of the LUC gene and an internal control REN driven by the CaMV 35S promoter was used as the reporter. For another set of constructs for another transcriptional activity assay, the coding sequences of SlERF.F12 and the truncated fragment SlERF.F12-ΔEAR2 (lacking the C-terminal EAR motif) were cloned into pGreenII 62sk vector as effectors; five copies of the GCC box and 35S minimal promoter were cloned into pGreenII 0800-LUC vector upstream of firefly LUC.
For the transactivation assays to test the regulation of SlERF.F12 and TPL on the ACS2, ACS4, PG2a, and PL promoters, each relevant promoter fragment was cloned into pGreenII 0800-LUC, while the coding sequences of SlERF.F12, SlTPL2, SlTPL4, and SlERF.F12ΔEAR2 were individually cloned into the pGreenII 62sk vector as effectors.
Mesophyll protoplasts were isolated from N. benthamiana leaves; the effector and reporter constructs were co-transfected into protoplasts by polyethylene glycol-mediated transfection as described by Huang et al. (2013). At ∼10–16 h after transfection, LUC and REN activities were measured using a dual LUC assay kit (Promega, Madison, WI, USA; E1910) according to the manufacturer’s instructions. The results are expressed as the ratio between LUC and REN activity from six independent biological replicates.
Agrobacterium-mediated transient gene expression in tomato fruits
The SlTPL2 coding sequence was cloned into the pBI121 vector to generate the SlTPL2 overexpression construct. To generate the SlTPL2-RNAi construct, a 380-bp SlTPL2 fragment and its reverse complement sequence were cloned into pBI121 on either side of an intron from ACTIN2 (At3g18780). The resulting plasmids were introduced into Agrobacterium strain GV3101. Agrobacterium-mediated transient expression was performed by infiltrating the bacteria into the pericarp of 35-DPA fruits at the fruit shoulder. The infiltrated fruits were collected after staying on-vine in a greenhouse at 22–25°C for 5 days. RT-qPCR was performed as described by Pirrello et al. (2006) and all primers are listed in Supplemental Data Set 4.
Y2H library screening
Total RNA was isolated from a mix of tomato fruits at different developmental and ripening stages; the cDNA library was constructed using SMART cDNA Library Construction Kit (Clontech, Mountain View, CA, USA; 634901) by Protein Interaction Ltd., Wuhan, China. The coding sequence of SlERF.F12 was cloned into the bait vector pGBKT7. The resulting bait vector pGBKT7-SlERF.F12 was transformed in the yeast Y2HGold strain (Clontech; 630498) and plated onto synthetic-defined (SD) medium lacking Trp at 30°C for 3 days. Competent cells were prepared from positive clones and transformed with the prey Y2H AD library. The co-transformants were then grown on SD medium lacking Trp, Leu, His, and Ade at 30°C for 5 days. Clones with a diameter >2 mm were restreaked onto selective SD medium containing X-α-gal to validate the interaction between SlERF.F12 and potential interactors. Blue clones were then grown in liquid SD medium lacking Trp and Leu for 16 h; plasmids isolation from yeast cells was performed according to the Yeast Plasmid Extraction Kit (Protein Interaction; PT1176). The inserts in each prey plasmid were identified by PCR using primers T7 and 3′AD, sequenced and analyzed by BLAST against the tomato genome (https://solgenomics.net/tools/blast/).
Y2H assay
Y2H and β-galactosidase activity assays were performed according to the procedure of the Matchmaker Gold Y2H System (Clontech). The coding sequences for SlERF.F12 and truncated fragments derived from SlERF.F12 including SlERF.F12ΔAP2 (73-154 amino acids without the AP2 domain), SlERF.F12-AP2 (1–72 amino acids), SlERF.F12-EAR2 (92–154 amino acids), SlERF.F12-ΔEAR2 (1–91 amino acids), SlERF.F12mEAR1 (mutation of the first EAR motif), and SlERF.F12mEAR2 (mutation of the second, C-terminal EAR motif) were cloned into pGADT7 as prey constructs. Similarly, the coding sequences for SlTPL1, SlTPL2, SlTPL3, SlTPL4, SlTPL5, and SlTPL6 were cloned into pGBKT7 to generate the bait constructs. Different pairs of bait and prey constructs were co-transformed into yeast strain AH109 and grown on SD medium lacking Leu and Trp (SD–Leu–Trp) for 2 days. The yeast cultures were tested on SD medium lacking Leu, Trp, His, and Ade and containing 4-mg/mL X-α-Gal for blue color development.
EMSA
EMSA was performed as described by Han et al. (2016). The sequences encoding full-length or the N-terminus of SlERF.F12 were cloned into pGEX-4T-1; the resulting glutathione S-transferase (GST) fusion proteins were produced in BM Rosetta (DE3) Competent Cells by induction with 0.5-mM isopropyl-β-d-1-thiogalactopyranoside for 5 h at 30°C. The recombinant protein was purified with Glutathione Sepharose 4B (GE Healthcare, Chicago, IL, USA) according to the manufacturer’s instructions. EMSA was performed using an EMSA kit (Thermo Fisher; 20148) according to the manufacturer’s instructions. The probes containing a GCC box or DRE/CRT motif derived from the PG2a or PL promoters were labeled with biotin using a DNA 3ʹ-End Biotinylation Kit (Thermo Fisher; 89818) and annealed to form double-stranded oligonucleotides. The same unlabeled DNA fragment was used as an unlabeled competitor, while probes in which the GCC box or DRE/CRT motif was changed to AAAAAA or AAAAA were used as mutated probes. The probes were incubated with the fusion protein at room temperature for 30 min in binding buffer (10 mM Tris–HCl, pH 8.0, 10-mM MgCl2, 5-mM DTT, 10% [v/v] glycerol and 50-ng/µL Poly [dI•dC] as nonspecific competitor). The reaction products were analyzed by 5% (w/v) native polyacrylamide gel electrophoresis and transferred to nylon membranes for chemiluminescent detection (Thermo Fisher; 20158).
Co-IP assays
Synthetic pBTEX-HA and pBTEX-FLAG vectors were kindly provided by Dr Fangming Xiao (Miao et al., 2014). The full-length coding sequences of SlERF.F12, HDA1/3, TPL2/4, and the sequences of SlERF.F12mEAR1 and SlERF.F12mEAR2 were amplified and cloned into appropriate restriction sites in pBTEX-HA or pBTEX-FLAG vectors. The pBTEX-SlTPL2/4-FLAG vector was co-infiltrated with pBTEX-SlERF.F12-HA or pBTEX-SlHDA1/3-HA into N. benthamiana leaves via Agrobacterium (strain GV2260)-mediated transient expression. Similarly, the pBTEX-SlHDA1/3-HA vectors were transiently co-infiltrated with pBTEX-SlERF.F12-FLAG in N. benthamiana leaves or with pBTEX-GFP-FLAG as the controls. Protein extraction was performed as described by Huang et al. (2013). Anti-FLAG M2 magnetic beads (Sigma, St Louis, MO, USA; M8823) were co-incubated with the protein extracts for 12–16 h at 4°C. Protein-bound beads were washed with IP wash buffer (0.15-M NaCl, 50-mM Tris–HCl, pH 7.5, 10% [v/v] glycerol, 0.1-mM phenylmethylsulfonyl fluoride (PMSF), pH = 7.5) 6 times. The isolated protein extracts were boiled for 5 min in 2 × SDS loading buffer, resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked in blocking buffer (1× Tris-buffered saline with 0.05% [v/v] Tween-20 [TBST] with 5% [w/v] nonfat milk) for 2 h at room temperature, followed by incubation with mouse anti-HA (Cell Signaling Technologies, Danvers, MA, USA; #2367) or rabbit anti-FLAG (Cell Signaling Technologies; #14793) antibody for 1–2 h. After three washes in 1× TBST buffer for 15 min each, the membranes were incubated with secondary antibodies (Cell Signaling Technologies, #14709 and #7074) for 1 h, washed 3 times for 15 min each with 1× TBST buffer, and visualized using an Immobilon Western Chemiluminescent HRP Substrate (Millipore, Burlington, MA, USA; WBKLS0100).
DNA pull-down assay
DNA pull-down assay was performed as described by Oh et al. (2014). Recombinant GST, GST-SlERF.F12 (from vector pGEX-4t-1), and SlTPL2-HIS, SlHDA1-HIS (from vector pET28a) proteins were produced in BL21 codon plus Escherichia coli cells and purified. The PG2a promoter fragment was amplified by PCR using 5′-biotin-labeled primers (Supplementary Data Set 4). SlTPL2-HIS (40 μg) and SlHDA1-HIS (40 μg) were incubated with the biotin-labeled DNA together with GST (40 μg) or GST-SlERF.F12 (40 μg) protein in HKMG buffer (10-mM HEPES, pH 7.9, 100-mM KCl, 5-mM MgCl2, 10% [v/v] glycerol, 1-mM DTT, and 0.5% [v/v] NP-40) containing protease and phosphatase inhibitors overnight. DNA-binding proteins were then pulled down with streptavidin agarose beads (Sigma; 16-126) and analyzed by immunoblotting using anti-GST antibody (Cell Signaling Technologies; #2622) and anti-HIS antibody (Cell Signaling Technologies; #9991).
Nuclei enrichment and immunoblotting
Enrichment of nuclei was performed as described Wang et al. (2021a). Fruits from the WT, SlHDA1-OE, and SlHDA1-RNAi lines at the 41-DPA stage were collected for nuclei isolation. Fruit pericarp was ground into a fine powder in liquid nitrogen and resuspended in buffer 1 (0.4-M sucrose, 10-mM Tris–HCl, pH 8.0, 5-mM β-mercaptoethanol, and 1-mM PMSF). The homogenates were filtered through two layers of Miracloth and centrifuged at 5,000g for 30 min at 4°C. The pellets were then resuspended in buffer 2 (0.25-M sucrose, 10-mM Tris–HCl, pH 8.0, 5-mM β-mercaptoethanol, and 1-mM PMSF) and placed in an ice bath for 30 min. After centrifugation at 12,000g for 10 min at 4°C, the pellets were boiled in 5× SDS–PAGE loading buffer (250-mM Tris–HCl pH6.8, 10% [w/v] SDS, 0.5% [w/v] bromophenol blue, 50% [v/v] glycerol, 5% [v/v] β-mercaptoethanol, pH 6.8) for 5 min. The boiled samples were resolved on 12% (w/v) SDS–PAGE and transferred to PVDF membranes. The membranes were blocked in blocking buffer (1× TBST with 5% [w/v] nonfat milk) for 2 h at room temperature and incubated with antibodies (Cell Signaling Technologies; #4499, #9649, and #8173) against H3, H3K9Ac, or H3K27Ac, respectively, for another 2 h. The membranes were then washed 3 times for 10 min each with 1×TBST and incubated for 1 h with the secondary antibody (Cell Signaling Technologies; #7074). The immune reactions were visualized using Immobilon Western HRP Chemiluminescent Substrate (Millipore; WBKLS0100) and ChemiDoc XRS+ System (BIO-RAD, Hercules, CA, USA).
ChIP assay
ChIP was performed as described by Zhang et al. (2015). Chromatin extracts were obtained from fruits of the WT, SlERF.F12-OE, SlERF.F12-RNAi, and SlERF.F12-FLAG lines at different stages. All fruits were fixed with 1% (v/v) formaldehyde. The chromatin was sonicated with a Bioruptor system (Qsonica, Newtown, CT, USA; Q800R2) with 30% power setting for 10 s “ON” and 10 s “OFF” for 11 min to shear DNA to an average length of 200–800 bp. Chromatin was then immunoprecipitated by the addition of protein A agarose beads and rabbit antibodies against FLAG (Cell Signaling Technologies; #14793), H3K9Ac (Cell Signaling Technologies; #9649), and H3K27Ac (Cell Signaling Technologies; #8173) and incubated at 4°C overnight. After the incubation, the beads were washed in the following buffers: once in low salt buffer (150-mM NaCl, 0.1% [w/v] SDS, 1% [v/v] Triton X-100, 2-mM EDTA, 20-mM Tris–HCl pH 8.0), once in high salt buffer (500-mM NaCl, 0.1% [w/v] SDS, 1% [v/v] Triton X-100, 2-mM EDTA, 20-mM Tris–HCl pH 8.0), once in LiCl buffer (250-mM LiCl, 1% [w/v] sodium deoxycholate, 1% [v/v] NP-40, 1-mM EDTA, 10-mM Tris–HCl pH 8.0), and twice in TE buffer (1-mM EDTA, 10-mM Tris–HCl pH 8.0). ChIP assays were performed with six biological replicates. The crosslinking was then reversed by incubating at 65°C overnight and then incubating with 50 -µg/mL Proteinase K (Cell Signaling Technologies; #10012S) at 65°C for 3 h. The immunoprecipitated chromatin was analyzed by qPCR.
TSA treatment
TSA was dissolved in DMSO and diluted to 10 μM in half-strength MS liquid medium without sucrose. The diluted solution was then infiltrated into the pericarp from the fruit shoulder at the 36-DPA stage. The fruits were collected for RNA isolation and RT-qPCR analysis after 1-week infiltration.
Identification of SlERF.F12 homologs in multiple climacteric fruits
The following reference genomes were used for this analysis: the tomato reference genome sequence SL4.0; the apple reference genome GDDH13 version 1.1; the banana genome version DH-Pahang version 2; the pear genome Pbr_v1.0; and the kiwifruit genome Red5_PS1_1.69.0. The protein-coding genes of each species were compared to infer orthology using OrthoFinder (Emms and Kelly, 2015). The phylogenetic tree of SlERF.F12 homologs was generated by MEGA X software (Kumar et al., 2018).
RNA-seq reads and processed data were obtained from public databases (Zhang et al., 2016; Hu et al., 2017; Li et al., 2020a, 2020b; Liu et al., 2021; Wang et al., 2021b) and retrieved from the NCBI database repository (https://www.ncbi.nlm.nih.gov/bioproject/) or the Genome Sequence Archive at the National Genomics Data Center (http://bigd.big.ac.cn/). For data analysis, paired reads were mapped to the tomato reference genome (SL4.0) using HISAT 2 with default parameters (Kim et al., 2015). The number of reads with a mapping quality (MAPQ) value >20 was collected for further analysis. The mapped fragments for each gene were counted with featureCounts (Liao et al., 2014) and transcripts per million were calculated.
Accession numbers
Sequence data from this article can be found in the Tomato Genome Protein Sequences (ITAG release 4.0) database under the following accession numbers: SlERF.F12 (Solyc02g077840), SlERF.F1 (Solyc10g006130), SlERF.F2 (Solyc07g064890), SLERF.F3 (Solyc07g049490), SlERF.F4 (Solyc07g053740), SlERF.F5 (Solyc10g009110), SlERF.F6 (Solyc12g005960), SlERF.F7(Solyc03g006320), SlERF.F8 (Solyc01g067540), SlERF.F9 (Solyc05g013540), SlERF.F10 (Solyc11g045680), SlERF.F11 (Solyc11g045690), SlERF.F13 (Solyc04g072300), E4 (Solyc03g111720), E8 (Solyc09g089580), ACO1 (Solyc07g049530), ACS2 (Solyc01g095080), ACS4 (Solyc05g050010), PG2a (Solyc10g080210), PL (Solyc03g111690), PSY1 (Solyc03g031860), β-LCY1 (Solyc04g040190), RIN (Solyc05g012020), NOR (Solyc10g006880), CNR (Solyc02g077920), SlTPL1 (Solyc03g117360), SlTPL2 (Solyc08g076030), SlTPL3 (Solyc01g100050), SlTPL4 (Solyc03g116750), SlTPL5 (Solyc07g008040), SlTPL6 (Solyc08g029050), SlHDA1 (Solyc09g091440), SlHDA3 (Solyc06g071680), and SlActin (Solyc11g005330). The RNA-seq data are available from the Genome Sequence Archive in the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, under the accession number CRA004377 and is publicly accessible at http://bigd.big.ac.cn/. Sequence alignments and machine-readable tree files have been placed in the Dryad database and are publicly accessible at https://datadryad.org/stash/share/gjm79KPtOChdOfMn49mt2vqB_JlDpsBded7PNXgjTjE.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of tomato SlERF.F subfamily members in tomato.
Supplemental Figure S2. Relative expression of ERF.F12 in vegetative tissues in WT plants treated with ethylene or 1-MCP.
Supplemental Figure S3. Relative expression levels of ERF.F subfamily genes in ERF.F12-RNAi fruits at 41 DPA.
Supplemental Figure S4. ERF.F12 ko mutants represses ethylene production, fruit softening, and ripening-related genes expression in fruits.
Supplemental Figure S5. Plant size and flowering phenotypes of the WT, ERF.F12-OE, and ERF.F12-RNAi lines.
Supplemental Figure S6. Dark-induced leaf senescence in the WT, ERF.F12-OE and ERF.F12-RNAi lines.
Supplemental Figure S7. Ethylene triple response in WT, ERF.F12-OE, and ERF.F12-RNAi seedlings.
Supplemental Figure S8. Ethylene triple response in etiolated seedlings of the WT and erf.f12 mutants.
Supplemental Figure S9. Phenotype of plant size and flowering time in the WT and erf.f12 ko mutants.
Supplemental Figure S10. Homologs of SlERF.F12 in different species with climacteric fruit are downregulated during the transition from unripe to ripe stages.
Supplemental File S1. Alignment of the tomato ERF.F family members shown in Supplemental Figure S1.
Supplemental File S2. Newick file format of the alignment shown in Supplemental Figure S1.
Supplemental File S3. Alignment of the homologs to tomato ERF.F12 shown in Supplemental Figure S10A.
Supplemental File S4. Newick file format of the alignment shown in Supplemental Figure S10A.
Supplemental Table S1. SlERF.F subfamily members.
Supplemental Table S2. List of the proteins interacting with SlERF.F12 identified by Y2H screening.
Supplemental Table S3. Off-target analysis in erf.f12-1 and erf.f12-2 mutants.
Supplemental Table S4. Description of mutants and transgenic lines used in this study.
Supplemental Data Set 1. Expression levels of DEGs between the WT and SlERF.F12-OE lines.
Supplemental Data Set 2. Expression levels of DEGs between the WT and SlERF.F12-RNAi lines.
Supplemental Data Set 3. Expression levels of genes upregulated in SlERF.F12-RNAi fruits but downregulated in SlERF.F12-OE lines, or genes downregulated in SlERF.F12-RNAi fruits but upregulated in SlERF.F12-OE lines.
Supplemental Data Set 4. List of primers used in this study.
Supplemental Data Set 5. Summary of statistical tests.
Supplementary Material
Acknowledgments
We would like to thank Zhangjun Fei (Boyce Thompson Institute) and Xin Wang (Boyce Thompson Institute) for insightful conversations.
Funding
This research was supported by the National Natural Science Foundation of China (No. 31901742, No. 32172271, No. 31772372, and No. 32172643), the National Key R&D Program of China (No. 2016YFD0400100), and this project is also supported by the Fundamental Research Funds for the Central Universities (No. SCU2021D006).
Conflict of interest statement. The authors declare that they have no conflict of interest.
These authors contributed equally (H.D. and Y.C.).
M.L. and H.D. planned and designed the research; H.D., Y.C., Zi.L., Zh.L., and R.W. performed experiments. P.S., Y.H., D.S., J.P., Z.L., and Y.L. analyzed data. M.L. and M.B. wrote the manuscript and J.G. and D.G. helped improve the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Mingchun Liu (mcliu@scu.edu.cn).
References
- Alexander L, Grierson D (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53: 2039–2055 [DOI] [PubMed] [Google Scholar]
- Benavente LM, Alonso JM (2006) Molecular mechanisms of ethylene signaling in Arabidopsis. Mol BioSyst 2: 165. [DOI] [PubMed] [Google Scholar]
- Burg SP, Burg EA (1962) Postharvest ripening of avocados. Nature 194: 398–399 [Google Scholar]
- Cao Y, Song F, Goodman RM, Zheng Z (2006) Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J Plant Physiol 163: 1167–1178 [DOI] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CJ, Liu JY (2010) The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol 10: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Cheng Y, Wu J, Cheng Q, Li W, Fan S, Jiang L, Xu Z, Kong F, Zhang D, et al. (2015) Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J Exp Bot 66: 2635–2647 [DOI] [PubMed] [Google Scholar]
- Dong W, Ai X, Xu F, Quan T, Liu S, Xia G (2012) Isolation and characterization of a bread wheat salinity responsive ERF transcription factor. Gene 511: 38–45 [DOI] [PubMed] [Google Scholar]
- Ecarnot M, Bączyk P, Tessarotto L, Chervin C (2013) Rapid phenotyping of the tomato fruit model, Micro-Tom, with a portable VIS-NIR spectrometer. Plant Physiol Biochem 70: 159–163 [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S (2015) OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy.Genome Biol 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Chen H, Gao H, Han Y (2019) Histone deacetylase CpHDA3 is functionally associated with CpERF9 in suppression of CpPME1/2 and CpPG5 genes during papaya fruit ripening. J Agric Food Chem 67:8919–8925 [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J, Nguyen C, Ampofo B, Zhong S, Fei Z (2017) The epigenome and transcriptional dynamics of fruit ripening. Ann Rev Plant Biol 68: 61–84 [DOI] [PubMed] [Google Scholar]
- Grierson D (2013) Ethylene and the control of fruit ripening. InSeymour GB, Poole M, Giovannoni JJ, Tucker GA, eds, The Molecular Biology and Biochemistry of Fruit Ripening, Blackwell Publishing Ltd., Oxford, pp 43–73 [Google Scholar]
- Gu C, Guo ZH, Hao PP, Wang GM, Jin ZM, Zhang SL (2017) Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot Stud 58: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JE, Hu Z, Yu X, Li A, Li F, Wang Y, Tian S, Chen G (2018) A histone deacetylase gene, SlHDA3, acts as a negative regulator of fruit ripening and carotenoid accumulation. Plant Cell Rep 37: 125–135 [DOI] [PubMed] [Google Scholar]
- Guo JE, Hu Z, Zhu M, Li F, Zhu Z, Lu Y, Chen G (2017) The tomato histone deacetylase SlHDA1 contributes to the repression of fruit ripening and carotenoid accumulation. Sci Rep 7: 7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YC, Kuang JF, Chen JY (2016) Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol 171: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Yamasaki K, Sarai A, Ohme-Takagi M (2002) Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry 41: 4202–4208 [DOI] [PubMed] [Google Scholar]
- Hao Y, Hu G, Breitel D, Liu M, Mila I, Frasse P, Fu Y, Aharoni A, Bouzayen M, Zouine M (2015) Auxin response factor SlARF2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PLoS Genet 11: e1005649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Wang H, Perry SE (2008) A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J 53: 172–185 [DOI] [PubMed] [Google Scholar]
- Hu W, Yan Y, Shi H, Liu J, Miao H, Tie W, Ding Z, Ding X, Wu C, Liu Y, et al. (2017) The core regulatory network of the abscisic acid pathway in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. BMC Plant Biol 17:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Miao M, Kud J, Niu X, Ouyang B, Zhang J, Ye Z, Kuhl JC, Liu Y, Xiao F (2013) SlNAC1, a stress-related transcription factor, is fine-tuned on both the transcriptional and the post-translational level. New Phytologist 197: 1214–1224 [DOI] [PubMed] [Google Scholar]
- Ju C, Chang C (2015) Mechanistic insights in ethylene perception and signal transduction. Plant Physiol 169: 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K (2011) EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Chapman N, David K, Angenent GC, Seymour GB, de Maagd RA (2014) Transcriptional control of fleshy fruit development and ripening. J Exp Bot 65: 4527–4541 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Shim D, Moon S, Lee J, Bae W, Choi H, Kim K, Ryu H (2019) Transcriptional network regulation of the brassinosteroid signaling pathway by the BES1-TPL-HDA19 co-repressor complex. Planta 250: 1371–1377 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Ann Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kumar S, , Stecher G,, Li M,, Knyaz C,, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Z, Wang Y, Tang K, Tang D, Datsenka T, Cheng J, Zhang Y, Handa AK, Zhu JK (2017) Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc Natl Acad Sci U S A 114: E4511–E4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Lelièvre JM, Tichit L, Dao P, Fillion L, Nam YW, Pech JC, Latché A (1997) Effects of chilling on the expression of ethylene biosynthetic genes in Passe-Crassane pear (Pyrus communis L.) fruits . Plant Mol Biol 33: 847–855 [DOI] [PubMed] [Google Scholar]
- Li S, Chen K, Grierson D (2021) Molecular and hormonal mechanisms regulating fleshy fruit ripening. Cells 10:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen T, Li Y, Wang Z, Cao H, Chen F, Li Y, Soppe WJJ, Li W, Liu Y (2019) ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression. Plant Cell 31: 832–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen Y, Zhou L, You S, Deng H, Chen Y, Alseekh S, Yuan Y, Fu R, Zhang Z, et al. (2020b) MicroTom metabolic network: rewiring tomato metabolic regulatory network throughout the growth cycle. Mol Plant 13:1203–1218 [DOI] [PubMed] [Google Scholar]
- Li S, Zhu B, Pirrello J, Xu C, Zhang B, Bouzayen M, Chen K, Grierson D (2020a) Roles of RIN and ethylene in tomato fruit ripening and ripening‐associated traits. New Phytologist 226:460–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhu B, Xu W, Zhu H, Chen A, Xie Y, Shao Y, Luo Y (2007) LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep 26: 1999–2008 [DOI] [PubMed] [Google Scholar]
- Liang Q, Deng H, Li Y, Liu Z, Shu P, Fu R, Zhang Y, Pirrello J, Zhang Y, Grierson D, et al. (2020) Like heterochromatin protein 1b represses fruit ripening via regulating the H3K27me3 levels in ripening-related genes in tomato. New Phytologist 227:485–497 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930 [DOI] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D (2009) Recent advances in ethylene research. J Exp Bot 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Liu M, , Pirrello J,, Kesari R,, Mila I,, Roustan J-P,, Li Z,, Latché A,, Pech J-C,, BouzayenM, , Regad F (2013) A dominant repressor version of the tomato Sl-ERF.B3 gene confers ethylene hypersensitivity via feedback regulation of ethylene signaling and response components. Plant J 76: 406–419 [DOI] [PubMed] [Google Scholar]
- Liu M, Diretto G, Pirrello J, Roustan JP, Li Z, Giuliano G, Regad F, Bouzayen M (2014) The chimeric repressor version of an Ethylene Response Factor (ERF) family member, Sl-ERF.B3, shows contrasting effects on tomato fruit ripening. New Phytologist 203: 206–218 [DOI] [PubMed] [Google Scholar]
- Liu M, Gomes BL, Mila I, Purgatto E, Peres LE, Frasse P, Maza E, Zouine M, Roustan JP, Bouzayen M, et al. (2016) Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol 170: 1732–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Pirrello J, Chervin C, Roustan JP, Bouzayen M (2015a) Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol 169: 2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, How-Kit A, Stammitti L, Teyssier E, Rolin D, Mortain-Bertrand A, Halle S, Liu M, Kong J, Wu C, et al. (2015b) A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc Natl Acad Sci USA 112: 10804– 10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hao N, Feng R, Meng Z, Li Y, Zhao Z (2021) Transcriptome and metabolite profiling analyses provide insight into volatile compounds of the apple cultivar ‘Ruixue’ and its parents during fruit development. BMC Plant Biol 21:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Lu J, Ju H, Zhou G, Zhu C, Erb M, Wang X, Wang P, Lou Y (2011) An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J 68: 583–596 [DOI] [PubMed] [Google Scholar]
- Lü P, Yu S, Zhu N, Chen YR, Zhou B, Pan Y, Tzeng D, Fabi JP (2018) Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat Plants 4: 784–791 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Y, Turečková V, Xue GP, Fernie AR, Mueller-Roeber B, Balazadeh S (2018) The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol 177: 1286–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor-and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurchie EJ, McGlasson WB, Eaks IL (1972) Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 237: 235–236 [DOI] [PubMed] [Google Scholar]
- Miao M, Zhu Y, Qiao M, Tang X, Zhao W, Xiao F, Liu Y (2014) The tomato DWD motif-containing protein DDI1 interacts with the CUL4-DDB1-based ubiquitin ligase and plays a pivotal role in abiotic stress responses. Biochem Biophys Res Commun 450: 1439–1445 [DOI] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. (2021) Pfam: the protein families database in 2021. Nucleic Acids Res 49: 412–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Munné-Bosch S (2015) Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol 169: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Ryu H, Hwang I, Wang ZY (2014) TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat Commun 5: 4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan IC, Li CW, Su RC, Cheng CP, Lin CS, Chan MT (2010) Ectopic expression of an EAR motif deletion mutant of SlERF3 enhances tolerance to salt stress and Ralstonia solanacearum in tomato. Planta 232: 1075–1086 [DOI] [PubMed] [Google Scholar]
- Pirrello J, Jaimes-Miranda F, Sanchez-Ballesta MT, Tournier B, Khalil-Ahmad Q, Regad F, Latché A, Pech JC, Bouzayen M (2006) Sl-ERF2, a tomato ethylene response factor involved in ethylene response and seed germination. Plant Cell Physiol 47: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Pirrello J, Prasad BC, Zhang W, Chen K, Mila I, Zouine M, Latche A, Pech JC, Ohme-Takagi M, Regad F, et al. (2012) Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol 12: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Ryu H, Cho H, Bae W, Hwang I (2014) Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat Commun 5: 4138– 4138 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Chapman NH, Chew BL, Rose JK (2013) Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol J 11: 269–278 [DOI] [PubMed] [Google Scholar]
- Shin JH, Mila I, Liu M, Rodrigues MA, Vernoux T, Pirrello J (2019) The RIN-regulated Small Auxin-Up RNA SAUR69 is involved in the unripe-to-ripe phase transition of tomato fruit via enhancement of the sensitivity to ethylene. New Phytologist 222:820–836 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CP, Galbraith DW (2006) AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol Biol 60: 241–257 [DOI] [PubMed] [Google Scholar]
- Stéphane A, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Sun Y, Liang B, Wang J, Kai W, Chen P, Jiang L, Du Y, Leng P (2018) SlPti4 affects regulation of fruit ripening, seed germination and stress responses by modulating ABA signaling in tomato. Plant Cell Physiol 59: 1956–1965 [DOI] [PubMed] [Google Scholar]
- Trujillo LE, Sotolongo M, Menéndez C, Ochogavía ME, Coll Y, Hernández I, Borrás-Hidalgo O, Thomma BP, Vera P, Hernández L (2008) SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant Cell Physiol 49: 512–525 [DOI] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci 110: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang P, Li X, Wang Y, Tian S, Qin G (2021a) The transcription factor SlHY5 regulates the ripening of tomato fruit at both the transcriptional and translational levels. Hortic Res 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Shu P, Zhang C, Zhang J, Chen Y, Zhang Y, Du K, Xie Y, Li M, Ma T, et al. (2021b) Integrative analyses of metabolome and genome-wide transcriptome reveal the regulatory network governing flavor formation in kiwifruit (Actinidia chinensis). New Phytologist doi: 10.1111/nph.17618. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
- Yin XR, Allan AC, Chen K, Ferguson IB (2010) Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol 153: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16: 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Chen M, Chen X, Xu Z, Li L, Guo J, Ma Y (2010) Isolation and characterization of a novel EAR-motif-containing gene GmERF4 from soybean (Glycine max L.). Mol Biol Rep 37: 809–818 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Alseekh S, Tohge T, Rallapalli G, Luo J, Kawar PG, Hill L, Santino A, Fernie AR, et al. (2015) Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat Commun 6: 8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MY, Xue C, Xu L, Sun H, Qin MF, Zhang S, Wu J (2016) Distinct transcriptome profiles reveal gene expression patterns during fruit development and maturation in five main cultivated species of pear (Pyrus L.). Sci Rep 6:28130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31: 154–159 [DOI] [PubMed] [Google Scholar]
- Zhou L, Tian S, Qin G (2019) RNA methylomes reveal the m(6)A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol 20: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.