Abstract
Background
Accurate CNS tumor diagnosis can be challenging, and methylation profiling can serve as an adjunct to classify diagnostically difficult cases.
Methods
An integrated diagnostic approach was employed for a consecutive series of 1258 surgical neuropathology samples obtained primarily in a consultation practice over 2-year period. DNA methylation profiling and classification using the DKFZ/Heidelberg CNS tumor classifier was performed, as well as unsupervised analyses of methylation data. Ancillary testing, where relevant, was performed.
Results
Among the received cases in consultation, a high-confidence methylation classifier score (>0.84) was reached in 66.4% of cases. The classifier impacted the diagnosis in 46.7% of these high-confidence classifier score cases, including a substantially new diagnosis in 26.9% cases. Among the 289 cases received with only a descriptive diagnosis, methylation was able to resolve approximately half (144, 49.8%) with high-confidence scores. Additional methods were able to resolve diagnostic uncertainty in 41.6% of the low-score cases. Tumor purity was significantly associated with classifier score (P = 1.15e−11). Deconvolution demonstrated that suspected glioblastomas (GBMs) matching as control/inflammatory brain tissue could be resolved into GBM methylation profiles, which provided a proof-of-concept approach to resolve tumor classification in the setting of low tumor purity.
Conclusions
This work assesses the impact of a methylation classifier and additional methods in a consultative practice by defining the proportions with concordant vs change in diagnosis in a set of diagnostically challenging CNS tumors. We address approaches to low-confidence scores and confounding issues of low tumor purity.
Keywords: brain tumor classification, deconvolution, DNA methylation profile, neuropathology, tumor purity
Key Points.
1. The methylation classifier has an impact on the diagnosis on a substantial proportion of cases received in a consultation practice, especially with the confidence score is high.
2. We describe the use of additional methods to resolve a substantial proportion of low-confidence-score cases.
3. We provide a proof-of-concept using deconvolution to classify low tumor-purity samples.
Importance of the Study.
Precision and accuracy are longstanding goals of CNS tumor diagnostics. The DKFZ/Heidelberg CNS tumor methylation classifier contributes to these goals, but more experience is required to understand its impact. We utilized the classifier and integrated diagnosis on a large cohort of >1000 cases, most of which were received from outside consultation for the purpose of methylation. The resultant integrated diagnosis was changed in 39% of these cases. The methylation classifier, however, does not reach high-confidence score (>0.84) on approximately one-third of cases, and we describe the use of additional methods to resolve 41.6% of these low-score cases. In addition, we identified tumor purity correlated with deceased performance of the methylation classifier and provided a proof-of-concept by using deconvolution to resolve of methylation profile in low purity. We identified 2 candidate novel tumor subtypes: a DMG-K27 subtype and an RB1 loss glioma class. Our experience provides a needed and practical guide toward the impact and future use of the methylation classifier in routine CNS tumor diagnostics.
Appropriate treatment for patients with central nervous system (CNS) tumors ultimately depends on accurate diagnosis. Traditional diagnosis is based on histology and advanced by molecular markers including point mutation, gene fusion, copy number variation (CNV), and other genomic alterations. Epigenetic profiling, especially DNA methylation arrays, is innovative in classifying both tissue-of-origin1–4 and tumor epigenetic phenotypes present in IDH-mutant (IDHmut) gliomas.5
In practical use, diagnostic interpretation of the DKFZ/Heidelberg DNA methylation-based classifier is based on determination of a class along with the calibrated score, where a high score (>0.90 or >0.84 as reported cutoffs) is indicative of high confidence.6,7 However, high-confidence scores are achieved in only 50%-70% in real-world studies.7–9 These studies also pointed to the utility of the methylation classifier to confirm, refine, and at times to substantially change diagnoses that were made based on histologic examination alone, while also highlighting challenges when the classifier confidence scores are below the cutoff. In addition to technical reasons, such as low-quality DNA, several reasons for low classifier scores exist: (1) the tumor type under question may not be represented in the classifier; (2) the tumor type may exist in the classifier but is an outlier at the boundary of the classifier, due to insufficient machine learning power or sample diversity; or (3) the tumor tissue may be in low tumor purity and was dominated by nonneoplastic tissues. Such cases may benefit from methods that are complementary and orthogonal to the classifier, such as unsupervised analyses, and identification of orthogonal tumor type-specific genomic alterations or immunohistochemical findings.
Here we evaluate the impact of the methylation classifier on CNS tumor diagnosis in 1258 CNS tumor cases, of which 1045 were received for consultation from outside institutions. Specific cases were analyzed by additional methods using nearest-neighbors assisted unsupervised analysis, CNV analysis, immunohistochemistry (IHC), or targeted DNA sequencing for further characterization, especially for more challenging cases as surrogate in diagnosis. We explored the potential factors associated with variation in classifier scores. We pay particular attention to the patterns of diagnostic changes as a result of the methylation classifier, as well as the relationships of tumor purity, classifier score, and diagnostic classification. Unsupervised analysis was performed to explore candidate new tumor types.
Materials and Methods
Patient Sample Collection
Patient materials and clinical data of the evaluated cohort (n = 1258) were collected from outside institutions (n = 1045) and consultation cases internal to the National Cancer Institute (NCI, n = 213). Appropriate institutional review board approval was obtained with a waiver of informed consent. Histological slides were evaluated to estimate tumor content and classification according to the current WHO criteria. Areas with highest tumor cell content (≥60% when possible) were marked and used for further DNA extraction.
Molecular Profiling
Immunohistochemical staining and sequencing analyses were performed according to standard methods appropriate for clinical testing. Methylation profiling was performed as previously described.10 The DKFZ/Heidelberg classifier was used and generated tumor classification with a calibrated score and CNV plot. Details are indicated in the Supplementary Information.
Unsupervised Analysis and Methylation Nearest-Neighbor Analysis
We performed t-distributed stochastic neighbor embedding (t-SNE)11 and uniform manifold approximation and projection (UMAP) using additional R code by integrating the Capper et al reference raw methylation IDAT files (n = 2801)6 and consultative cases. In order to measure the distance of consultation cases to reference tumor entities, we queried methylation nearest-neighbors in the reference data using PC distance. To interrogate the distance of a new case to the reference entities, 15 neighbors were first retrieved, and distance thresholds were applied as detailed in Supplementary Information. We integrated the classifier result and our additional methylation analysis in an R pipeline which generates an HTML report for every case with all the described analyses (an example in Supplementary Figure S1). The code is available by request.
Integrated Diagnosis
An integrated diagnosis for tumor cases was rendered by neuropathologists (K.A., M.Q., and D.P.) during consensus conference to facilitate diagnosis.12 Details, as well as a description of unsupervised analyses and tumor purity estimation, are described in Supplementary Tables S1 and S2 and Supplementary Information.
Tumor Impurity Adjustment
To deconvolve bulk tumor methylation profile for gliomas, we estimated cell fractions using methylCIBERSORT13 using a signature derived from public profiles of high-grade glioma, H3-K27M-mutant cell lines, and nonneoplastic cell lines including glia, neurons, neutrophils, B- and T cells, NK cells, monocytes, and endothelial cells (Supplementary Table S3; Supplementary Information). Adjustment of methylation beta values was performed using InfiniumPurify R package.14 Highly variable probes (SD > 0.23 for both unadjusted and adjusted datasets) were used for t-SNE analysis.
Results
Integrated Diagnosis for CNS Tumor in a Consultative Practice
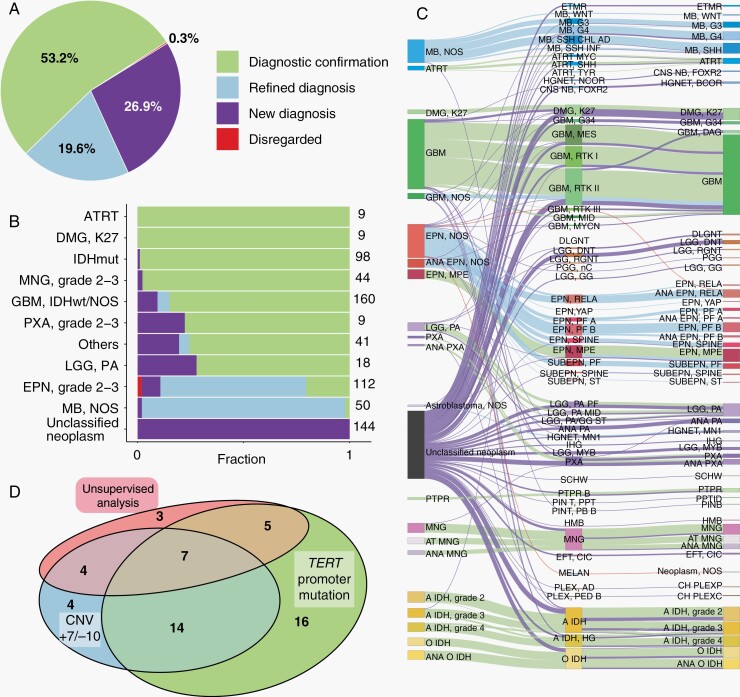
We evaluated a consecutive series of 1258 surgical neuropathology cases in a predominantly consultative practice in the period between 2018 and 2020, of which 1045 were received from outside institutions for consultation. We focused on these 1045 cases and found that approximately 2/3 of the cases received a high-confidence score (>0.84), a proportion similar to previously reported studies.8,9,15 Within this subset of cases with high-confidence scores, we compared the integrated diagnosis with the pre-classifier impression. The results are summarized as (A) concordant/unchanged; (B) concordant/subtype determined; (C) new/changed diagnoses; or (D) descriptive diagnoses (Table 1). Overall, approximately half (53.2%) of the results were concordant/unchanged. Considering the cases where methylation had an impact on the diagnosis, in 19.7% of cases in the total cohort, the methylation classifier was consistent with the original histopathological diagnosis but provided additional, clinically relevant subtyping information. Importantly, a new diagnosis was made in 26.9% of the cases as a result of the classifier. Patterns of change between the original (“pre-classifier”) diagnosis and post-classifier/integrated diagnosis for cases with high classifier scores are shown in Figure 1, where Figure 1b indicates the proportion and type of change in diagnosis (if any), and Figure 1c and Supplementary Table S4 provide a detailed view of the pre-classifier impression, methylation classification, and integrated diagnosis for high-confidence score cases. For cases that received suggestive (≤0.84 and >0.30) and “no-match” (≤0.30) scores, the classifier was contributory, but to a lesser extent, as a definitive diagnosis could be less frequently achieved in these situations (66.2% and 31.1%, respectively, Table 1).
Table 1.
Summary of Classifier Score and Change in Diagnosis on 1045 Consultation Cases
| # | Classifier Score | Final vs Initial Diagnosis | Number | % Within-score | % Total |
|---|---|---|---|---|---|
| 1A | >0.84 | Concordant | 369 | 53.2 | 35.3 |
| 1B | Concordant, subtyped | 137 | 19.7 | 13.1 | |
| 1C | New diagnosis | 187 | 26.9 | 17.9 | |
| 1D | Descriptive | 1 | 0.1 | 0.1 | |
| 2A | 0.3-0.84 | Concordant | 65 | 30.1 | 6.2 |
| 2B | Concordant, subtyped | 25 | 11.6 | 2.4 | |
| 2C | New diagnosis | 53 | 24.5 | 5.1 | |
| 2D | Descriptive | 73 | 33.8 | 7.0 | |
| 3A | ≤0.3 | Concordant | 31 | 23.0 | 3.0 |
| 3B | Concordant, subtyped | 3 | 2.2 | 0.3 | |
| 3C | New diagnosis | 8 | 5.9 | 0.8 | |
| 3D | Descriptive | 93 | 68.9 | 8.9 |
Cases are grouped into 3 classifier score categories (>0.84, 0.3-0.84, and ≤0.3) and 4 categories of impact (A-D) as indicated.
Fig. 1.
Impact of methylation classifier on CNS tumor diagnosis. (a) The impact of methylation profiling on 694 high-score referral cases is shown proportionally as: Diagnostic confirmation (light green); Refined diagnosis (light blue); New diagnosis (purple); and Disregarded (red). (b) Impact of the classifier across different histopathologically diagnosed tumor types. From top to bottom: 9 atypical teratoid/rhabdoid tumors (ATRT), 9 K27M mutated diffuse midline gliomas (DMG-K27), 98 IDH-mutant gliomas (IDHmut), 44 meningiomas (MNG), 160 glioblastomas (GBM) or GBM-NOS, 9 pleomorphic xanthoastrocytomas (PXA), 41 less common tumors (Others, other tumors with <6 samples in this cohort), 18 pilocytic astrocytomas (LGG-PA), 112 WHO grade 2-3 ependymomas (EPN, grade 2-3), 50 medulloblastoma-NOS (MB-NOS), and 144 unclassified neoplasm. (c) Sankey diagram illustrating the diagnosis of the high-score cases. From left to right are pre-classifier diagnosis, methylation classification, and final integrated diagnosis. Lines are colored by classifier impact. (d) Venn diagram showing the utilization of additional methods used for GBM diagnosis.
We examined the impact of the classifier across different histologically diagnosed tumor types (Figure 1b and c), where we observed distinct patterns based on the initial histologic (pre-classifier) impression. First, some pre-classifier diagnostic classes (ATRT, DMG-K27, IDHmut gliomas, and meningiomas) were as a group, largely confirmed by methylation, and a diagnostic change was rarely found in these cases. As one example, a diagnostic change of pre-classifier “IDHmut astrocytoma” to glioblastoma (GBM) (IDH wildtype) was observed on case #V587, where such change was firstly induced by the classifier with high confidence and supported by additional analyses (Supplementary Figure S2). Second, in some pre-classifier diagnostic classes, methylation was largely confirmatory but also resulted in a proportion of new diagnoses, including GBM/GBM-NOS, PXA, and LGG-PA. Third, the impact of the classifier on histologically diagnosed ependymoma and medulloblastoma was primarily by diagnostic refinement/subtyping. Last and importantly, a large proportion of cases (n = 289) were received without a definitive diagnosis. These were received with descriptive histologic diagnoses (eg, “glioma, NOS”, “glioneuronal tumor”, “embryonal tumor”) in which definitive diagnoses were not rendered on the contributing pathology report. The classifier gave high classifier scores in 144 (49.8%) of these cases (Figure 1b). As seen in Figure 1c, these 144 cases were resolved into a large variety of entities, which included high-grade tumors (medulloblastoma, grade 4 diffuse gliomas) as well and lower grade circumscribed glial/glioneuronal entities (eg, LGG-PA, PXA) and IDHmut gliomas, highlighting the utility of methylation classifier in resolving highly challenging cases. Details are described in Supplementary Information and Supplementary Table S4.
Practical Integrated Diagnosis for Histologically Diagnosed GBMs
To illustrate the diagnostic procedure and role of methylation profiling on a common CNS tumor type, we detail our integrated diagnostic experience on GBM, the most common adult intrinsic CNS tumor. Two hundred thirty-nine cases were initially designated as GBM (GBM-IDH-wt) or GBM-NOS (GBM without IDH status specified) by the submitting institution prior to methylation. The classifier gave high-score (>0.84) classifications to 66.9% (n = 160) of these cases, with diagnostic confirmation of GBM-IDH-wt in 90.6% (n = 145) of these 160 cases (Figure 1b and c). We used additional methods to interrogate the diagnosis of the 15 cases which received a high score for a classification other than GBM and were able to confirm these changes: 6 cases of DMG-K27 (H3-K27M-mutated), 4 IDHmut astrocytoma (IDH1-mutated), 2 ANA-PXA (CDKN2A/B loss was identified in both cases; BRAF-V600E mutation was identified in one case), and single cases of GBM-G34 (supportive by H3 G34-mutant-specific IHC) ANA-PA (supported by classifier score and the copy number result) and HGNET-BCOR. Thus, among pre-classifier histopathologically diagnosed GBMs which received high classifier scores, most (~90%) were confirmed as GBM, but the remaining 10% were found to be alternative entities by the methylation classifier.
We then turned our attention to the 79 pre-classifier “GBM” cases that did not receive a high classifier score, where 19.6% (n = 47) received a suggestive (≤0.84 and >0.3) score and 13.4% (n = 32) cases received a no-match (≤0.3) score. Additional methods were used to interrogate these 79 cases and a final diagnosis of GBM was given to 70.9% (n = 56) of these. Specifically, unsupervised analysis showed co-embedding with GBM tumors in some cases (n = 19). Others showed the canonical chromosome 7 gain and 10 loss (+7/−10, n = 29) common to adult GBM and/or TERT promoter mutation (n = 42) which supported the diagnosis of GBM. Interestingly, 43.3% (n = 23) cases were supported by only one of the three methods (Figure 1d). Nine other cases that received low scores were diagnosed as non-GBM tumors after interrogation, and the remainder received a descriptive diagnosis. Overall, the results indicate a substantial contribution of the methylation classifier, as well as additional methods when necessary, to histologically diagnosed GBM.
Interrogating Challenging Cases Using Additional Diagnostic Methods
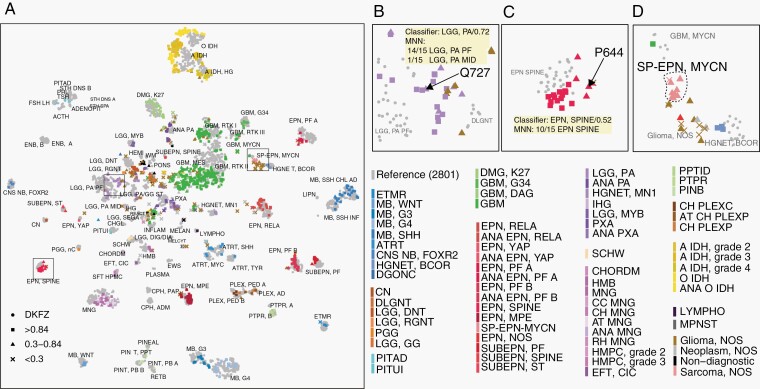
The methylation classifier provided “suggestive” (0.30-0.84) score to 20.7% (n = 216) and a “noncontributory” (≤0.3) score to 12.9% (n = 135) of the cases. Additional methods were employed where possible and enabled definitive diagnoses to be rendered for 41.6% (n = 146) of these cases. When compared to the original histologies diagnosis, the impact of methylation was (1) diagnostic confirmation in 78 cases; (2) diagnostic refinement/subtyping in 26 cases; and (3) a new/changed diagnosis in 42 cases. For such cases without high-confidence classifier scores, unsupervised analysis/dimension reduction of methylation profiles assisted in resolving a diagnosis for 76 cases. While we found individual cases near specific groups in the t-SNE analysis (Figure 2a), it is known that the displayed distance between samples on the t-SNE plot is not necessarily proportional to their similarity. To address this, we therefore incorporated nearest-neighbors to assist the unsupervised analysis. In one example, case #Q727 was initially diagnosed as pilocytic astrocytoma (LGG-PA) by histopathology, and the methylation classifier gave classification as “LGG-PA posterior fossa” (LGG-PA-PF) but with a score of 0.73. We found this case to be located near the border of LGG-PA-PF group on the t-SNE (Figure 2b). The 15 neighbors for this case in the reference cohort all passed the distance threshold and belong to LGG-PA-PF or LGG-PA-MID which increased the confidence in determination of these samples as LGG-PA. A second case (#P644) was initially diagnosed as ependymoma, and methylation suggested it as ependymoma-SPINE with a score of 0.52. This case was embedded with ependymoma-SPINE in UMAP (not shown) and located near the ependymoma-SPINE group on t-SNE (Figure 2c). All of the top 15 nearest-neighbors for this case were ependymoma-SPINE, which supported a diagnosis of ependymoma, spinal subtype. In addition, for tumor entities that were not present in the classifier such as the recently discovered spinal ependymoma with MYCN amplification (SP-EPN-MYCN) subtype, nearest-neighbor, and t-SNE analyses could not map the tumors into a specific entity group in the reference data. t-SNE analysis showed that SP-EPN-MYCN tumors located as an independent group in an open space region (Figure 2d) and interestingly nearest-neighbor frequently identified a few GBM-MYCN as nearest-neighbors in the reference data. Methylation array-based CNV analysis identified high level of MYCN amplification for all these ependymomas as a strong evidence for their defining diagnosis. As an additional method to resolve low-score cases, we used orthogonal data, including CNV and mutation analysis, which identified definitive/supportive mutations for 106 cases (Supplementary Tables S1 and S2). For example, CNV of +7/−10 provided supportive information for diagnosis of 37 GBMs, and 1p/19q co-deletion derived from the arrays provided the required information for the diagnosis of 48 IDHmut oligodendrogliomas.
Fig. 2.
Nearest-neighbor assisted unsupervised interrogation of the current cohort cases. (a) T-distributed stochastic neighbor embedding (t-SNE) analysis of in-house 1258 cases together projected on Capper et al. 2801 samples (reference set). Reference tumors are shown in gray dots, the 1258 consultative cases are colored by final integrated diagnosis of tumor entities and shaped by their classifier scores: high-confidence scores >0.84 (square), suggestive scores between 0.3 and 0.84 (triangle), and not contributary scores <0.3 (cross). (b–d) Case study of low classification score cases Q727 (b) and P644 (c) as well as tumor of a class that is absent from the classifier: spinal ependymoma with MYCN amplification (SP-EPN-MYCN) (d).
Unsupervised Analysis Identifies Potential New Tumor Subtypes
To evaluate the role of unsupervised analysis identifies for new tumor subtype determination, we projected our 1258 methylation cases on the DKFZ reference dataset using t-SNE (Figure 2a). Most of the diagnostically resolved cases co-embedded with known specific entities in the reference set, suggesting overall, that our external dataset was comparable to the reference set. That said, we did observe that some entities (eg, IDHmut gliomas and GBMs) did not always overlap with the reference set, most likely because of technical issues (eg, batch effects) (Supplementary Figure S3) and real-world tumor heterogeneity. The recently discovered new entity SP-EPN-MYCN formed a single group. Several GBM-MYCN samples are co-localized with SP-EPN-MYCN (Figure 2a) suggesting similar methylation profiles of a subset of MYCN-activated CNS tumors.
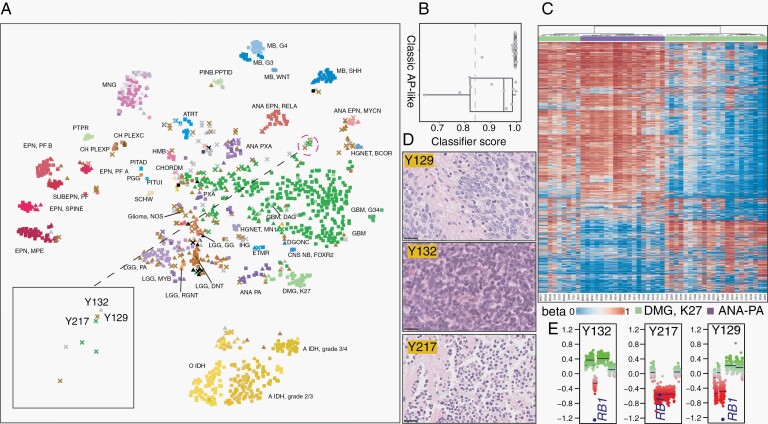
The methylation class DMG-K27 was found to separate into 2 groups (Figure 3a): one co-embedded with the reference DMG-K27 tumors (classic DMG-K27), and a new group near ANA-PA (AP-like DMG-K27). The classifier scores for the 2 groups were both high, except for several AP-like DMG-K27 cases (Figure 3b). To further explore their relationship, we clustered these cases with 18 ANA-PAs using unsupervised hierarchical clustering and also obtained the 2 DMG-K27 clusters (Figure 3c). The methylation profile of AP-like DMG-K27 was more similar to ANA-PA than classic DMG-K27 and showed global hypermethylation relative to classic DMG-K27, suggesting potentially a new tumor subtype. We then explored the possibility of an additional tumor entity among a set of cases with low-confidence scores who received descriptive diagnoses. The t-SNE plot revealed 3 cases grouped together at the open space (Figure 3a). These patients were male and aged 61 to 69 with “small round blue cell” histopathology (Figure 3d). CNV analysis identified loss of tumor suppressor gene RB1 in these cases (Figure 3e), suggesting a possible molecular commonality as a CNS tumor subtype, perhaps linked to prior studies on RB1-altered GBMs and low-grade glioma.16–18
Fig. 3.
Unsupervised analysis of the methylation-profiled cohort. (a) t-SNE analysis of 1258 methylation-profiled tumor cases colored by final diagnosis and shaped by classification scores as indicated in Figure 2a. (b) The classifier scores of the cases diagnosed as DMG-K27 tumors in 2 groups: classic DMG-K27 and AP-like DMG-K27. (c) Cluster heatmap of in-house 18 ANA-PA and 31 DMG-K27 tumors indicating 2 groups of DMG-K27. Tumor types are colored on the top of heatmap, light green: DMG-K27, purple: ANA-PA. (d) H&E stains of Y132, Y217, and Y129 with 25 µm scale bars on bottom left (e) CNV plots on chromosome 13 revealing the common RB1 loss in Y132, Y217, and Y129.
DNA Input Amount and Tumor Purity
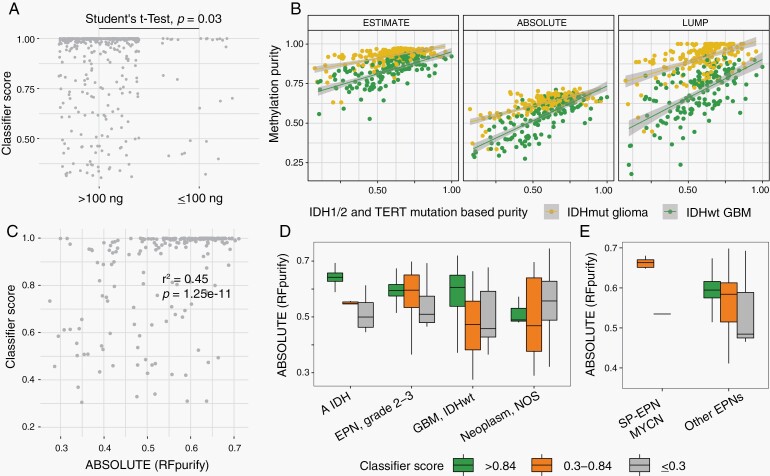
Low DNA input and tumor purity were assessed as factors related to confidence score. While we aimed for the recommended 250 ng DNA input amount, methylation profiling was also performed when only a lower amount of DNA was available. As expected, we observed a significantly lower mean classifier score for low DNA input (<100 ng) samples compared with samples of higher DNA amounts (Figure 4a, P = .03, Student’s t test), suggesting that DNA amount explained a proportion of diminished methylation performance.
Fig. 4.
Tumor purity is significantly associated with classifier classification confidence score. (a) Comparison of the classifier classification score with the DNA input amount used for methylation profiling. (b) Comparison of IDH1/2 and TERT mutation allele/variant frequency-based tumor purity with 3 methylation-based tumor purity estimation methods from RF_purity in IDH-mutated gliomas and TERT-mutated glioblastomas. (c) Correlation of the tumor purity and the classifier classification score in pre-classifier GBMs. Tumor purity is estimated using RF_purity-ABSOLUTE method. (d) Tumor purity in different classification score groups across different tumor entities. (e) Tumor purity and classifier score groups of the recently discovered spinal ependymoma with MYCN amplification subtype (SP-EPN-MYCN), which is unrepresented in the classifier, compared with other ependymoma subtypes represented in the classifier.
To investigate the impact of tumor purity on classifier score, we estimated tumor purity using 3 methods derived from the methylation profiles.19,20 We compared these purity estimations with the variant allele frequency (VAF)-based tumor purity approximations, using IDH1/2 mutations in IDHmut gliomas and TERT promoter in GBM. Methylation-based tumor purity estimation methods were all significantly correlated with VAF-based purity (Figure 4b). We then used purity estimations from RF_Purify-ABSOLUTE method for downstream analysis, which showed the highest correlation with VAF-based purity. As expected, tumor purity correlated with classifier score in histopathologically diagnosed GBM (r2 = 0.45, P = 1.25e−11) (Figure 4c). To further explore this, we analyzed additional well-represented tumor types and found that cases which received high scores tended to be higher purity tumors (Figure 4d).
An analysis of variables associated with classifier scores including tumor purity, tumor type, and DNA amount showed that tumor purity was most significantly associated with classifier score (P = 1.15e−11, generalized logistic regression). An additional reason for a low score was that the tumor type in question is not in the DKFZ reference set used for comparison. Accordingly, we compared a recently identified subtype of SP-EPN-MYCN,21–23 which is not represented in the DKFZ reference set, with other ependymoma subtypes (which are included in the reference set). In general, ependymoma subtypes with high classifier scores were of high purity. As expected, none of the high purity SP-EPN-MYCN cases received high-confidence score (Figure 4e). Overall, the analysis demonstrated that low tumor purity, low DNA starting amount, and the absence of the corresponding entities (known or undiscovered entities) in the classifier are the common reasons for low classifier scores.
In Silico Methylation Purity Adjustment Improves Classification of Low-Purity Glioma Specimens
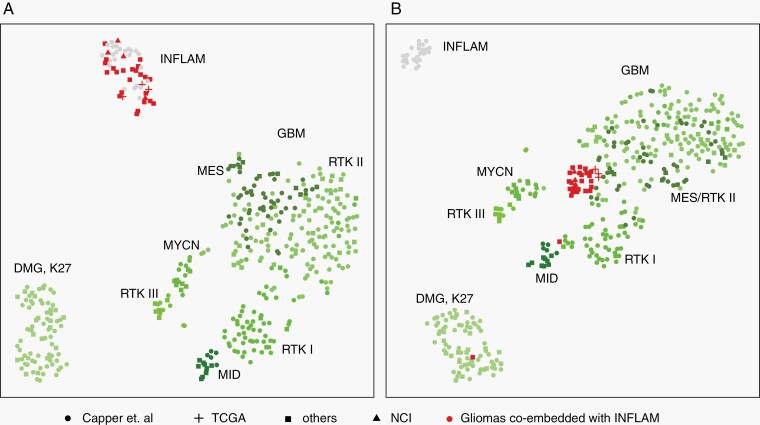
We then examined whether purity adjustment of the methylation profile could improve classification and diagnostic confidence. For example, GBM tumors formed a dispersed group with low-purity cases admixed with low-grade gliomas in t-SNE analysis (Figure 3a; Supplementary Figure S4). These low-purity GBM cases received lower classifier scores, which was in line with the significant association between tumor purity and classifier score. In a collection of in-house and previously published GBM and DMG-K27 tumors (Supplementary Table S3), we observed several tumors co-embedded with inflammatory tissue on t-SNE (Figure 5a), suggesting high immune cell infiltration. Recent computational methods showed promising results for deconvolving methylation profile that adjust the tumor purity to obtain the neoplastic profile.24,25 To test if tumor purity adjustment could improve methylation classification, we performed deconvolution for neoplastic samples in Figure 5a. Following methylation profile deconvolution, the low-purity samples showed co-embedding with the appropriate tumor types (Figure 5b), which suggested a proof-of-concept method for improving classification confidence after estimating and accounting for the nonneoplastic cellular components from bulk methylation data, upon suspicion of a specific tumor type.
Fig. 5.
Tumor purity adjustment for inflammatory infiltrated gliomas improved tumor classification. (a) t-SNE plot of GBM, DMG-K27, control inflammatory tissue (INFLAM, gray dots), and a collection of gliomas co-embedded with inflammatory tissue (red) before (a) and after (b) tumor purity adjustment. Tumor purity was adjusted using the InfiniumPurify R package. Circle dot, cross, square, and triangle represent samples from Capper et al, TCGA, other public resources and our in-house samples (Supplementary Table S3).
Discussion
DNA methylation has been demonstrated as tissue/cell-type-specific and abnormal methylation plays an important role in cancer development.1–4,26,27 It has been widely used in tumor type/subtype classification, prognosis prediction, and biomarker identification. Recent experience with the CNS tumor classifier has revealed the importance of this technique in tumor diagnosis and classification.6,7 However, additional experience is required to better understand the role of methylation profiling in daily practice, in the context of an integrated diagnosis. In the present study, we have integrated DNA methylation profiling for tumor diagnosis with the implementation of methylation-based classifier in 1258 CNS tumors, with a focus on diagnostically challenging cases.
We assessed the impact of the integrated diagnosis by comparing the pre-classifier histopathology-based diagnosis, methylation classification, and final integrated diagnosis by using 4 types of impact and 3 classifier score categories (high score, suggestive score, and noncontributory score). We found that approximately two-thirds of the cases received a high score, a proportion similar to previous studies.8,9,15 The impact of this process varied across different tumor types (Figure 1; Supplementary Table S4). Broadly speaking, among cases received in consultation, this approach was directly confirmatory in approximately half (53.2%) of the high-score cases (Table 1). For an additional 19.7% of the high-score cases (mostly medulloblastomas and ependymomas), methylation provided additional clinically relevant subtyping information. Importantly, the classifier gave high-confidence score classifications that led to a substantially new diagnosis in 26.9% of the high-score cases. Most of these cases were received with either a descriptive diagnosis (eg, “glial neoplasm,” embryonal neoplasm,” or similar) or were simply received without a histopathological diagnosis, showing the value of methylation to help classify tumors that elude definitive diagnosis by conventional means. Experience from this work provides insights regarding the types of cases that may specifically benefit from methylation profiling. For example (Supplementary Table S4), most (94%) histologically diagnosed meningiomas were confirmed by methylation without a change in diagnosis, as were IDHmut gliomas (99%). However, a proportion that was higher than might have been expected (20%) of histologically diagnosed GBM cases were changed following methylation profiling. Additional cases that may benefit from the classifier include histologically diagnosed PXA and PA as well as those that elude a specific diagnosis using routine histopathology. Methylation could resolve a specific diagnosis in approximately half of the cases that elude a definite diagnosis by current practice standards, suggesting its value in that clinical setting. Finally, tumors such as medulloblastoma and ependymoma, while often confirmed in these categories benefit from the diagnostic requirement of subtyping that is a component of the methylation classifier.
Less understood is how to approach cases with “suggestive” scores (0.3-0.84) received from the classifier, which were observed in 23.2% of cases (n = 242). This issue has been previously recognized but remains an important problem in interpretation.8–10,15 Such cases are challenging to interpret and the reasons for such scores below the cutoff cannot always be determined with certainty. One reason for such scores is due to low tumor content/purity, although this determination is sometimes difficult using histopathological interpretation. Other reasons include the possibility that the case in question is not represented in the reference set. In cases of low classifier scores, supportive ancillary data (specific mutational profiles, fusions, and/or stereotypic copy number changes) can lead to a definitive diagnosis in the right setting. In this work, we endeavored to use dimension reduction (t-SNE and UMAP) as well as nearest-neighbor analysis to provide additional diagnostic information as an unsupervised approach. Nearest-neighbor analysis combined with t-SNE/UMAP identifies reference samples closest to the interrogated sample as a surrogate for distance measurement and often provided a clue as to a possible diagnosis, which could then be confirmed with additional tests. As a practical matter, the routine inclusion of UMAP, t-SNE, and nearest-neighbor analysis provided a platform to evaluate the concordance of methylation-based classification. While it is difficult to quantify, we conservatively estimate that UMAP/t-SNE as well as nearest-neighbor analysis contributed to a definite diagnosis of a specific tumor entity in 35.2% (n = 76) of the suggestive-score cases. Additional molecular marker testing was also used for cases with specific genomic alterations in suspicion. In summary, nearest-neighbor-assisted unsupervised analysis resolved 76 of 216 outside cases which received suggestive scores from the classifier, and together with ancillary molecular testing, 112 cases were resolved by combining these 2 methods. For cases that received noncontributory/no-match scores molecular testing and/or unsupervised embedding of methylation profiles provided supportive information (although to a lesser extent). Overall the results suggest an important role for the methylation classifier, when placed in context with alternative unsupervised methods to examine methylation data, as well as supportive molecular information when relevant.
One hundred sixty-nine cases (13.4%) were left unresolved and reasons for this, including the high diversity of CNS tumor types as well as low tumor purity, a common reason for low classifier score and/or a classifier score of control/reactive brain tissue. We examined tumor purity in some detail and found that it was a significant factor (P = 1.15e−11) that was associated with classifier score groups (high, suggestive, and low scores). We adjusted for low tumor purity in a set of suspected high-grade gliomas that mapped to control/inflammatory tissue in a t-SNE analysis. Adjustment for inflammatory infiltrated gliomas using a deconvolution approach correctly embedded low-purity tumor samples. Though this purity adjustment required prior knowledge of tumor entity, it provided a proof-of-concept in accounting for nonneoplastic components in bulk methylation profile. Development of a pan-CNS tumor deconvolution method to address low tumor purity samples is a goal for future studies and which, if successful, could improve diagnostic confidence for tumors with low purity.
In summary, experience with a large cohort of CNS tumors, primarily referred due to diagnostic difficulty, methylation profiling and integrated diagnosis led to a new or refined diagnosis in nearly 40% of the cases where a high classifier score could be achieved (Table 1). The classifier was helpful, but to a lesser degree, when scores were below the 0.84 cutoff. Ancillary and complementary methods were helpful to resolve a diagnosis, especially when classifier scores were low. Specific findings in this study include an estimation of the performance of the classifier for cases (n = 289) that could not be determined diagnostically through conventional means, where methylation was able to diagnostically resolve approximately half (144/289) of these cases. Additional noteworthy results included a proportion of histologically diagnosed GBMs that were reclassified into alternative tumor types as a result of the classifier, as well as some specific initial histologic diagnoses (eg, PA and PXA) that were frequently changed following methylation. Based on these results, we conclude that methylation profiling should be considered, at minimum, for cases where (1) the diagnosis is unclear or in question; or (2) there is a need for clinically relevant subtype information (eg, ependymomas, medulloblastomas). We further show the utility of additional analytic techniques in cases of low-confidence scores and suggest an approach to informatically define a tumor methylation class in a set of low-purity GBM cases. Additional work to address tumor purity will likely assist to understand and resolve some low-confidence score cases and help to distinguish these from new or rare tumor entities. While the methylation array incurs a cost, we believe that this cost is a good value and is comparable to a potentially large IHC panel that can be performed on a diagnostically challenging tumor. Additional work and experience will likely lead to improvements in diagnostics by methylation, as this modality is increasingly incorporated into the classification of CNS tumors and into clinical practice.
Supplementary Material
Acknowledgments
This work utilized the computational resources of the NIH HPC Biowulf cluster. We thank all the patients and clinicians involved in the submitted case material for expert interpretation analyzed in this study.
Contributor Information
Zhichao Wu, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Zied Abdullaev, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Drew Pratt, Department of Pathology, University of Michigan, Ann Arbor, Michigan, USA.
Hye-Jung Chung, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Shannon Skarshaug, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Valerie Zgonc, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Candice Perry, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Svetlana Pack, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Lola Saidkhodjaeva, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Sushma Nagaraj, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Manoj Tyagi, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Vineela Gangalapudi, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Kristin Valdez, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Rust Turakulov, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Liqiang Xi, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Mark Raffeld, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Antonios Papanicolau-Sengos, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Kayla O’Donnell, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Michael Newford, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Mark R Gilbert, Neuro-Oncology Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Felix Sahm, Department of Neuropathology, Institute of Pathology, University Hospital of Heidelberg, Heidelberg, Germany.
Abigail K Suwala, Department of Neuropathology, Institute of Pathology, University Hospital of Heidelberg, Heidelberg, Germany.
Andreas von Deimling, Department of Neuropathology, Institute of Pathology, University Hospital of Heidelberg, Heidelberg, Germany.
Yasin Mamatjan, Division of Neurosurgery, Department of Surgery, University of Toronto, Toronto, Ontario, Canada.
Shirin Karimi, Division of Neurosurgery, Department of Surgery, University of Toronto, Toronto, Ontario, Canada.
Farshad Nassiri, Division of Neurosurgery, Department of Surgery, University of Toronto, Toronto, Ontario, Canada.
Gelareh Zadeh, Division of Neurosurgery, Department of Surgery, University of Toronto, Toronto, Ontario, Canada.
Eytan Ruppin, Cancer Data Science Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Martha Quezado, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Kenneth Aldape, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Funding
This research is supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute (NCI), and Center for Cancer Research (CCR).
Conflict of interest statement. The authors declare that they have no competing interests.
Authorship statement. All authors listed in the manuscript contributed significantly to this manuscript. All authors were also involved in the writing of the manuscript and have read and approved the manuscript.
Data Availability Statement
The raw methylation IDAT files and code are available for research by request.
References
- 1. Byun HM, Siegmund KD, Pan F, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18(24):4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagae G, Isagawa T, Shiraki N, et al. Tissue-specific demethylation in CpG-poor promoters during cellular differentiation. Hum Mol Genet. 2011;20(14):2710–2721. [DOI] [PubMed] [Google Scholar]
- 3. Yuen RK, Neumann SM, Fok AK, et al. Extensive epigenetic reprogramming in human somatic tissues between fetus and adult. Epigenetics Chromatin. 2011;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fernandez AF, Assenov Y, Martin-Subero JI, et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012;22(2):407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noushmehr H, Weisenberger DJ, Diefes K, et al. ; Cancer Genome Atlas Research Network . Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capper D, Stichel D, Sahm F, et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 2018;136(2):181–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pickles JC, Fairchild AR, Stone TJ, et al. DNA methylation-based profiling for paediatric CNS tumour diagnosis and treatment: a population-based study. Lancet Child Adolesc Health. 2020;4(2):121–130. [DOI] [PubMed] [Google Scholar]
- 9. Priesterbach-Ackley LP, Boldt HB, Petersen JK, et al. Brain tumour diagnostics using a DNA methylation-based classifier as a diagnostic support tool. Neuropathol Appl Neurobiol. 2020;46(5):478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karimi S, Zuccato JA, Mamatjan Y, et al. The central nervous system tumor methylation classifier changes neuro-oncology practice for challenging brain tumor diagnoses and directly impacts patient care. Clin Epigenetics. 2019;11(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 12. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 13. Chakravarthy A, Furness A, Joshi K, et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat Commun. 2018;9:3220–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng X, Zhang N, Wu HJ, Wu H. Estimating and accounting for tumor purity in the analysis of DNA methylation data from cancer studies. Genome Biol. 2017;18(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jaunmuktane Z, Capper D, Jones DTW, et al. Methylation array profiling of adult brain tumours: diagnostic outcomes in a large, single centre. Acta Neuropathol Commun. 2019;7(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim YH, Lachuer J, Mittelbronn M, et al. Alterations in the RB1 pathway in low-grade diffuse gliomas lacking common genetic alterations. Brain Pathol. 2011;21(6):645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldhoff P, Clarke J, Smirnov I, et al. Clinical stratification of glioblastoma based on alterations in retinoblastoma tumor suppressor protein (RB1) and association with the proneural subtype. J Neuropathol Exp Neurol. 2012;71(1):83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suwala AK, Stichel D, Schrimpf D, et al. Glioblastomas with primitive neuronal component harbor a distinct methylation and copy-number profile with inactivation of TP53, PTEN, and RB1. Acta Neuropathol. 2021;142(1):179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johann PD, Jäger N, Pfister SM, Sill M. RF_Purify: a novel tool for comprehensive analysis of tumor-purity in methylation array data based on random forest regression. BMC Bioinformatics. 2019;20(1):428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Swanson AA, Raghunathan A, Jenkins RB, et al. Spinal cord ependymomas with MYCN amplification show aggressive clinical behavior. J Neuropathol Exp Neurol. 2019;78(9):791–797. [DOI] [PubMed] [Google Scholar]
- 22. Ghasemi DR, Sill M, Okonechnikov K, et al. MYCN amplification drives an aggressive form of spinal ependymoma. Acta Neuropathol. 2019;138(6):1075–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raffeld M, Abdullaev Z, Pack SD, et al. High level MYCN amplification and distinct methylation signature define an aggressive subtype of spinal cord ependymoma. Acta Neuropathol Commun. 2020;8(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lutsik P, Slawski M, Gasparoni G, Vedeneev N, Hein M, Walter J. MeDeCom: discovery and quantification of latent components of heterogeneous methylomes. Genome Biol. 2017;18:55–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rahmani E, Schweiger R, Rhead B, et al. Cell-type-specific resolution epigenetics without the need for cell sorting or single-cell biology. Nat Commun. 2019;10:3417–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. [DOI] [PubMed] [Google Scholar]
- 27. Baylin SB, Höppener JW, de Bustros A, Steenbergh PH, Lips CJ, Nelkin BD. DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer Res. 1986;46(6):2917–2922. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw methylation IDAT files and code are available for research by request.