When looking at a leaf surface under a microscope, you will see a continuous array of puzzle-shaped cells, called pavement cells, which provide a structural barrier against external mechanical insults. The barrier is interrupted by small pores surrounded by specialized guard cells, forming stomata. This apparently simple organization, however, influences many plant functions including gas exchange, photosynthesis, water loss and nutrient transport, general growth, organogenesis, morphogenesis, defense against pathogens, and more.
The phenotypic plasticity of pavement cells and stomata allows plants to optimize growth and resource allocation (Pillitteri and Torii, 2012; Liu et al., 2021). The precise segmentation, a process to partition an image into distinct regions/objects, and the quantification of cell characteristics (e.g., cell count, cell size, and length/width ratio) are critical for understanding this plasticity, but have been a challenge for computational biologists across a variety of plant research disciplines (Chickarmane et al., 2010). The “ground truth” method—manual segmentation of cell shape and counting—is unfortunately very laborious, time consuming, and subject to bias by the experimenter. For large-scale genetic studies and breeding programs, manual quantification becomes practically impossible. A few automatic, high-throughput image processing pipelines have been reported, each limited by their own preferred image input formats. Generally, these high-throughput methods have performed poorly at segmenting simple bright-field images, which are the cheapest and most readily available. To develop an “universal” program that can handle images taken by bright-field microscopy as well as other methods from a wide range of species, Shaopeng Li, Linmao Li, and colleagues (Li et al., 2022) present LeafNet, an integrated program capable of precisely detecting stomata and segmenting pavement cells from complex dataset. The toolkit is available as a web server (https://leafnet.whu.edu.cn/), a standalone program with graphical interface and a command line program, making it widely accessible to users with different computational background.
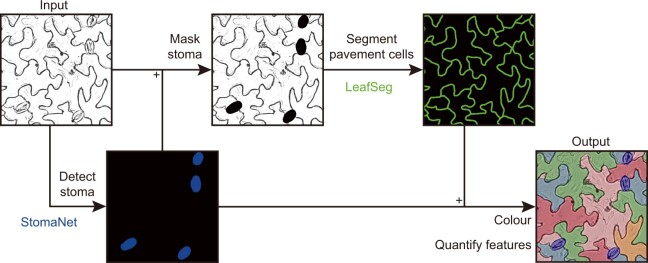
The key strategy employed is a hierarchical image processing pipeline (see Figure). First, stomata are detected based on a deep neural network and then masked out. Pavement cell contours are then detected by a region merging algorithm. The gross outcome is that segmentation of both stomata and pavement cells in the same image can be resolved with minimal interference. Noise due to incidental dots, lines, dirty background, and uneven lighting can be minimized. It is also important to recognize that LeafNet generates segmentation output, which allows quantification of dozens of cell features not assessed/released by other methods. These segmentation datasets will be valuable to cross-check with different methods and to serve as training/test samples to facilitate future development of new image processing tools.
Figure.
Graphical illustration for hierarchical segmentation of stomata and pavement cells using LeafNet. StomaNet module for detecting stomata and LeafSeg module for segmenting pavement cells on stomata-masked input. Adopted from Li et al. (2022), Figure 1E.
LeafNet was originally designed to analyze bright-field images taken by light microscopes, a difficult task in plant image processing. The authors then demonstrate that LeafNet is flexible and performs well with confocal and fluorescence images as well as a wide sampling dataset containing a random mix of differential interference contrast, scanning electron microscopy, and bright-field images across plant taxa. Biological differences between Arabidopsis genotypes can also be resolved, making it a promising tool for high-throughput analytical studies.
Imaging and image processing are the foundation for studying plant morphology and function. It will be exciting and challenging to further take into consideration time and space. In the case of stomata function, physiological output (e.g. CO2 assimilation) and the number of stomata are not always correlated, underlying the importance of cell dynamics. Stomatal opening and closure are tightly regulated processes in response to various environmental cues, and high-throughput quantification of stomatal aperture is another great challenge. The work of Li et al. might also serve as inspiration for the development of additional high-throughput imaging tools such as this in the near future.
References
- Chickarmane V, Roeder AHK, Tarr PT, Cunha A, Tobin C, Meyerowitz EM (2010) Computational morphodynamics: A modeling framework to understand plant growth. Annu Rev Plant Biol 61: 65–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li L, Fan W, Ma S, Zhang C, Kim JC, Wang K, Russinova E, Zhu Y, Zhou Y (2022) LeafNet: A Tool for Segmenting and Quantifying Stomata and Pavement Cells. The Plant Cell 34: 1171--1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Jobert F, Rahneshan Z, Doyle SM, Robert S (2021) Solving the puzzle of shape regulation in plant epidermal pavement cells. In Merchant SS, ed., Annual Review of Plant Biology, Vol 72, pp 525–550 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU (2012) Mechanisms of stomatal development. In Merchant SS, ed., Annual Review of Plant Biology, Vol 63, pp 591–614 [DOI] [PubMed] [Google Scholar]