Abstract
Background
Extracellular Vesicle (EV)-based therapy has been identified as a leading alternative approach in several disease models. EV derived from the Olfactory Ensheathing Cell (OEC) has been documented for its strong neuro-regenerative capacity. However, no information on its cargo that may contribute to its therapeutic effect has been available.
Objective
To report the first miRNA profile of human OEC (hOEC) -EV, and investigate the neuroprotective effects.
Methods
hOEC-EV was isolated and sequenced. We established in vitro experiments to assess the therapeutic potential of hOEC-EVs with respect to insulted neural progenitor cells (NPCs), and the angiogenesis effect. Secondary post-injury insults were imitated using t-BHP-mediated oxidative stress.
Results
We noted a strong abundance of hOEC-EV-miRNAs, including hsa-miR148a-3p, has-miR151a-3p and several members of let-7 family. The common targets of 15 miRNAs among the top 20 miRNAs were thrombospondin 1 and cyclin dependent kinase 6. We demonstrated that hOEC-EVs promote normal NPC proliferation and differentiation to neuron-like morphologies with prolonged axons. hOEC-EVs protect cells from t-BHP mediated apoptosis. We also found that the migration rate of either NPCs or endothelial cells significantly improved with hOEC-EVs. Furthermore, in vitro tube formation assays indicated that angiogenesis, an important process for tissue repair, was significantly enhanced in human umbilical vein endothelial cells exposed to hOEC-EVs.
Conclusion
Our results revealed that hOEC-EVs exert neuroprotective effects by protecting cells from apoptosis and promoting in vitro biological processes that are important to neural tissue repair, including neural cell proliferation, axonal growth, and cell migration, in addition to enhancing angiogenesis.
Keywords: Human olfactory ensheathing cell, extracellular vesicle, miRNA, neural progenitor, neuro-regeneration, in vitro angiogenesis
1. INTRODUCTION
Olfactory Ensheathing Cells (OECs) are specialized glia found in the lamina propria of the olfactory mucosa [1]. Their exceptional properties include active lifelong neurogenesis and olfactory neuron guidance from peripheral regions to the Central Nervous System (CNS) [2-4]. Furthermore, OECs have been shown to promote axonal regeneration and functional recovery in a Spinal Cord Injury (SCI) model [5-7]. In the demyelinated spinal cord model rat, transplantation of OECs or Schwann cells results in re-myelination [8]. Furthermore, the acute transplantation of OECs into the spinal cords of transected rats showed that OECs can limit activation and infiltration of immune cells, diminish inhibitory molecules in the lesion, and create a favorable environment for axon regeneration and neuron survival [9]. A previous study also demonstrated that OECs not only promote neuron sprouting through direct contact with the damaged neurons but also act as a physical substrate for neuron regeneration after injury. OECs also secrete soluble factors that assist in post-injury neurite sprouting [10], including several important molecules for axon regeneration, such as neuron growth factor neurotropic factors [11-13] and extracellular matrix molecules [10, 14, 15]. For these special characteristics, which are critical for neurogenesis and neural regeneration, OECs have been recognized as an alternative promising transplantation tool for repairing a defective CNS, such as in SCI [16-18], peripheral nerve injury (PNI) [19-22], glioma [23], and Parkinson’s disease [24]. However, the outcomes of cell transplantation-based therapies are variable and depend not only on the immunological response but also on cell survival after transplantation, and the integration of transplanted cells [25].
Recently, extracellular vesicles (EVs) have attracted attention and been medically applied as a cell-free therapy and drug delivery system. EVs are membrane-bound nanovesicles released from the cell into the extracellular space [26]. These nanovesicles carry lipids, proteins, and various functional RNA species such as microRNA (miRNA), contributing to intercellular communication [27]. The communication between neural cells is essential for nervous system activity. EVs from several cell types, such as astrocytes [28], microglia [29], and oligodendrocytes [30], modulate cell-to- cell or neuron-glia communication in both, healthy and diseased brains [31]. EVs have emerged as essential instruments of neurogenesis and regeneration. EVs from mesenchymal stem cells (MSCs) and Schwann cells enhance peripheral nerve regeneration [32]. In a rat stroke model [33], systemic administration of MSC-derived exosomes could enhance neurogenesis, neurite remodeling, and angiogenesis and improve functional recovery. Recently, neural stem cell-derived EVs were found to potentially attenuate neuronal apoptosis, inhibit neuroinflammation, and enhance functional recovery in SCI rats [34]. Exosomes from bone MSCs are also capable of suppressing neuroinflammation, attenuating neuron apoptosis, and enhancing angiogenesis after traumatic SCI [35]. Similarly, EVs of mesenchymal stromal cells can promote primary cortical neuron axonal growth [36]. Furthermore, fibroblast-derived EVs promote neurite outgrowth, and their application can enhance optic nerve regeneration after injury [37].
Despite the long-term application of OEC transplantation to neuron regenerative therapy, and the evidence of neurological function recovery [38], the effects of OEC-EVs on neurogenesis or neural regeneration are still not well understood. The first available report of OEC-derived EVs and their effect on neuro-regeneration was a study conducted on a rat model [39]. Rat OEC-derived EVs were found to promote the axonal growth of the dorsal root ganglion and improve axon regeneration in the damaged sciatic nerve. Previously, we reported the very first characterization of human OEC (hOEC)-derived EVs [40]. However, their potential effect on neuro-regeneration for further applications remains to be investigated. Overall, the evidence demonstrated that miRNAs were enriched in EV from various cells that carry potential effects for neuro-regeneration [32, 41]. To better understand the function and application of hOEC-EV in neuro-regeneration, enriched miRNA profiling is essential. To date, the miRNA profile of hOEC-EV has not been uncovered. Therefore, the aim of this study was to characterize miRNA profile of hOEC-EV and investigate the effect of hOEC-EVs on neurogenesis and repair potential based on an in vitro model of neuron progenitor damage.
2. MATERIALS AND METHODS
2.1. OEC Cell Cultivation
hOECs were isolated from the Olfactory Mucosa (OM) tissue. The OM was endoscopically collected under general anesthesia after signed informed consent was obtained from the donor. OEC primary culture was performed as described previously [36]. Briefly, the collected OM was first placed in cold Phosphate Buffered Saline (PBS). The tissue was washed three times in cold PBS and incubated in a dispase II enzyme solution at 37°C for 20 min. The lamina propria was dissected under a microscope and cut into small pieces with sterile instruments. The suspension was centrifuged at 200 × g for 3 min to pellet the dissociated cells. The pellet was resuspended in Dulbecco’s modified Eagle’s medium (DMEM/F12; Gibco®, ThermoFisher Scientific) plus supplements and cultured at 37°C in an incubator with 5% CO2. The fresh medium was replaced every other day. Before conditional medium was collected for EV isolation, cells were cultured in the serum-free medium.
2.2. Immunocytochemistry
Cells (2 × 104) were seeded in each well of a 48-well plate and maintained under optimized conditions. Once cell density reached 80% confluence, the culture medium was discarded. The cells were rinsed in PBS and fixed with 4% paraformaldehyde (PFA) for 15 min. Cells were washed three times in PBS prior to permeabilization by incubating them in 0.1% tritonX-100 for 15 min at room temperature. After three washes in PBS, cells were blocked with 3% bovine serum albumin (BSA) for 1 h. Cells were incubated overnight with an optimized dilution of primary antibody in 1% BSA at 4°C. As OEC characteristic markers, S100β (1:250; ab52642, Abcam), Vimentin (1:250; ab8978, Abcam), and p75NTR (1:100; ab52987, Abcam) antibodies were used in this study. For NPC labelling, a Tuj1 (1:500; ab7751, Abcam) antibody was utilized. On the second day, cells were washed three times in 1% BSA and incubated for 1 h with secondary antibody (goat anti-rabbit AlexaFluor® 488; ab150081, Abcam or Goat anti-mouse AlexaFuor® 594; ab150116, Abcam). DAPI mounting medium was used to label the cell nucleus. Cell imaging was performed with an inverted fluorescence microscope (Olympus, CKX53).
2.3. EVs Isolation and Characterization
hOEC-EVs were isolated by sequential ultracentrifugation. Briefly, the culture medium was decanted. The cells were rinsed in PBS and then cultured in the serum-free medium (13 mL/T75 flask) for 72 h. The culture medium was harvested into 50-mL centrifuge tubes and centrifuged at 2,000 × g for 10 min at 4°C to first remove large cell debris. The supernatant was then transferred to a new tube and centrifuged at 10,000 × g for 10 min to remove the remaining small cell debris. The supernatant was then transferred to an ultracentrifuge tube and centrifuged at 100,000 × g for 70 min at 4°C. The supernatant was discarded, and the EV pellet was washed in PBS with final centrifugation at 100,000 × g for 70 min at 4°C. The isolated EVs were resuspended in PBS for further characterization and subsequent cell experiments. For western blot-based EVs characterization, lysis buffer was directly added to the EV pellet. The EVs were immediately used or stored at 4°C for <1 week until use or at −80°C for longer storage. The morphology of the isolated hOEC-EVs was characterized with a transmission electron microscope (TEM; Hitachi HT7700 (100 kV)). The TEM analysis was performed by MA-tek Material Analysis Technology Inc. The nanovesicle size distribution was analyzed by Nanoparticle Tracking Analysis (NTA) (Zetaview Particle Metrix, Germany). The expression of EV surface proteins was identified by western blotting.
2.4. miRNA Sequencing and Data Analysis
Total RNA was collected from hOEC-EVs by adding TRIzolTM reagent (Invitrogen) directly to the EV pellet after PBS washing. The solution was then stored at -80°C. Three replicates of RNA samples were pooled and sent for RNA extraction, RNA library preparation and sequencing by TopGen Biotechnology Co., Ltd. (Taiwan). After raw sequencing data was obtained, the adaptor and primer sequences were trimmed using CLIP v1.0.3. Filter FASTQ v1.1.5 was used as the filter platform. The sequence data in fastq file format was quality checked by FastQC v0.11.8. miRNA sequences mapped and annotated on miRbase v.22.1 [42]. DIANA-mirPath v3 [43] was utilized to identify enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and gene ontology (GO) categories. Enrichment p-values were corrected for false discovery rate (FDR), and were considered significant for values of p ≤ 0.05. To predict the putative mRNA target and related pathway of the top abundant miRNAs, different algorithms (experimentally supported Tarbase v7.0 and predicted interaction microT-CSD) provided in mirPath v3 package were considered. We predicted the interaction network of identified miRNAs and their targets using miRNet, a miRNA-centric network visual analytics platform [44].
2.5. Western Blot Analysis
Lysis buffer containing radioimmunoprecipitation assay buffer (PierceTM RIPA buffer, Invitrogen) plus 1× protease inhibitor (HaltTM Protease and Phosphatase Inhibitor cocktail, Invitrogen) was applied to harvest total protein from cells. To extract total exosomal protein from EV pellets, the lysis buffer was added directly to the ultracentrifuge tube after washing with PBS. The dissolved EV or cell pellet was vortexed and centrifuged at 13,000 × g for 5 min. The supernatant was then transferred to a new tube. The protein concentration was quantified using the BCA kit (Pierce BCA protein assay, Invitrogen), and 25 µg of isolated protein was loaded into each well for SDS-PAGE. Proteins on the gel were transferred onto polyvinylidene difluoride membranes. After blocking the membrane in 5% skim milk in TBST buffer (tris-buffered saline, 0.1% Tween 20) for 1 h at room temperature, the membrane was incubated with primary antibodies at 4°C overnight with gentle shaking. Then, it was washed three times in TBST and incubated for 1 h with horseradish peroxidase-conjugated secondary antibodies at room temperature with gentle shaking. Finally, three washes in TBST were then conducted. Enhanced chemiluminescence reagent was used to detect immunoreactive bands according to the manufacturer’s instructions (Millipore). Primary antibodies used in this study were as follows: CD81 (1:1000; GTX31381, GeneTex, Inc.), CD9 (1:1000; GTX66709, GeneTex, Inc.), CD63 (1:1000; EXOAB-CD63A-1, System Biosciences, Inc.), TSG101 (1:1000; GTX70255, GeneTex, Inc.), BCl-2 (1:1000; GTX100064, GeneTex, Inc.), Bax (1:1000; GTX109683, Genetex, Inc.) caspase 3 cleaved Asp175 (1:800; GTX86952, GeneTex, Inc.), and beta-actin (1:20000; GTX109639, GeneTex, Inc.).
2.6. Induced Pluripotent Stem Cell (iPSC)-NPC Differentiation and Cultivation
We differentiated iPSCs to neural progenitor cells (NPC) following a previously described protocol with minimum modifications [37]. In summary, iPSCs were reprogrammed from fibroblasts via transfection with viral vectors expressing the transcription factors Sox2, c-Myc, Klf4, and Oct4. The generated iPSCs were then maintained in mTeSRTM (Stem Cell Technologies). Selected iPSC colonies were then transferred to a new Matrigel pre-coated plate and cultured in mTeSRTM1 medium. To differentiate NPCs, iPSC colonies were cultured in a PSC induction medium, containing Neurobasal® medium and 2% neural induction supplement, for 4-6 days. Thereafter, the PSC selection medium was replaced with a neural expansion medium containing 1:1 Neurobasal:DMEM/F12 and 2% neural induction supplement (Gibco® Neural Induction Medium, ThermoFisher Scientific).
2.7. Cell Viability Assay (CCK-8)
Cell proliferation of NPCs was evaluated using a Cell Counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.) following the manufacturer’s instructions. In brief, 1 × 104 cells were seeded in a 96-well plate with 100 µL medium/well and incubated for 24 or 48 h with or without hOEC-EVs. Each well was then incubated with CCK-8 working solution at 37°C for 3 h. Optical density at 490 nm was measured on a microplate reader (BioTek® 800™ TS Absorbance Reader).
2.8. Tert-Butyl Hydroperoxide (t-BHP) Treatment and Lactate Dehydrogenase (LDH) Cytotoxicity Assay
NPCs were exposed to 150 µM t-BHP for 1.5 h. Next, the supernatant was discarded, cells were washed with PBS, and replated with fresh medium with or without hOEC-EVs. Cell cytotoxicity or injury was quantified by performing an LDH assay (Dojindo Molecular Technologies, Inc.) according to the manufacturer’s instructions.
2.9. Apoptosis (Terminal Deoxynucleotidyl Transferase dUTP Nick End Labelling (TUNEL) Assay)
NPCs (2.5 × 104 per well) were seeded in a 48-well plate and treated with t-BHP as described previously herein. After treatment, cells were fixed in 4% PFA for 10 min. Cell apoptosis was evaluated using a TUNEL assay kit HRP-DAB (abcam206386, Abcam) according to the manufacturer’s instructions. The brown stained cells were evaluated as apoptotic cells under a microscope.
2.10. Migration Assay
The silicone inserts (2-well culture-insert, ibidi®) were utilized for cell migration assay. NPCs or Human Umbilical Vein Endothelial Cells (HUVECs) were diluted to 7 × 105 cells/mL, and 70 μL of cell suspension was plated per insert chamber. After a 24-h incubation, the inserts were removed, and the cells were rinsed in PBS. Medium with or without hOEC-EVs was supplied to each well, and cells were incubated for 24 h. Images around the gaps were acquired at 0, 12, and 24 h of incubation. The gap width and number of migrating cells were quantified from the images.
2.11. Invasion (Transwell Assay)
NPCs or HUVECs were seeded in 12-well transwell inserts, 0.8-µm pore size (JET Biofil®), with 300 µL serum-free NPC or HUVEC medium. The lower chamber was filled with 750 µL of 10% FBS-containing medium. hOEC-Evs or PBS were added to each insert well. Cells were cultured for 24 h, the medium was discarded, and the lower surfaces of inserts were dipped in 4% PFA for 10 min. After washing by dipping in PBS, the lower surfaces of inserts were dipped in crystal violet solution (0.5%) for 30 s. The inner surfaces of inserts were gently cleaned with cotton swabs. The lower surfaces of inserts were placed on a glass slide and observed under a microscope.
2.12. In vitro Angiogenesis (Tube Formation Assay)
The Cultrex® In vitro Angiogenesis Tube Formation Assay kit (Trevigen Inc.) was used in this study. Here, 50 µL of gel was coated on each well of a pre-cooled 96-well plate and incubated at 37°C for 30 min. HUVECs were plated at a cell density of 2 × 104 cells/100 µL/well. hOEC-Evs or PBS were added into each well, and tube formation was observed at 0, 6, 12, and 24 h after seeding.
2.13. Statistical Analysis
Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, Inc. San Diego, CA, USA). To compare the differences between groups, one-way or two-way ANOVA was utilized. Statistical significance was presented as *p < 0.05, **p < 0.01, and ***p < 0.001. Error bars represent the standard error of the mean.
3. RESULTS
3.1. hOEC-EV Isolation, Characterization, and Quantification
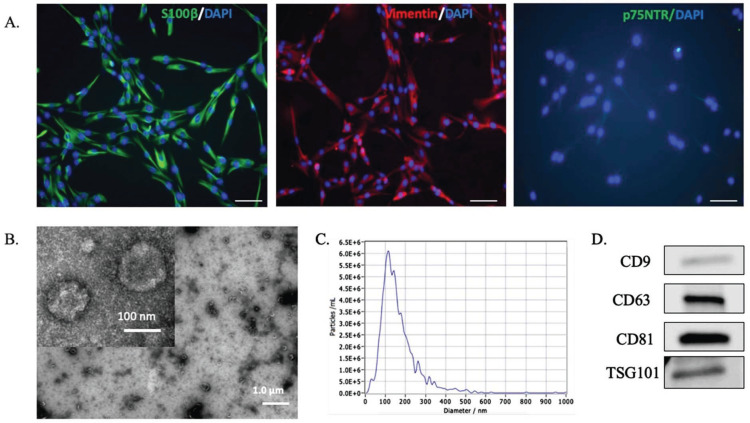
Primary hOECs were characterized by immunocytochemistry (Fig. 1). Primary hOECs extensively expressed S100β and Vimentin, while p75NTR, which is recognized as an OEC-specific marker in several species, was undetectable in this study (Fig. 1A). hOECs were incubated in the serum-free medium for 72 h prior to conditioned medium harvesting. The isolated EVs were characterized based on TEM images, western blotting, and NTA. TEM images revealed the cup-shaped structure typical of EVs (Fig. 1B). The size distribution of hOEC-Evs measured by NTA showed a peak at 114.2 nm, indicating the size of the major detected population (Fig. 1C). Western blot analysis indicated the expression of EV markers, specifically CD9, CD63, CD81, and TSG101 (Fig. 1D). Prior to EV treatment analysis, the concentration of isolated EVs was quantified by ELISA. Consistent with western blot results, CD9, CD63, and CD81 were detected. For the further ELISA-based quantification of hOEC-EVs isolated from different batches before the next cell treatment, we employed CD63-based analysis.
Fig. (1).
Human olfactory ensheathing cell (hOEC)- EV characterization. (A) Immunocytochemistry for specific OEC markers (S100β, Vimentin, p75NTR) in primary cultured hOECs at passage 1. Scale bars: 100 µm. (B) Transmission electron microscopy (TEM) images of isolated hOEC-EVs (scale bar, 500 nm.) (C) Nanoparticle Tracking Analysis results demonstrating the particle size distribution of the isolated hOEC-EVs. (D) Western blot results indicating the expression of EV surface markers (CD9, CD63, CD81, and TSG101) on hOEC-EVs. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.2. miRNA Sequencing Result
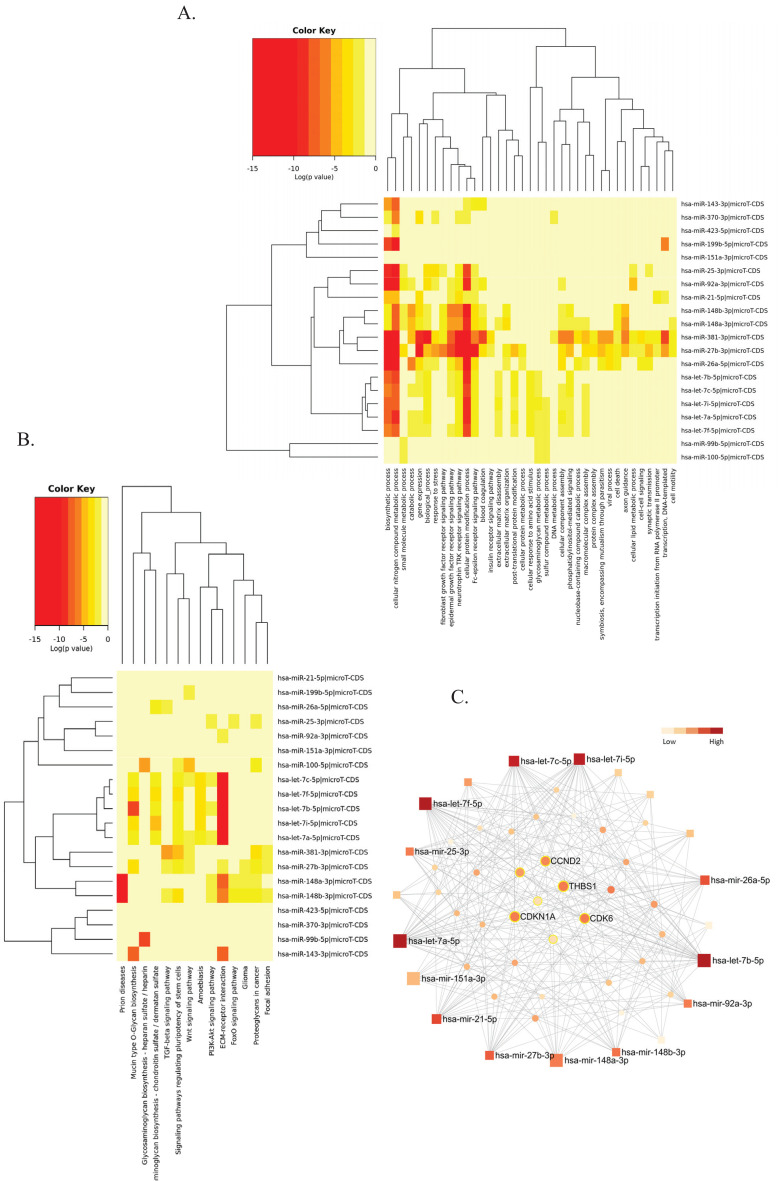
We sought to uncover novel information on hOEC-EV to further investigate its therapeutic potential. Here, we provide the first report of the miRNA profile of hOEC-EVs. Next-generation sequencing data showed a total of 6,280,260 qualified reads, of which 234,191 reads were mapped to a reference miRNA database (miRBase v22.1) and 145 miRNAs were identified. The identified miRNAs profile was provided as a supplementary file. The five most enriched miRNAs were hsa-miR-148a-3p, hsa-miR-151-3p, hsa-let-7b-5p, hsa-let-7i-5p and hsa-miR-143-3p. Five members of the let-7 family (let-7a-5p, let-7b-5p, let-7c-5p, let-7f-5p and let-7i-5p) were identified among the top 20 miRNAs. In this list, we identified the miRNA related to neurogenesis and differentiation or neuro-regeneration as indicated in previous studies, namely, let-7b and miR-25 [45], miR-21 [46], miR-27 [47], miR-148b, let-7i, and miR-26a [48].
A GO analysis using a microT-CSD algorithm suggested that the top 20 miRNA targets were enriched in several biological processes, including but not limited to cellular nitrogen compound, biosynthetic process, cellular protein modification process, and the neurotrophin tropomyosin-related kinase (TRK) receptor signaling pathway. Cellular nitrogen compounds and the neurotrophin-TRK receptor signaling pathway were predicted by both microT-CSD and Tarbase algorithms as high p-value GO categories. A KEGG pathway analysis using a microT-CSD algorithm indicated that these abundant miRNAs of hOEC-EC were involved in extracellular matrix (ECM)-receptor interaction, prion disease, mucin type O-glycan biosynthesis, signaling pathways regulating pluripotency of stem cells, and proteoglycans in cancer. Proteoglycans in cancer, ECM-receptor interaction, and prion diseases were both outcomes of the microT-CSD and Tarbase predictions of the roles of the top 20 miRNAs. Heatmaps of the GO annotation and KEGG pathway analyses of the top 20 abundant miRNAs by microT-CSD algorithms of mirPath v3.0 are presented in Fig. (2A and B), respectively.
Fig. (2).
Pathway analysis of hOEC-EV- miRNA profile. (A) Gene Ontology categories of top 20 hOEC-EV-miRNAs constructed in the DIANA-mirPath v3. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of top 20 hOEC-EV-miRNAs constructed in the DIANA-mirPath v3. (C) The minimum interaction network of top 20 hOEC-EV-miRNAs and their targets built in the miRNET 2.0. Square nodes represented the miRNA queries. Circle nodes represented predicted targets. The color key bar indicated the degree of interaction. The highly connected targets (high degree of interaction) were located in the middle of the network. The nodes of targets related to the p53 pathway were labelled with a yellow line (A higher resolution / colour version of this figure is available in the electronic copy of the article).
We constructed the miRNA-target network of these 20 miRNAs to observe common interactions that may play a key role in hOEC-EV function by using miRNet 2.0. The network revealed that the highest degree of interaction was in the let-7 family (28-34 degree). The common targets (extracted from Tarbase v8.0 and miRTarBase v8.0) with the three highest degrees of interaction included thrombospondin 1 (THBS1), cyclin dependent kinase 6 (CDK6), CDK inhibitor 1A (CDKN1A), and cyclin D2 (CCND2) (Fig. 2C). Most of these genes were related to the p53 signaling pathway or cell cycle.
3.3. hOEC-EVs Enhance NPC Proliferation and Axonal Growth
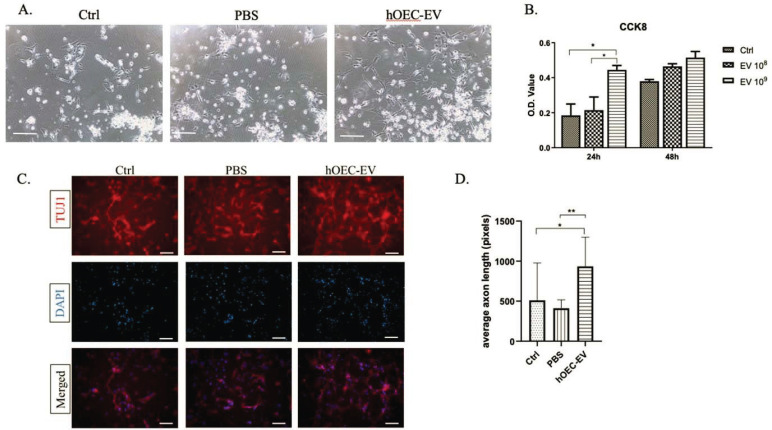
In our previous study [40], we demonstrated that hOEC-EVs could significantly stimulate NPCs’ proliferation after 72-h incubation. However, we suspected that the concentration of hOEC-EVs might affect their potential. In the present study, we incubated NPCs with various concentrations of EVs (108 or 109 particles/mL) for 24 or 48 h (Fig. 3A). CCK-8 assay results indicated that hOEC-EVs promoted cell proliferation/survival in a dose-dependent manner (Fig. 3B). EVs at 109 particles/mL significantly enhanced NPC’s proliferation rate after 24 h incubation. However, at 48 h of incubation, the proliferation stimulating effect was insignificant. A recent report revealed that rat OEC-derived EVs promote axonal growth of the dorsal root ganglion [39]. In this study, we sought to determine whether hOEC-EV could enhance neurogenesis by promoting NPC differentiation to neurons with axon elongation. We performed immunostaining to label NPCs with an antibody for TUJ1, a specific neuron marker (Fig. 3C). Axon length was measured and averaged per random field. We showed that the average axon length of NPCs exposed to hOEC-EVs was significantly longer than that of controls (Fig. 3D). These results confirmed that hOEC-EVs enhance NPC proliferation and differentiation to neuron-like morphology with elongated axons.
Fig. (3).
Human olfactory ensheathing cell (hOEC)-EVs stimulate neural progenitor cell (NPC) proliferation and axon elongation. (A) Image of NPCs after incubation for 24 h in medium with hOEC-EV supplementation or PBS, or of mock controls. Scale bars: 200 µm. (B) CCK-8 results in the bar graph are based on optical density (O.D.) values at 24 or 48 h of incubation. hOEC-EVs at either 108 or 109 particles/mL (*p<0.05). (C) Immunostaining for Tuj1, a neuron marker, to label cell body and axon. (D) Axon length was measured using Image J. The average axon lengths from two random fields (40×) of three replications each are presented in the graph. Significantly increased average axon length was found in the hOEC-EV-treated group (*p<0.05, ** p<0.01) (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.4. hOEC-EVs Ameliorate NPC Apoptosis
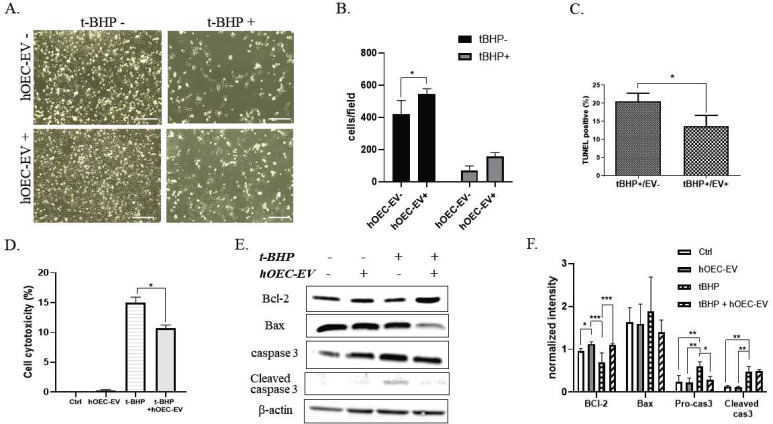
Traumatic injuries to the nervous system, such as SCI, consist of two mechanisms, including a primary mechanical injury and secondary insults associated with inflammation and apoptosis. Long-term neurological deficits due to these secondary insults lead to the expansion of damage and worsen paralysis. We sought to establish new approaches to prevent or treat neuronal inflammation and apoptosis using hOEC-EVs. NPCs were exposed to 150 µM t-BHP to activate oxidative stress damage. After 1.5 h incubation, a fresh medium was added and hOEC-EVs were administered to the cells. Notably, we found that the cell number was drastically decreased in t-BHP-treated groups, indicating that t-BHP causes cell death and detachment (Fig. 4A). However, the number of surviving cells was increased in the EV-treated group, suggesting that hOEC-EVs protect the cells from t-BHP-induced cell death (Fig. 4A and B). We further employed TUNEL staining to evaluate cell death. TUNEL-positive (apoptotic) cells were abundant in the t-BHP-treated group and significantly decreased in the hOEC-EV-treated group (p<0.05) (Fig. 4C). LDH assay results indicated that NPC cytotoxicity was remarkably increased after t-BHP treatment. This result is consistent with the low cell viability and decreased cell number observed in Fig. (4A). However, cell cytotoxicity mediated by t-BHP was significantly abolished in the hOEC-EV treatment group (p<0.05) (Fig. 4D). To determine the effect of hOEC-EVs on NPC’s apoptotic protein expression, western blot analysis was performed. The representative blotting results and quantification of blot densitometry are depicted in Fig. (4E and F) and showed that hOEC-EV administration was able to upregulate expre administration was able to upregulate expression of the anti-apoptotic protein BCl-2 in NPCs. Treatment with t-BHP reduced BCl-2 expression, an effect significantly attenuated by hOEC-EVs (p<0.01). Expression of pro-apoptotic protein Bax and cleavage form of caspase 3 was reduced by hOEC-EV in t-BHP treated groups. Furthermore, Pro-caspase 3 in NPCs was significantly induced by t-BHP treatment and ameliorated by hOEC-EV administration. Overall, this result revealed that t-BHP activates NPC apoptosis, whereas hOEC-EV treatment mitigates the effect.
Fig. (4).
Human olfactory ensheathing cell (hOEC)-EVs protect neural progenitor cells (NPCs) from apoptosis. (A) Appearance of NPCs with different treatments. t-BHP caused a large amount of cell death. Scale bars: 200 µm. (B) The cell number was counted to evaluate cell damage after t-BHP treatment. (C) TUNEL assays were performed to count apoptotic cells after treatment. Quantification of TUNEL-positive cells was calculated as a percentage. (D) Lactate dehydrogenase (LDH) assay indicating cytotoxicity after t-BHP treatment and the alleviating effect of hOEC-EVs. (E) Representative western blot images of apoptotic protein expression. (F) Densitometric bar graph quantified from the three western blot replicates. *p<0.05, **p<0.01, ***p<0.001 (A higher resolution / colour version of this figure is available in the electronic copy of the article).
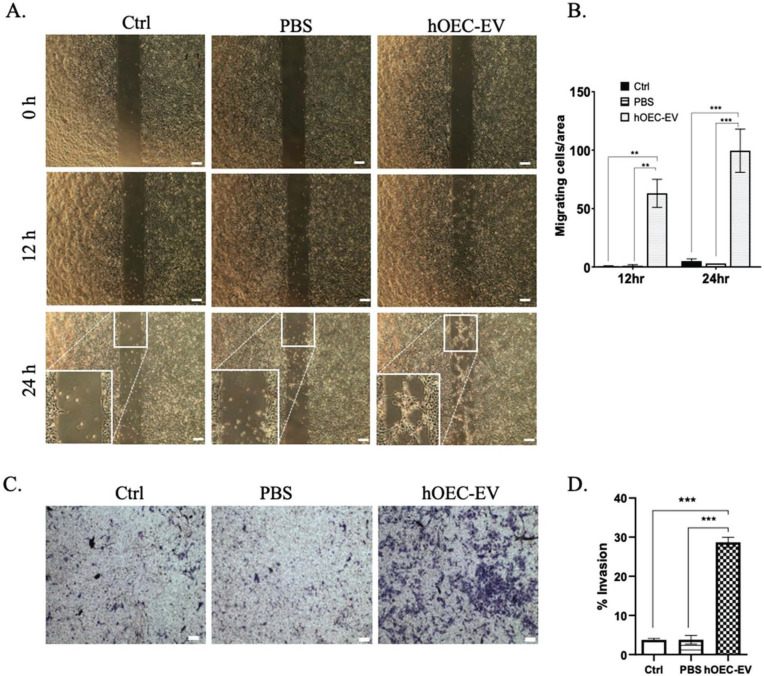
3.5. hOEC-EVs Stimulate NPC Migration and Invasion
The migration of neuron precursor cells was previously shown to be an important process during wound healing in SCI repair in zebrafish [49]. Herein, we sought to investigate the effect of hOEC-EVs on the migration ability of NPC. We evaluated the migration ability of NPCs based on the culture-insert 2-well method. We observed the migrating cell under a microscope at 0, 12, 24 h incubation. The number of cells exhibiting migration activity was counted per 4× magnification field. Consistently, we demonstrated that NPCs cultured in a medium with hOEC-EV showed significantly higher migration rates at 12 h (p<0.01) and 24 h (p<0.001) (Fig. 5A and B). Furthermore, we evaluated the invasion ability of NPCs. Here, NPCs were cultured on the upper surface of a transwell insert and incubated in a medium with or without hOEC-EVs. We found that NPCs cultured in a medium with hOEC-EVs migrated to the lower surface of the insert at a significantly higher rate (p-value < 0.001) than those in the other groups (Fig. 5C and D). This evidence consistently supported the hypothesis that hOEC-EVs can probably promote the repair of injured neural tissue by enhancing neural cell migration and wound healing.
Fig. (5).
Human olfactory ensheathing cell (hOEC)-EVs stimulate the migration of neural progenitor cells (NPCs). (A) Cell migration ability confirmed by the culture-insert method. hOEC-EVs significantly promoted NPCs’ cell migration ability. (B) The average number of migrating cells per area was counted and is shown in the bar graph. (C) NPCs’ invasion ability was assessed using the transwell method. hOEC-EVs significantly increased NPCs’ invasion ability. (D) The number of invading cells in each group was counted and averaged and is depicted in the bar graph. **p<0.01, ***p<0.001. Scale bars: 200 µm (A higher resolution / colour version of this figure is available in the electronic copy of the article).
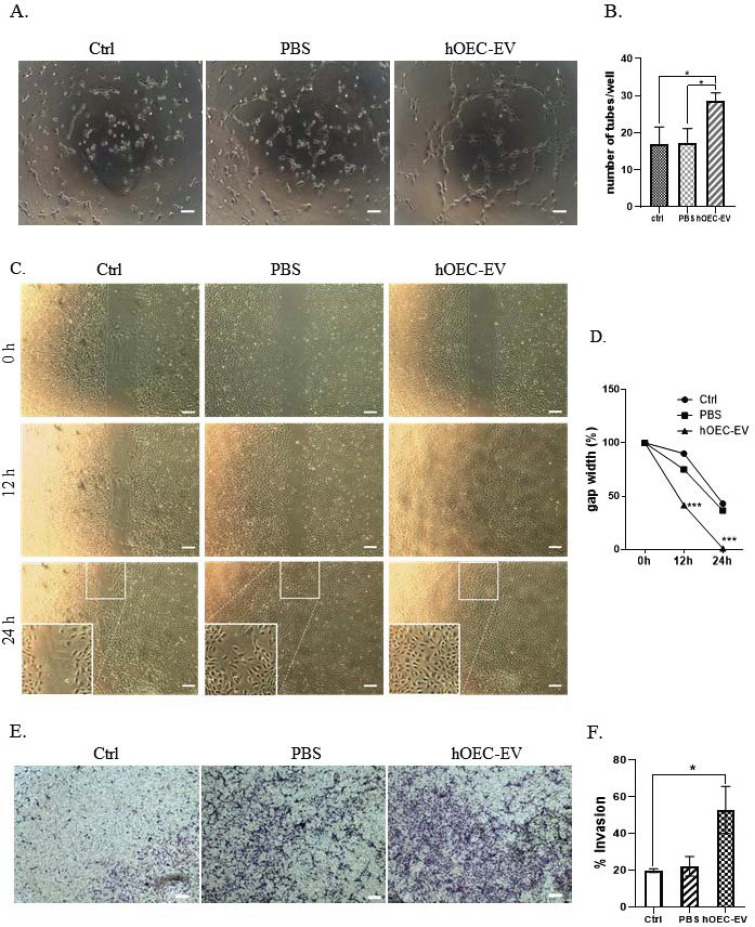
3.6. hOEC-EVs Promote In vitro Angiogenesis and HUVECs’ Migration
Angiogenesis is another important process required for damaged tissue repair. A previous study demonstrated that EVs derived from bone MSCs stimulate in vitro angiogenesis and in vivo vascularization [35]. Therefore, we conducted in vitro angiogenesis tests. HUVECs were seeded in gel-pre-coated 96-well plates and incubated with PBS or hOEC-EVs. Tube formation was observed and the number of tubes per well was directly counted under a microscope. Representative images of tube formation at 6 h are shown in Fig. (6A). We demonstrated that hOEC-EV-treated cells formed significantly more complete tubes per well than cells of the other groups (Fig. 6B). At 24 h, formed tubes partially disconnected in every group. However, the number of remaining tubes in the hOEC-EV-treated group was higher than in the others. Furthermore, HUVECs’ migration ability was also evaluated by the aforementioned 2-well culture-insert method. Here, the gap was only completely filled by HUVECs exposed to hOEC-EVs after 24-h incubation (Fig. 6C and D). This result indicated that hOEC-EV-treated HUVECs display a higher migration rate. Finally, similarly to NPCs, the invasion ability of HUVECs was also determined using transwells. We proved that HUVECs exposed to hOEC-EVs migrated to the lower surface of transwells at a higher rate than control cells (Fig. 6E and F). In summary, hOEC-EVs stimulate in vitro vessel formation, and endothelial cell migration and invasion, which are hallmarks of angiogenesis.
Fig. (6).
Human olfactory ensheathing cell (hOEC)-EVs promote in vitro angiogenesis and HUVECs’ migration. (A) In vitro angiogenesis assay showing the tube formation ability of HUVECs at 6 h with different treatments. (B) Average number of tubes formed in each group. (C) HUVEC migration ability at different time points evaluated by culture-inserts. (D) Gap width as a percentage of the initial size. (E) HUVEC invasion ability assessed using the transwell method. (F) Quantification of the invasion assay. *p<0.05, ***p<0.001. Scale bars: 200 µm (A higher resolution / colour version of this figure is available in the electronic copy of the article).
4. DISCUSSION
In this study, we focused on characterizing hOEC-EVs, miRNA profiling, and their effect on neuroprotection or neuro-regeneration. We first isolated OECs from human nasal tissue. We demonstrated that these cells exclusively expressed OEC characteristic markers, such as S100β, Vimentin, GFAP, and SOX10 both here and in our preliminary report [40]. However, p75NTR, expressed in OECs of several species, including rats [39], was not positively immunostained in our study. This is consistent with a previous report revealing that human OECs do not express P75NTR, whereas the perineural olfactory nerve fibers that surround OECs do [50, 51]. Characterization by TEM, NTA, and western blot analysis confirmed that hOEC-derived EVs were successfully prepared.
In this study, we reported the first miRNA profile of hOEC-EV. The most abundant miRNA in hOEC-EV was hsa-miR-148a-3p, hsa-miR-151a-3p, and let-7 family. Recently, miR-148a/b-3p has been reported as an angiogenesis regulator [52] and a tumor suppressor in several cancers, including ovarian cancer [53]. hsa-miR-151a-3p, the second enriched miRNA in this study, was found to be upregulated in patients with Alzheimer’s disease (AD) [54]. Inconsistently, it was reported for its decrease in the plasma EV of AD patient respect to those of control [55]. Another study showed that miR-151a-3p enhances anti-apoptotic, anti-oxidative, and neuroprotective effects of dexmedetomidine [56]. Our miRNA profile demonstrated that there were several members of the let-7 family enriched in hOEC-EV, including let-7b-5p and let-7i-5p. Let-7 family is well-recognized as a tumor suppressor [57]. A previous report showed that let-7 miRNA is highly expressed in new-born olfactory bulb neurons, and its suppression impairs the radial migration [58]. Let-7b can promote neurogenesis, regulating neural stem cell proliferation and differentiation through targeting nuclear receptor and cell cycle regulator cyclin D1 [59]. Let-7i, as well as miR-148b, is also indicated as miRNA specifically related to neuron cell generation [48].
GO category and KEGG pathway analyses revealed that these abundant miRNAs are involved in several biological processes, such as the cellular nitrogen compound metabolic process and ECM-receptor interaction. Some evidence has demonstrated the association of nitrogen with neurogenesis. Meanwhile, the ECM pathway has been widely mentioned in the context of neural development. The ECM provides structural support, regulates proliferation, promotes the differentiation of neural progenitors, and regulates neuronal migration [60]. Neurotrophin-TRK receptor signaling was also predicted as a pathway related to the hOEC-EV-miRNAs. The neurotrophin-TRK receptor interaction is critical for the long-distance communication between axon terminals and cell bodies, in addition to neural circuit maintenance [61]. These prediction results pointed to the potential of hOEC-EVs in light of neurological applications.
Based on the predicted miRNA-target interaction network in miRnet 2.0, THBS1, CDK6, CDN1A, and CCND2, were highly connected target genes. THBS1 is found in the brain and is primarily expressed by astrocytes [62]. THBS1 has been reported as a controller of synaptogenesis [63], related to angiogenesis [64, 65], inflammation [66], and cell- type specific axon regeneration [67]. Elevated THBS1 is found in glioma and its silencing inhibits tumor growth [68]. CDK6 is an essential protein regulating the cell cycle in the G1 phrase; CDK6 controls the switching between cell proliferation and neuronal differentiation [69]. CDKN1A or p21 is a cell cycle inhibitor and downstream protein of p53. The inhibition of p21 improves the cell proliferation ability of intestinal progenitor cells and the self-renewal of hematopoietic stem cells of mice with dysfunctional telomeres without accelerating cancer development [70]. A part of the cyclin family, CCND2, is mainly involved in the regulation of G1/S transition [71]. Most of these common targets were linked to the p53 signaling pathway, an important pathway regulating cell cycle arrest, senescence or apoptosis. By an overall miRNA profile analysis, these identified miRNAs probably contribute to the neuroregulatory effect of hOEC-EV by promoting or inhibiting target cell proliferation, neuronal differentiation, migration, and apoptosis.
While rat OEC-EVs were proven to have the ability to promote axonal growth of dorsal root ganglion (DRG) neurons [39], our study is the first showing that hOEC-EVs significantly augmented cell proliferation and axon growth of NPC. In vertebrate mammals, NPCs are found lining along the spinal cord canal in a quiescent state [72], and in rodent models, NPCs increase active proliferation and differentiation, and migrate to the lesion after SCI [73]. The manipulation of endogenous NPC activity may offer potential benefits for neuron regeneration after SCI [74]. Accumulating evidence has also shown that transplanted NPCs could extend large numbers of axons into the caudal part of a host spinal lesion, and the host axons may also regenerate into the cell grafts to form new synapses and improve neural function [75-78]. Thus, our finding suggested that hOEC-EV may support the neuron regeneration after injury by activating NPC proliferation and axon growth.
Our data also suggested that hOEC-EVs protect NPCs from apoptosis in t-BHP-mediated oxidative stress. We demonstrated that the anti-apoptotic protein BCl-2 was upregulated in hOEC-EV-treated cells either under normal or t-BHP-mediated stress conditions. Expression of cleaved caspase 3 and Bax was reduced insignificantly after hOEC-EV treatment under t-BHP-mediated stress condition. Therefore, the anti-apoptotic activity of hOEC-EVs might be partially related to BCl-2 upregulation and pro-caspase 3 downregulation. Despite the insignificant effect of hOEC-EVs on Bax and cleaved caspase 3 expression, the number of surviving cells in the t-BHP+EV treatment group was higher than in the t-BHP only group. Furthermore, the number of TUNEL-positive cells in the hOEC-EVs treated group was significantly lower. Consistently, LDH assay results demonstrated the lower cytotoxicity and increased number of surviving cells in the EV-treated group. Taken together, hOEC-EVs had a protective effect on NPC viability under t-BHP-mediated stress by BCl-2 upregulation. Similar results have been recently reported in several studies using different EV sources. EVs derived from neural stem cells can attenuate apoptosis after traumatic SCI and in vitro glutamate-induced neurotoxicity by reducing cleaved caspase 3 and elevating BCl-2 expression [34]. Bone MSC-derived EVs could exert anti-apoptotic effects on in vitro glutamate-induced apoptosis and in an in vivo SCI model [35]. Astrocyte-derived EVs are also capable of inhibiting neuron apoptosis in vitro and in vivo [79]. The characterization of these different EV cargos, such as RNA sequencing, may provide informative data to identify the essential miRNAs that regulate the anti-apoptotic effect, thereby informing the further development of EV-based therapy.
One of our most significant findings was the effect of hOEC-EVs on cell migration. We demonstrated that hOECs stimulated the migration ability of either NPCs or HUVECs. hOEC-EV treatment remarkably stimulated NPC migration in a 2-well culture-insert method. Furthermore, transwell methods confirmed that both the migration and invasion abilities of NPCs were significantly improved by hOEC-EVs. In zebrafish, the early migration of neuron precursors initiates neuron regeneration following SCI [49]. Furthermore, previous research indicates that NSPC migration is essential for CNS development and neurogenesis [80, 81]. In addition, NSPC migration to the injured CNS sites is an adaptive response to limit and repair damage in various types of brain injury, such as those involving ischemia and trauma [82-85]. NSPC proliferation can be activated after injury, and NSPCs might differentiate into neurons or glia [86]. Thus, NSPCs’ migration to the lesion is critical to promote tissue repair. Consequently, we think that, by activating neuron precursor migration, hOEC-EVs probably have potential benefits for the repair and regeneration of neuronal tissue after traumatic injury.
Previous evidence indicates that SCI causes vascular deconditioning below the injured area, including vascular destruction and disorganization, microcirculation loss, endothelial cell death, and vascular remodeling [87]. The devastated vasculature consequently enhances secondary injury following the primary mechanical injury to the spinal cord. Given that blood supply is critical for cell/tissue survival and functionality, inadequate blood supply due to ischemic conditions exacerbates neuronal cell death [88, 89]. In an SCI rat model, the delivery of angiogenic microspheres into the injury site promotes angiogenesis and recruitment of neural precursors, stimulates axon growth, reduces white matter volume loss, and enhances neurologic recovery [90]. Thus, revascularization is another important process for tissue repair. In the present report, we demonstrated that hOEC-EVs can stimulate HUVECs’ in vitro tube formation. Furthermore, they also increased their migration and invasion ability, important processes involved in vascularization. These results are similar to those of a previous study showing that EVs from bone MSCs can promote angiogenesis in vitro and in vivo in an SCI model [35]. Our study is the first report highlighting hOEC-EVs’ pro-angiogenic functions and their possible capacity of stimulating tissue repair through revascularization. However, in vivo study is required to validate the potential of hOEC-EVs in neuron regeneration and revascularization after injury.
CONCLUSION
In conclusion, we isolated, characterized and reported the first miRNA profile of hOEC-EVs. hsa-miR-148a-3b, hsa-miR-151a-3p, hsa-miR143-3p, and let-7 family were found abundantly in hOEC-EVs. The most common targets connected among the 20 most enriched miRNAs were THBS1 and CDK6, both of which are related to the p53 signaling pathway. The miRNA data provided useful information for the future study of hOEC-EV applications. The novel miRNAs profiled in this study require validation of their specific function in specific target cell types. We revealed that isolated hOEC-EVs harbored promising potential for the protection and repair of damaged NPCs. Specifically, hOEC-EVs stimulated NPC proliferation and axon elongation. Under t-BHP-mediated oxidative stress, hOEC-EVs protected NPCs from apoptosis by upregulating the anti-apoptotic protein BCl-2, while suppressing pro-caspase 3 expression. In this study, we uncovered the significant effect of hOEC-EVs on NPC and HUVEC cell migration and invasion, both of which represent important processes of neuron tissue repair. Furthermore, HUVEC in vitro angiogenesis was enhanced by hOEC-EV treatment. Taken together, hOEC-EVs are a promising therapeutic tool to alleviate neuronal damage due to secondary insults such as oxidative stress, while promoting biological processes related to tissue repair. Since our study was an in vitro study, the effect of hOEC-EVs in neurodegenerative diseases, such as traumatic brain injury or SCI, remains to be verified in vivo.
ACKNOWLEDGEMENTS
Declared none.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All the study protocols were approved by the Hospital, I-Shou University, Taiwan (Approval no. IRB: EMRP45107N).
HUMAN AND ANIMAL RIGHTS
No animals were used for studies that are the base of this research. All the humans procedure were followed in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013 (http:// ethics.iit.edu/ecodes/node/3931).
CONSENT FOR PUBLICATION
All participants signed written informed consent.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author [YKT] upon reasonable request.
FUNDING
We thank the grants from the Ministry of Science and Technology (MOST) of the Taiwan Government (MOST 109-2314-B-650-002-MY2; MOST 109-2221-E-650-001-MY2).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Doucette J.R. The glial cells in the nerve fiber layer of the rat olfactory bulb. Anat. Rec. 1984;210(2):385–391. doi: 10.1002/ar.1092100214. [DOI] [PubMed] [Google Scholar]
- 2.Doucette R. Glial influences on axonal growth in the primary olfactory system. Glia. 1990;3(6):433–449. doi: 10.1002/glia.440030602. [DOI] [PubMed] [Google Scholar]
- 3.Graziadei P.P., Monti Graziadei G.A. Neurogenesis and neuron regeneration in the olfactory system of mammals. III. Deafferentation and reinnervation of the olfactory bulb following section of the fila olfactoria in rat. J. Neurocytol. 1980;9(2):145–162. doi: 10.1007/BF01205155. [DOI] [PubMed] [Google Scholar]
- 4.Raisman G. Specialized neuroglial arrangement may explain the capacity of vomeronasal axons to reinnervate central neurons. Neuroscience. 1985;14(1):237–254. doi: 10.1016/0306-4522(85)90176-9. [DOI] [PubMed] [Google Scholar]
- 5.Raisman G. Olfactory ensheathing cells - another miracle cure for spinal cord injury? Nat. Rev. Neurosci. 2001;2(5):369–375. doi: 10.1038/35072576. [DOI] [PubMed] [Google Scholar]
- 6.Ramón-Cueto A., Santos-Benito F.F. Cell therapy to repair injured spinal cords: olfactory ensheathing glia transplantation. Restor. Neurol. Neurosci. 2001;19(1-2):149–156. [PubMed] [Google Scholar]
- 7.Ramón-Cueto A., Nieto-Sampedro M. Regeneration into the spinal cord of transected dorsal root axons is promoted by ensheathing glia transplants. Exp. Neurol. 1994;127(2):232–244. doi: 10.1006/exnr.1994.1099. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama Y., Lankford K., Radtke C., Greer C.A., Kocsis J.D. Remyelination of spinal cord axons by olfactory ensheathing cells and Schwann cells derived from a transgenic rat expressing alkaline phosphatase marker gene. Neuron Glia Biol. 2004;1(1):47–55. doi: 10.1017/S1740925X04000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khankan R.R., Griffis K.G., Haggerty-Skeans J.R., Zhong H., Roy R.R., Edgerton V.R., Phelps P.E. Olfactory ensheathing cell transplantation after a complete spinal cord transection mediates neuroprotective and immunomodulatory mechanisms to facilitate regeneration. J. Neurosci. 2016;36(23):6269–6286. doi: 10.1523/JNEUROSCI.0085-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung R.S., Woodhouse A., Fung S., Dickson T.C., West A.K., Vickers J.C., Chuah M.I. Olfactory ensheathing cells promote neurite sprouting of injured axons in vitro by direct cellular contact and secretion of soluble factors. Cell. Mol. Life Sci. 2004;61(10):1238–1245. doi: 10.1007/s00018-004-4026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipson A.C., Widenfalk J., Lindqvist E., Ebendal T., Olson L. Neurotrophic properties of olfactory ensheathing glia. Exp. Neurol. 2003;180(2):167–171. doi: 10.1016/S0014-4886(02)00058-4. [DOI] [PubMed] [Google Scholar]
- 12.Wewetzer K., Grothe C., Claus P. In vitro expression and regulation of ciliary neurotrophic factor and its alpha receptor subunit in neonatal rat olfactory ensheathing cells. Neurosci. Lett. 2001;306(3):165–168. doi: 10.1016/S0304-3940(01)01891-2. [DOI] [PubMed] [Google Scholar]
- 13.Woodhall E., West A.K., Chuah M.I. Cultured olfactory ensheathing cells express nerve growth factor, brain-derived neurotrophic factor, glia cell line-derived neurotrophic factor and their receptors. Brain Res. Mol. Brain Res. 2001;88(1-2):203–213. doi: 10.1016/S0169-328X(01)00044-4. [DOI] [PubMed] [Google Scholar]
- 14.Guérout N., Derambure C., Drouot L., Bon-Mardion N., Duclos C., Boyer O., Marie J.P. Comparative gene expression profiling of olfactory ensheathing cells from olfactory bulb and olfactory mucosa. Glia. 2010;58(13):1570–1580. doi: 10.1002/glia.21030. [DOI] [PubMed] [Google Scholar]
- 15.Marçal H., Sarris M., Raftery M.J., Bhasin V., McFarland C., Mahler S.M. Expression proteomics of olfactory ensheathing cells. J. Chem. Technol. Biotechnol. 2008;83(4):473–481. doi: 10.1002/jctb.1900. [DOI] [Google Scholar]
- 16.Assinck P., Duncan G.J., Hilton B.J., Plemel J.R., Tetzlaff W. Cell transplantation therapy for spinal cord injury. Nat. Neurosci. 2017;20(5):637–647. doi: 10.1038/nn.4541. [DOI] [PubMed] [Google Scholar]
- 17.Gomes E.D., Mendes S.S., Assunção-Silva R.C., Teixeira F.G., Pires A.O., Anjo S.I., Manadas B., Leite-Almeida H., Gimble J.M., Sousa N., Lepore A.C., Silva N.A., Salgado A.J. Co-transplantation of adipose tissue-derived stromal cells and olfactory ensheathing cells for spinal cord injury repair. Stem Cells. 2018;36(5):696–708. doi: 10.1002/stem.2785. [DOI] [PubMed] [Google Scholar]
- 18.Gómez R.M., Sánchez M.Y., Portela-Lomba M., Ghotme K., Barreto G.E., Sierra J., Moreno-Flores M.T. Cell therapy for spinal cord injury with olfactory ensheathing glia cells (OECs). Glia. 2018;66(7):1267–1301. doi: 10.1002/glia.23282. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L., Li B., Liu B., Dong Z. Co-transplantation of epidermal neural crest stem cells and olfactory ensheathing cells repairs sciatic nerve defects in rats. Front. Cell. Neurosci. 2019;13:253. doi: 10.3389/fncel.2019.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Wang W-T., Gong C-R., Li C., Shi M. Combination of olfactory ensheathing cells and human umbilical cord mesenchymal stem cell-derived exosomes promotes sciatic nerve regeneration. Neural Regen. Res. 2020;15(10):1903–1911. doi: 10.4103/1673-5374.280330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radtke C., Aizer A.A., Agulian S.K., Lankford K.L., Vogt P.M., Kocsis J.D. Transplantation of olfactory ensheathing cells enhances peripheral nerve regeneration after microsurgical nerve repair. Brain Res. 2009;1254:10–17. doi: 10.1016/j.brainres.2008.11.036. [DOI] [PubMed] [Google Scholar]
- 22.Guérout N., Duclos C., Drouot L., Abramovici O., Bon-Mardion N., Lacoume Y., Jean L., Boyer O., Marie J.P. Transplantation of olfactory ensheathing cells promotes axonal regeneration and functional recovery of peripheral nerve lesion in rats. Muscle Nerve. 2011;43(4):543–551. doi: 10.1002/mus.21907. [DOI] [PubMed] [Google Scholar]
- 23.Carvalho L.A., Teng J., Fleming R.L., Tabet E.I., Zinter M., de Melo Reis R.A., Tannous B.A. Olfactory ensheathing cells: a trojan horse for glioma gene therapy. J. Natl. Cancer Inst. 2019;111(3):283–291. doi: 10.1093/jnci/djy138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q., Qin Q., Sun H., Zhong D., An R., Tian Y., Chen H., Jin J., Wang H., Li G. Neuroprotective effect of olfactory ensheathing cells co-transfected with Nurr1 and Ngn2 in both in vitro and in vivo models of Parkinson’s disease. Life Sci. 2018;194:168–176. doi: 10.1016/j.lfs.2017.12.038. [DOI] [PubMed] [Google Scholar]
- 25.Reshamwala R., Shah M., St John J., Ekberg J. Survival and integration of transplanted olfactory ensheathing cells are crucial for spinal cord injury repair: Insights from the last 10 years of animal mdel studies. Cell Transplant. 2019;28(1_suppl):132S–159S. doi: 10.1177/0963689719883823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaborowski M.P., Balaj L., Breakefield X.O., Lai C.P. Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience. 2015;65(8):783–797. doi: 10.1093/biosci/biv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caruso Bavisotto C., Scalia F., Marino Gammazza A., Carlisi D., Bucchieri F., Conway de Macario E., Macario A.J.L., Cappello F., Campanella C. Extracellular vesicle-mediated cell-cell communication in the nervous system: focus on neurological diseases. Int. J. Mol. Sci. 2019;20(2):434. doi: 10.3390/ijms20020434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frühbeis C., Fröhlich D., Krämer-Albers E-M. Emerging roles of exosomes in neuron-glia communication. Front. Physiol. 2012;3:119–119. doi: 10.3389/fphys.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potolicchio I., Carven G.J., Xu X., Stipp C., Riese R.J., Stern L.J., Santambrogio L. Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J. Immunol. 2005;175(4):2237–2243. doi: 10.4049/jimmunol.175.4.2237. [DOI] [PubMed] [Google Scholar]
- 30.Krämer-Albers E.M., Bretz N., Tenzer S., Winterstein C., Möbius W., Berger H., Nave K.A., Schild H., Trotter J. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: Trophic support for axons? Proteomics Clin. Appl. 2007;1(11):1446–1461. doi: 10.1002/prca.200700522. [DOI] [PubMed] [Google Scholar]
- 31.Frühbeis C., Fröhlich D., Kuo W.P., Krämer-Albers E.M. Extracellular vesicles as mediators of neuron-glia communication. Front. Cell. Neurosci. 2013;7:182. doi: 10.3389/fncel.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pegtel D.M., Peferoen L., Amor S. Extracellular vesicles as modulators of cell-to-cell communication in the healthy and diseased brain. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369(1652):369. doi: 10.1098/rstb.2013.0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xin H., Li Y., Cui Y., Yang J.J., Zhang Z.G., Chopp M. Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. J. Cereb. Blood Flow Metab. 2013;33(11):1711–1715. doi: 10.1038/jcbfm.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rong Y., Liu W., Wang J., Fan J., Luo Y., Li L., Kong F., Chen J., Tang P., Cai W. Neural stem cell-derived small extracellular vesicles attenuate apoptosis and neuroinflammation after traumatic spinal cord injury by activating autophagy. Cell Death Dis. 2019;10(5):340. doi: 10.1038/s41419-019-1571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W., Wang Y., Gong F., Rong Y., Luo Y., Tang P., Zhou Z., Zhou Z., Xu T., Jiang T., Yang S., Yin G., Chen J., Fan J., Cai W. Exosomes derived from bone mesenchymal stem cells repair traumatic spinal cord injury by suppressing the activation of a1 neurotoxic reactive astrocytes. J. Neurotrauma. 2019;36(3):469–484. doi: 10.1089/neu.2018.5835. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Chopp M., Liu X.S., Katakowski M., Wang X., Tian X., Wu D., Zhang Z.G. Exosomes derived from mesenchymal stromal cells promote axonal growth of cortical neurons. Mol. Neurobiol. 2017;54(4):2659–2673. doi: 10.1007/s12035-016-9851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tassew N.G., Charish J., Shabanzadeh A.P., Luga V., Harada H., Farhani N., D’Onofrio P., Choi B., Ellabban A., Nickerson P.E.B., Wallace V.A., Koeberle P.D., Wrana J.L., Monnier P.P. Exosomes mediate mobilization of autocrine Wnt10b to promote axonal regeneration in the injured CNS. Cell Rep. 2017;20(1):99–111. doi: 10.1016/j.celrep.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Huang H., Xi H., Chen L., Zhang F., Liu Y. Long-term outcome of olfactory ensheathing cell therapy for patients with complete chronic spinal cord injury. Cell Transplant. 2012;21(Suppl. 1):S23–S31. doi: 10.3727/096368912X633734. [DOI] [PubMed] [Google Scholar]
- 39.Xia B., Gao J., Li S., Huang L., Ma T., Zhao L., Yang Y., Huang J., Luo Z. Extracellular vesicles derived from olfactory ensheathing cells promote peripheral nerve regeneration in rats. Front. Cell. Neurosci. 2019;13:548. doi: 10.3389/fncel.2019.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu Y-K., Hsueh Y-H. Extracellular vesicles isolated from human olfactory ensheathing cells enhance the viability of neural progenitor cells. Neurol. Res. 2020;42(11):959–967. doi: 10.1080/01616412.2020.1794371. [DOI] [PubMed] [Google Scholar]
- 41.Ge X., Guo M., Hu T., Li W., Huang S., Yin Z., Li Y., Chen F., Zhu L., Kang C., Jiang R., Lei P., Zhang J. Increased microglial exosomal miR-124-3p alleviates neurodegeneration and improves cognitive outcome after rmTBI. Mol. Ther. 2020;28(2):503–522. doi: 10.1016/j.ymthe.2019.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460-6. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang L., Zhou G., Soufan O., Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48(W1):W244–W251. doi: 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji F., Lv X., Jiao J. The role of microRNAs in neural stem cells and neurogenesis. J. Genet. Genomics. 2013;40(2):61–66. doi: 10.1016/j.jgg.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Ning X-J., Lu X-H., Luo J-C., Chen C., Gao Q., Li Z-Y., Wang H. Molecular mechanism of microRNA-21 promoting Schwann cell proliferation and axon regeneration during injured nerve repair. RNA Biol. 2020;17(10):1508–1519. doi: 10.1080/15476286.2020.1777767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z., Yuan Y., Zhang Z., Ding K. Inhibition of miRNA-27b enhances neurogenesis via AMPK activation in a mouse ischemic stroke model. FEBS Open Bio. 2019;9(5):859–869. doi: 10.1002/2211-5463.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho J.A., Park H., Lim E.H., Lee K.W. MicroRNA expression profiling in neurogenesis of adipose tissue-derived stem cells. J. Genet. 2011;90(1):81–93. doi: 10.1007/s12041-011-0041-6. [DOI] [PubMed] [Google Scholar]
- 49.Vandestadt C., Vanwalleghem G.C., Castillo H.A., Li M., Schulze K., Khabooshan M., Don E., Anko M-L., Scott E.K., Kaslin J. Early migration of precursor neurons initiates cellular and functional regeneration after spinal cord injury in zebrafish. BioRxiv. 2019:539940. doi: 10.1101/539940. [DOI]
- 50.Oprych K., Cotfas D., Choi D. Common olfactory ensheathing glial markers in the developing human olfactory system. Brain Struct. Funct. 2017;222(4):1877–1895. doi: 10.1007/s00429-016-1313-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holbrook E.H., Rebeiz L., Schwob J.E. Office-based olfactory mucosa biopsies. Int. Forum Allergy Rhinol. 2016;6(6):646–653. doi: 10.1002/alr.21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim H., Ko Y., Park H., Zhang H., Jeong Y., Kim Y., Noh M., Park S., Kim Y-M., Kwon Y-G. MicroRNA-148a/b-3p regulates angiogenesis by targeting neuropilin-1 in endothelial cells. Exp. Mol. Med. 2019;51(11):1–11. doi: 10.1038/s12276-019-0344-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang W., Dong J., Wang M., Yao S., Tian X., Cui X., Fu S., Zhang S. miR-148a-3p suppresses epithelial ovarian cancer progression primarily by targeting c-Met. Oncol. Lett. 2018;15(5):6131–6136. doi: 10.3892/ol.2018.8110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Leidinger P., Backes C., Deutscher S., Schmitt K., Mueller S.C., Frese K., Haas J., Ruprecht K., Paul F., Stähler C., Lang C.J.G., Meder B., Bartfai T., Meese E., Keller A. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013;14(7):R78–R78. doi: 10.1186/gb-2013-14-7-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gámez-Valero A., Campdelacreu J., Vilas D., Ispierto L., Reñé R., Álvarez R., Armengol M.P., Borràs F.E., Beyer K. Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl. Neurodegener. 2019;8(1):31. doi: 10.1186/s40035-019-0169-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Y., Wu Y., Li N., Wang Z. Up-regulation of miRNA-151-3p enhanced the neuroprotective effect of dexmedetomidine against β-amyloid by targeting DAPK-1 and TP53. Exp. Mol. Pathol. 2021;118:104587. doi: 10.1016/j.yexmp.2020.104587. [DOI] [PubMed] [Google Scholar]
- 57.Xu H., Liu C., Zhang Y., Guo X., Liu Z., Luo Z., Chang Y., Liu S., Sun Z., Wang X. Let-7b-5p regulates proliferation and apoptosis in multiple myeloma by targeting IGF1R. Acta Biochim. Biophys. Sin. (Shanghai) 2014;46(11):965–972. doi: 10.1093/abbs/gmu089. [DOI] [PubMed] [Google Scholar]
- 58.Petri R., Pircs K., Jönsson M.E., Åkerblom M., Brattås P.L., Klussendorf T., Jakobsson J. let-7 regulates radial migration of new-born neurons through positive regulation of autophagy. EMBO J. 2017;36(10):1379–1391. doi: 10.15252/embj.201695235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao C., Sun G., Li S., Lang M.F., Yang S., Li W., Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. USA. 2010;107(5):1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Long K.R., Huttner W.B. How the extracellular matrix shapes neural development. Open Biol. 2019;9(1):180216–180216. doi: 10.1098/rsob.180216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scott-Solomon E., Kuruvilla R. Mechanisms of neurotrophin trafficking via Trk receptors. Mol. Cell. Neurosci. 2018;91:25–33. doi: 10.1016/j.mcn.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eroglu C. The role of astrocyte-secreted matricellular proteins in central nervous system development and function. J. Cell Commun. Signal. 2009;3(3-4):167–176. doi: 10.1007/s12079-009-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Christopherson K.S., Ullian E.M., Stokes C.C., Mullowney C.E., Hell J.W., Agah A., Lawler J., Mosher D.F., Bornstein P., Barres B.A. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120(3):421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 64.Lawler J. Thrombospondin-1 as an endogenous inhibitor of angiogenesis and tumor growth. J. Cell. Mol. Med. 2002;6(1):1–12. doi: 10.1111/j.1582-4934.2002.tb00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bender H.R., Campbell G.E., Aytoda P., Mathiesen A.H., Duffy D.M. Thrombospondin 1 (THBS1) promotes follicular angiogenesis, luteinization, and ovulation in primates. Front. Endocrinol. (Lausanne) 2019;10:727–727. doi: 10.3389/fendo.2019.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stein E.V., Miller T.W., Ivins-O’Keefe K., Kaur S., Roberts D.D. Secreted thrombospondin-1 regulates macrophage interleukin-1β production and activation through CD47. Sci. Rep. 2016;6(1):19684. doi: 10.1038/srep19684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bray E.R., Yungher B.J., Levay K., Ribeiro M., Dvoryanchikov G., Ayupe A.C., Thakor K., Marks V., Randolph M., Danzi M.C., Schmidt T.M., Chaudhari N., Lemmon V.P., Hattar S., Park K.K. Thrombospondin-1 mediates axon regeneration in retinal ganglion cells. Neuron. 2019;103(4):642–657.e7. doi: 10.1016/j.neuron.2019.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daubon T., Léon C., Clarke K., Andrique L., Salabert L., Darbo E., Pineau R., Guérit S., Maitre M., Dedieu S., Jeanne A., Bailly S., Feige J-J., Miletic H., Rossi M., Bello L., Falciani F., Bjerkvig R., Bikfalvi A. Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat. Commun. 2019;10(1):1146. doi: 10.1038/s41467-019-08480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beukelaers P., Vandenbosch R., Caron N., Nguyen L., Belachew S., Moonen G., Kiyokawa H., Barbacid M., Santamaria D., Malgrange B. Cdk6-dependent regulation of G(1) length controls adult neurogenesis. Stem Cells. 2011;29(4):713–724. doi: 10.1002/stem.616. [DOI] [PubMed] [Google Scholar]
- 70.Choudhury A.R., Ju Z., Djojosubroto M.W., Schienke A., Lechel A., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J., Lee H-W., von Zglinicki T., Ganser A., Schirmacher P., Nakauchi H., Rudolph K.L. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat. Genet. 2007;39(1):99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- 71.Sherr C.J. D-type cyclins. Trends Biochem. Sci. 1995;20(5):187–190. doi: 10.1016/S0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 72.Hamilton L.K., Truong M.K., Bednarczyk M.R., Aumont A., Fernandes K.J. Cellular organization of the central canal ependymal zone, a niche of latent neural stem cells in the adult mammalian spinal cord. Neuroscience. 2009;164(3):1044–1056. doi: 10.1016/j.neuroscience.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 73.Mothe A.J., Tator C.H. Proliferation, migration, and differentiation of endogenous ependymal region stem/progenitor cells following minimal spinal cord injury in the adult rat. Neuroscience. 2005;131(1):177–187. doi: 10.1016/j.neuroscience.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 74.Mao Y., Nguyen T., Sutherland T., Gorrie C.A. Endogenous neural progenitor cells in the repair of the injured spinal cord. Neural Regen. Res. 2016;11(7):1075–1076. doi: 10.4103/1673-5374.187035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu P., Woodruff G., Wang Y., Graham L., Hunt M., Wu D., Boehle E., Ahmad R., Poplawski G., Brock J., Goldstein L.S., Tuszynski M.H. Long-distance axonal growth from human induced pluripotent stem cells after spinal cord injury. Neuron. 2014;83(4):789–796. doi: 10.1016/j.neuron.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bonner J.F., Connors T.M., Silverman W.F., Kowalski D.P., Lemay M.A., Fischer I. Grafted neural progenitors integrate and restore synaptic connectivity across the injured spinal cord. J. Neurosci. 2011;31(12):4675–4686. doi: 10.1523/JNEUROSCI.4130-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kadoya K., Lu P., Nguyen K., Lee-Kubli C., Kumamaru H., Yao L., Knackert J., Poplawski G., Dulin J.N., Strobl H., Takashima Y., Biane J., Conner J., Zhang S.C., Tuszynski M.H. Spinal cord reconstitution with homologous neural grafts enables robust corticospinal regeneration. Nat. Med. 2016;22(5):479–487. doi: 10.1038/nm.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kumamaru H., Kadoya K., Adler A.F., Takashima Y., Graham L., Coppola G., Tuszynski M.H. Generation and post-injury integration of human spinal cord neural stem cells. Nat. Methods. 2018;15(9):723–731. doi: 10.1038/s41592-018-0074-3. [DOI] [PubMed] [Google Scholar]
- 79.Pei X., Li Y., Zhu L., Zhou Z. Astrocyte-derived exosomes suppress autophagy and ameliorate neuronal damage in experimental ischemic stroke. Exp. Cell Res. 2019;382(2):111474. doi: 10.1016/j.yexcr.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 80.Gage F.H. Neurogenesis in the adult brain. J. Neurosci. 2002;22(3):612–613. doi: 10.1523/JNEUROSCI.22-03-00612.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hatten M.E. Central nervous system neuronal migration. Annu. Rev. Neurosci. 1999;22(1):511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- 82.Arvidsson A., Collin T., Kirik D., Kokaia Z., Lindvall O. Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat. Med. 2002;8(9):963–970. doi: 10.1038/nm747. [DOI] [PubMed] [Google Scholar]
- 83.Iwai M., Sato K., Kamada H., Omori N., Nagano I., Shoji M., Abe K. Temporal profile of stem cell division, migration, and differentiation from subventricular zone to olfactory bulb after transient forebrain ischemia in gerbils. J. Cereb. Blood Flow Metab. 2003;23(3):331–341. doi: 10.1097/01.WCB.0000050060.57184.E7. [DOI] [PubMed] [Google Scholar]
- 84.Fricker R.A., Carpenter M.K., Winkler C., Greco C., Gates M.A., Björklund A. Site-specific migration and neuronal differentiation of human neural progenitor cells after transplantation in the adult rat brain. J. Neurosci. 1999;19(14):5990–6005. doi: 10.1523/JNEUROSCI.19-14-05990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Z., Kato T., Kawagishi K., Fukushima N., Yokouchi K., Moriizumi T. Cell dynamics of calretinin-immunoreactive neurons in the rostral migratory stream after ibotenate-induced lesions in the forebrain. Neurosci. Res. 2002;42(2):123–132. doi: 10.1016/S0168-0102(01)00314-5. [DOI] [PubMed] [Google Scholar]
- 86.Yao L., Li Y. The role of direct current electric field-guided stem cell migration in neural regeneration. Stem Cell Rev. Rep. 2016;12(3):365–375. doi: 10.1007/s12015-016-9654-8. [DOI] [PubMed] [Google Scholar]
- 87.Tator C.H., Koyanagi I. Vascular mechanisms in the pathophysiology of human spinal cord injury. J. Neurosurg. 1997;86(3):483–492. doi: 10.3171/jns.1997.86.3.0483. [DOI] [PubMed] [Google Scholar]
- 88.Lou J., Lenke L.G., Ludwig F.J., O’Brien M.F. Apoptosis as a mechanism of neuronal cell death following acute experimental spinal cord injury. Spinal Cord. 1998;36(10):683–690. doi: 10.1038/sj.sc.3100632. [DOI] [PubMed] [Google Scholar]
- 89.Liu X.Z., Xu X.M., Hu R., Du C., Zhang S.X., McDonald J.W., Dong H.X., Wu Y.J., Fan G.S., Jacquin M.F., Hsu C.Y., Choi D.W. Neuronal and glial apoptosis after traumatic spinal cord injury. J. Neurosci. 1997;17(14):5395–5406. doi: 10.1523/JNEUROSCI.17-14-05395.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu S., Yao S., Wen Y., Wang Y., Wang H., Xu Q. Angiogenic microspheres promote neural regeneration and motor function recovery after spinal cord injury in rats. Sci. Rep. 2016;6(1):33428. doi: 10.1038/srep33428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author [YKT] upon reasonable request.