Abstract
Shoot apical meristem (SAM) and root apical meristem (RAM) homeostasis is tightly regulated by CLAVATA3 (CLV3)/EMBRYO SURROUNDING REGION-related (CLE) peptide signaling. However, the intracellular signaling components after CLV3 is perceived by the CLV1–CLV3-INSENSITIVE KINASE (CIK) receptor complex and CLE25/26/45 are sensed by the BARELY ANY MERISTEM (BAM)–CIK receptor complex are unknown. Here, we report that PBS1-LIKE34/35/36 (PBL34/35/36), a clade of receptor-like cytoplasmic kinases, are required for both CLV3-mediated signaling in the SAM and CLE25/26/45-mediated signaling in the RAM. Physiological assays showed that the SAM and RAM of pbl34 pbl35 pbl36 were resistant to CLV3 and CLE25/26/45 treatment, respectively. Genetic analyses indicated that pbl34 pbl35 pbl36 greatly enhanced the SAM defects of clv2 and rpk2 but not clv1, and did not show additive effects with bam3 and cik2 in the RAM. Further biochemical assays revealed that PBL34/35/36 interacted with CLV1, BAM1/3, and CIKs, and were phosphorylated by CLV1 and BAM1. All these results suggest that PBL34/35/36 act downstream of CLV1 and BAM1/3 to mediate the CLV3 and CLE25/26/45 signals in maintaining SAM and RAM homeostasis, respectively. Our findings shed light on how CLE signals are transmitted intracellularly after being perceived by cell surface receptor complexes.
Introduction
Plant cells constantly communicate with their neighboring cells, allowing normal development and responses to ever-changing environmental conditions. Peptide hormones and cell surface-localized receptor-like protein kinases (RLKs) are crucial mediators of these communications (Olsson et al., 2019). The shoot apical meristem (SAM) and root apical meristem (RAM), which are responsible for morphogenesis of the aboveground and underground plant parts, respectively, also recruit peptide–RLK receptor pairs to maintain their homeostasis (Kitagawa and Jackson, 2019).
In the SAM, the stem cell-derived peptide CLAVATA 3 (CLV3) diffuses into the organization center (OC) to repress the expression of WUSCHEL (WUS; Brand et al., 2000; Schoof et al., 2000). WUS, a key regulator maintaining the shoot stem cell niche, acts in a non-cell-autonomous manner to activate CLV3 expression in a threshold-dependent manner (Perales et al., 2016; Rodriguez et al., 2016). Thus, the CLV–WUS negative-feedback loop directs the balance between cell self-renewal and cell differentiation (Somssich et al., 2016). CLV3 has been proposed to be perceived by three different receptors, including CLV1, CLV2, and RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2; Clark et al., 1997; Kayes and Clark, 1998; Guo et al., 2010; Kinoshita et al., 2010). All of these receptors employ CLV3-INSENSITIVE KINASES (CIKs) as co-receptors to limit SAM size (Hu et al., 2018; Gou and Li, 2020). BARELY ANY MERISTEMS (BAMs), the homologs of CLV1, are ectopically expressed in the rib meristem (RM) in the clv1 mutant to partially compensate for the loss-of-function of CLV1, indicating that BAMs and CLV1 act as redundant receptors in regulating SAM homeostasis (Deyoung and Clark, 2008; Nimchuk et al., 2015).
Genetic evidence indicates that CLV1, CLV2, and RPK2 act in three parallel pathways transducing the CLV3 signal to regulate SAM homeostasis, implying that these three receptors may employ different downstream components to transduce the CLV3 signal into the cytoplasm (Kinoshita et al., 2010). CORYNE (CRN), a receptor-like cytoplasmic kinase (RLCK) that lacks an extracellular domain (Müller et al., 2008), can interact with CLV2, a leucine-rich repeat receptor-like protein (LRR-RLP), via their transmembrane domains to form a complex (Bleckmann et al., 2010). The loss-of-function of CRN can strongly enhance the SAM defects of clv1 and rpk2, but not clv2, indicating that CRN specifically functions in the CLV2 pathway (Müller et al., 2008). Heterotrimeric G proteins play key roles in transmitting signals from RLK receptors into the cytoplasm (Pandey, 2019). Additional mutations of GTP BINDING PROTEIN BETA 1 (AGB1, Gβ) or ARABIDOPSIS G-PROTEIN GAMMA-SUBUNIT 1/2 (AGG1/2, Gγ) do not alter SAM defects of the rpk2 mutant, but significantly enhance the SAM defects of clv1 and clv2. Further biochemical evidence shows that AGB1 physically interacts with RPK2. Therefore, it was proposed that Gβγ and RPK2 regulate SAM maintenance through a common pathway (Ishida et al., 2014). However, the immediate events after CLV3 is recognized by the CLV1–CIK complex are not clear in Arabidopsis.
The CLV–WUS feedback signaling pathway appears to be conserved across angiosperms (Somssich et al., 2016). Maize (Zea mays) THICK TASSEL DWARF 1 (TD1) and FASCIATED EAR 2 (FEA2) are orthologs of Arabidopsis CLV1 and CLV2 (Taguchi-Shiobara et al., 2001; Bommert et al., 2005). FEA3, another LRR-RLP, is expressed in the central region of the SAM to perceive ZmFON2-LIKE CLE PROTEIN 1 (ZmFCP1), the CLV3 ortholog in maize, to regulate SAM homeostasis (Je et al., 2016). Similar to CLV1 and CLV2 in Arabidopsis, TD1 functions in parallel with FEA2 and FEA3 in maize (Je et al., 2016). FEA2 also employs ZmCRN as a downstream effector transducing the ZmFCP1 signal to regulate stem cell niche in maize (Je et al., 2018). Interestingly, maize COMPACT PLANT 2 (CT2, Gα) and ZmGB1 (Gβ) also act downstream of FEA2 to maintain SAM homeostasis by perceiving ZmCLE7, another CLV3 ortholog in maize (Bommert et al., 2013b; Wu et al., 2020). The downstream module of TD1 in maize is still undiscovered.
In addition to the aforementioned regulators, several other essential components involved in stem cell maintenance have been identified. POLTERGEIST (POL) and POL-Like 1 (PLL1), two PP2C phosphatases, function upstream of WUS as negative regulators in the CLV3 signaling pathway. The loss-of-function of POL and PLL1 suppresses SAM defects of the weak clv1 and clv2 mutants, and the pol pll1 double mutant shows phenotypes similar to the wus mutant (Song and Clark, 2005; Song et al., 2006). A recent work showed that POL and PLLs directly interact with and dephosphorylate CLV1 and BAMs (DeFalco et al., 2021). Mitogen-activated protein kinase (MAPK) cascades, which have been proposed to mediate multiple RLK signals in the cytoplasm (Wang and Gou, 2020), are also involved in CLV3-mediated SAM regulation (Betsuyaku et al., 2011b). The CLV3-activated phosphorylation of MPK3 and MPK6 is dependent on CLV1 and BAM1. In addition, the SAM of a conditional mpk3 mpk6 double mutant is insensitive to CLV3 treatment (Lee et al., 2019). However, the MAPK kinases (MKKs) and MKK kinases (MKKKs) acting upstream of MPK3/6, and the signaling elements connecting MKKKs and CLV1/BAM1 receptors in the SAM need further investigation.
Besides functioning in the SAM, CLE peptide-triggered signaling is also crucial for RAM homeostasis. Exogenous applications of Groups A and B CLEs greatly decrease the activity of the proximal root meristem (Ito et al., 2006; Betsuyaku et al., 2011a), possibly due to the suppression of protophloem differentiation (Depuydt et al., 2013; Hazak et al., 2017). Furthermore, many key regulators in the SAM also exhibit conserved functions in the proximal root meristem (Zhu et al., 2020). BAM1/3, the homologs of CLV1, and CIKs form a receptor complex in a ligand-induced manner to regulate CLE25/26/45-mediated suppression of protophloem differentiation and consumption of the proximal root meristem (Depuydt et al., 2013; Anne et al., 2018; Ren et al., 2019; Hu et al., 2022). In addition, CLV2/CRN, a heteromer of CLV2 and CRN, and RPK2 are involved in CLE signaling to regulate homeostasis of the proximal root meristem (Fiers et al., 2005; Müller et al., 2008; Bleckmann et al., 2010; Kinoshita et al., 2010; Gujas et al., 2020). RPK2 also forms a heteromeric complex with BAM1, which is involved in CLE peptide-dependent shrinkage of the proximal root meristem (Shimizu et al., 2015). Although several receptors have been demonstrated to mediate CLE signaling in the RAM, more signaling elements that directly interact with these receptors to transmit the signals into the cytoplasm have not been characterized yet.
To date, it has been revealed that RLCKs are employed as the immediate signaling components acting downstream of RLKs to transduce the corresponding ligand signals. For example, CONSTITUTIVE DIFFERENTIAL GROWTH 1 (CDG1) and the BRASSINOSTEROID (BR)-SIGNALING KINASE (BSK) family members are phosphorylated by BRASSINOSTEROID-INSENSITIVE 1 (BRI1), an LRR type RLK functioning as a major BR receptor, to modulate BR signaling (Tang et al., 2008; Kim et al., 2011; Sreeramulu et al., 2013); BOTRYTIS-INDUCED KINASE1 (BIK1) and its homolog PBS1-LIKE 1 (PBL1) directly interact with various pattern recognition receptors, such as FLAGELLIN SENSING 2 (FLS2), ELONGATION FACTOR-TU (EF-Tu) RECEPTOR (EFR), PEP RECEPTORs (PEPRs), and LYSINE MOTIF RECEPTOR KINASE 5 (LYK5), to regulate plant innate immune responses (Lu et al., 2010a; Zhang et al., 2010; Liu et al., 2013; Kadota et al., 2014; Ranf et al., 2014); RPM1-INDUCED PROTEIN KINASE (RIPK) is required for RAPID ALKALINIZATION FACTOR 1 (RALF1) signaling and functions downstream of FERONIA (FER) to regulate primary root and root hair growth (Du et al., 2016); MARIS (MRI) acts as a key downstream component of ANXUR 1/2 (ANX1/2) and FER to monitor cell wall integrity in pollen tube and root hair (Boisson-Dernier et al., 2015; Liao et al., 2016); CAST AWAY (CST) interacts with both HAESA (HAE) and SUPPRESSOR OF BIR 1 (SOBIR1)/EVERSHED (EVR) to inhibit organ abscission (Burr et al., 2011). It has been accepted as a paradigm that RLK-regulated biological processes require RLCKs to transduce the corresponding signals to downstream signaling components (Liang and Zhou, 2018). However, whether CLV1 and BAM1/3 also need RLCKs to transduce the CLV3 and CLE25/26/45 signals in the SAM and RAM has not been elucidated.
Here, we report that PBL34/35/36, three members of the RLCK VII subfamily required for LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION (LORE)-mediated plant immunity regulation (Luo et al., 2020), play key roles in CLV3-mediated SAM regulation and CLE25/26/45-mediated RAM regulation through acting directly downstream of CLV1 and BAM1/3, respectively. The root length and protophloem of pbl34 pbl35 pbl36 show resistance to CLE45 treatment, which is similar to observations for bam3 and cik2 (Depuydt et al., 2013; Anne et al., 2018; Ren et al., 2019). Moreover, the pbl34 pbl35 pbl36 triple mutants generate enlarged SAMs and extra flower organs, similar to those observed for clv1 (Clark et al., 1997). The loss-of-function of PBL34/35/36 greatly enhances the SAM defects of clv2 and rpk2, but not clv1. Furthermore, PBL34/35/36 can interact with CLV1, BAM1/3, and CIKs both in vivo and in vitro, and can be phosphorylated by CLV1 and BAM1 in vitro. Our studies reveal that PBL34/35/36 act as conserved regulators downstream of CLV1 and BAM1/3 to maintain homeostasis of both the SAM and RAM.
Results
PBL34 may sense CLE45 to regulate root growth
Increasing lines of evidence indicate that RLCKs act as direct components downstream of RLKs to regulate diverse biological processes (Liang and Zhou, 2018), implying that CLV1-mediated SAM homeostasis and BAM1/3-mediated RAM homeostasis may need such RLCKs as well. There are 149 RLCKs that were divided into 17 subgroups based on their sequence homology in Arabidopsis (Shiu et al., 2004). Although the functions of most RLCKs have not been elucidated, some members of the RLCK VII subfamily have been clarified to be involved in plant immunity and development. Several of them act downstream of RLKs to transduce peptide signals including INFLORESCENCE DEFICIENT IN ABSCISSION (IDA; Burr et al., 2011), whose mature peptide is similar to CLE (Zhang et al., 2020). We wondered whether the RLCK VII subfamily members are also required for CLE peptide signaling.
To this end, 28 homozygous T‐DNA insertion lines of the RLCK VII subfamily genes were treated with 20-nM CLE45 on half-strength Murashige–Skoog (MS) medium, and the primary root length was examined at 5 days after germination. We found that the relative root length of pbl34 was significantly longer than that of other rlck vii mutants upon 20-nM CLE45 treatment. To further confirm that pbl34 was resistant to CLE45, different concentrations of CLE45 were applied to pbl34 seedlings. Root growth of the pbl34 single mutant was also less sensitive than the wild-type control to treatment with 100-nM CLE45, indicating that PBL34 may be involved in CLE45 signaling to regulate root growth (Supplemental Figure S1A).
PBL34/35/36 function redundantly to sense root-active CLE peptides
Despite the root of pbl34 seedlings being less sensitive than the wild-type control to CLE45, the treatment still resulted in significantly shortened pbl34 root when compared with the mock treatment (Supplemental Figure S1A), indicating that CLE45 signaling was partially retained in the pbl34 mutant and the homologs of PBL34 may function redundantly to regulate root growth. PBL34 belongs to the RLCK VII-5 subclade that contains two homologs PBL35 and PBL36 (Supplemental Figure S1B and Supplemental Data Set S1; Rao et al., 2018).
To explore the potential functional redundancy of PBL35 and PBL36 with PBL34 in CLE45 signaling, we examined the expression patterns of these PBLs in the RAM using the ProPBL:gPBL-YFP reporter lines. PBL34 is mainly expressed in the stem cell niche and stele in the RAM; PBL35 is mainly expressed in the proximal root meristem, and PBL36 is ubiquitously expressed throughout the RAM (Supplemental Figure S2). Taken together, all three PBL genes are expressed in the RAM, indicating that they may function redundantly to regulate the proximal root meristem.
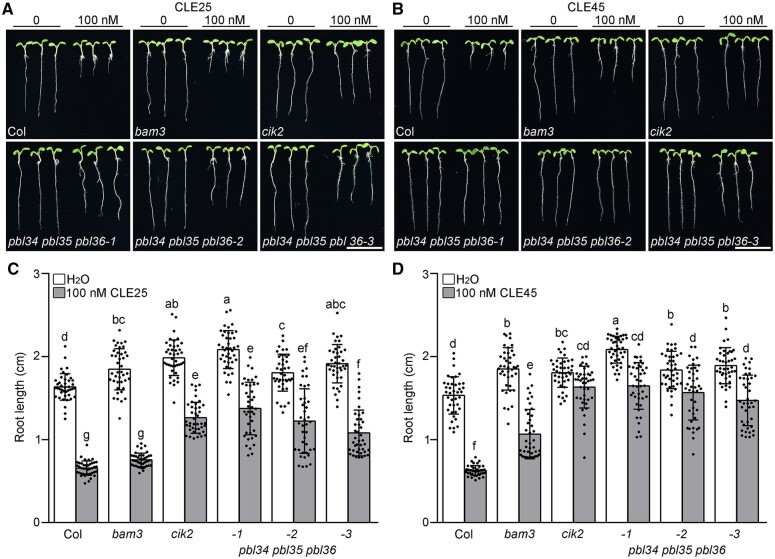
Therefore, the pbl34 pbl36 and pbl35 pbl36 double mutants and three independent pbl34 pbl35 pbl36 triple mutants (Supplemental Figure S3) were generated by genetic cross or clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR‐associate protein 9 (Cas9)-based gene editing for analyzing their responses to CLE treatment. The wild-type plants grown on 1/2 MS medium with 100-nM CLE25 or CLE45 showed significantly shortened roots with a decreased proximal root meristem (Figure 1; Supplemental Figures S1A and S4). When treated with CLE45, the pbl34 pbl36 and pbl35 pbl36 double mutants showed reduced sensitivity than the wild-type control, which was similar to that of pbl34, while the pbl34 pbl35 pbl36-1 (hereafter named pbl34 pbl35 pbl36 for brevity) triple mutants were less sensitive than the other pbl mutants tested (Supplemental Figure S1A), suggesting that PBL34, PBL35, and PBL36 function redundantly to sense CLE signals in regulating root growth and maintaining the proximal root meristem, and that PBL34 plays the most important role in this process. Two other independent pbl34 pbl35 pbl36 triple mutants showed similarly reduced sensitivity when treated with 100-nM CLE25 or CLE45 (Figure 1; Supplemental Figure S4). The complementation of pbl34 pbl35 pbl36-2 by PBL34, PBL35, and PBL36 under a UBQ10 promoter further confirmed that PBL34/35/36 function in CLE-mediated proximal root meristem maintenance and root growth (Supplemental Figure S5A).
Figure 1.
PBL34/35/36 are required for sensing CLE25 and CLE45. A, B, Five-day-old seedlings of Col, bam3, cik2, pbl34 pbl35 pbl36-1, pbl34 pbl35 pbl36-2, and pbl34 pbl35 pbl36-3 grown on 1/2 MS medium with 0-nM, 100-nM CLE25 (A) or CLE45 (B) peptide. Roots of cik2, pbl34 pbl35 pbl36-1, pbl34 pbl35 pbl36-2, and pbl34 pbl35 pbl36-3 showed reduced sensitivity to CLE25 or CLE45 when compared with the wild-type plants. Scale bar: 1 cm. C, D, Quantitative analyses of root length of the indicated genotypes under the conditions shown in (A) and (B). Data are shown as mean ± sd (n ≥ 40). Different lowercase letters indicate statistically significant differences based on two-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test (P < 0.05).
As BAM1/3 and CIKs are required for the sensing of all root-active CLE peptides (Figure 1; Anne et al., 2018; Hu et al., 2018, 2022), which prompted us to investigate whether PBL34/35/36 are also required for the sensing of various CLEs. The results showed that the pbl34 pbl35 pbl36 mutants were not only partially resistant to CLE45, but also partially resistant to all the root-active CLE peptides (Supplemental Figure S6). Taken together, these data demonstrate that PBL34/35/36 function redundantly to regulate CLE-mediated proximal root meristem maintenance.
PBL34/35/36 are required for CLE-mediated suppression of protophloem differentiation in the root
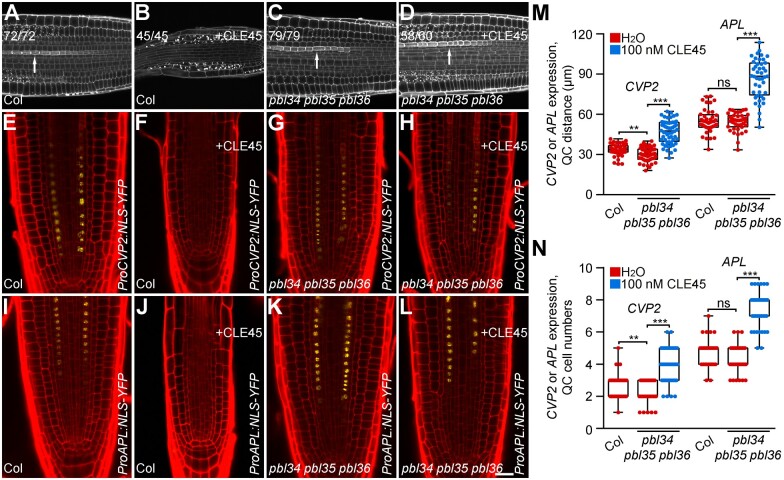
Exogenous application of CLE peptides suppresses the differentiation of protophloem to affect maintenance of the proximal root meristem (Hazak et al., 2017). Protophloem precursor cell differentiation is associated with a thickening of the protophloem cell wall, which can be strongly stained by propidium iodide (PI; Truernit et al., 2008). When the wild-type seedlings were treated with CLE45, the typical cell wall staining of the protophloem cells was abolished. However, the strongly stained cells were clearly observed in almost all roots of pbl34 pbl35 pbl36 (Figure 2, A–D), suggesting that the protophloem cells can be properly differentiated upon CLE45 treatment when the PBL34/35/36-mediated signaling is blocked.
Figure 2.
PBL34/35/36 are required for CLE45-mediated suppression of protophloem differentiation. A–D, The protophloem of 5-day-old seedlings was analyzed by mPS-PI staining. Representative roots of Col (A and B) and pbl34 pbl35 pbl36 (C and D) grown on 1/2 MS medium without (A and C) or with (B and D) 100-nM CLE45 peptide are shown. E–L, Expression of CVP2 and APL was detected by ProCVP2:NLS-YFP and ProAPL:NLS-YFP reporter lines, respectively, in Col (E, F, I, and J) and pbl34 pbl35 pbl36 (G, H, K, and L) treated with (F, H, J, and L) or without (E, G, I, and K) 100-nM CLE45. The YFP fluorescence signals (yellow) in 5-day-old seedlings were observed using confocal laser scanning microscopy after PI staining (red). Scale bar: 50 µm. M and N, Quantitative analyses of the distance and cell number between the first cell with detectable CVP2 or APL expression and the QC position in the indicated genotypes. Box limits indicate 25th and 75th percentiles; horizontal line is the median; whiskers display minimum and maximum values. Dots represent individual measurements from more than 40 samples per group. ***P < 0.001, **P < 0.01, ns represents no significant difference (two-tailed Student’s t test).
To further confirm that the protophloem is still retained in the pbl34 pbl35 pbl36 mutants, promoters of COTYLEDON VASCULAR PATTERN 2 (CVP2; Rodriguez-Villalon et al., 2015) and ALTERED PHLOEM DEVELOPMENT (APL; Bonke et al., 2003), two protophloem markers, were cloned to drive the expression of a nuclear-localized yellow fluorescent protein (YFP) in the wild-type and pbl34 pbl35 pbl36 mutant background. Interestingly, CVP2 was expressed earlier in the pbl34 pbl35 pbl36 mutants than in the wild-type plants, indicating that the protophloem differentiation may be premature in pbl34 pbl35 pbl36 (Figure 2, E, G, M, and N). Consistent with the previous results, the expression of CVP2 and APL was not detected in the wild-type plants upon CLE45 treatment, while the fluorescence signal was still observed in the pbl34 pbl35 pbl36 mutants upon CLE45 treatment (Figure 2, E–L), indicating that PBL34/35/36 are essential for CLE45-mediated suppression of protophloem differentiation. Although CVP2 and APL are still expressed in the pbl34 pbl35 pbl36 mutants upon CLE45 treatment, the distance between cells with CVP2 or APL expression and the quiescent center (QC) is markedly farther (Figure 2, E–N). These results suggest that CLE signaling is greatly impaired but not completely blocked in the pbl34 pbl35 pbl36 mutants. It is possible that, besides PBL34/35/36, other PBL homologs can partially compensate for the loss-of-function of PBL34/35/36.
PBL34/35/36 act in the CLE–BAM–CIK signaling pathway to regulate proximal root meristem homeostasis
It was reported that the CLE–BAM–CIK signaling module is involved in regulating protophloem differentiation and proximal root meristem (Hu et al., 2022). As described above, since PBL34/35/36 are required for CLE sensing, we wondered whether PBL34/35/36 act in the same genetic pathway with BAM–CIK. Therefore, we compared the CLE resistances of bam3 pbl34 pbl35 pbl36 with bam3 and pbl34 pbl35 pbl36, and cik2 pbl34 pbl35 pbl36 with cik2 and pbl34 pbl35 pbl36. The results showed that bam3 pbl34 pbl35 pbl36 and cik2 pbl34 pbl35 pbl36 did not show significantly additive effects when compared with the corresponding single mutants and pbl34 pbl35 pbl36, suggesting that PBL34/35/36 might act in the same pathway with BAM–CIK (Supplemental Figure S7).
BREVIS RADIX (BRX) and OCTOPUS (OPS) have been reported to be required for proper differentiation of protophloem and maintenance of the proximal root meristem (Truernit et al., 2012; Rodriguez-Villalon et al., 2014). The short-root phenotype of brx and ops, which may be due to the amplified CLE signaling, can be largely rescued by bam3 and cik2 cik3 cik4 cik5 cik6 (Depuydt et al., 2013; Rodriguez-Villalon et al., 2014; Breda et al., 2019; Hu et al., 2022). To further investigate the roles of PBL34/35/36 in CLE25/26/45 signaling, additional genetic analyses were carried out to elucidate whether PBL34/35/36 function together with BRX and OPS. The results showed that pbl34 pbl35 pbl36 partially rescued the short-root phenotype of brx and ops since the brx pbl34 pbl35 pbl36 and ops pbl34 pbl35 pbl36 quadruple mutants exhibited roots that were slightly longer than those of the brx and ops mutants (Supplemental Figure S8), suggesting that PBL34/35/36 may be involved in BRX and OPS signaling. It is notable that the bam3 and cik2 cik3 cik4 cik5 cik6 mutations can greatly rescue the short-root phenotype of the brx and ops mutants (Depuydt et al., 2013; Rodriguez-Villalon et al., 2014), which is similar to but more significant than the effect of the pbl34 pbl35 pbl36 triple mutant, suggesting that additional homologs of PBL34/35/36 may still function to mediate the root-active CLE signals. Taken together, these results suggest that PBL34/35/36, possibly with their homologs, may function in the BAM–CIK signaling pathway to regulate the proximal root meristem.
SAM homeostasis is disturbed in the pbl34 pbl35 pbl36 mutant
It was reported that SAM and RAM share many common or similar regulators to maintain their homeostasis (Zhu et al., 2020). To explore whether PBL34/35/36 also regulate SAM homeostasis, we first detected the expression patterns of PBL34/35/36 in the SAM using the ProPBL:gPBL-YFP reporter lines. The fluorescent reporters revealed that PBL34/35/36 are widely expressed in the inflorescence, including the inflorescence meristems (IMs), flower primordia, and flower meristems. In the IM, PBL34/35/36 are expressed in the central zone (CZ), peripheral zone, and OC (Supplemental Figure S9), overlapping with the expression of CLV1 and CIKs (Hu et al., 2018; Blümke et al., 2021).
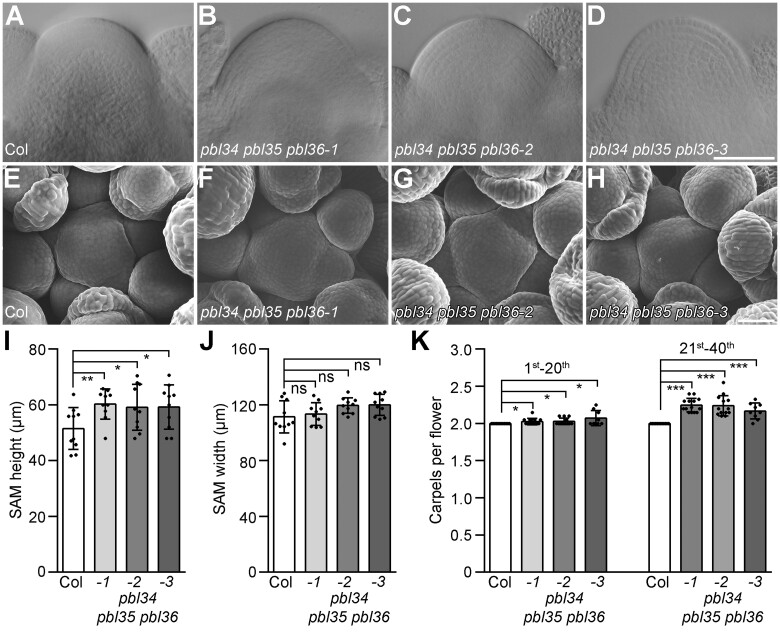
We therefore examined the SAM size, IM morphology, and carpel numbers of the pbl34 pbl35 pbl36 mutants. Results showed that the SAM heights of three independent pbl34 pbl35 pbl36 triple mutants are slightly greater than those of the wild-type plants, and the SAM widths are indistinguishable from those of the wild-type plants (Figure 3, A–D, I, and J). Although the IM morphology is not strongly disturbed in the pbl34 pbl35 pbl36 mutants (Figure 3, E–H), three independent pbl34 pbl35 pbl36 triple mutants develop flowers with increased numbers of carpels relative to the wild-type flower, which produces two carpels. Reintroduction of PBL34, PBL35, and PBL36 into the pbl34 pbl35 pbl36-2 mutants can completely suppress the extra carpels of the pbl34 pbl35 pbl36-2 mutants (Supplemental Figure S5B). Interestingly, siliques at the top part of the inflorescence have more carpels than those at the basal part (Figure 3K) indicating that SAM homeostasis is disrupted and the defects are enhanced over time in the pbl34 pbl35 pbl36 mutants.
Figure 3.
The pbl34 pbl35 pbl36 triple mutant shows an enlarged SAM. A–D, DIC images of SAM from 19-day-old seedlings of Col (A), pbl34 pbl35 pbl36-1 (B), pbl34 pbl35 pbl36-2 (C), and pbl34 pbl35 pbl36-3 (D). Scale bar: 50 µm. E–H, SEM images of 5-week-old IM in Col (E), pbl34 pbl35 pbl36-1 (F), pbl34 pbl35 pbl36-2 (G), and pbl34 pbl35 pbl36-3 (H). Scale bar: 50 µm. I–K, Quantitative analysis of SAM height (I), width (J), and carpel number in the 1st–20th and 21st–40th siliques (K). Data are shown as mean ± sd (n = 10 independent plants, each sample represents the average carpel number of the first (left) or second (right) 20 siliques of each plant.). ***P < 0.001, **P < 0.01, *P < 0.05, ns represents no significant difference (two-tailed Student’s t test).
To further confirm that SAM homeostasis is disrupted in the pbl34 pbl35 pbl36 mutants, the expression patterns of CLV3 and WUS, two marker genes expressed in the stem cells and OC of the SAM, respectively, were analyzed by RNA in situ hybridization. In the wild-type plants, CLV3 is exclusively expressed as a small triangle in the CZ. However, the expression domain of CLV3 is obviously expanded in the SAM in all three independent pbl34 pbl35 pbl36 triple mutants. The expression levels of CLV3 are also dramatically increased in the pbl34 pbl35 pbl36 mutants (Figure 4, A–D). In the wild-type plants, WUS is expressed in a small group of cells (11.1 ± 1.2, n = 8). In the pbl34 pbl35 pbl36 mutants, the expression of WUS is markedly expanded into more cells (17 ± 2.5, n = 7), although the expression level is not significantly increased (Figure 4, E–H). These SAM defects in the pbl34 pbl35 pbl36 mutants, including enlarged SAM, siliques with extra carpels, and enhanced and expanded expression of CLV3 and WUS, are reminiscent of the clv mutants (Clark et al., 1997; Kayes and Clark, 1998), suggesting that the CLV–WUS negative-feedback loop is disrupted in the pbl34 pbl35 pbl36 mutant.
Figure 4.
PBL34/35/36 function in the CLV3-WUS pathway to maintain SAM homeostasis. A–H, RNA in situ hybridization showing the expression patterns of CLV3 (A–D) and WUS (E–H) in the IMs of Col (A and E), pbl34 pbl35 pbl36-1 (B and F), pbl34 pbl35 pbl36-2 (C and G), and pbl34 pbl35 pbl36-3 (D and H). Scale bar: 30 µm. The cells that were marked by antisense probe of WUS are indicated with dotted black lines. I–L, Four-week-old seedlings of Col (I and J) and pbl34 pbl35 pbl36 (K and L) grown on MS medium without (I and K) or with (J and L) 5 µM CLV3. Scale bar: 5 mm. White arrows indicate the bolting inflorescence stem, and the red arrow indicates the terminated SAM. M–P, SEM images of 30-day-old SAMs from Col (M), pbl34 pbl35 pbl36 (N), wus (O), and wus pbl34 pbl35 pbl36 (P). Scale bars: 100 µm (M and N) and 500 µm (O and P).
PBL34/35/36 are involved in the CLV3–WUS pathway to maintain SAM homeostasis
The above results indicated that PBL34/35/36 are involved in regulating SAM homeostasis possibly through the CLV–WUS pathway. Since PBL34/35/36 are members of the RLCK family, they may be involved in sensing the CLV3 signal in the SAM. Therefore, both the wild-type plants and the pbl34 pbl35 pbl36 mutants were treated with 5-µM synthesized CLV3 to examine their sensitivities. Consistent with previous reports, the wild-type seedlings displayed terminated SAMs and did not generate inflorescence stems when they were grown on MS medium containing 5-µM CLV3 for 4 weeks. However, about 60% of the pbl34 pbl35 pbl36 mutants still retained the SAM and generated an inflorescence stem under the same growth conditions (Figure 4, I–L; Supplemental Figure S10). To elucidate the differential contributions of each PBL for maintaining SAM homeostasis, all single and double mutants of PBL34/35/36 were further examined to analyze their sensitivities to CLV3. The results showed that all single mutants exhibited responses similar to the wild-type plants, while about 3% of the pbl35 pbl36 mutants, 6% of the pbl34 pbl36 mutants, and 24% of the pbl34 pbl35 mutants exhibited resistance to CLV3 treatment and generated inflorescence stems (Supplemental Figure S10). Taken together, these results demonstrate that PBL34/35/36 are redundantly required for CLV3-mediated SAM homeostasis, and that PBL34 may play a major role in this process.
WUS, which acts downstream of CLV3, is necessary and sufficient to determine stem cell identity (Gallois et al., 2004). Disruption of WUS function leads to the loss of stem cells in the SAM (Mayer et al., 1998). To determine the genetic relationship between PBL34/35/36 and WUS, a wus pbl34 pbl35 pbl36 quadruple mutant was generated to examine its phenotypes in the meristem. We found that the wus pbl34 pbl35 pbl36 mutant exhibits premature meristem termination, which is similar to the wus mutant (Figure 4, M–P), suggesting that PBL34/35/36 act upstream of WUS to maintain SAM homeostasis.
PBL34/35/36 function in the CLV1 pathway to regulate SAM homeostasis
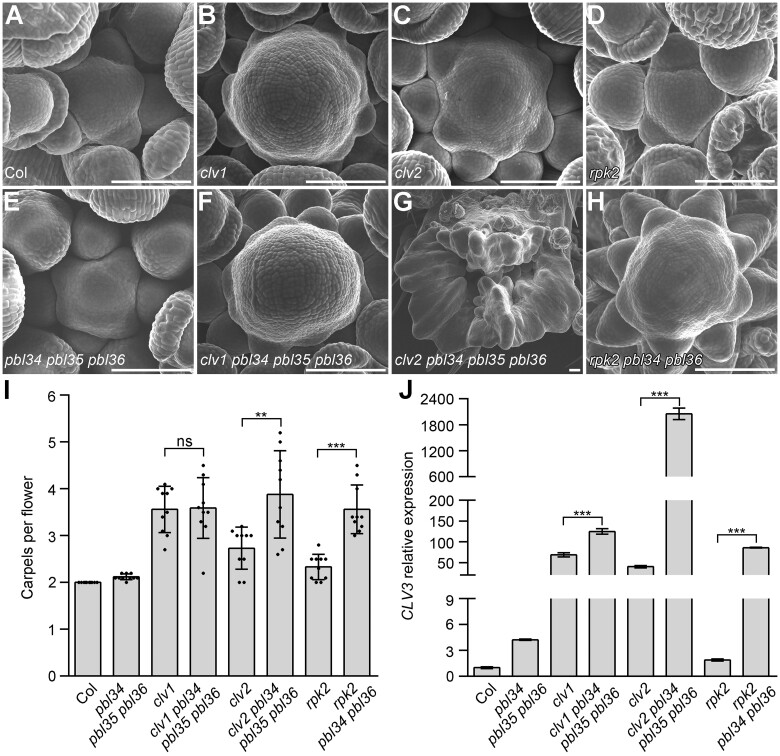
It has been demonstrated that the CLV3 signal is transduced by three parallel pathways mediated by CLV1, CLV2/CRN, and RPK2 to maintain SAM homeostasis (Clark et al., 1997; Kayes and Clark, 1998; Müller et al., 2008; Kinoshita et al., 2010; Kitagawa and Jackson, 2019). Our results suggest that PBL34/35/36 are involved in CLV3-mediated SAM regulation. To investigate whether PBL34/35/36 are required by these three signaling pathways to regulate SAM homeostasis, we created the quadruple mutants clv1 pbl34 pbl35 pbl36, clv2 pbl34 pbl35 pbl36, and triple mutant rpk2 pbl34 pbl36 due to the genetic linkage between RPK2 and PBL35, to examine the SAM-related phenotypes. Surprisingly, rosettes of the clv1 pbl34 pbl35 pbl36 quadruple mutants do not display significant morphological alterations when compared with those of pbl34 pbl35 pbl36 and clv1 (Supplemental Figure S11, B, E, and F). In addition, the clv1 pbl34 pbl35 pbl36 quadruple mutant and the clv1 single mutant show discontinuous flower outgrowth indistinguishable from each other (Supplemental Figure S11, J and N). However, the clv2 pbl34 pbl35 pbl36 and rpk2 pbl34 pbl36 mutants produce significantly more rosette leaves when compared with clv2 and rpk2, respectively (Supplemental Figure S11, C, D, G, and H). Moreover, the clv2 pbl34 pbl35 pbl36 quadruple mutants produce a fasciated inflorescence stem with many more flowers than the clv2 and pbl34 pbl35 pbl36 mutants (Supplemental Figure S11, K, M, and O). The rpk2 pbl34 pbl36 mutant displays a discontinuous flower outgrowth, which resembles that of clv1 and is much more severe than that of the rpk2 and pbl34 pbl35 pbl36 mutants (Supplemental Figure S11, L, M, and P).
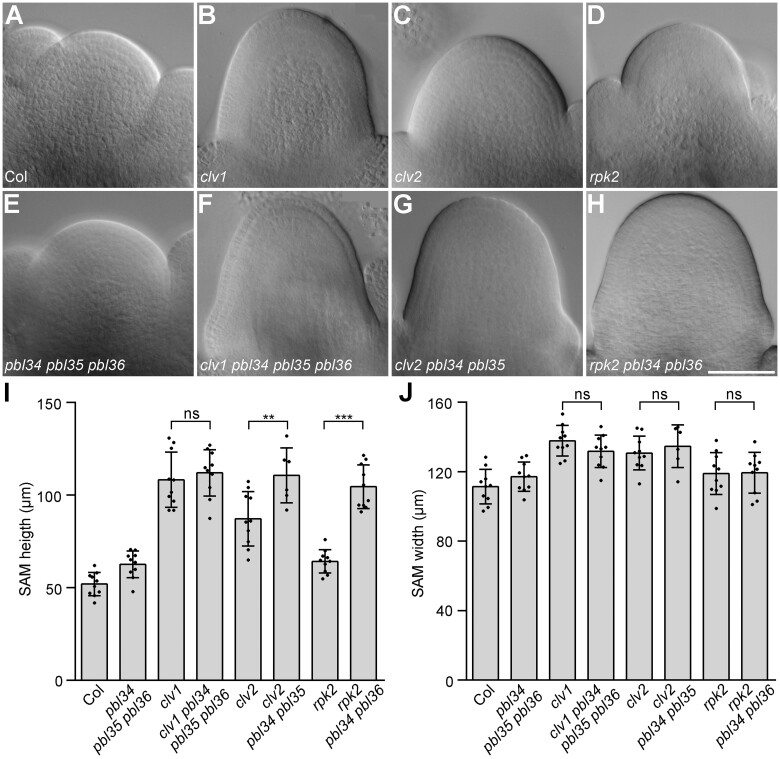
The SAMs of clv1 pbl34 pbl35 pbl36, clv2 pbl34 pbl35, and rpk2 pbl34 pbl36 were further analyzed. The results showed that the SAM of clv1 pbl34 pbl35 pbl36 is higher than that of the wild-type and the pbl34 pbl35 pbl36 mutant, but is indistinguishable from that of the clv1 mutant (Figure 5, A, B, F, and I). Due to the extremely irregular SAM of clv2 pbl34 pbl35 pbl36, the SAM of the clv2 pbl34 pbl35 triple mutant was analyzed. Both clv2 pbl34 pbl35 and rpk2 pbl34 pbl36 generate a significantly higher SAM when compared with the corresponding single mutants clv2 and rpk2, and the pbl34 pbl35 pbl36 triple mutant, respectively (Figure 5, C, D, and G–I). To further confirm the genetic effect of pbl34 pbl35 pbl36 in the clv1, clv2, and rpk2 background, we analyzed the IM morphology, carpel number, and CLV3 expression level of these mutants. The results showed that the clv1 pbl34 pbl35 pbl36 mutant produces an enlarged IM, which is similar to that of clv1 (Figure 6, B and F). In addition, clv1 pbl34 pbl35 pbl36 and clv1 produce siliques with similar carpel numbers (Figure 6I). Consistent with the higher vegetative SAM than clv2, clv2 pbl34 pbl35 pbl36 exhibits a dramatically enlarged irregular IM and significantly increased carpel numbers when compared with the clv2 single mutant (Figure 6, C, G, and I). Similarly, rpk2 pbl34 pbl36 also shows an enlarged IM and significantly increased carpel numbers when compared with the rpk2 single mutant (Figure 6, D, H, and I). Consistent with the IM size, the CLV3 expression level in the inflorescence of clv1 pbl34 pbl35 pbl36 is just slightly upregulated when compared with clv1, whereas both clv2 pbl34 pbl35 pbl36 and rpk2 pbl34 pbl36 exhibit more than 40-fold upregulated expression of CLV3 when compared with clv2 and rpk2, respectively (Figure 6J).
Figure 5.
pbl34 pbl35 pbl36 cannot enhance the SAM defects of clv1. A–H, DIC images of SAMs from 19-day-old seedlings of Col (A), clv1 (B), clv2 (C), rpk2 (D), pbl34 pbl35 pbl36 (E), clv1 pbl34 pbl35 pbl36 (F), clv2 pbl34 pbl35 (G), and rpk2 pbl34 pbl36 (H). Scale bar: 50 µm. I, J, Quantitative analysis of SAM height (I) and width (J). Data are shown as mean ± sd (n = 10 independent plants). ***P < 0.001, **P < 0.01, ns represents no significant difference (two-tailed Student’s t test).
Figure 6.
PBL34/35/36 function in the CLV1 pathway to transduce the CLV3 signal. A–H, SEM images of IMs in Col (A), clv1 (B), clv2 (C), rpk2 (D), pbl34 pbl35 pbl36 (E), clv1 pbl34 pbl35 pbl36 (F), clv2 pbl34 pbl35 pbl36 (G), and rpk2 pbl34 pbl36 (H). Scale bars: 100 µm. I, Quantitative analyses of carpels per flower of the indicated genotypes. Data are shown as mean ± sd (n = 10 independent plants; each sample represents the average carpel number of each plant.). ***P < 0.001, **P < 0.01, ns represents no significant difference (two-tailed Student’s t test). J, Relative expression of CLV3 in the indicated genotypes was quantified by RT-qPCR. Data are shown as mean ± sd (n = 3 technical replicates, two biological replicates showed similar results). ***P < 0.001 (two-tailed Student’s t test).
Taken together, these data indicate that mutations of PBL34/35/36 cannot enhance the SAM defects of clv1, but greatly enhance the SAM defects of clv2 and rpk2. In other words, our genetic data suggest that PBL34/35/36 may function in the CLV1 signaling pathway, and possibly act parallel to CLV2/CRN and RPK2 in mediating the CLV3 signal for maintaining SAM homeostasis.
PBL34/35/36 interact with CLV1, BAM1/3, and CIKs
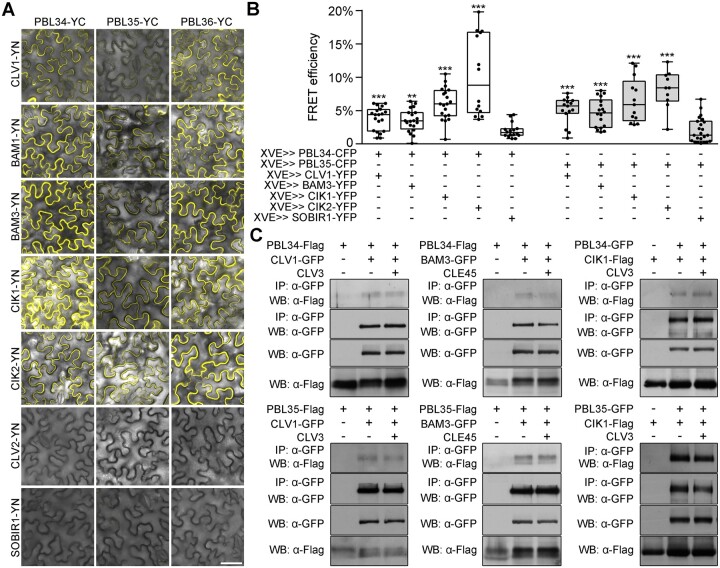
Our results strongly indicate that PBL34/35/36 play roles in both CLE-mediated signaling in the proximal root meristem and CLV3-mediated signaling in the SAM. In addition, the genetic data suggest that PBL34/35/36 act together with BAM3 and CIK2 to sense CLE45 in the RAM (Supplemental Figure S7), and act together with CLV1 to transduce the CLV3 signal in the SAM (Figures 5 and 6). On the other hand, CIKs have been shown to function as coreceptors of CLV1 and BAMs in both the SAM and RAM (Hu et al., 2018, 2022). Therefore, we next investigated whether PBL34/35/36 can interact with CLV1, BAMs, and CIKs. Results of yeast two-hybrid analysis demonstrated that PBL34/35/36 can interact with CLV1, BAM3, CIK1, and CIK2, but not SOBIR1 (Supplemental Figure S12A). A split YFP-based bimolecular fluorescence complementation (BiFC) assay also confirmed the interactions between PBL34/35/36 and CLV1, BAM1, BAM3, CIK1, and CIK2 (Figure 7A). Consistent with the genetic results, PBL34/35/36 did not interact with CLV2, which is different from CRN, indicating that PBL34/35/36 may act in parallel with CLV2/CRN. After isolating mesophyll protoplasts of Nicotiana benthamiana, these interactions were shown to occur at the plasma membrane, which is consistent with the notion that these proteins are membrane-localized (Supplemental Figure S12B). Moreover, the fluorescence resonance energy transfer (FRET) assays between PBL34/35 and CLV1, BAM3, or CIK1/2 were performed with inducible expression, and the results also showed that PBL34/35 interact with CLV1, BAM3, and CIK1/2 (Figure 7B).
Figure 7.
PBL34/35/36 interact with CLV1, BAM1/3, and CIKs. A, BiFC assay in N. benthamiana leaves to show interactions between PBL34/35/36 and CLV1, BAM1, BAM3, CIK1, CIK2, CLV2, and SOBIR1. PBLs were fused to the C-terminal part of YFP; CLV1, BAM1, BAM3, CIK1, CIK2, CLV2, and SOBIR1 were fused to the N-terminal part of YFP. Scale bar: 50 µm. B, Interactions of PBL34/35 with CLV1, BAM3, CIK1, CIK2, and SOBIR1 were detected by FRET in N. benthamiana leaves. All these proteins were transiently expressed under the control of the XVE≫oLexA-35S estradiol-inducible system. The FRET efficiencies between PBL34/35 and CLV1, BAM3, or CIKs were significantly higher than those with the control SOBIR1. Box limits indicate 25th and 75th percentiles; horizontal line is the median; whiskers display minimum and maximum values. Dots represent individual measurements from 10 to 23 samples per group. ***P < 0.001, **P < 0.01 (two-tailed Student’s t test). C, In vivo co-IP of PBL34/35-Flag and CIK1-Flag by CLV1/BAM3-GFP or PBL34/35-GFP, respectively, in stable transgenic plants. Ten-day-old stable transgenic seedlings harboring PBL34/35-Flag CLV1/BAM3-GFP or CIK1-Flag PBL34/35-GFP were harvested for protein extraction after treatment with or without 10-µM CLV3 or CLE45 for 10 min. Protein extracts were immunoprecipitated with an α-GFP antibody (IP: α-GFP), and then immunoblotted with an α-Flag (WB: α-Flag) or an α-GFP antibody (WB: α-GFP). Protein extracts before immunoprecipitation were immunoblotted with either α-Flag or α-GFP antibody as input.
Stable double transgenic plants harboring PBL-Flag and CLV1-GFP, or PBL-Flag and BAM3-GFP, or PBL-GFP and CIK1-Flag were further used for co-immunoprecipitation (co-IP) assays. Consistent with the above results, PBL34/35 interact with CLV1, BAM3, and CIK1 in vivo. However, the interactions between PBL34/35 and CLV1, BAM3, or CIK1 in planta cannot be enhanced by CLV3 or CLE45 treatment (Figure 7C). Taken together, these results demonstrate that PBL34/35/36 associate with the CLV–CIK complex to control SAM homeostasis, and with the BAM–CIK complex to control proximal root meristem homeostasis.
PBL34/35 can be phosphorylated by CLV1 and BAM1 in vitro
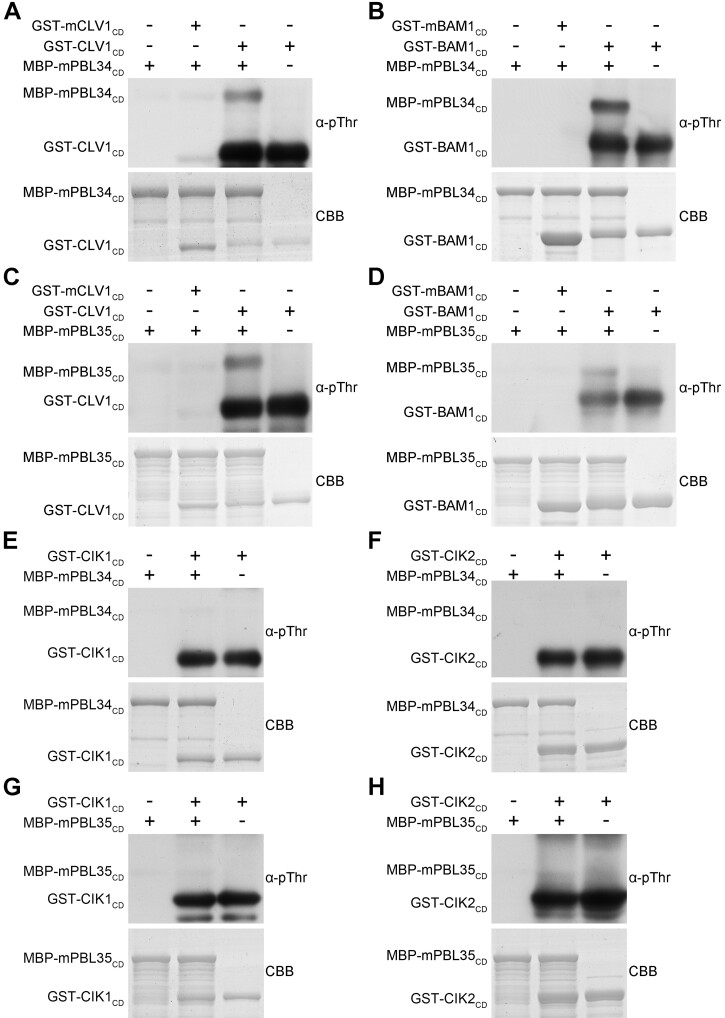
Our previous work demonstrated that the CLV3 and CLE25/26/45 signals are transmitted through phosphorylation (Hu et al., 2018, 2022). Therefore, we speculated that PBL34/35/36 might be phosphorylated by CLV1, BAMs, or CIKs directly. In vitro kinase assays using recombinant proteins purified from Escherichia coli were performed to determine whether PBL34/35 can be phosphorylated by CLV1, BAMs, or CIKs. As we unable to obtain the fusion protein of the BAM3 cytoplasmic domain, the cytoplasmic domain of CLV1, BAM1, and CIK1/2 was used (Shimizu et al., 2015). Immunoblotting assays using an α-pThr antibody revealed that GST-CLV1CD, GST-BAM1CD, GST-CIK1CD, and GST-CIK2CD possess strong autophosphorylation activities, whereas GST-mCLV1CD, GST-mBAM1CD, MBP-mPBL34CD, and MBP-mPBL35CD, in which a conserved lysine residue involved in ATP binding in their kinase domain was mutated, do not show any detectable autophosphorylation activities. It is apparent that GST-CLV1CD and GST-BAM1CD can directly phosphorylate MBP-mPBL34CD and MBP-mPBL35CD, but GST-CIK1CD and GST-CIK2CD cannot (Figure 8). These data indicate that PBL34/35 act as direct substrates of CLV1 and BAM1/3 to maintain homeostasis of the SAM and RAM.
Figure 8.
CLV1 and BAM1 phosphorylate PBL34 and PBL35 in vitro. A–H, In vitro kinase assays were performed by incubating GST-CLV1CD or GST-mCLV1CD, GST-BAM1CD or GST-mBAM1CD, GST-CIK1CD or GST-mCIK1CD, and GST-CIK2CD or GST-mCIK2CD with MBP-mPBL34CD or MBP-mPBL35CD, respectively. The phosphorylation levels were detected by α-pThr antibody. CBB, Coomassie brilliant blue staining.
Discussion
PBL34/35/36 function directly downstream of BAM1/3 to control CLE25/26/45-mediated proximal root meristem maintenance and protophloem differentiation
RLCKs act as essential players in RLK-mediated signaling pathways to connect the activated RLKs with downstream signaling components in a variety of plant defense responses and growth processes (Liang and Zhou, 2018). Although BAM1/3 and CIKs were found to be involved in proximal root meristem maintenance and protophloem differentiation (Depuydt et al., 2013; Anne et al., 2018; Ren et al., 2019; Hu et al., 2022), the direct downstream effectors of the BAM–CIK complex in this process have not been identified. Here, we show that the root length, proximal root meristem, and protophloem of the pbl34 pbl35 pbl36 triple mutants are resistant to the root-active CLE peptides (Figures 1 and 2; Supplemental Figures S4 and S6). Similar results were recently reported in a bioRxiv preprint (DeFalco et al., 2021). Genetic analysis also revealed that bam3 pbl34 pbl35 pbl36 and cik2 pbl34 pbl35 pbl36 do not show any additive defects when compared with bam3, cik2, and pbl34 pbl35 pbl36 (Supplemental Figure S7). Further biochemical assays show that PBL34/35/36 interact with BAM1/3 and CIKs both in vivo and in vitro (Figure 7). These results demonstrate that PBL34/35/36 act downstream of BAM1/3 and CIKs, which mediate CLE signaling to regulate the proximal root meristem and protophloem differentiation.
However, the loss-of-function of PBL34/35/36 can only slightly rescue the short-root phenotype of brx and ops (Supplemental Figure S8), which may be caused by the functional redundancy of PBL homologs. A similar scenario was found for cik2, which is partially resistant to all the root-active CLE peptides, but cannot suppress the root defects of brx, and can only slightly rescue the root defects of ops. Furthermore, the cik2 cik3 cik4 cik5 cik6 mutation largely rescues the root defects of both brx and ops (Anne et al., 2018; Ren et al., 2019; Hu et al., 2022). Therefore, in brx pbl34 pbl35 pbl36 and ops pbl34 pbl35 pbl36, other PBL members may compensate for the loss-of-function of PBL34/35/36 to transduce CLE signals. Generating and analyzing higher-order pbl mutants may provide insight into this possibility.
Interestingly, although PBL34/35/36 interact with both BAMs and CIKs, they can only be phosphorylated by BAM1, but not by CIKs in vitro (Figure 8). These results indicate that PBL34/35/36 may function as the direct substrates of BAMs to transduce CLE signals from the cell surface into the cytoplasm. Similarly, BSKs are the direct phosphorylation substrates of BRI1 to activate the downstream BR signaling components, whereas BSKs cannot be phosphorylated by BAK1 although BAK1 shows dramatically enhanced phosphorylation levels upon BR perception (Wang et al., 2005; Tang et al., 2008). Plants may employ a similar strategy by forming an RLK–RLCK complex that transduces extracellular signals into the cytoplasm.
PBL34/35/36 exhibit the CLV1 pathway-specific function to maintain SAM homeostasis
The SAM and RAM are evolutionarily related structures, and they employ several common or related regulators to maintain stem cell niches. CLV1 is involved in regulating both SAM and the distal root meristem (Clark et al., 1997; Stahl et al., 2013); CLV2 and CRN are required to maintain the homeostasis of both the SAM and proximal root meristem (Kayes and Clark, 1998; Müller et al., 2008; Hazak et al., 2017); CIKs function redundantly in all these meristems (Hu et al., 2018; Zhu et al., 2021). In this study, our findings indicate that PBL34/35/36 also function as conserved regulators in both the SAM and RAM.
The pbl34 pbl35 pbl36 mutants exhibit enlarged SAMs relative to the wild-type plants (Figure 3), and show reduced sensitivity to exogenously applied CLV3 (Figure 4; Supplemental Figure S10). Genetic analysis shows that additional mutations of PBL34/35/36 greatly enhance the phenotype of clv2 and rpk2, but not clv1, suggesting that PBL34/35/36 function in the CLV1 pathway, and act independently of CLV2 and RPK2 in maintaining SAM homeostasis. Biochemical assays show that PBL34/35/36 interact with CLV1 and CIKs both in vivo and in vitro, and they can be phosphorylated by CLV1, but not by CIKs in vitro, which is similar to the scenario that is found in PBL34/35/36-regulated RAM maintenance. The current findings support the notion that PBL34/35/36 may specifically act downstream of CLV1 in mediating the CLV3 signal to regulate SAM homeostasis.
We noticed that the pbl34 pbl35 pbl36 mutant exhibits weaker SAM defects when compared with the clv1, clv3, and cik mutants (Figure 3; Hu et al., 2018). It is very possible that other PBL homologs also play a role in maintaining SAM homeostasis, similar to the situation found in PBL34/35/36-mediated RAM regulation. Alternatively, it cannot be ruled out that additional pathways parallel to PBL34/35/36 may also function downstream of CLV1 to mediate the CLV3 signal. Distinct effectors downstream of the same receptor may exist to transduce the same or different signals. For instance, FEA2 employs two different downstream effectors, CT2 (Gα) and ZmCRN, to maintain SAM homeostasis in maize by transducing the ZmFCP1 and ZmCLE7 signals, respectively (Je et al., 2018).
Potential applications of PBL34/35/36 in agriculture
The CLV–WUS negative-feedback loop is conserved across angiosperms. Various clv mutants have been identified in rice (Oryza sativa), maize, and tomato (Solanum lycopersicum). The appropriately enlarged IMs and floral meristems of these clv mutants can result in more and bigger fruits (Somssich et al., 2016). Usually, strong clv mutants generate extremely flattened or split meristems, giving rise to very disorganized inflorescences that have poor seed yield, such as the null td1 and fea2 mutants in maize (Taguchi-Shiobara et al., 2001; Bommert et al., 2005). In contrast, meristems with slightly increased size in the weak clv mutants in crops will not lead to dramatically disorganized inflorescences, such as the weak alleles of fea2, fea3, Zmcle7, Zmfcp1, and Zmcle1e5 mutants in maize, and can significantly improve crop productivity (Bommert et al., 2013a; Je et al., 2016; Liu et al., 2021). Actually, the homologs of PBL34/35/36 are widely spread in various species. Loss-of-function of BnaA09g22690D and BnaA09g22670D, two homologs of PBL34/35/36 in Brassica napus, results in a fasciated stem, ultrahigh branching, and more flowers (Li et al., 2020), suggesting that PBLs possess conserved functions in maintaining SAM homeostasis across species. As pbl34 pbl35 pbl36 is a weak mutant of the CLV signaling pathway, these three genes, and possibly their potential homologs, are appropriate candidate targets that can be used in modern breeding to improve crop productivity.
High-yield breeding requires not only good yield traits but also proper tolerance to various stresses since plants constantly cope with adverse environments. In general, turning on defense signaling often leads to decreased growth and yield, and turning off defense signaling will result in infected plants by pathogens. This defense–growth tradeoff is the biggest challenge for modern crop breeding (Wu et al., 2020). Therefore, a comprehensive understanding of the cross-talk between developmental and immune signaling is critical for providing essentially basic knowledge to maximize crop productivity. In addition to the functions in RAM and SAM discussed in this study, PBL34/35/36 have also been demonstrated to be downstream signaling components of LORE that recognizes medium-chain 3-hydroxy fatty acids in the plant immune response (Luo et al., 2020), indicating that PBL34/35/36 act as critical regulators in both SAM maintenance and the immune response. Therefore, PBL34/35/36 have the potential to balance the trade-off between defense response and development of inflorescences, branches, and flowers to improve crop productivity. Especially, the differentially phosphorylated sites of PBL34/35/36 by LORE and CLV1 may be engineered as potential targets to improve crop yield, but not affect the immune response.
Briefly, the results presented in this report demonstrate that CLV1- and BAM1/3-mediated CLE peptide signaling in the SAM and RAM require PBL34/35/36 functioning as downstream components to transduce the signals into the cytoplasm. Our findings not only provide essential insights into the molecular mechanisms controlling meristem homeostasis but also further substantiate the conservative strategy employed by RLK-mediated signaling pathways. In addition, pbl34 pbl35 pbl36 mutants show reduced sensitivity to all of those CLE peptides that can consume the proximal root meristem, suggesting that PBL34/35/36 probably mediate multiple CLE signals to regulate plant growth and development.
Materials and methods
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col), various mutants, and transgenic plants in the Col background were used in this study. The T-DNA insertion lines, including pbl34-1 (SALK_067743), pbl34-2 (SALK_126209), pbl35-1 (SALK_039402), pbl36-1 (SAIL_885_B03), pbl36-2 (SALK_074901), pbl36-3 (SALK_058890), bam3 (SALK_044433), cik2 (SALK_066568), clv1-20 (SALK_008670), clv2-101 (GK686A09), rpk2-1 (SALK_062412), and wus-101 (GK870H12), were obtained from the Arabidopsis Biological Resource Center (ABRC). brx-2 and ops-1 were used in previous studies (Li et al., 2009; Truernit et al., 2012). The pbl34-3, pbl35-2, and pbl35-3 mutants were generated by CRISPR/Cas9-based gene editing (Wang et al., 2015). The corresponding single guide RNA (sgRNA) sequences are listed in Supplemental Table S1. The pbl34-1, pbl35-1, and pbl36-1 mutants were used to generate the corresponding pbl double mutants and the pbl34 pbl35 pbl36-1 triple mutant, which were employed for physiological and genetic analysis. The pbl34 pbl35 pbl36-1 triple mutant was named pbl34 pbl35 pbl36 for brevity in the text. The pbl34-2, pbl35-2, and pbl36-2 mutants were used to create the pbl34 pbl35 pbl36-2 mutant. The pbl34 pbl35 pbl36-3 mutant was generated by crossing the pbl34-3, pbl35-3, and pbl36-3 mutants. Seeds were sown in the soil after vernalization at 4°C for 3 days and then cultivated in a greenhouse with 16-h light at 20°C–22°C until the plants were used. For seedling analysis, surface-sterilized seeds were vernalized for 3 days and grown on half-strength MS medium plates containing 1% (w/v) sucrose and 0.8% (w/v) agar in a growth chamber with 16-h light (120 µmol s−1 m−2 light intensity provided by white LED lamps) and 8-h dark at 22°C. Peptide assays were performed as described previously (Hu et al., 2018; Zhu et al., 2021).
Phylogenetic analysis
Amino acid sequences of PBL34/35/36 and BIK1 were downloaded from The Arabidopsis Information Resource. The multiple sequence alignment was performed using MEGA6 (https://www.megasoftware.net). The phylogenetic tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates. The evolutionary distances were computed using the Poisson correction method, and are in the units of the number of amino acid substitutions per site.
Creation of constructs and transgenic plants
To analyze the expression patterns of PBL34/35/36, the genomic fragments that encompass the promoter sequence and the coding sequence without the stop codon of each PBL were cloned into the pBIB-BASTA-GWR-YFP vector. To analyze protophloem differentiation, the promoters of CVP2 and APL (Bonke et al., 2003; Scacchi et al., 2010) were cloned into the pFYTAG destination vector (Zhang et al., 2005) using Gateway technology.
The coding sequences of PBL34/35/36, CIK1, CLV1, and BAM3 were cloned into the pUBQ10-GWR-Flag and pH35GWG destination vectors using Gateway technology. Double transgenic plants expressing PBL-Flag and CLV1/BAM3-GFP or PBL-GFP and CIK1-Flag were then generated for co-IP analyses. All transgenic plants were generated by the floral dip method (Clough and Bent, 1998) using the Agrobacterium tumefaciens GV3101 strain.
For BiFC assays, PBL34/35/36 were fused with the C-terminal part of YFP (YC) in the pEarleygate201-YC vector, and CLV1, BAM1, BAM3, CIK1, CIK2, CLV2, and SOBIR1 were fused with the N-terminal part of YFP (YN) in the pEarleygate202-YN vector (Lu et al., 2010b).
For FRET analyses with inducible vectors, PBL34/35 were fused with CFP in the pER8-CFP vector, and CLV1, BAM3, CIK1, CIK2, and SOBIR1 were fused with YFP in the pER8-YFP vector. Both pER8-CFP and pER8-YFP were modified by adding CFP or YFP into the binary destination vector pER8 (Zuo et al., 2000) that was linearized by XhoI and SpeI. The Gateway cassette and CFP/YFP were amplified from pBIB-BASTA-GWR-CFP/YFP.
For protein purification, the kinase-inactive cytoplasmic domains of PBL34/35 were fused with an N-terminal MBP-tag using the pMal-cRi vector, and the wild-type or kinase-inactive cytoplasmic domains of CLV1, BAM1, CIK1, and CIK2 were fused with an N-terminal GST tag using the pDEST15 vector.
Phenotypic analysis
The siliques were examined under a dissection microscope to count carpel number. The carpel number of each genotype was determined by averaging the number of carpels from 10 plants. The carpel number of each plant was determined by averaging carpel numbers from 20 or all siliques of the plant. For SAM analysis, leaves of 19-day-old seedlings were removed to expose the SAM under a dissection microscope, then the SAM was cleared in a mixture of chloral hydrate, glycerol, and water (4:1:2, w/v/v) on a glass slide for several minutes. Differential interference contrast (DIC) images of the SAM were taken using a microscope (Leica DM6000B) equipped with a DFC420C digital camera. The base of the SAM was defined as the location of leaf primordium, and the distance from the top of a SAM to the base was measured as the height. To observe the IM, flowers were removed from the inflorescence to expose the IM. The prepared samples were stuck to the stage of a microscope and were then frozen in liquid nitrogen for about 30 s (keeping the samples above the liquid nitrogen). The samples were subsequently examined by a Hitachi 4700 scanning electron microscope.
Microscopy analysis
PI staining and mPS-PI assays were performed as described previously (Zhu et al., 2021). Samples were observed and photographed using a confocal laser scanning microscope (LSM-880, ZEISS and A1R + Ti2-E, Nikon). Confocal imaging of fluorescence reporters in the IM of living plants was performed by using a Nikon (A1R + Ti2-E) confocal microscope. NIS-Elements AR (Nikon software) was used to reconstruct the Z-stacks for a longitudinal view. To image the YFP and PI signals simultaneously in the IM, the multitracking mode of the Nikon confocal microscope was used. YFP was excited at 487 nm and the signals at 500–550 nm were collected. PI was excited using a 560 nm laser and the signals at 570–620 nm were collected.
FRET experiments were conducted using a Zeiss LSM 880. CFP donor fluorophores were excited at 458 nm, and the signals were collected at 463–510 nm. YFP acceptor fluorophores were excited at 514 nm, and the signals were collected at 520–620 nm. To perform acceptor photobleaching, the selected regions of interest were photobleached by defined parameters. The prebleach and postbleach images were collected, and the fluorescence intensity of CFP and YFP were recorded before and after photobleaching.
RNA in situ hybridization and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
RNA in situ hybridization of CLV3 and WUS was performed as described previously (Hu et al., 2018). Total RNA of Col, pbl34 pbl35 pbl36, clv1, clv1 pbl34 pbl35 pbl36, clv2, clv2 pbl34 pbl35 pbl36, rpk2, and rpk2 pbl34 pbl36 was extracted from 5-week-old IMs with young flowers using an RNA extraction kit (TIANGEN, DP432). Five micrograms of total RNA of each sample was used to synthesize cDNA using HiScript II Q RT SuperMix (Vazyme Biotech, R223). Quantitative RT-PCR analysis was performed with a StepOnePlus Real-time PCR System (Applied Biosystems) using Hieff qPCR SYBR Green Master Mix (YEASEN, 11203ES03) to examine the relative expression of CLV3. Primers used in this study are listed in Supplemental Table S1.
Biochemical assays
BiFC, mbSUS yeast two-hybrid, co-IP, and in vitro phosphorylation assays were performed as described previously (Cui et al., 2018; Hu et al., 2018). Commercial antibodies of Flag (α-Flag; Abmart, M20008L, lot 334077; 1:3,000 dilution), GFP (α-GFP; Roche; 11814460001, lot 47859600; 1:3,000 dilution), and phosphothreonine (α-pThr; Cell Signaling Technology; 9381, lot 25; 1:2,500 dilution) were used for immunoblot analyses. For the mbSUS yeast two-hybrid assay, full-length coding sequences of PBL34, PBL35, and PBL36 were co-transferred into the yeast strain THY.AP5 with the vector pX-NubWTgate linearized by EcoRI and SmaI, and full-length coding sequences of CLV1, BAM3, CIK1, CIK2, and SOBIR1 were co-transferred into the yeast strain THY.AP4 with the vector pMetYCgate linearized by PstI and HindIII.
For isolating mesophyll protoplasts of N. benthamiana, the infiltrated leaves were cut into small pieces and then incubated with 5-mL protoplast Enzyme Solution (1.5% [w/v] cellulase R10, and 0.4% [w/v] macerozyme R10, 0.4-M mannitol, 20-mM KCl, 20-mM MES, pH 5.7, 10-mM CaCl2, 1.83-µL 2-mercaptoethanol, 0.1% [w/v] BSA) in a vacuum for 30 min. The samples were then transferred into darkness for 4 h with gentle shaking. After an equal volume of W5 Solution (2-mM MES, pH 5.7, 154-mM NaCl, 125-mM CaCl2, 5-mM KCl) was added to each sample, the precipitates were collected by centrifuging at 100g for 2 min at room temperature and then resuspended with 0.5-mL washing and incubation solution (0.5-M mannitol, 4-mM MES, pH 5.7, 20-mM KCl).
For inducible FRET, the transiently transformed N. benthamiana leaves under the control of an estradiol-inducible promoter (Zuo et al., 2000) were grown for 36 h and then sprayed with 20-µM β-estradiol 6 h prior to imaging. The FRET efficiencies were quantified by measuring the intensity increase of the donor fluorophore (CFP) after bleaching of the acceptor according to the following formula (Weidtkamp-Peters and Stahl, 2017):
For protein purification, the expression of MBP-tagged kinase-inactive cytoplasmic domains of PBL34/35 was induced with 0.2-mM IPTG for 6 h at 28°C and the expression of GST-tagged cytoplasmic domains of CLV1, BAM1, CIK1, and CIK2 was induced with 0.2-mM IPTG for 16 h at 18°C.
Quantification and statistical analysis
Data for quantification analyses are presented as mean ± sd. Statistical analyses were performed using GraphPad Prism software (version 6.0) and Microsoft Excel 2010. Results of all statistical analyses are presented in Supplemental Data Set S2.
Accession numbers
Sequence data from this article can be found in the Arabidopsis TAIR database (www.arabidopsis.org) under the following accession numbers: APL (AT1G79430), BAM1 (AT5G65700), BAM3 (AT4G20270), BRX (AT1G31880), CIK1 (AT1G60800), CIK2/CLERK (AT2G23950), CLE25 (AT3G28455), CLE45 (AT1G69588), CLV1 (AT1G75820), CLV2 (AT1G65380), CLV3 (AT2G27250), CVP2 (AT1G05470), OPS (AT3G09070), PBL34 (AT5g15080), PBL35 (AT3G01300), PBL36 (AT3g28690), RPK2 (AT3G02130), SOBIR1 (AT2G31880), WUS (AT2G17950).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. pbl34 is less sensitive to CLE45 treatment.
Supplemental Figure S2. PBL34/35/36 are expressed in the RAM.
Supplemental Figure S3. Gene structures of PBL34/35/36 and location of T-DNA insertions and CRISPR/Cas9-mediated mutations.
Supplemental Figure S4. PBL34/35/36 are required for CLE45-mediated proximal root meristem consumption.
Supplemental Figure S5. Complementation of PBL34/35/36 can rescue the defects of pbl34 pbl35 pbl36.
Supplemental Figure S6. pbl34 pbl35 pbl36 showed resistance to root-active CLEs.
Supplemental Figure S7. PBL34/35/36 function in a common pathway with BAM3 and CIK2.
Supplemental Figure S8. pbl34 pbl35 pbl36 partially rescues the short root of brx and ops.
Supplemental Figure S9. PBL34/35/36 are expressed in the SAM.
Supplemental Figure S10. PBL34/35/36 are involved in the CLV3-mediated signaling pathway.
Supplemental Figure S11. pbl34 pbl35 pbl36 and pbl34 pbl36 enhance the phenotype of clv2 and rpk2, respectively.
Supplemental Figure S12. PBL34/35/36 interact with CLV1, BAM3, CIK1, and CIK2 on the plasma membrane.
Supplemental Table S1. Primers used in this study.
Supplemental Data Set S1. Alignments used to generate the phylogeny presented in Supplemental Figure S1B.
Supplemental Data Set S2. Summary of statistical analyses.
Supplementary Material
Acknowledgments
We thank the ABRC for the T-DNA insertion lines used in this study. We thank Dr. Jun Liu (Institute of Microbiology, Chinese Academy of Sciences) for nicely providing us with the MBP-mPBL34CD construct. We are grateful to Dr Chun‐Ming Liu (Peking University) for kindly providing us with the brx-2 and ops-1 mutants. We are grateful to Ms Liang Peng, Ms Haiyan Li, Ms Liping Guan, Dr Yahu Gao, and Dr Yang Zhao (Core Facility for Life Science Research, Lanzhou University) for technical assistance.
Funding
This work was supported by National Natural Science Foundation of China (31970339, 32170332, 31770312, 31900166, and 32000594), the 111 Project (B16022), China Postdoctoral Science Foundation (BX20180133), Fundamental Research Funds for the Central Universities (lzujbky-2021-kb05, lzujbky-2022-kb05), and Lanzhou City’s Scientific Research Funding Subsidy to Lanzhou University.
Conflict of interest statement. The authors declare no competing interests.
Contributor Information
Wenping Wang, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Chong Hu, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Xiaonan Li, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Yafen Zhu, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Liang Tao, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Yanwei Cui, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Dingqian Deng, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Xiaoxuan Fan, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Hong Zhang, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Jia Li, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China; School of Life Sciences, Guangzhou University, Guangzhou 510006, China.
Xiaoping Gou, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Jing Yi, Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
W.W., C.H., X.G., and J.Y. conceived the project, designed all experiments, analyzed the data, and wrote the manuscript. X.G. and J.Y. supervised the study. W.W. and C.H. performed most of the experiments and prepared the data. X.L. and L.T. performed the RNA in situ hybridization analyses and in vivo fluorescence imaging in the SAM. Y.Z. conducted the in vitro phosphorylation assays. Y.C., D.D., X.F., and H.Z. contributed to the generation and analysis of transgenic plants and high-order mutants. J.L. helped prepare the manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Xiaoping Gou (gouxp@ lzu.edu.cn) and Jing Yi (yijing@lzu.edu.cn).
References
- Anne P, Amiguet-Vercher A, Brandt B, Kalmbach L, Geldner N, Hothorn M, Hardtke CS (2018) CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis. Development 145: dev162354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku S, Sawa S, Yamada M (2011a) The function of the CLE peptides in plant development and plant-microbe interactions. Arabidopsis Book 9: e0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku S, Takahashi F, Kinoshita A, Miwa H, Shinozaki K, Fukuda H, Sawa S (2011b) Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol 52: 14–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R (2010) Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol 152: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blümke P, Schlegel J, Gonzalez-Ferrer C, Becher S, Pinto KG, Monaghan J, Simon R (2021) Receptor-like cytoplasmic kinase MAZZA mediates developmental processes with CLAVATA1-family receptors in Arabidopsis. J Exp Bot 72: 4853–4870 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Franck CM, Lituiev DS, Grossniklaus U (2015) Receptor-like cytoplasmic kinase MARIS functions downstream of CrRLK1L-dependent signaling during tip growth. Proc Natl Acad Sci USA 112: 12211–12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Nagasawa NS, Jackson D (2013a) Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat Genet 45: 334–337 [DOI] [PubMed] [Google Scholar]
- Bommert P, Je BI, Goldshmidt A, Jackson D (2013b) The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558 [DOI] [PubMed] [Google Scholar]
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr W (2005) THICK TASSEL DWARF1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mahonen AP, Hauser MT, Helariutta Y (2003) APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Breda AS, Hazak O, Schultz P, Anne P, Graeff M, Simon R, Hardtke CS (2019) A cellular insulator against CLE45 peptide signaling. Curr Biol 29: 2501–2508 [DOI] [PubMed] [Google Scholar]
- Burr CA, Leslie ME, Orlowski SK, Chen I, Wright CE, Daniels MJ, Liljegren SJ (2011) CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol 156: 1837–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui Y, Hu C, Zhu Y, Cheng K, Li X, Wei Z, Xue L, Lin F, Shi H, Yi J, et al. (2018). CIK receptor kinases determine cell fate specification during early anther development in Arabidopsis. Plant Cell 30: 2383–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco TA, Anne P, James SR, Willoughby A, Johanndrees O, Genolet Y, Pullen A-M, Zipfel C, Hardtke CS, Nimchuk ZL (2021) A conserved regulatory module regulates receptor kinase signaling in immunity and development. bioRxiv preprint. doi: 10.1101/2021.01.19.427293 (January 20, 2021) [DOI] [Google Scholar]
- Depuydt S, Rodriguez-Villalon A, Santuari L, Wyser-Rmili C, Ragni L, Hardtke CS (2013) Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc Natl Acad Sci USA 110: 7074–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyoung BJ, Clark SE (2008) BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Li X, Chen J, Chen W, Li B, Li C, Wang L, Li J, Zhao X, Lin J, et al. (2016). Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc Natl Acad Sci USA 113: 8326–8334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM (2005) The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois JL, Nora FR, Mizukami Y, Sablowski R (2004) WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev 18: 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, Li J (2020) Paired receptor and coreceptor kinases perceive extracellular signals to control plant development. Plant Physiol 182: 1667–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujas B, Kastanaki E, Sturchler A, Cruz TMD, Ruiz-Sola MA, Dreos R, Eicke S, Truernit E, Rodriguez-Villalon A (2020) A reservoir of pluripotent phloem cells safeguards the linear developmental trajectory of protophloem sieve elements. Curr Biol 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Guo Y, Han L, Hymes M, Denver R, Clark SE (2010) CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J 63: 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O, Brandt B, Cattaneo P, Santiago J, Rodriguez-Villalon A, Hothorn M, Hardtke CS (2017) Perception of root-active CLE peptides requires CORYNE function in the phloem vasculature. EMBO Rep 18: 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Zhu Y, Cui Y, Cheng K, Liang W, Wei Z, Zhu M, Yin H, Zeng L, Xiao Y, et al. (2018) A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat Plants 4: 205–211 [DOI] [PubMed] [Google Scholar]
- Hu C, Zhu Y, Cui Y, Zeng L, Li S, Meng F, Huang S, Wang W, Kui H, Yi J, et al. (2022) A CLE-BAM-CIK signalling module controls root protophloem differentiation in Arabidopsis. New Phytol 233: 282–296 [DOI] [PubMed] [Google Scholar]
- Ishida T, Tabata R, Yamada M, Aida M, Mitsumasu K, Fujiwara M, Yamaguchi K, Shigenobu S, Higuchi M, Tsuji H, et al. (2014). Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep 15: 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Je BI, Xu F, Wu Q, Liu L, Meeley R, Gallagher JP, Corcilius L, Payne RJ, Bartlett ME, Jackson D (2018) The CLAVATA receptor FASCIATED EAR2 responds to distinct CLE peptides by signaling through two downstream effectors. eLife 7: e35673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je BI, Gruel J, Lee YK, Bommert P, Arevalo ED, Eveland AL, Wu Q, Goldshmidt A, Meeley R, Bartlett M, et al. (2016). Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat Genet 48: 785–791 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Kayes JM, Clark SE (1998) CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125: 3843–3851 [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, Wang ZY (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S (2010) RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Jackson D (2019) Control of meristem size. Annu Rev Plant Biol 70: 269–291 [DOI] [PubMed] [Google Scholar]
- Lee H, Jun YS, Cha OK, Sheen J (2019) Mitogen-activated protein kinases MPK3 and MPK6 are required for stem cell maintenance in the Arabidopsis shoot apical meristem. Plant Cell Rep 38: 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gao J, Chen J, Wang Z, Shen W, Yi B, Wen J, Ma C, Shen J, Fu T, Tu J (2020) Identification and fine mapping of a major locus controlling branching in Brassica napus. Theor Appl Genet 133: 771–783 [DOI] [PubMed] [Google Scholar]
- Li J, Mo X, Wang J, Chen N, Fan H, Dai C, Wu P (2009) BREVIS RADIX is involved in cytokinin-mediated inhibition of lateral root initiation in Arabidopsis. Planta 229: 593–603 [DOI] [PubMed] [Google Scholar]
- Liang X, Zhou JM (2018) Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu Rev Plant Biol 69: 267–299 [DOI] [PubMed] [Google Scholar]
- Liao HZ, Zhu MM, Cui HH, Du XY, Tang Y, Chen LQ, Ye Z, Zhang XQ (2016) MARIS plays important roles in Arabidopsis pollen tube and root hair growth. J Integr Plant Biol 58: 927–940 [DOI] [PubMed] [Google Scholar]
- Liu L, Gallagher J, Arevalo ED, Chen R, Skopelitis T, Wu Q, Bartlett M, Jackson D (2021) Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat Plants 7: 287–294 [DOI] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou JM (2013) BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci USA 110: 6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010a) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, et al. (2010b). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61: 259–270 [DOI] [PubMed] [Google Scholar]
- Luo X, Wu W, Liang Y, Xu N, Wang Z, Zou H, Liu J (2020) Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J 39: e102856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Zhou Y, Tarr PT, Peterson BA, Meyerowitz EM (2015) Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson V, Joos L, Zhu S, Gevaert K, Butenko MA, De Smet I (2019) Look closely, the beautiful may be small: precursor-derived peptides in plants. Annu Rev Plant Biol 70: 153–186 [DOI] [PubMed] [Google Scholar]
- Pandey S (2019) Heterotrimeric G-protein signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 70: 213–238 [DOI] [PubMed] [Google Scholar]
- Perales M, Rodriguez K, Snipes S, Yadav RK, Diaz-Mendoza M, Reddy GV (2016) Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc Natl Acad Sci USA 113: 6298–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Frohlich K, Westphal L, Scheel D, Lee J (2014) Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol 14: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Zhou Z, Miao P, Bi G, Hu M, Wu Y, Feng F, Zhang X, Zhou JM (2018) Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol 177: 1679–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren SC, Song XF, Chen WQ, Lu R, Lucas WJ, Liu CM (2019) CLE25 peptide regulates phloem initiation in Arabidopsis through a CLERK-CLV2 receptor complex. J Integr Plant Biol 61: 1043–1061 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Villalon A, Gujas B, van Wijk R, Munnik T, Hardtke CS (2015) Primary root protophloem differentiation requires balanced phosphatidylinositol-4,5-biphosphate levels and systemically affects root branching. Development 142: 1437–1446 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Villalon A, Gujas B, Kang YH, Breda AS, Cattaneo P, Depuydt S, Hardtke CS (2014) Molecular genetic framework for protophloem formation. Proc Natl Acad Sci USA 111: 11551–11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez K, Perales M, Snipes S, Yadav RK, Diaz-Mendoza M, Reddy GV (2016) DNA-dependent homodimerization, sub-cellular partitioning, and protein destabilization control WUSCHEL levels and spatial patterning. Proc Natl Acad Sci USA 113: 6307–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacchi E, Salinas P, Gujas B, Santuari L, Krogan N, Ragni L, Berleth T, Hardtke CS (2010) Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proc Natl Acad Sci USA 107: 22734–22739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jurgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Shimizu N, Ishida T, Yamada M, Shigenobu S, Tabata R, Kinoshita A, Yamaguchi K, Hasebe M, Mitsumasu K, Sawa S (2015) BAM 1 and RECEPTOR-LIKE PROTEIN KINASE 2 constitute a signaling pathway and modulate CLE peptide-triggered growth inhibition in Arabidopsis root. New Phytol 208: 1104–1113 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich M, Je BI, Simon R, Jackson D (2016) CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143: 3238–3248 [DOI] [PubMed] [Google Scholar]
- Song SK, Clark SE (2005) POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev Biol 285: 272–284 [DOI] [PubMed] [Google Scholar]
- Song SK, Lee MM, Clark SE (2006) POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 133: 4691–4698 [DOI] [PubMed] [Google Scholar]
- Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D, Hayun LB, Gruetter C, Rauh D, Ori N, et al. (2013) BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J 74: 905–919 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, Kuhnemuth R, Weidtkamp-Peters S, Pinto KG, Kirschner GK, Schmid JB, Wink RH, Hulsewede A, et al. (2013) Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol 23: 362–371 [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The FASCIATED EAR2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Belcram K, Barthelemy J, Palauqui JC (2012) OCTOPUS, a polarly localised membrane-associated protein, regulates phloem differentiation entry in Arabidopsis thaliana. Development 139: 1306–1315 [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthelemy J, Palauqui JC (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gou X (2020) Receptor-like protein kinases function upstream of MAPKs in regulating plant development. Int J Mol Sci 21: 7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Stahl Y (2017) The use of FRET/FLIM to study proteins interacting with plant receptor kinases. Methods Mol Biol 1621: 163–175 [DOI] [PubMed] [Google Scholar]
- Wu Q, Xu F, Liu L, Char SN, Ding Y, Je BI, Schmelz E, Yang B, Jackson D (2020) The maize heterotrimeric G protein beta subunit controls shoot meristem development and immune responses. Proc Natl Acad Sci USA 117: 1799–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Gong FC, Lambert GM, Galbraith DW (2005) Cell type-specific characterization of nuclear DNA contents within complex tissues and organs. Plant Methods 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liu L, Kucukoglu M, Tian D, Larkin RM, Shi X, Zheng B (2020) Predicting and clustering plant CLE genes with a new method developed specifically for short amino acid sequences. BMC Genomics 21: 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Hu C, Gou X (2020) Receptor-like protein kinase-mediated signaling in controlling root meristem homeostasis. aBIOTECH 1: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Hu C, Cui Y, Zeng L, Li S, Zhu M, Meng F, Huang S, Long L, Yi J, et al. (2021). Conserved and differentiated functions of cik receptor kinases in modulating stem cell signaling in Arabidopsis. Mol Plant 14: 1119–1134 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.