Abstract
Background
Tumor-associated macrophages/microglia (TAMs) are prominent microenvironment components in human glioblastoma (GBM) that are potential targets for anti-tumor therapy. However, TAM depletion by CSF1R inhibition showed mixed results in clinical trials. We hypothesized that GBM subtype-specific tumor microenvironment (TME) conveys distinct sensitivities to TAM targeting.
Methods
We generated syngeneic PDGFB- and RAS-driven GBM models that resemble proneural-like and mesenchymal-like gliomas, and determined the effect of TAM targeting by CSF1R inhibitor PLX3397 on glioma growth. We also investigated the co-targeting of TAMs and angiogenesis on PLX3397-resistant RAS-driven GBM. Using single-cell transcriptomic profiling, we further explored differences in TME cellular compositions and functions in PDGFB- and RAS-driven gliomas.
Results
We found that growth of PDGFB-driven tumors was markedly inhibited by PLX3397. In contrast, depletion of TAMs at the early phase accelerated RAS-driven tumor growth and had no effects on other proneural and mesenchymal GBM models. In addition, PLX3397-resistant RAS-driven tumors did not respond to PI3K signaling inhibition. Single-cell transcriptomic profiling revealed that PDGFB-driven gliomas induced expansion and activation of pro-tumor microglia, whereas TAMs in mesenchymal RAS-driven GBM were enriched in pro-inflammatory and angiogenic signaling. Co-targeting of TAMs and angiogenesis decreased cell proliferation and changed the morphology of RAS-driven gliomas.
Conclusions
Our work identifies functionally distinct TAM subpopulations in the growth of different glioma subtypes. Notably, we uncover a potential responsiveness of resistant mesenchymal-like gliomas to combined anti-angiogenic therapy and CSF1R inhibition. These data highlight the importance of characterization of the microenvironment landscape in order to optimally stratify patients for TAM-targeted therapy.
Keywords: angiogenesis, CSF1R inhibition, glioblastoma subtypes, single-cell transcriptomics, tumor-associated microglia and macrophages
Key Points.
CSF1R inhibition blocks the growth of PDGFB-driven but not RAS-driven murine gliomas.
TAMs in PDGFB- or RAS-driven gliomas exhibit functionally distinct characteristics.
Co-targeting of TAM and angiogenesis reduces proliferation and alters tumor phenotype.
Importance of the Study.
Recent studies indicate tumor-associated microglia/macrophages (TAMs) are highly heterogeneous in human gliomas. CSF1R inhibition that targets tumor-associated macrophages has been applied for anti-tumor therapy in other cancers; however, clinical trials of the CSF1R inhibitor PLX3397 in recurrent human GBMs have shown no population-wide response. Thus, the significance of TAM functions in GBM growth and progression remains to be defined. Here we identify distinct roles of TAM subpopulations in the growth of different glioma subtypes. TAM targeting inhibits the growth of PDGFB-driven but not RAS-driven gliomas. In contrast to pro-tumor TAMs in PDGFB-driven gliomas, TAMs in mesenchymal-like RAS-driven gliomas are enriched in pro-inflammatory and angiogenic signaling, leading to the resistance to CSF1R inhibition. We further find that co-targeting anti-angiogenic therapy with CSFR1 inhibition reduces cell proliferation and alters tumor morphology. Our work may help clinicians better identify GBM patients who may benefit from TAM targeting.
Glioblastoma (GBM) is a highly malignant brain tumor with poor prognosis. Despite aggressive multimodal therapies including radiation and temozolomide,1 median survival of GBM patients is a dismal 14 months. Based on transcriptional profiling, human GBMs have been classified into at least 3 subtypes including a proneural subtype associated with oligodendrocyte lineage signatures that often exhibits PDGFRA amplification, TP53, loss and increased phosphoinositide 3-kinase (PI3K) signaling; a classical subtype enriched in astrocytic signatures characterized by EGFR gain of function; and a mesenchymal subtype associated with immune infiltration and the loss of NF1 and TP53.2 However, clinical trials targeting subtype-specific signaling pathways in GBMs have not been successful.1
Individual tumors display significant molecular and cellular heterogeneity with diverse co-existing cell types including tumor cells, immune cells, and vascular cells.3,4 Single-cell profiling has demonstrated that individual tumor cells exist on a continuum between 4 cell lineages: astrocyte-like, oligodendrocyte progenitor cell-like, neural progenitor cell-like, and mesenchymal-like subtypes.5 Brain tumor cells have been shown to recruit immune cells and vascular networks to support their growth.4 How the tumor microenvironment (TME) influences the growth of genetically and phenotypically distinct glioma subtypes remains poorly understood. Therapies targeting the TME represent a potential approach to anti-glioma therapy due to the mutational stabilities of these cells.4 Tumor-associated macrophages/microglia (TAMs) are the most prevalent microenvironmental cells in GBM,4 especially in the mesenchymal subtype of GBM.2 TAMs in the central nervous system (CNS) include microglia, CNS border-associated macrophages (BAMs), and monocyte-derived macrophages.6 Resident microglia in the brain play important roles in brain development, homeostasis, maintenance of neuronal networks, and local inflammation.7,8 BAMs present at CNS interfaces, such as the perivascular and perimeningeal spaces,8 participate in antigen presentation to infiltrating T cells.9 Macrophages localize to the tumor core of mouse and human gliomas, support angiogenesis and tumor growth, and are associated with poor prognosis in GBM.10–12
Targeting of TAMs has been explored through evaluation of small molecular inhibitors of macrophage colony-stimulating factor 1 receptor (CSF1R) signaling, which is involved in recruitment, proliferation, and polarization of TAMs.4 CSF1R inhibition in mouse models of glioma has revealed that TAMs function in tumor growth, immunosuppression, regulation of vascular networks, and regulation of invasion, depending on the tumor model.4,10,11 Treatment of a mouse PDGFB-driven glioma model with CSF1R inhibitor BLZ945 promoted durable regression and dramatically improved long-term survival of tumor-bearing mice.13 However, clinical trials of CSF1R inhibitor PLX3397 in recurrent human GBMs showed no population-wide response, while 2 patients in the trial showed an extended survival response,14 suggesting that glioma subtypes vary in their responses to CSF1R-dependent TAM targeting.
To determine the responsiveness of glioma subtypes to TAM targeting, we examined different animal models of gliomas for sensitivity to CSF1R inhibition. Similar to the previous report,15 we found that the CSF1R inhibitor hindered tumor growth in a proneural-like PDGFB-DNp53-induced glioma model. In contrast, CSF1R inhibition in oncogenic RAS-driven mesenchymal-like gliomas led to acceleration of tumor growth at the early phase of tumor progression. Single-cell transcriptomic profiling revealed that programs associated with inflammation and immunosuppression are upregulated in TAMs in the microenvironment of RAS-driven tumors compared with those in the PDGFB-driven TME. In RAS-driven gliomas, interactions between TAMs and the vasculature are prevalent. Further, co-targeting of TAMs and angiogenesis reduced proliferation and altered tumor morphology in RAS-driven gliomas. Together, our studies suggest that the microenvironment landscapes elicited by molecularly distinct GBM subtypes dramatically impact the efficacy of anti-microenvironment therapies.
Methods
Animal Use
Immunodeficient NOD SCID gamma (NSG) mice and immunocompetent Boy/J mice, a congenic C57BL6/J strain with a CD45.1 variant, were provided by Cincinnati Children’s Hospital Medical Center Cancer Core. The animal studies and procedures were approved by the IACUC (Institutional Animal Care and Use Committees) of the Cincinnati Children’s Hospital Medical Center, USA.
Cloning of HRASV12-dnp53 Viral Vector
The coding sequence from pWZL-hygro-HRASV12 (Addgene #18749) was ligated into pQCXIX-IRES-dnp53 destination vector.16 See Supplementary Methods for more details.
Virus Production
Retroviruses were packaged using calcium phosphate transfection of pQCXIX-PDGFB-IRES-dnp53 or pQCXIX-HRASV12-IRES-dnp53 into 293T cells. See Supplementary Methods for more details.
Stereotactic Injections
Retrovirus was injected in the subventricular zone (−1.5, 2.0, −2.3) or subgranular zone (−2, 1.5, −2.3). Tumor cells were injected in the mouse cortex (1, 0.5, −2) or striatum (0.2, 2.2, −3). See Supplementary Methods for more details.
Cell Culture
Human GSC262 and GSC20, and murine tumor cell lines were grown in serum-free DMEM/F12 media. Cell proliferation was assessed using a WST-1 cell viability kit (Takara, Bio MK400, Mountain View, CA, USA). See Supplementary Methods for more information.
Primary Tumor Cell Line Isolation
Tumor tissue was isolated and digested using Accutase and cell suspension was plated in the appropriate media above. See Supplementary Methods for more information.
In Vivo Drug Treatments
PLX3397 hydrochloride and BKM120 (MedKoo Biosciences, Morrisville, NC, USA), and cediranib (LC Laboratories, Woburn, MA, USA) were used for drug treatments in vivo. Mice were sacrificed when they developed neurologic symptoms. See Supplementary Methods for more information.
Tissue Sectioning and Staining
Brains were processed for vibratome and paraffin sectioning as previously described.16 See Supplementary Methods for more information.
Image Quantification
All other quantifications were done using the multipoint tool in ImageJ. See Supplementary Methods for more information.
RNA Sequencing and Analysis
Analysis of differential expression genes (FDR <0.05 and fold changes log2 > 2) was performed using the DESeq2 package from R 4.0.2 using raw gene counts as an input. See Supplementary Methods for more information.
Single-Cell RNA Sequencing and Analysis
Primary tumors were dissociated and analyzed using Drop-Seq or 10× Chromium 3v3 Profiling as previously described.16 See Supplementary Methods for more information.
Statistical Analysis
Detailed information of all the statistical analyses performed is described in the Supplementary Methods.
Results
Targeting TAMs Blocks Tumor Growth in Murine PDGFB-Driven Gliomas But Not Human Proneural GBMs
To determine the function of TAMs in glioma growth, we used primary tumor cells from an animal model of proneural GBM16 that overexpressed human PDGFB and dominant-negative p53 (dnp53). We implanted these cells in immunocompetent adult mice to establish a syngeneic GBM model in which recipient mice develop full-blown tumors at around 14 days’ post-implantation. Recipient tumors exhibit pathological and morphological characteristics of human GBM.16
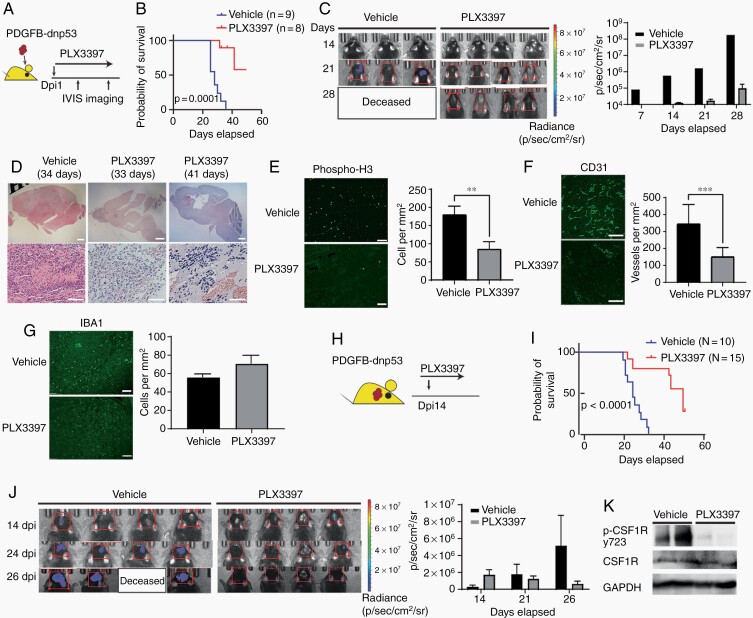
To inhibit macrophage activity, we treated mice daily with PLX3397, an FDA-approved oral brain-penetrant CSF1R inhibitor that causes macrophage depletion or reprogramming, starting the day after tumor implantation (Figure 1A). In contrast to vehicle-treated mice that died around 32 days, the treatment with PLX3397 blocked the growth of the transplanted tumor cells and significantly (P = .0001) extended animal lifespan (Figure 1B and C). There were no apparent tumor masses in the PLX3397-treated mice that had been transplanted with PDGFB-driven GBM cells (Figure 1D). Mice that died while being treated showed small residual tumors with high degrees of hemorrhage (Figure 1D).
Fig. 1.
Targeting TAMs blocks tumor growth in murine PDGFB-driven proneural-like gliomas. (A) Experimental design of PLX3397 treatment in PDGFB-driven glioma. Animals were treated every day post-tumor implant (dpi) and monitored by bioluminescent imaging. (B) Kaplan-Meier survival curve of mice treated with PLX3397 or vehicle. Log-rank test. (C) Representative bioluminescent images of mice treated with vehicle or PLX3397. Bar graph: average photon flux at each time point. (D) Representative H&E-stained sections of PDGFB-driven tumors treated with vehicle or PLX3397. Low power scale: 1 mm, High power scale: 300 µM. (E–G) Left: Representative immunostaining for p-H3 (E), CD31 (F), or IBA1 (G) in PDGFB-driven GBM treated with vehicle or PLX3397 for 3 days post-tumor implant. Right: Quantification of label-positive cells per unit area (n = 3 mice/group); **P < .01; ***P < .001, unpaired t test. Scale bars in E and F, 100 µM; G, 300 µM. (H) Experimental design of PLX3397 treatment of established PDGFB-driven tumors. Mice were treated daily with PLX3397 or vehicle beginning 14 days after tumor implantation. (I) Kaplan-Meier survival curve of mice. Log-rank test. (J) Representative bioluminescent images. Bar graph of average photon flux at 14, 21, and 26 days. (K) Western blot of phospho-CSF1R from mouse PDGFB tumors (n = 2 samples per group and repeated twice) treated with vehicle or 100 mg/kg PLX3397 for 2 days from day 14 to 16. Abbreviations: CSF1R, colony-stimulating factor 1 receptor; GBM, glioblastoma; PDGFB, platelet-derived growth factor subunit B; TAMs, tumor-associated macrophages/microglia.
Examination of tumor tissues after 3 days of PLX3397 treatment showed a dramatic decrease in the number of proliferative phospho-H3+ cells and in CD31+ blood vessel density (Figure 1E and F). Despite the reduction of CSF1R activity, the number of IBA1+ TAMs was not altered by PLX3397 treatment (Figure 1G). This suggests that TAMs are reprogrammed rather than depleted by PLX3397 in the PDGFB-driven GBM model. This is consistent with a previous report in a similar PDGFB-induced glioma model (RCAS-hPDGFB/Nestin-Tv-a; Ink4a/Arf−/−).13,17
To assess whether targeting TAMs with the CSF1R inhibitor extended survival in established tumors, we treated established PDGFB-driven GBM tumor-bearing mice with PLX3397 at 14 days after tumor transplant (Figure 1H). PLX3397 treatment extended mouse survival (Figure 1I) and inhibited the outgrowth of established tumors (Figure 1J). CSF1R phosphorylation decreased in tumors after PLX3397 treatment (Figure 1K). In contrast to the strong effects of PLX3397 on tumors in vivo, treatment of tumor cells with PLX3397 in vitro did not reduce tumor cell viability (Supplementary Figure S1A). Similarly, other murine glioma cell lines and human GSC lines showed minimal responsiveness to PLX3397 in vitro (Supplementary Figure S1B–E). Furthermore, the phosphorylation level of PDGFRb, a potential target of PLX3397, was not substantially altered in PDGFB-driven tumors in vivo after treatment (Supplementary Figure S1F). These observations suggest that CSF1R inhibition targets TAMs rather than tumor cells to block initiation and progression of PDGFB-driven GBM tumors.
To investigate if TAMs are important in the growth of human proneural GBMs, we used a patient-derived xenograft (PDX) GBM model transplanted with proneural glioma stem-like cells (GSC262)18—a cell line with EGFR amplification, TP53 mutation, and CDKN2A deletion. We treated these mice with PLX3397 after the tumor had been established in the brain. In this model, CSF1R inhibition did not have significant effects on tumor growth, histology, or animal survival (Supplementary Figure S2). This suggested that, unlike the proneural-like PDGFB-driven syngeneic mice, human proneural GBM PDX tumors derived from GSC262 are not responsive to CSF1R inhibition.
RAS-Driven Mesenchymal-Like Gliomas Are Resistant to CSF1R Inhibition
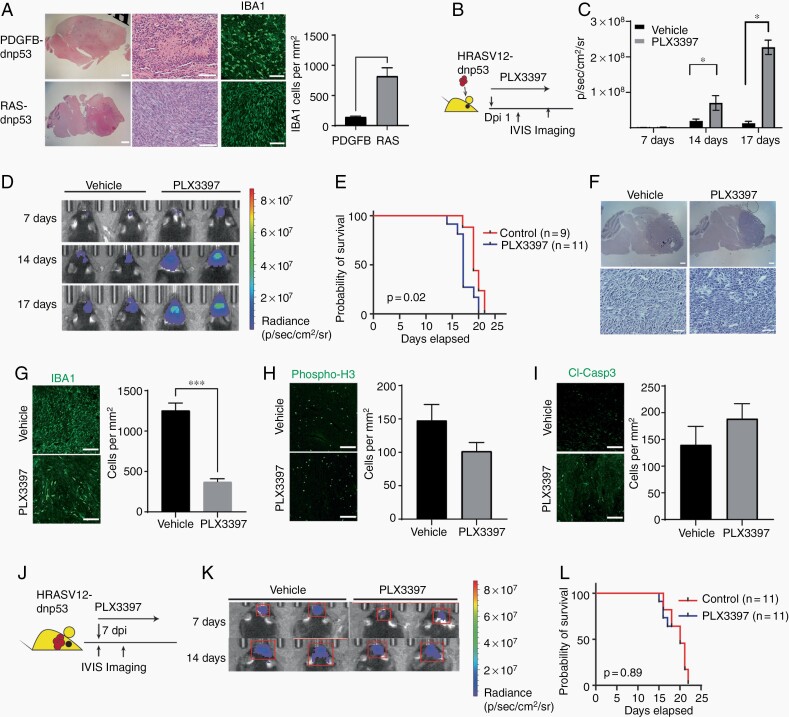
GBM tumors of the mesenchymal subtype are driven by activation of RAS signaling due to NF1 loss and are densely infiltrated by TAMs.2 To determine whether CSF1R inhibition could suppress tumor growth of mesenchymal GBM, we developed an animal model of mesenchymal-like gliomas by expressing oncogenic human HRASV12 and dnp53, which mimic the loss of NF1 and TP53 (designated as RAS-driven GBM). Expression of HRASV12 and dnp53 has previously been shown to induce the mesenchymal-like GBM in mice.19 Adult mice transduced with HRASV12-dnp53 retrovirus in the subventricular zone developed GBMs as early as 30 days’ post-transduction (Supplementary Figure S3A and B). Secondary tumors derived from transplantation of the RAS-driven GBM cells resulted in animal death within 25 days. Compared to corresponding endpoint tumors of PDGFB-driven GBMs, RAS-driven GBMs in implanted mice appeared to have higher cell density and intense IBA1+ TAM infiltration (Figure 2A) as well as pro-inflammatory MHCII+ TAMs (Supplementary Figure S3C and D. To verify the mesenchymal identify of RAS-driven GBM lines, we performed transcriptional profiling of cell lines isolated from RAS-driven GBM tumors. We observed enrichment of terms related to mesenchymal GBM, epithelial-mesenchymal transition and Hippo signaling, and upregulation of YAP/TAZ, the characteristic features of mesenchymal GBM phenotype (Supplementary Figure S3E and F).
Fig. 2.
RAS-driven mesenchymal-like mouse gliomas are resistant to CSF1R inhibition. (A) Representative images of PDGFB- and RAS-driven tumors stained with H&E and for IBA1 and quantification (right). Scale (low power, 1 mm; high power, 300 µM). (B) Experimental design of early treatment of RAS-driven gliomas with PLX3397. (C) Average photon flux from RAS-driven tumors at indicated time points in mice treated with PLX3397 or vehicle. *P < .05, t test. (D) Representative bioluminescent images of mice with RAS-driven tumors treated with vehicle or PLX3397 beginning 1 day after implantation. (E) Kaplan-Meier survival curve of mice with RAS-driven tumors treated with vehicle or PLX3397 starting 1 day after tumor transplantation. P-value calculated using log-rank test. (F) Representative images of H&E-stained tumors from mice with RAS-driven tumors treated with vehicle or PLX3397. Scale bars: upper, 300 µM, and lower, 100 µM. (G–I) Left: Representative images of tumors stained for IBA1 (G), phospho-H3 (H), or cleaved caspase 3 (I) from mice with RAS-driven tumors treated with vehicle or PLX3397. Scale: 100 µM. Right: Quantification of label+ cells per unit area (n = 3 mice/group, ***P < .001, unpaired t test). (J) Experimental design for CSF1R inhibition in established RAS-driven tumors. (K) Representative bioluminescent images of mice with established RAS-driven tumors treated with vehicle or PLX3397. (L) Kaplan-Meier survival curves of mice with RAS-driven tumors treated with vehicle or PLX3397 after tumors were established. Log-rank test. Abbreviations: CSF1R, colony-stimulating factor 1 receptor; PDGFB, platelet-derived growth factor subunit B.
To determine the effect of CSF1R inhibition on mesenchymal-like glioma growth, we treated the RAS-driven GBM syngeneic mice with PLX3397 daily starting the day after tumor cell transplantation (Figure 2B). Surprisingly, we found that PLX3397 treatment at the early phase of tumorigenesis accelerated tumor growth (Figure 2C and D). This resulted in a significant shorter animal life span in the syngeneic RAS-driven GBM mice (Figure 2E). Tumor morphology was not altered substantially following PLX3397 treatment (Figure 2F), but PLX3397 treatment did cause a decrease in the number of IBA1+ TAMs in RAS-driven GBM tumors (Figure 2G). This contrasts to the maintenance of these cells in PDGFB-driven GBM tumors after PLX3397 treatment (Figure 1G).13 In addition, cell proliferation and apoptosis assayed by phospo-H3 and cleaved caspase 3 immunostaining were not significantly changed by CSF1R inhibition in the RAS-driven GBM tumors (Figure 2H and I).
We further investigated the effect of TAM inhibition on the tumor progression of established RAS-driven GBM tumors. In contrast to increased tumor growth when PLX3397 treatment was initiated immediately after implantation, PLX3397 administration in animals with established RAS-driven GBM tumors did not have a substantial effect on tumor progression or animal survival compared with vehicle-treated mice (Figure 2J–L).
To explore whether primary resistance to CSF1R inhibition is a general characteristic of mesenchymal glioma, we examined 2 additional mesenchymal GBM models. In a model with tumors derived from neural progenitors overexpressing PDGFRA-D842V and dnp53,20 PLX3397 treatment did not inhibit tumor progression in the syngeneic mice with established GBM tumors (Supplementary Figure S4). Similarly, PDX mice orthotopically transplanted with human mesenchymal GSC2018 cells also failed to show responses to PLX3397 (Supplementary Figure S5). These observations suggest that targeting of TAMs with PLX3397 does not effectively inhibit tumor growth in genetically distinct mesenchymal subtype glioma models.
Targeting Intrinsic PI3K Signaling Does Not Sensitize RAS-Driven Gliomas to PLX3397 Inhibition
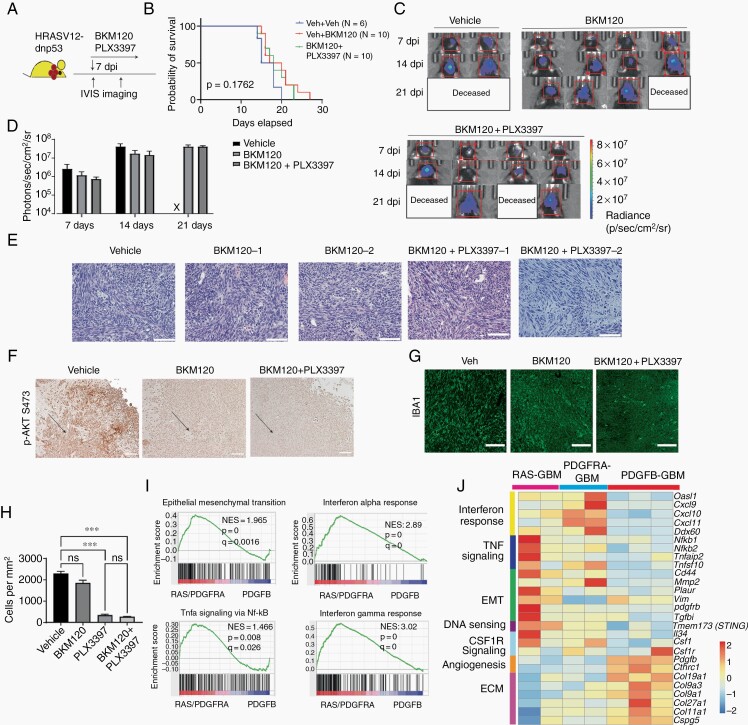
Activation of PI3K signaling has been associated with resistance to PLX3397 in PDGFB-driven glioma models.17 To determine whether inhibition of PI3K signaling sensitizes RAS-driven tumors to CSF1R inhibition, we treated RAS-driven gliomas with a brain-penetrant reversible PI3K inhibitor BKM120,17 or the combination of BKM120 and PLX3397 starting 7 days after tumor transplantation (Figure 3A). In contrast to previous studies in the PDGFB-driven relapsed PDGFB-driven glioma model,17 individual or combination treatment did not lead to significant improvement in animal survival (Figure 3B). Tumor growth and morphology were similar among treatment groups (Figure 3C–E). Mice treated with BKM120 singly or in combination with PLX3397 showed lower phospho-AKT (p-Akt) staining (Figure 3F). Inhibition of PI3K signaling alone did not have a significant influence on the number of IBA1+ cells detected, but the combination of BKM120 with PLX3397 did decrease the number of IBA1+ TAMs in tumor tissues (Figure 3G and H). Similarly, the combination treatment did not impact tumor growth in a PDGFRA-driven mesenchymal-like glioma model20 (Supplementary Figure S5). These observations suggest that inhibition of PI3K-mediated signaling is insufficient to sensitize RAS- or PDGFRA-driven gliomas to CSF1R inhibition.
Fig. 3.
Tumor-intrinsic PI3K signaling does not explain the resistance of RAS-driven GBM to PLX3397 inhibition. (A) Experimental design of trial of PI3K inhibitor BKM120 (20 mg/kg) in RAS-driven gliomas. (B) Kaplan-Meier survival curves of mice with RAS-driven tumors with different treatments. Log-rank t test. (C) Bioluminescent imaging of mice with RAS-driven gliomas treated with BKM120 or combination therapy. (D) Quantification of average flux from RAS-driven tumors in mice treated with vehicle, BKM120, or combination therapy. All vehicle mice were deceased at 21 days. (E) Representative histology of RAS-driven tumors from mice treated with vehicle, PI3K inhibitor, or combination therapy. Scale 300 µM. (F, G) Representative images of tumors stained for phospho-Akt S473 (F, scale 1 mm.) or IBA1 (G, scale 100 µM) in RAS-driven tumors treated with vehicle, BKM120, or combination therapy. (H) Quantification of IBA1+ cells per unit area in each treatment group (n = 3 mice/treatment group). One-way ANOVA and multiple comparisons (***P < .001). (I) Hallmark terms enriched in transcriptomes of CSF1R-resistant tumors (RAS-driven and PDGFRA-dnp53) compared to those of CSF1R-sensitive tumors (PDGFB tumors). (J) Heatmap of genes enriched in CSF1R-resistant tumor cells (RAS-driven and PDGFRA-dnp53) when compared to PDGFB gliomas. Abbreviations: CSF1R, colony-stimulating factor 1 receptor; GBM, glioblastoma ;PDGFB, platelet-derived growth factor subunit B.
To identify other potential tumor-intrinsic regulators of resistance to CSF1R inhibition, we performed transcriptome profiling of primary cell lines from PDGFB-driven GBM, RAS-driven GBM, and PDGFRA-driven GBM tumors.20 As expected due to their mesenchymal identities, PDGFRA- and RAS-driven GBM cell lines showed enrichment of genes associated with epithelial-mesenchymal transition (Figure 3I), such as Cd44, Vim, Pdgfrb, Tgfbi, Loxl1, and Mmp2 (Figure 3J). In addition, we found that the CSF1R inhibition-resistant GBM lines (RAS- and PDGFRA-driven GBM lines) were enriched in transcripts involved in pro-inflammatory signaling pathways such as the interferon response and TNF-mediated signaling compared to CSF1R inhibition-sensitive PDGFB-driven GBM gliomas (Figure 3I and J).
Bone Marrow-Derived Macrophages Do Not Have a Pro-Growth Effect on PDGFB- or RAS-Driven Tumors
Macrophage ontogeny has been proposed as an important regulator of TAM function in GBM.12 Bone marrow-derived macrophages can infiltrate tumors and produce growth factors to drive the immune-suppressive phenotype and growth of GBMs.11,12 The chemokine receptor CCR2 is critical for the migration of bone marrow-derived macrophages into the CNS.11,12 To determine whether infiltration of myeloid-derived cells into PDGFB-driven tumors could lead to the responsiveness to CSF1R inhibition, we examined tumor growth in Ccr2-knockout mice, wherein the Ccr2 gene is disrupted by RFP insertion.21 Intriguingly, the loss of Ccr2 did not affect tumor growth or animal survival when PDGFB-driven GBM cells were transplanted into Ccr2-knockout mice (Supplementary Figure S7A and B). There was very little co-localization between RFP and IBA1, suggesting that the IBA1+ TAM compartment is not continuously replenished by CCR2+ bone marrow-derived cells (Supplementary Figure S7C). Accordingly, CCR2 knockout led to only modest changes in the infiltration of IBA1+ cells (Supplementary Figure S7E). While the numbers of CCR2-RFP+ cells only modestly decreased, we detected an increase in a population of RFP+ cells expressing a T cell marker CD3 (Supplementary Figure S7F and G), which is consistent with a report that CCR2 deficiency leads to an increase in CCR2+ CD4+ T cells in glioma tissues.22 Similarly, the growth of RAS-driven tumors was not altered in Ccr2-knockout mice (Supplementary Figure S7H). In addition, PLX3397 treatment did not appear to result in a significant difference in survival outcomes between control and Ccr2-knockout mice bearing either RAS- or PDGFB-driven tumors (Supplementary Figure 7I and J). Given the lack or reduction of marrow-derived macrophages in Ccr2-knockout mice, our data suggest that bone marrow-derived macrophages may not contribute substantially to the growth of PDGFB- or RAS-driven gliomas.
Single-Cell RNA-Seq Identifies Differences in TAM Populations Between RAS- and PDGFB-Driven Gliomas
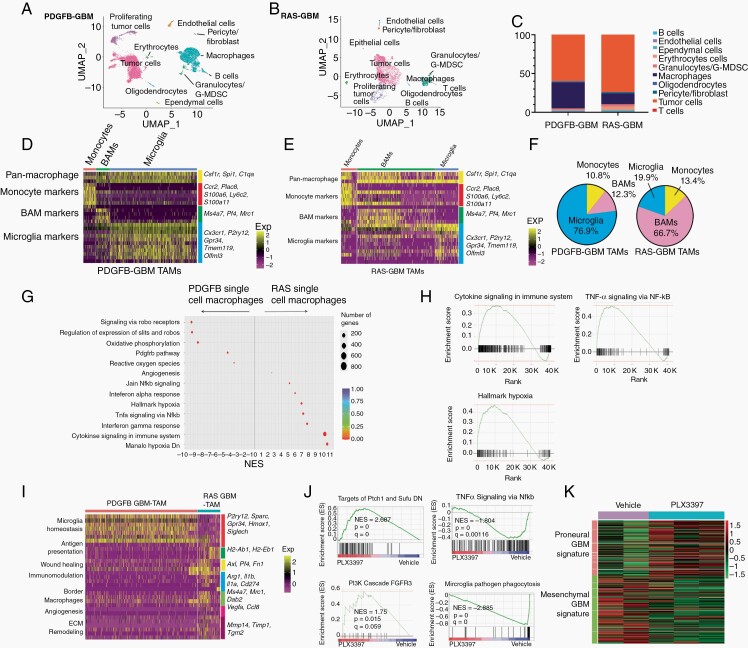
To dissect the cellular compositions and functions of TAMs in RAS- and PDGFB-driven tumors, we performed 10× Chromium single-cell RNA-seq (scRNA-seq) analysis of primary RAS-driven gliomas and compared it with single-cell transcriptomic data from PDGFB-driven gliomas induced by PDGFB and dnp53.16 Transcriptome data were analyzed from 15 565 cells from RAS-driven GBM and 16 773 cells from PDGFB-driven GBM. Unsupervised clustering analysis identified 8 different clusters with distinct gene expression signatures (Figure 4A and B). The clusters included tumor cells and immune cell populations (eg, microglia, microphages, T cells, B cells, dendritic cells, granulocytes, pericytes, and endothelial cells). The scRNA-seq analysis was also performed using the highly parallel droplet-based single-cell transcriptomics (Drop-seq) platform,23 and similar cellular clusters were identified (Supplementary Figure S8).
Fig. 4.
scRNA-seq reveals differences in TAM populations in RAS- and PDGFB-driven GBM. (A, B) UMAP plot of clusters derived from 10× Genomics profiling of end-stage PDGFB-driven tumors (A) and RAS-driven glioma (B). (C) Bar graph of distribution of cell types in the two glioma models. (D, E) Heatmap of expression of marker genes for monocytes, BAMs, and microglia in PDGFB-driven gliomas (D) and RAS-driven gliomas (E). (F) Pie chart showing subdivision of macrophage populations in the two GBM models. (G) Rich plot of differentially enriched terms between RAS-driven and PDGFB-driven glioma macrophages. (H) Plots of terms enriched in transcriptomes of macrophages from RAS-driven vs PDGFB-driven tumors. (I) Heatmap of differentially expressed genes in macrophages from PDGFB- and RAS-driven tumors. (J) Gene set enrichment analysis of gene expression between vehicle and PLX3397 treated RAS-GBM (n = 3 samples/group). (K) Expression of proneural and mesenchymal signature genes from RAS-driven tumors treated with vehicle or PLX3397. Abbreviations: BAMs, border-associated macrophages; GBM, glioblastoma; PDGFB, platelet-derived growth factor subunit B; TAMs, tumor-associated macrophages/microglia; UMAP, Universal Manifold Approximation and Projection.
To assess the heterogeneity of TAMs, we identified populations of TAMs expressing pan-macrophage markers such as C1qa, Csf1r, and Spi1 in PDGFB- and RAS-driven tumors. In PDGFB-driven tumors, 26% of the cells were TAMs, whereas in RAS-driven tumors, 5.7% of cells were TAMs (Figure 4C). Clustering of TAMs based on gene expression patterns identified cellular characteristics of cells previously reported to be present in normal brain and in brain tumors24 including BAMs expressing Ms4a7, F13a1, Pf4, and Dab2, microglial TAMs (Gpr34, Sall1, and Sparc), and monocytic TAMs (Ccr2, Ifitm3, Ly6c2, and Ifitm2) (Figure 4D and E). Brain microglial cells were the predominant TAM (76% of total) in PDGFB-driven gliomas (Figure 4D and F), while the remainder were perivascular TAMs (12%) or monocyte-derived TAMs (11%). In contrast, in Ras-driven glioma, 65.5%, expressed markers of BAMs, 15.6% were brain microglial cells, and 18.9% were monocyte-derived macrophages (Figure 4E and F).
TAM Characteristics Differ in RAS-Driven and PDGFB-Driven Tumors
We next compared gene expression in TAMs from RAS-driven and PDGFB-driven tumors and identified pathways that are differentially activated. The macrophages in both RAS-driven and PDGFB-driven tumors did not show strong expression of classical M1 or M2 macrophage marker genes or increase M1 or M2 signatures after PLX3397 treatment (Supplementary Figure S9A–C). Unbiased gene set enrichment analysis (GSEA) revealed enrichment of pathways including Slit-Robo signaling, oxidative phosphorylation, PDGFRB, and neuroactive signaling in TAMs from PDGFB-driven tumors (Figure 4G). The predominant TAMs in PDGFB-driven GBM were characterized by relatively high expression of brain microglia-enriched genes (P2ry12, Sparc, Gpr34, and Siglech). TAMs from PDGFB-driven tumors treated with PLX3397 exhibited a downregulation of genes associated with cell proliferation and antigen presentation, while an increase in genes associated with regulation of vascular stability and Hippo signaling (Supplementary Figure S9D), suggesting transcriptional reprogramming in TAMs. In contrast, in TAMs from RAS-driven tumors, enrichment in inflammatory signaling such as TNF, interferon, NF-κb, and hypoxia pathways were observed (Figure 4G and H). Strikingly, in TAMs from RAS-driven tumors, there was upregulation of transcripts involved in antigen presentation (eg, H2-Ab1- and H2-Eb1-encoding MHCII proteins), inflammation (IL1b), angiogenesis (Vegfa and Ccl8), and immunosuppression (Arg1 and Cd274, the latter encoding checkpoint protein PD-L1) (Figure 4I).
To confirm the pro-inflammatory nature of TAMs from RAS-driven tumors, we performed bulk transcriptomic profiling of endpoint RAS-driven tumors treated with vehicle or PLX3397. GSEA analysis revealed that PLX3397-treated RAS-driven tumors had lower levels of transcripts related to NF-κb signaling and phagocytosis, suggesting a decrease in tumor inflammation (Figure 4J). There was a concomitant upregulation of pro-growth signaling pathways such as Hedgehog and PI3K-FGFR signaling following macrophage depletion (Figure 4J), consistent with accelerated tumor growth after CSF1R inhibition in RAS-driven tumors at early stages. PLX3397-treated RAS-driven tumors also showed an enrichment in the proneural signature of GBM, whereas tumors from vehicle-treated mice showed an enrichment of the mesenchymal GBM signature (Figure 4K). This suggests that TAM targeting resulted in a mesenchymal to proneural transition in RAS-driven tumors, consistent with TAMs as a driver of mesenchymal GBM identity.25
TAM Subtypes in Murine GBMs Resemble Their Human Counterparts
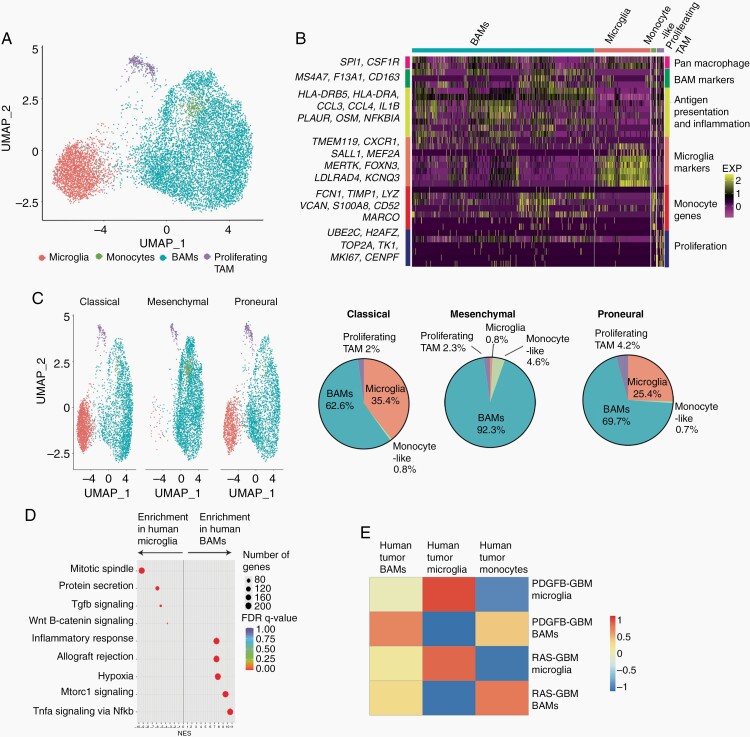
To compare the compositions of TMEs in human GBM subtypes, we examined single-cell transcriptome data of GBM tumors from publicly available datasets.5,26,27 Human TAMs segregated into 4 distinct clusters (Figure 5A). All TAM clusters showed expression of pan-macrophage markers SPI1 and CSF1R (Figure 5A and B). One cluster included canonical microglia as indicated by the expression of TMEM119, SALL1, and MEF2A. Another cluster appears to be similar to BAMs (MS4A7, CD163, and F13A1), and a third cluster expressed monocyte markers FCN1, VCAN, and LYZ (Figure 5A and B). The final cluster includes proliferative TAMs that express cell proliferation markers. We found that mesenchymal subtype tumors had a higher fraction of the BAM cluster cells than the other subtypes (Figure 5C) and that mesenchymal subtype tumors were enriched in genes associated with inflammatory responses, hypoxia, and NF-κb signaling (Figure 5D). Human microglia TAM and BAMs showed enrichment of signatures corresponding to the PDGFB- and RAS-driven counterparts, respectively (Figure 5E), suggesting that TAM subtypes in murine GBMs resemble those of the human counterparts.
Fig. 5.
TAMs in human GBMs segregate based on compartment of origin. (A) UMAP plot of 10× Genomics scRNA-seq and snRNA-seq of TAMs from human GBM datasets clustered using Seuratv3. (B) Heatmap of marker genes for BAMs, monocyte-like cells, microglia, and proliferating TAMs from human GBMs. (C) UMAP plots of TAM clusters and percentages of BAMs, monocyte-like cells, microglia, and proliferating TAMs in different human GBM subtypes. (D) Rich plot of Hallmark gene sets enrichment in human BAMs vs microglia. (E) Correlation of mouse RAS-BAM and PDGFB-microglia signatures with TAMs in human GBMs. Abbreviations: BAMs, border-associated macrophages; GBM, glioblastoma; PDGFB, platelet-derived growth factor subunit B; TAMs, tumor-associated macrophages/microglia; UMAP, Universal Manifold Approximation and Projection.
Combination of Angiogenesis and CSF1R Inhibition Decreases Cell Proliferation in RAS-Driven GBM
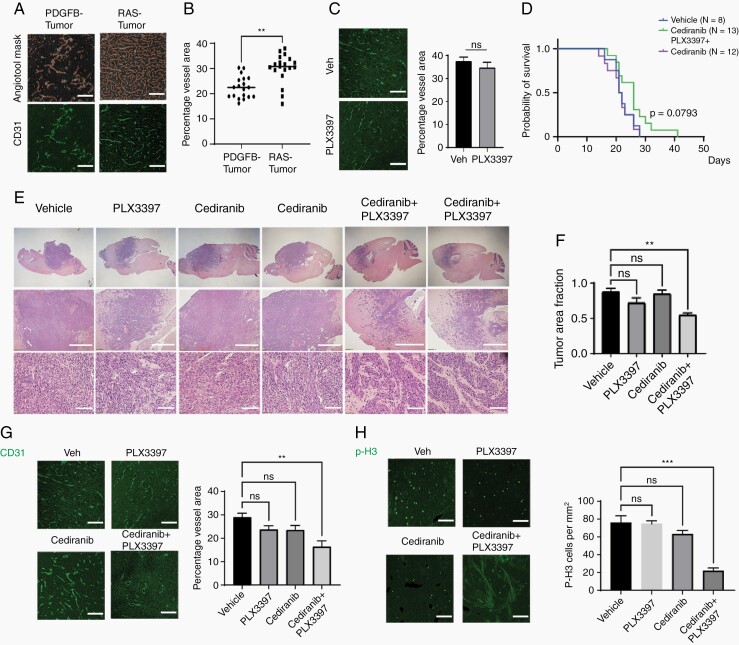
To further understand how TME cells communicate with tumor cells in PDGFB- and RAS-driven GBM, we performed receptor ligand profiling using the CellChat pipeline.28 We found that RAS-driven tumors had more extensive TME communication and increased TAM-to-vascular signaling than PDGFB-driven GBM (Supplementary Figure S10A and B). In PDGFB-driven GBM, endothelial cells VEGF signaling from pericytes (Supplementary Figure S10C and D). In contrast, RAS-driven GBM endothelial cells receive angiogenic signals from multiple cell types including macrophages, granulocytic cells, tumor cells, and pericytes (Supplementary Figure S10C and D). Consistent with the predicted multiple overlapping angiogenic systems, RAS-driven GBMs exhibited a higher tumor vessel density when compared to PDGFB-driven GBMs (Figure 6A and B). Notably, PLX3397 treatment caused a decrease in CD31+ vessels in PDGFB-driven tumors (Figure 1), whereas vascular density was not affected by PLX3397 treatment when tumors were driven by RAS (Figure 6C).
Fig. 6.
Combination of anti-angiogenic therapy with CSF1R inhibition decreases proliferation in RAS-driven GBM but does not improve survival outcomes. (A) Representative images of PDGFB- and RAS-driven tumors stained for CD31. Scale 100 µM. (B) Vessel density assayed by CD31 staining in PDGFB- and RAS-driven tumors by Angiotool (n = 4 samples/group, ***P < .001). (C) Representative images of CD31+ vessels from Ras-driven tumors treated with vehicle or PLX3397, scale 100 µM. Vessel area measured by Angiotool (ns, not significant). (D) Kaplan-Meier survival curve of mice with RAS-driven tumors treated with cediranib (6 mg/kg daily), vehicle, or the combination of cediranib with PLX3397 (100 mg/kg daily). (E) Representative tissue histology of tumors treated with vehicle, PLX3397, cediranib, or combination therapy. Scale: middle (1 mm) and lower panels (300 µM). (F) Quantification of tumor area fraction for tumors receiving vehicle, PLX3397, cediranib, or combination therapy (n = 3 tumors/group). **P < .01, 1-way ANOVA with multiple comparisons. ns, not significant. (G–H) Left: Representative images of CD31+ blood vessels (G) and p-H3+ cells (H) in endpoint RAS-driven tumors. Scale 100 µM. Right: Quantification of label+ cells in all treatment groups (n = 3 mice/group). **P < .01, ***P < .001, 1-way ANOVA with multiple comparisons. ns, not significant. Abbreviations: CSF1R, colony-stimulating factor 1 receptor; GBM, glioblastoma; PDGFB, platelet-derived growth factor subunit B.
To examine whether co-targeting of TAMs and angiogenesis would impact the phenotype and growth of RAS-driven GBM, we treated mice implanted with RAS-driven GBM cells with PLX3397, cediranib (a VEGFR2 inhibitor), or both. Treatment with cediranib, PLX3397, or both yielded similar survival curves in RAS-driven GBM models (Figure 6D). The combination treatment resulted in less dense tumor lesions, which appeared to be infiltrative (Figure 6E and F). We observed that vessel density was decreased in tumors from mice treated with the combination of cediranib and PLX3397, but not individual agents alone (Figure 6G). In addition, the combination treatment but not either agent alone, reduced cell proliferation in RAS-driven GBM tumors as measured by staining for the proliferative marker p-H3 (Figure 6H). Cediranib treatment led to an increase in infiltration of CD3+ and Granzyme B (Gzmb+) T cells but did not affect the fraction of PD-1+ exhaustion T cells or the number of NK1.1+ NK cells (Supplementary Figure S11A–C and F), consistent with a trend toward improved survival in RAS-driven glioma (Figure 6D). However, the increase in active T cell infiltration was suppressed by depletion of TAMs in combination treatment (Supplementary Figure S11A–D). In addition, cediranib treatment increased the number of intratumoral regulatory T cells (FoxP3+ Treg cells) (Supplementary Figure S11A and D), which might limit the effects of the influx of cytolytic T cells on tumor growth.
We next investigated the effect of combination therapy in other PDGFB- and PDGFRA-driven glioma models. In the PDGFB-driven model, neither cediranib nor combination therapy with CSF1R inhibition improved survival or altered tumor morphologies compared with PLX3397 monotherapy (Supplementary Figure S12A and B), suggesting an absence of the synergy of combination therapy in PDGFB-driven tumors. Combination therapy in PDGFRA-driven tumors tended to have a worse outcome, similar to RAS-driven tumors but did not increase tumor cell invasiveness (Supplementary Figure S12C and D). These observations suggest a model-specific response to PLX3397 treatment among different glioma models.
Discussion
The growth of GBMs is controlled not only by genetic alterations in tumor cells but also by microenvironment fitness. The brain has a unique microenvironment that has evolved to dampen immunity. How genetically distinct gliomas reprogram this microenvironment to support cancer progression remains poorly understood. A better understanding of the role of the TME will facilitate the development of innovative and efficacious strategies for targeting brain tumors. Current therapies focus mostly on targeting the glioma tumor cells without accounting for TME constituents. In this study, we show that targeting unique microenvironmental TAMs present in disparate glioma subtypes can act as a double-edged sword, contributing to the suppression of cancer cells in the PDGFB-driven model but promoting their growth and survival in other glioma models. Thus, our data reveal a context-dependent role of TME on glioma growth and progression.
Early characterization of glioma-associated macrophages classified these cells into tumor-inhibiting M1 and tumor-promoting M2-like phenotypes.4 Recent profiling studies indicate that both human and rodent TAMs are highly heterogenous, transcriptionally diverse populations with both pro- and anti-tumorigenic characteristics distinct from M1 or M2 macrophages.10,29 CSF1R is the receptor that mediates signaling that regulates production, differentiation, and function of macrophages. CSF1R inhibition of TAMs has been proposed as a therapeutic strategy for targeting the growth of gliomas.15 Consistent with a previous study in a preclinical model of gliomas with PDGFB expression,13 we showed that CSF1R inhibition blocked tumor growth and progression in a PDGFB-driven glioma model. However, the CSF1R inhibitor did not significantly slow growth of mesenchymal GBM models (RAS-driven GBM and PDGFRA-driven GBM), despite a mesenchymal to proneural transition in RAS-driven tumors after TAM targeting. Phase II clinical trials of the CSF1R inhibitor PLX3397 did not show efficacy in patients with recurrent GBM,14 suggesting a tumor subtype-specific response to CSF1R inhibition. While the different tumor models such as cell transplantation-based allografts and endogenously developed tumors may result in the different prevalence of TAMs, the underlying mechanisms remain to be determined. Nonetheless, TAMs from both allograft and endogenous tumor models such as RAS-driven gliomas exhibit the similar phenotypes or properties. Intriguingly, PDX models with human proneural or mesenchymal GBM failed to show any growth response or survival benefit following PLX3397 treatment, suggesting that GBM phenotype alone is not sufficient to predict tumor responsiveness to CSF1R inhibition. This may be due to a consequence of mismatch between human cytokines and mouse receptors or intrinsic difference between human and mouse glioma stem-like cells.
We identified 2 main functionally distinct TAM populations from bulk and single-cell transcriptomic profiles in different animal models of GBMs. TAMs in proneural-like PDGFB-driven GBM tumors are enriched in tumor-associated microglia that are critical for driving tumor growth. In contrast, in mesenchymal-like RAS-driven GBM tumors, TAMs exhibit expression profiles characteristic of BAMs or perivascular-associated macrophages with enrichment of genes involved in inflammation and antigen presentation. Targeting the BAM population may result in a removal of the potential brake of antigen presentation on tumor growth, which might lead to accelerated tumor growth after depletion at an early tumorigenic stage in RAS-driven tumors. In our murine GBM models, the predominant TAMs correspond to CNS-resident microglia or macrophages. Inhibition of peripheral macrophage migration into the CNS by CCR2 deletion did not alter tumor growth, suggesting a minimal impact of monocyte-derived macrophages on GBM growth in these models.
TAMs were reprogrammed by PLX3397 treatment in the PDGFB-driven GBM model, consistent with a previous report.13 In contrast, CSF1R inhibition resulted in depletion of TAMs in the RAS-driven model. The mechanisms underlying the differential effects of CSF1R inhibition on TAM reprogramming and growth remain to be defined. Nonetheless, our observations suggest a context-dependent role of TAMs in the outgrowth of distinct gliomas, indicating that the responses of GBM to CSF1R inhibitors depend on both microenvironmental TAMs and glioma subtypes.
Due to the lack of efficacy of CSF1R inhibition in GBM clinical trials, there has been a renewed focus on development of combination therapies to improve response. A previous study showed that there was activation of PI3K signaling upon tumor relapse in PDGFB-driven tumors treated with CSF1R inhibitor BLZ945 and that co-targeting of CSF1R with PI3K inhibition was sufficient to re-sensitize relapsed tumors to CSF1R inhibition.17 However, we found that RAS-driven tumors are not responsive to inhibition of both PI3K and CSF1R signaling, suggesting that additional mechanisms or signaling pathways confer intrinsic resistance to PI3K and CSF1R inhibition. In contrast to PDGFB-driven gliomas, the RAS-driven GBM tumors exhibit high tumor vessel density.
The fact that only combined targeting of VEGFR signaling and TAMs was able to reduce vascular density suggests that TAMs may produce additional pro-angiogenic molecules, such as Fgf2, Pdgfb, Cxcl2, and Wnt7b30 to maintain the vasculature after anti-angiogenic therapy. Co-targeting of tumor VEGFR and TAMs reduced cell proliferation in RAS-driven tumors, resulting in less dense tumor phenotypes, although the combined treatment did not improve animal lifespan. The exact mechanism underlying the lack of enhanced survival remains unknown. Treatment-related side effects, unidentified tumor growth regulators, or increased tumor cell infiltration may counteract the reduction in cell proliferation. Nonetheless, our data suggest that combination inhibition of angiogenesis and TAMs may improve treatment response in mesenchymal-like PLX3397-resistant gliomas.
Our data indicate that CSF1R inhibition blocks the growth of PDGFB-overexpressing glioma but not the growth of other genetically distinct gliomas including RAS- and PDGFRA-driven tumors, indicating that there are tumor subtype- or microenvironment signaling-specific responses to TAM targeting, although the intrinsic and extrinsic mechanisms underlying differential responsiveness to CSF1R inhibition remain to be defined. This highlights the need to distinguish between different TME landscapes in order to use targeted therapies most effectively. PDGF mutational events such as PDGFA amplification or overexpression drive initial tumorigenic events in GBM patients.31 Our data suggest that proneural gliomas with PDGFB overexpression are likely sensitive to CSF1R inhibition, whereas mesenchymal GBMs are resistant. There are approximately 1% of human GBMs that have PDGFB overexpression, for instance, a subset of human diffuse midline pediatric gliomas show high PDGFB expression and microglia/macrophage infiltration.32 However, it remains to be determined if they are responsive to CSF1R inhibition. Future efforts could broaden the spectrum of sensitive proneural tumors by reprogramming GBMs into a “PDGFB-driven” state. The presence of an inflammatory subset of TAMs in mesenchymal-like RAS-driven GBM tumors might potentially offer a therapeutic target for immunotherapy with macrophage checkpoint inhibitors such as CD47 that would augment the phagocytic activity of the TAMs. Combination therapy targeting different tumor compartments has the potential to lead to more robust or durable responses.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Katherine Yutzey for the gift of the CCR2-KO mouse strain and Dr Matthew Garrett for reading the manuscript.
Contributor Information
Rohit Rao, Brain Tumor Center, Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA; Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
Rong Han, Brain Tumor Center, Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
Sean Ogurek, Brain Tumor Center, Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
Chengbin Xue, Brain Tumor Center, Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
Lai Man Wu, Brain Tumor Center, Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
Liguo Zhang, Brain Tumor Center, Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
Li Zhang, Department of Environmental and Public Health Sciences, University of Cincinnati, Cincinnati, Ohio, USA.
Jian Hu, Department of Cancer Biology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Timothy N Phoenix, Division of Pharmaceutical Sciences, James L. Winkle College of Pharmacy, University of Cincinnati, Cincinnati, Ohio, USA.
Stephen N Waggoner, Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA; Center for Autoimmune Genomics and Etiology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
Q Richard Lu, Brain Tumor Center, Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA; Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
Funding
This work was funded in part by CancerFreeKids Foundation, TeamConnor Foundation, and Pray-Hope Believe Foundation to Q.R.L. and Albert Ryan fellowship to R.R.
Conflict of interest statement. The authors declare no conflict of interest.
Authorship statement. Conceptualization: R.R. and Q.R.L.; Investigation: R.R., S.O., R.H., C.X., L.M.W., Lig Z., and Li Z.; Writing: R.R., Q.R.L., and R.H.; Resource: J.H.; Review and editing: T.N.P., S.N.W., and Q.R.L.; Funding acquisition: Q.R.L.
Data and software availability. All the high-throughput data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GEO: GSE185420.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group . Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32(1):42–56.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31(3):326–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mrdjen D, Pavlovic A, Hartmann FJ, et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity. 2018;48(2):380–395.e6. [DOI] [PubMed] [Google Scholar]
- 7. Paolicelli RC, Bolasco G, Pagani F, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333(6048):1456–1458. [DOI] [PubMed] [Google Scholar]
- 8. Jordao MJC, Sankowski R, Brendecke SM, et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363(6425):eaat7554. [DOI] [PubMed] [Google Scholar]
- 9. Kierdorf K, Masuda T, Jordão MJC, Prinz M. Macrophages at CNS interfaces: ontogeny and function in health and disease. Nat Rev Neurosci. 2019;20(9):547–562. [DOI] [PubMed] [Google Scholar]
- 10. Klemm F, Maas RR, Bowman RL, et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell. 2020;181(7):1643–1660.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bowman RL, Klemm F, Akkari L, et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 2016;17(9):2445–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Müller S, Kohanbash G, Liu SJ, et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 2017;18(1):234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pyonteck SM, Akkari L, Schuhmacher AJ, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19(10):1264–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Butowski N, Colman H, De Groot JF, et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro Oncol. 2016;18(4):557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan D, Kowal J, Akkari L, et al. Inhibition of colony stimulating factor-1 receptor abrogates microenvironment-mediated therapeutic resistance in gliomas. Oncogene. 2017;36(43):6049–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weng Q, Wang J, Wang J, et al. Single-cell transcriptomics uncovers glial progenitor diversity and cell fate determinants during development and gliomagenesis. Cell Stem Cell. 2019;24(5):707–723.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quail DF, Bowman RL, Akkari L, et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352(6288):aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang S, Zhao BS, Zhou A, et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017; 31(4):591–606.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Friedmann-Morvinski D, Narasimamurthy R, Xia Y, Myskiw C, Soda Y, Verma IM. Targeting NF-κB in glioblastoma: a therapeutic approach. Sci Adv. 2016;2(1):e1501292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patel SK, Hartley RM, Wei X, et al. Generation of diffuse intrinsic pontine glioma mouse models by brainstem-targeted in utero electroporation. Neuro Oncol. 2020;22(3):381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saederup N, Cardona AE, Croft K, et al. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS One. 2010;5(10):e13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flores-Toro JA, Luo D, Gopinath A, et al. CCR2 inhibition reduces tumor myeloid cells and unmasks a checkpoint inhibitor effect to slow progression of resistant murine gliomas. Proc Natl Acad Sci U S A. 2020;117(2):1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammond TR, Dufort C, Dissing-Olesen L, et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50(1):253–271.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sa JK, Chang N, Lee HW, et al. Transcriptional regulatory networks of tumor-associated macrophages that drive malignancy in mesenchymal glioblastoma. Genome Biol. 2020;21(1):216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yuan J, Levitin HM, Frattini V, et al. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 2018;10(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L, Babikir H, Müller S, et al. The phenotypes of proliferating glioblastoma cells reside on a single axis of variation. Cancer Discov. 2019;9(12):1708–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Szulzewsky F, Pelz A, Feng X, et al. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One. 2015;10(2):e0116644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin P, Gurevich DB. Macrophage regulation of angiogenesis in health and disease. Semin Cell Dev Biol. 2021. doi: 10.1016/j.semcdb.2021.06.010 [DOI] [PubMed] [Google Scholar]
- 31. Ozawa T, Riester M, Cheng YK, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26(2):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ross JL, Chen Z, Herting CJ, et al. Platelet-derived growth factor beta is a potent inflammatory driver in paediatric high-grade glioma. Brain. 2021;144(1):53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.