Abstract
Low temperature causes poor coloration of strawberry (Fragaria sp.) fruits, thus greatly reducing their commercial value. Strawberry fruits accumulate anthocyanins during ripening, but how low temperature modulates anthocyanin accumulation in plants remains largely unknown. We identified MITOGEN-ACTIVATED PROTEIN KINASE3 (FvMAPK3) as an important negative regulator of anthocyanin accumulation that mediates the poor coloration of strawberry fruits in response to low temperature. FvMAPK3 activity was itself induced by low temperature, leading to the repression of anthocyanin accumulation via two mechanisms. Activated FvMAPK3 acted as the downstream target of MAPK KINASE4 (FvMKK4) and SUCROSE NONFERMENTING1-RELATED KINASE2.6 (FvSnRK2.6) to phosphorylate the transcription factor FvMYB10 and reduce its transcriptional activity. In parallel, FvMAPK3 phosphorylated CHALCONE SYNTHASE1 (FvCHS1) to enhance its proteasome-mediated degradation. These results not only provide an important reference to elucidate the molecular mechanisms underlying low-temperature-mediated repression of anthocyanin accumulation in plants, but also offer valuable candidate genes for generating strawberry varieties with high tolerance to low temperature and good fruit quality.
FvMAPK3 regulates the transcriptional activity of FvMYB10 and the degradation of FvCHS1 to participate in low temperature caused poor coloration in strawberry fruit.
Introduction
Low-temperature stress can delay plant growth, reduce plant fertility, and decrease the yield and quality of crops (Yang et al., 2019). Current research on low-temperature stress mainly focuses on vegetative organs (Ding et al., 2015; Li et al., 2017; Zhao et al., 2017; Zhang et al., 2017b; Yang et al., 2019). However, compared to vegetative organs, reproductive organs such as flowers and fruits are more sensitive and vulnerable to low-temperature stress, and they are often severely damaged before vegetative organs exhibit any symptoms (Thakur et al., 2010). A thorough exploration of the mechanisms of response to low-temperature stress in reproductive organs should help reduce much of the economic loss caused by low temperatures.
Strawberry (Fragaria sp.) is an important horticultural crop with high nutritional, health, and commercial values (Taghavi et al., 2019). Recently, strawberry has emerged as a research model for members of the Rosaceae family as well as for nonclimacteric fruits (Kang et al., 2013; Castillejo et al., 2020; Gaston et al., 2020; Martín-Pizarro et al., 2021). The optimum temperature for strawberry growth is between 5°C and 30°C, while the optimum temperature for fruiting is around 15°C–25°C. In practice, a low temperature of 10°C can cause abnormal development and ripening of strawberry fruits, which can result in great economic loss to strawberry farmers (Davik et al., 2000; Koehler et al., 2015). The main consequences of low-temperature stress on strawberry fruits are delayed and abnormal ripening, which manifest as reduced pigment accumulation rates, low pigment contents, and variegated coloring. These easily detectable phenotypes make strawberry fruits an ideal material to study the regulatory mechanisms underlying low-temperature stress responses during fruit ripening. Identifying the specific molecular determinants responsible for poor strawberry fruit coloration at low temperatures would illuminate the underlying regulatory networks and provide valuable candidates for molecular breeding of lines with reduced low-temperature damage.
The main pigments in strawberry fruits are anthocyanins, which contribute to the fruit quality and nutritional value of fruits (He and Giusti, 2010; Jaakola, 2013). Anthocyanin biosynthesis is catalyzed by a series of enzymes encoded by the structural genes PHENYLALANINE AMMONIA-LYASE (PAL), CINNAMATE 4-HYDROXYLASE (C4H), CHALCONE SYNTHASE (CHS), CHALCONE ISOMERASE (CHI), FLAVONOID 3-HYDROXYLASE (F3H), DIHYDROFLAVONOL 4-REDUCTASE (DFR), ANTHOCYANIDIN SYNTHASE (ANS), and several GLYCOSYLTRANSFERASEs (UFGTs; Almeida et al., 2007; Xu et al., 2017). Among these, CHS is the rate-limiting enzyme in the biosynthetic pathway (Feinbaum and Ausubel, 1988). Transcriptional control of CHS expression has been extensively studied in many plants (Hoffmann et al., 2006; Yuan et al., 2009; Hosokawa et al., 2013; Xu et al., 2017; Nakayama et al., 2019). Recently, a posttranslational regulatory mechanism for CHS abundance was reported in Arabidopsis (Arabidopsis thaliana), whereby CHS protein levels are regulated by a Kelch domain-containing F-box (KFB) protein, which mediates the ubiquitination and degradation of CHS (Zhang et al., 2017a). Whether the KFBCHS–CHS module is conserved in different plant species and different organs and whether other types of protein modifications are involved in the regulation of CHS remain unknown.
Besides anthocyanin biosynthetic genes, previous studies have also identified a number of regulatory factors involved in modulating anthocyanin accumulation in strawberry fruits, including MYB10, MYB1, GAMYB (gibberellic acid-induced MYB), MYB9/MYB11, basic Helix–Loop–Helix3 (bHLH3), TRANSPARENT TESTA GLABRA1 (TTG1), RELATED TO ABI3/VP1 1 (RAV1), SNF1-Related Protein Kinase 2.6 (SnRK2.6), MADS1, MADS9, REDUCED ANTHOCYANINS IN PETIOLES (RAP), and RIPENING-INDUCING FACTOR (RIF; Aharoni et al., 2001; Seymour et al., 2011; Salvatierra et al., 2013; Schaart et al., 2013; Lin-Wang et al., 2014; Medina-Puche et al., 2014; Han et al., 2015; Vallarino et al., 2015; Lu et al., 2018; Vallarino et al., 2019; Wang et al., 2019; Castillejo et al., 2020; Gao et al., 2020; Zhang et al., 2020b; Martín-Pizarro et al., 2021). Most of these regulatory proteins are transcription factors, among which the R2R3-type FvMYB10/FaMYB10, the homolog of Arabidopsis MYB75, is considered to be a central activator of downstream structural genes in the anthocyanin biosynthesis pathway in both octoploid (Fragaria × ananassa) and diploid (Fragaria vesca) fruits (Lin-Wang et al., 2014; Medina-Puche et al., 2014; Castillejo et al., 2020). A single-nucleotide mutation in MYB10 was shown to be responsible for a change in the color of strawberry fruits; in addition, various alleles of MYB10 are associated with natural variation in skin and flesh color in different strawberry varieties (Hawkins et al., 2016; Castillejo et al., 2020; Zhang et al., 2020b). Although FvMYB10 is the key molecular switch controlling anthocyanin biosynthesis in strawberry fruits, it remains largely unclear whether and what upstream components might affect the transcriptional activation activity of FvMYB10 and how FvMYB10 senses environmental or phytohormonal signals to regulate anthocyanin accumulation (Zhang et al., 2020b).
Anthocyanin biosynthesis is highly affected by temperature, as illustrated by previous studies that demonstrated how low temperatures promote anthocyanin accumulation, while high temperatures reduce anthocyanin accumulation in vegetative organs (Steyn et al., 2002; Zhang et al., 2010). Notably, strawberry fruits respond to temperature in the opposite direction as vegetative organs, with low temperatures limiting pigment accumulation (Wang et al., 2002; Wang, 2006; Ledesma et al., 2008; Mirahmadi et al., 2012; Taghavi et al., 2019). Our knowledge of how cells in strawberry fruits sense and relay the temperature signal and how low temperatures regulate anthocyanin biosynthesis is incomplete.
In previous work, we showed that FaSnRK2.6 is regulated by low temperature and negatively regulates anthocyanin accumulation in strawberry fruits (Han et al., 2015). The Arabidopsis homolog of FaSnRK2.6, SnRK2.6 (also named OPEN STOMATA1 [OST1]), is a key regulator of low-temperature signal transduction (Ding et al., 2015, 2018). Besides OST1, several other members of MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) cascades, such as MAPK KINASE1 (MKK1), MKK2, MAPK3, MAPK4, and MAPK6, were also reported to be important for low-temperature signaling in Arabidopsis (Ding et al., 2019). In particular, the MAPK3–INDUCER OF C-REPEAT BINDING FACTOR (CBF) EXPRESSION 1 (ICE1) phosphorylation module acts as a central component of low-temperature signaling by regulating the expression of CBF and COLD-REGULATED (COR) genes in Arabidopsis or TREHALOSE-6-PHOSPHATE PHOSPHATASE1 (OsTPP1) in rice (Oryza sativa; Li et al., 2017; Zhao et al., 2017; Zhang et al., 2017b). However, MAPK3-mediated phosphorylation of ICE1 results in its degradation in Arabidopsis but higher stability of ICE1 in rice, indicating that although the signaling cascade might be conserved, MAPK3 acts as a negative regulator of low-temperature tolerance in Arabidopsis but is a positive regulator in rice (Li et al., 2017; Zhao et al., 2017; Zhang et al., 2017b). Therefore, despite its conservation, the exact regulatory mechanism of the MAPK3–ICE1 module may differ between species.
In general, MAPK cascades consist of three consecutive kinases (MAPK kinase kinases [MKKKs], MKKs, and MAPKs) that govern diverse biological functions in plants (Komis et al., 2018). Although MAPK cascade genes have been identified in strawberry (Wei et al., 2017; Zhou et al., 2017a), their precise biological function remains unknown, with the exception of FaMAPK19, which was reported to be involved in resistance to the fungus Botrytis cinerea in fruits (Zhang et al., 2020a). Notably, SnRK2 and MAPK cascades have been demonstrated to form a phosphorylation module that regulates osmotic stress in Arabidopsis (Droillard et al., 2002; Umezawa et al., 2013; Wang et al., 2013a). Whether FvSnRK2.6 participates in a functional module with MAPK cascades in strawberry and whether MAPK cascades are involved in the regulation of fruit ripening and low-temperature responses of fruits have not been explored.
Here, we report that FvMAPK3 is an important regulator of poor fruit coloration in response to low-temperature stress in strawberry. FvMAPK3 phosphorylated FvCHS1 through the FvMKK4/FvMAPK3 module while also phosphorylating FvMYB10 via both the FvSnRK2.6/FvMAPK3 and FvMKK4/FvMAPK3 modules. FvMAPK3 repressed anthocyanin accumulation at low temperatures, largely as a consequence of increasing FvKFB1-mediated degradation of FvCHS1 and decreasing the transcriptional activity of FvMYB10. Additionally, we determined that a phosphorylation-dead mutant of FvMYB10 (FvMYB104A) may have potential breeding value to allow strawberry fruits to maintain normal coloration under low-temperature stress.
Results
Low-temperature stress represses anthocyanin accumulation and increases FvMAPK3 activity in strawberry fruits
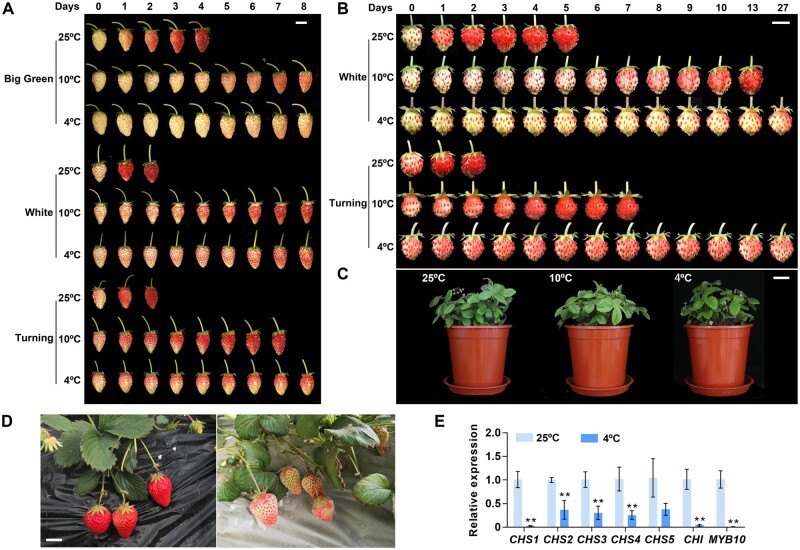
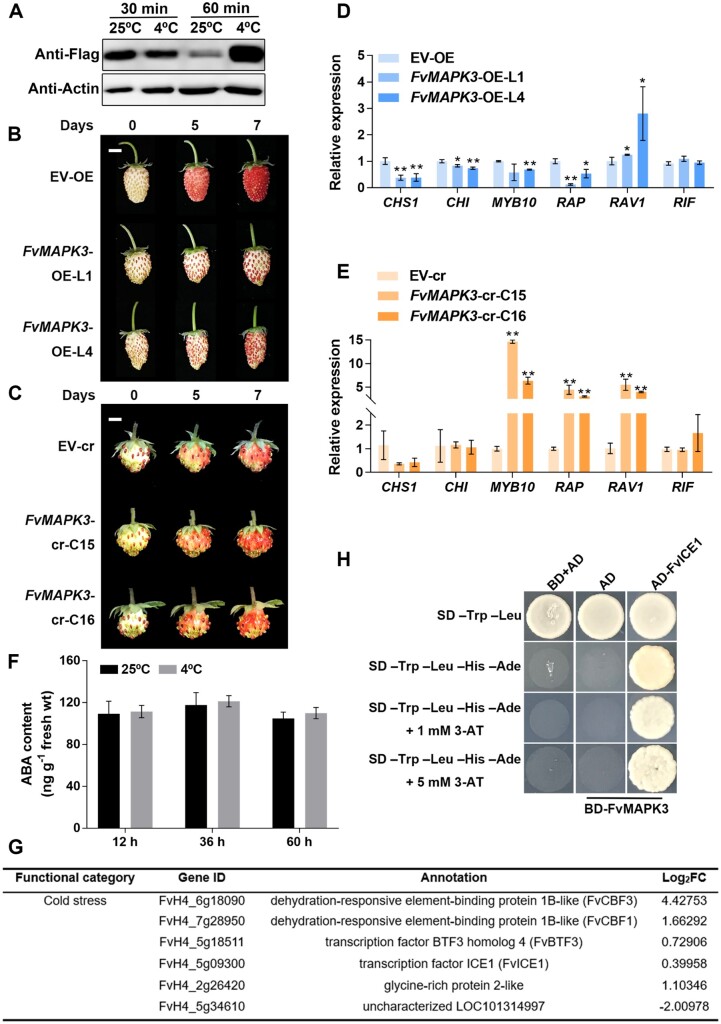
Low temperature delays the ripening of strawberry fruits, which is most easily seen as a delay in the accumulation of anthocyanins (Mirahmadi et al., 2012; Han et al., 2015; Taghavi et al., 2019). To better understand the effects of low temperature on anthocyanin accumulation in diploid strawberry (F. vesca) fruits, we followed the changes in pigment contents in both attached and detached fruits at different developmental stages when exposed to low temperatures. We determined that exposure to low temperatures of 4°C or 10°C equally delayed the rate of anthocyanin accumulation and decreased total anthocyanin contents in both attached and detached strawberry fruits (Figure 1, A and B). When strawberry plants with white fruits were cultivated at 10°C or 4°C for 8 days, their vegetative parts were comparable to those of plants maintained in the control growth conditions of 25°C (Figure 1C). However, their fruits largely failed to accumulate anthocyanins, suggesting that white fruits are more sensitive to low-temperature stress than turning fruits or mature leaves. The results also showed that white fruits are largely unable to accumulate anthocyanins even after 27 days of exposure to 4°C, indicating that this low-temperature condition has devastating effects on strawberry coloration (Figure 1B). The effect of low temperature on anthocyanin accumulation was not limited to white fruits, as large green fruits and turning stage fruits also showed a striking delay in pigmentation, indicating that strawberry fruits are highly sensitive to low-temperature stress throughout the ripening process (Figure 1A). These results are consistent with field phenotypes observed in octoploid strawberry (F. × ananassa) exposed to low-temperature stress at the white stage during greenhouse production (Figure 1D; Han et al., 2015).
Figure 1.
Low-temperature stress represses the accumulation of anthocyanins in strawberry fruits. A, Phenotypes of detached strawberry fruits under low-temperature stress. Diploid strawberry fruits (F. vesca, cv Fragola di Bosco) at the big green, white, and turning stages were fed with 88 mM sucrose at 25°C, 10°C, or 4°C. Photographs were taken every day, and representative images are shown. Bar, 1 cm. B, Strawberry plants carrying three white or turning fruits per plant were placed in a growth chamber set to 25°C (control conditions), 10°C, or 4°C. Photographs were taken every day, and representative images are shown. Bar, 1 cm. C, Phenotypes of strawberry plants carrying white fruits cultivated at 25°C, 10°C, or 4°C for 8 days. Bar, 5 cm. D, Fruits of the cultivar Benihoppe grown at ∼20°C (left) or 4°C (right) in the greenhouse. Bar, 2 cm. E, Relative expression levels of selected regulators involved in anthocyanin biosynthesis in white strawberry fruits at 25°C or subjected to 4°C treatment. Values are means ± standard error of mean (sem) of three biological replicates. Statistical significance was determined by Student’s t test (*P < 0.05, **P < 0.01).
The white fruit stage is the key stage during which strawberry plants initiate fruit ripening, and it determines fruit quality (Jia et al., 2013; Wei et al., 2018). To unravel the molecular regulatory mechanism underlying low-temperature-mediated repression of anthocyanin accumulation, we measured the relative transcript levels for genes involved in anthocyanin biosynthesis in white fruits. Low-temperature stress of 4°C caused a downregulation of FvCHS1, FvCHI, and FvMYB10 expression relative to the control condition of 25°C, suggesting that these genes contribute to low-temperature-mediated inhibition of anthocyanin accumulation (Figure 1E).
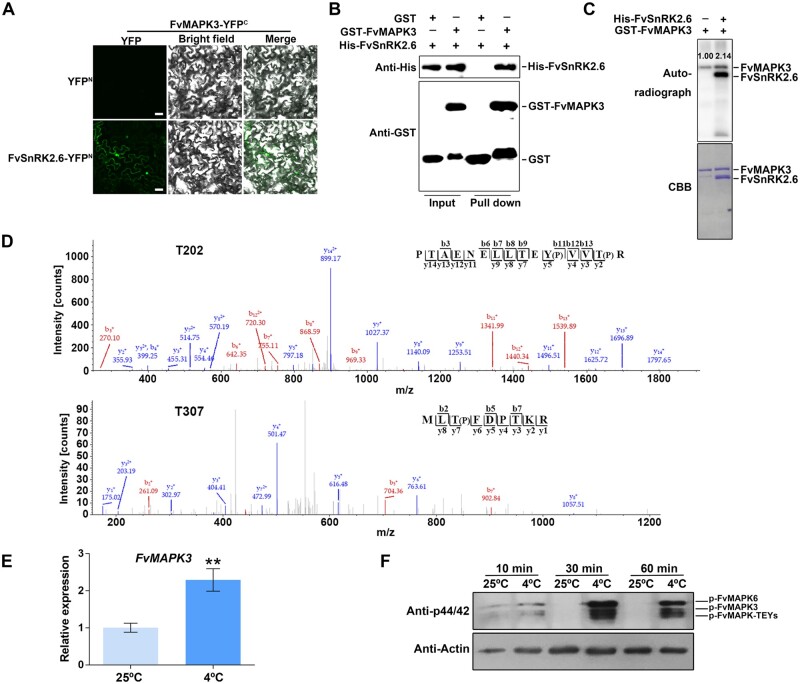
We showed previously that FaSnRK2.6 is involved in low-temperature-mediated anthocyanin accumulation in strawberry (Han et al., 2015). Given that MAPK cascades are a conserved component of low-temperature signaling in both Arabidopsis and rice, and because SnRK2 was reported to phosphorylate MAPKs (Droillard et al., 2002; Umezawa et al., 2013; Wang et al., 2013a; Ding et al., 2019), we hypothesized that FvSnRK2.6 might regulate low-temperature-mediated anthocyanin accumulation by targeting a MAPK cascade in strawberry fruits. To test this possibility, we first used yeast two-hybrid (Y2H) assays to determine the potential for interaction between FvSnRK2.6 and all 12 annotated FvMAPK proteins in the current version (version 4.0.a1) of the diploid strawberry genome (Wei et al., 2017; Zhou et al., 2017a; Edger et al., 2018). FvMAPK3 only weakly interacted with FvSnRK2.6 on synthetic defined (SD) medium lacking Trp, Leu, His, and Ade and did not interact on SD medium lacking Trp, Leu, His, and Ade medium and containing 3-amino-1,2,4-triazole (3-AT) (Supplemental Figure S1, A and B). However, the interaction of kinases with their partners and substrates may be affected by their phosphorylation status, which may not be fully activated in yeast cells (Li et al., 2016). We, therefore, tested this interaction via bimolecular fluorescence complementation (BiFC) assays between FvMAPK3 and FvSnRK2.6 in plant cells: We detected fluorescence from yellow fluorescent protein (YFP), indicative of an interaction between the two proteins in the Nicotiana benthamiana leaf cells (Figure 2A;Supplemental Figure S1C). To determine whether the interaction between FvMAPK3 and FvSnRK2.6 was dependent on the presence of other proteins, we performed pull-down assays with recombinant proteins, which showed that GST-FvMAPK3 and His-FvSnRK2.6 interact directly in vitro (Figure 2B).
Figure 2.
FvMAPK3 is activated by FvSnRK2.6 and low temperature. A, FvSnRK2.6 and FvMAPK3 interact, as seen by BiFC. FvMAPK3 was fused to the C terminus of YFP and FvSnRK2.6 to the N terminus of YFP. The encoding constructs were co-infiltrated into N. benthamiana leaves. FvMAPK3-YFPc and YFPn were used as negative control. Bars, 50 µm. B, In vitro pull-down assay showing that FvSnRK2.6 interacts with FvMAPK3. Recombinant GST or GST-FvMAPK3 bounded to Glutathione Sepharose beads was incubated with recombinant His-FvSnRK2.6 protein and immunoblotted with Anti-His antibody. C, In vitro kinase assay with FvSnRK2.6 and FvMAPK3. Purified recombinant proteins were separated by 10% SDS–PAGE after incubation in kinase reaction buffer for 30 min at 30°C. Phosphorylated FvMAPK3 and FvSnRK2.6 were visualized by autoradiography (top panel). Recombinant FvMAPK3 and FvSnRK2.6 were detected by CBB staining (bottom panel). D, Liquid chromatography tandem mass spectrometry (LC–MS/MS) detection of phosphorylated amino acids in FvMAPK3. Thr-202 and Thr-307 residues were both phosphorylated by FvSnRK2.6. b and y represent the N- and C-terminal ion of the peptide fragment with retained charge, respectively. The label numbers below indicate the location of the identified peptide. (P) stands for phosphorylated amino acids. E, Relative FvMAPK3 expression levels in white strawberry fruits at 25°C (control conditions) or after exposure to low-temperature stress at 4°C for 24 h. Values are means ± sem of three biological replicates. Statistical significance was determined by Student’s t test (*P < 0.05, **P < 0.01). F, FvMAPK3 activity is rapidly induced by low temperatures in strawberry fruits. Total proteins were extracted from white fruits cultivated at 25°C (control conditions) or transferred to 4°C for 10, 30, or 60 min. Total proteins were subjected to immunoblotting to detect the phosphorylation levels of FvMAPK-TEY kinases with anti-p44/42 antibody. Anti-actin antibody was used as loading control. FvMAPK6 was distinguished by its molecular weight, and FvMAPK3 was validated using Flag-FvMAPK3 transgenic plants and its molecular weight.
To test whether FvSnRK2.6 phosphorylates FvMAPK3, we then used recombinant GST-FvMAPK3 as a substrate for phosphorylation by recombinant His-FvSnRK2.6 to perform in vitro kinase activity assays: Both His-FvSnRK2.6 and GST-FvMAPK3 demonstrated autophosphorylation activity, and His-FvSnRK2.6 slightly increased the phosphorylation levels of GST-FvMAPK3 in vitro (Figure 2C). We identified amino acid residues Thr-202 and Thr-307 by mass spectrometry (MS) as the GST-FvMAPK3 phosphorylation sites from His-FvSnRK2.6 activity, while Thr-197, Tyr-199, and Thr-311 resulted from GST-FvMAPK3-mediated autophosphorylation (Figure 2D;Supplemental Figure S1D).
To place FvMAPK3 in the context of low-temperature responses during strawberry fruit ripening, we determined relative FvMAPK3 transcript levels after a 24-h exposure to 4°C, which revealed a 2.5-fold increase relative to the control temperature of 25°C (Figure 2E). Next, we detected phosphorylation levels of FvMAPK3 in white fruits upon transfer from 25°C to 4°C using a commercial Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204)-specific antibody. The Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204)-specific antibody recognizes the phosphorylated/active form of MAPK proteins that contain a TEY motif (p-TEpY; Li et al., 2016; Zhao et al., 2017; Zhang et al., 2017b; Guo et al., 2020), which is the case for FvMAPK1-7 in strawberry (Supplemental Figure S2). FvMAPK-TEY proteins are recognized by the p44/42 antibody as three bands in F. vesca and four bands in F. × ananassa (Figures 2, F and 3, B). Based on their molecular weights, the first band was assigned to phosphorylated FvMAPK6 (Figure 2F;Supplemental Figure S2B). We also used an anti-Flag antibody to determine the position of the FvMAPK3 protein in immunoblots on protein extracts from transgenic Flag-FvMAPK3 strawberry plants: FvMAPK3 corresponded to the second band (Supplemental Figure S2, B and D). As shown in Figure 2F, FvMAPK3 activity increased slightly after 10 min at 4°C and then rose sharply after 30 min. These results suggest that FvMAPK-TEY kinases are downstream signaling components that can rapidly respond to low-temperature stress at the posttranslational level.
Figure 3.
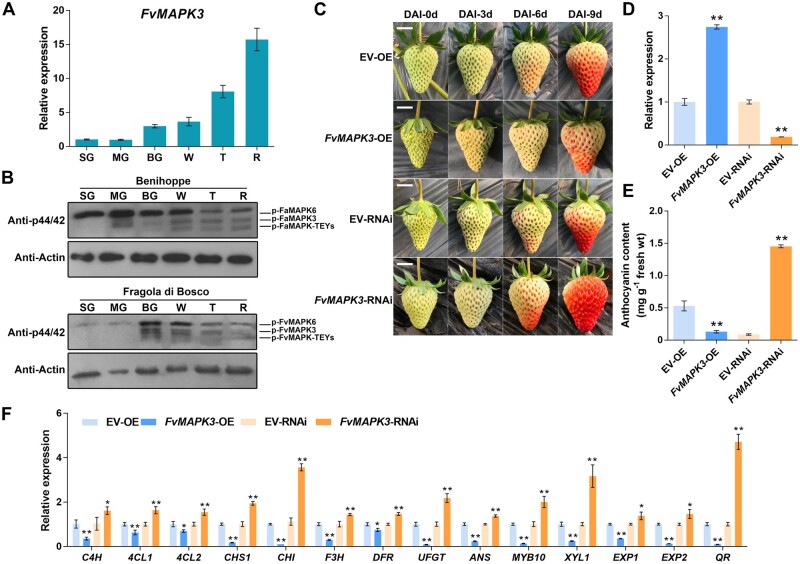
Transient transformation of strawberry fruits with FvMAPK3 modulates their ripening. A, Relative FvMAPK3 expression levels at different developmental stages in diploid strawberry fruits, as determined by RT-qPCR. SG, small green fruit; MG, middle green fruit; BG, big green fruit; W, white fruit; T, turning fruit; R, red fruit. The mean values ± sem from three biological replicates are shown. B, MAPK3, MAPK6, and other MAPK-TEY kinase activity at different developmental stages in octoploid (F. × ananassa Ducherne, cv Benihoppe) and diploid (F. vesca, cv Fragola di Bosco) strawberry fruits. Total proteins were extracted, and MAPK activity was analyzed by immunoblotting with anti-p44/42 antibody. Anti-actin antibody was used as loading control. C, Phenotypes associated with the transient OE (FvMAPK3-OE) and silencing of FvMAPK3 (FvMAPK3-RNAi) in Benihoppe strawberry fruits. DAI, days after injection. Bars, 1 cm. D, Relative FvMAPK3 expression levels in transiently transformed strawberry fruits, as determined by RT-qPCR. Values are means ± sem of three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05, **P < 0.01. E, Anthocyanin contents in FvMAPK3-OE and FvMAPK3-RNAi fruits. Error bars represent sem of three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05, **P < 0.01. F, Relative expression levels of ripening-related genes in FvMAPK3-OE and FvMAPK3-RNAi fruits, as determined by RT-qPCR. FvACTIN was used as internal reference. Values are means ± sem of three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05, **P < 0.01.
FvMAPK3 negatively regulates fruit ripening in strawberry fruits
To analyze the function of FvMAPK3 in strawberry fruits, we investigated the native changes of FvMAPK3 transcript levels during strawberry fruit ripening. FvMAPK3 expression gradually and continuously increased during strawberry fruit ripening, from the small green to the red stage (Figure 3A). Similarly, FaMAPK3/FvMAPK3 native kinase activity rose at the middle green or big green stages and then remained constant until the end of fruit ripening in diploid and octoploid strawberry fruits (Figure 3B). However, the native kinase activity of FaMAPK6/FvMAPK6 kinases showed a different pattern, suggesting that the biological function of FaMAPK6/FvMAPK6 may differ during strawberry fruit ripening.
We transiently overexpressed (FvMAPK3-OE) and silenced (via RNA interference; FvMAPK3-RNAi) FvMAPK3 in strawberry fruits from the “Benihoppe” cultivar. We observed a delay in fruit ripening in FvMAPK3-OE fruits and an acceleration in FvMAPK3-RNAi fruits (Figure 3C). We confirmed the higher (in FvMAPK3-OE) and lower (in FvMAPK3-RNAi) relative FvMAPK3 transcript levels in transiently transformed strawberry fruits by real time quantitative polymerase chain reaction (RT-qPCR) (Figure 3D). Consistent with these results, anthocyanin contents and the expression of the anthocyanin biosynthesis genes FvCHS1, FvCHI, FvF3H, FvDFR, FvUFGT, and FvANS were significantly reduced in FvMAPK3-OE fruits and increased in FvMAPK3-RNAi fruits (Figure 3, E and F). Notably, the expression of FvMYB10 was also affected in the same direction as anthocyanin biosynthesis structural genes in FvMAPK3 transgenic fruits (Figure 3F). In addition, a series of structural genes involved in the phenylpropanoid pathway were also regulated in transiently transformed strawberry fruits such as C4H and 4CL1 (Figure 3F). These results indicated that FvMAPK3 acts upstream of the regulation of FvMYB10, FvCHS1, and FvCHI transcription for anthocyanin accumulation in strawberry fruits. Moreover, the expression of softening-related and aroma production-related genes such as XYLOSIDASE1 (FvXYL1), EXPANSIN1 (FvEXP1), FvEXP2, and QUINONE OXIDOREDUCTASE (FvQR) followed the same pattern as FvMYB10 in the transgenic fruits, suggesting that FvMAPK3 has diverse functions in the regulation of fruit ripening (Figure 3F).
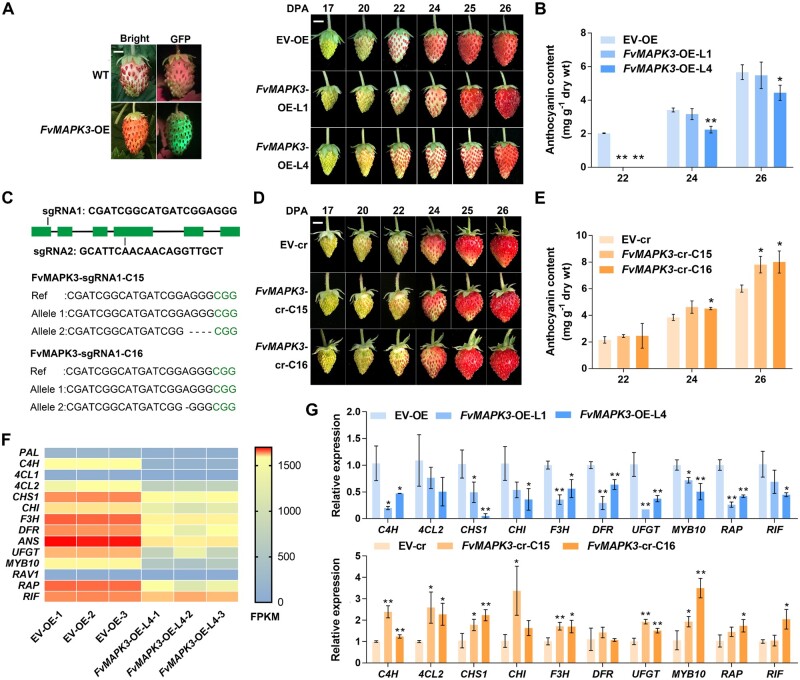
To fully explore the biological functions of FvMAPK3 in strawberry, we generated stable overexpression (OE) and genome-edited (cr) transgenic lines for FvMAPK3 in the diploid strawberry background (F. vesca). After transformation, we selected the two independent FvMAPK3-OE lines L1 and L4 from five detected lines for further experiments (Supplemental Figure S3A). Compared to control EV-OE strawberry lines transformed with empty vector (EV) only, FvMAPK3-OE-L1 and FvMAPK3-OE-L4 plants grew normally (Supplemental Figure S3B) but exhibited a delay in fruit anthocyanin accumulation (Figure 4, A and B). To obtain mutants edited at the FvMAPK3 locus, we selected 58 lines with high expression for a single-guide RNA (sgRNA) targeting FvMAPK3 and for the nuclease gene Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats [CRISPR]-associated nuclease) from individual antibiotic-resistant callus. We then genotyped all seedlings by PCR amplification over the edited site, followed by DNA sequencing, resulting in the identification of two heterozygous mutants in FvMAPK3 for sgRNA1. However, sgRNA2 did not generate an editing event in the selected seedlings (Figure 4C;Supplemental Figure S3C). FvMAPK3-cr line C15 harbored a 4-bp deletion, while line C16 carried a 1-bp deletion. Both gene-edited lines were easily distinguished from wild-type strawberry plants due to their enlarged, rounded, crinkly leaves with serrated margins, elongated fruiting branches, and oblate fruits; importantly, these phenotypes were stably inherited to the T1 generation (Supplemental Figure S3, D and E). However, we failed to identify homozygous mutants among all T1 plants, as only heterozygous mutants survived the antibiotic selection for the CRISPR construct, suggesting that FvMAPK3 may be essential for the development of strawberry embryos and/or may affect sexual reproduction.
Figure 4.
FvMAPK3 negatively regulates anthocyanin accumulation in strawberry fruits. A, Visualization of GFP accumulation in FvMAPK3-OE fruits (left) and ripening phenotypes of EV-OE and FvMAPK3-OE-L1 and FvMAPK3-OE-L4 transgenic strawberry fruits (right). Fruits at 17, 20, 22, 24, 25, and 26 DPA are shown. Bars: 0.5 cm. B, Anthocyanin contents in EV-OE and FvMAPK3-OE-L1 and FvMAPK3-OE-L4 fruits at 22, 24, and 26 DPA. Error bars represent sem of three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05, **P < 0.01. C, Diagram of the FvMAPK3 locus and target sites for designed sgRNAs. Green boxes, exons; lines, introns. sgRNA1 and sgRNA2 were used for genome editing. Sequencing revealed successful genome editing at FvMAPK3 in lines C15 and C16, obtained with sgRNA1. D, Fruit developmental phenotypes of EV-cr and FvMAPK3-cr-C15 and FvMAPK3-cr-C16 transgenic strawberry fruits. Bar: 0.5 cm. E, Anthocyanin contents in EV-cr and FvMAPK3-cr-C15 and FvMAPK3-cr-C16 fruits. Error bars represent sem of three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05, **P < 0.01. F, Expression profiles of anthocyanin biosynthesis genes differently expressed between EV-OE and FvMAPK3-OE-L4 fruits, based on RNA-seq analysis of three biological replicates. The color scale was used to indicate FPKM values. G, Validation of relative expression levels of anthocyanin biosynthesis genes in EV-OE and FvMAPK3-OE-L1 and -L4, and EV-cr and FvMAPK3-cr-C15 and -C16 strawberry fruits by RT-qPCR. FvACTIN was used as internal reference. Values are means ± sem of three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05, **P < 0.01.
Because the generation of transgenic plants may lead to changes in ploidy (Zhang et al., 2014), and since FvMAPK3-cr plants had enlarged leaves, a phenotype commonly associated with higher ploidy levels (Zhang et al., 2014), we determined the ploidy of FvMAPK3-cr plants by flow cytometry: FvMAPK3-cr plants were in fact tetraploid (Supplemental Figure S3F). As a suitable control for these lines, we analyzed more seedlings that had been transformed with the EV control (EV-cr) and identified one tetraploid EV-cr plant (Supplemental Figure S3, F and G). When compared to seeds from this tetraploid EV-cr line, FvMAPK3-cr seeds showed a lower germination rate (Supplemental Figure S3H). However, anthocyanin accumulation rates were comparable in EV-cr and FvMAPK3-cr fruits, although total anthocyanin contents in fully ripe FvMAPK3-cr fruits were significantly higher relative to those of EV-cr fruits (Figure 4, D and E).
To decipher the molecular mechanisms underlying FvMAPK3-mediated repression of anthocyanin accumulation in strawberry fruits, we performed transcriptome deep sequencing (RNA-seq) using EV-OE and FvMAPK3-OE-L4 fruits (Supplemental Figure S4; Supplemental Data Set 1). We also validated the expression of several genes of interest by RT-qPCR; in both cases, the expression of ripening-regulated transcription factor genes FvMYB10, FvRAP, and FvRIF and of most anthocyanin biosynthetic genes was severely downregulated in FvMAPK3-OE fruits relative to EV-OE fruits (Figure 4, F and G; Table 1; Supplemental Data Set 1). In addition to anthocyanin biosynthetic genes, the expression of several transcription factor genes belonging to the MYB, bHLH, and WRKY families and some important members of abscisic acid (ABA) and jasmonic acid (JA) signaling transduction was also significantly downregulated in FvMAPK3-OE fruits (Table 1; Supplemental Data Set 1). Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of differentially expressed genes that were selected by the criteria |Log2FC| ≥ 1 and pad j ≤ 0.05 between EV-OE and FvMAPK3-OE fruits revealed enrichment in several pathways that are closely associated with low-temperature responses such as plant hormone signal transduction (Thomashow, 1999; Lee et al., 2005), carbon metabolism, starch and sucrose metabolism, galactose metabolism (Kaplan and Guy, 2004), and the MAPK signaling pathway (Supplemental Figure S4B; Li et al., 2017; Zhang et al., 2017b; Zhao et al., 2017). Further analysis showed that FvMAPK3 is also co-expressed with JA and ABA signaling components such as JASMONATE ZIM-DOMAIN PROTEINs (JAZs), MYC2, PYRIBACTIN (PYR)-LIKEs (PYLs), SnRK2.2, ABA INSENSITIVE1 (ABI1), and ABI5 (Supplemental Figure S4D). Additionally, two genes that encode lignin biosynthetic enzymes were also identified as co-expressed genes with FvMAPK3 (Supplemental Figure S4D).
Table 1.
Selected differentially expressed genes (DEGs) in FvMAPK3-OE fruits compared to EV-OE fruits
| Functional category | Gene ID | Annotation | Log2FC |
|---|---|---|---|
| Flavonoid biosynthesis | FvH4_3g40570 | Trans-cinnamate 4-monooxygenase | −3.7 |
| FvH4_7g01160 | Polyketide synthase 1 | −3.7 | |
| FvH4_7g20870 | Chalcone--flavonone isomerase 1 | −2.7 | |
| FvH4_1g11810 | Naringenin, 2-oxoglutarate 3-dioxygenase | −3.0 | |
| FvH4_2g39520 | Bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase | −4.0 | |
| FvH4_5g01170 | Leucoanthocyanidin dioxygenase | −3.1 | |
| FvH4_7g33840 | Anthocyanidin 3-O-glucosyltransferase 2 | −4.1 | |
| FvH4_1g22020 | Transcription factor MYB114-like (FvMYB10) | −2.5 | |
| FvH4_1g27460 | Glutathione S-transferase F11-like (FvRAP) | −4.6 | |
| Plant hormone signal transduction | FvH4_4g09440 | ABA receptor PYL8-like | +4.6 |
| FvH4_5g22060 | ABA receptor PYR1-like | −3.4 | |
| FvH4_5g04500 | Probable protein phosphatase 2C 8 | −2.6 | |
| FvH4_3g24800 | ABA-INSENSITIVE 5-like protein 1 | −4.9 | |
| FvH4_6g18510 | Protein ABA-INSENSITIVE 5 | –5.8 | |
| FvH4_6g35140 | Protein TIFY 11A-like | +3.7 | |
| FvH4_1g09070 | Protein TIFY 10A | +3.8 | |
| FvH4_4g23000 | Protein TIFY 9 | +4.3 | |
| FvH4_7g17380 | Transcription factor MYC2 | +9.3 | |
| Transcription factors | |||
| bHLH family | FvH4_4g07090 | bHLH35 | +8.1 |
| FvH4_6g26360 | bHLH118-like | +8.1 | |
| FvH4_5g10850 | bHLH-MYC and R2R3-MYB transcription factors N-terminal | +7.7 | |
| FvH4_1g18740 | bHLH18-like | +4.9 | |
| FvH4_1g18930 | bHLH93-like | +2.8 | |
| FvH4_7g24720 | bHLH51 | +2.6 | |
| FvH4_7g10840 | UNE10 | –3.2 | |
| APETALA2 family | FvH4_4g26740 | Uncharacterized LOC101314095 | –4.5 |
| FvH4_7g10070 | Ethylene-responsive transcription factor ERF107-like | +8.1 | |
| FvH4_5g33180 | Ethylene-responsive transcription factor ERF020 | +5.8 | |
| FvH4_7g10080 | Ethylene-responsive transcription factor 13-like | +4.5 | |
| FvH4_6g18090 | Dehydration-responsive element-binding protein 1B-like | +4.4 | |
| FvH4_7g28960 | Ethylene-responsive transcription factor ERF027 | +4.3 | |
| FvH4_4g03450 | Ethylene-responsive transcription factor ERF096-like | +4.2 | |
| FvH4_3g08790 | Ethylene-responsive transcription factor 12 | +4.1 | |
| Functional category | Gene ID | Annotation | Log2FC |
| FvH4_2g06050 | Ethylene-responsive transcription factor ERF109-like | +2.8 | |
| FvH4_5g19840 | Ethylene-responsive transcription factor 2 | +2.6 | |
| FvH4_6g01400 | Dehydration-responsive element-binding protein 2D | –3.1 | |
| FvH4_7g24760 | Ethylene-responsive transcription factor ABI4 | –3.6 | |
| FvH4_6g34710 | AP2-like ethylene-responsive transcription factor AIL1 | –4.3 | |
| FvH4_2g38880 | Dehydration-responsive element-binding protein 2D-like | –4.3 | |
| MYB family | FvH4_2g35010 | MYB44-like | +7.2 |
| FvH4_2g31090 | MYB6-like | +4.8 | |
| FvH4_7g16990 | MYB12-like | +4.4 | |
| FvH4_3g28890 | MYB46 | +4.2 | |
| FvH4_4g31190 | myb-like protein Q | +3.9 | |
| FvH4_5g11930 | MYB108-like | +3.4 | |
| FvH4_6g44340 | myb-related protein Myb4-like | +3.0 | |
| FvH4_2g21500 | myb-related protein Myb4 | +2.6 | |
| FvH4_1g22020 | MYB114-like | –2.5 | |
| FvH4_5g37310 | MYB39 | –2.7 | |
| FvH4_5g17111 | MYB3-like (FvMYB1) | –3.3 | |
| FvH4_5g03100 | MYB86 | –4.7 | |
| FvH4_3g15320 | myb-related protein 330 | –4.8 | |
| FvH4_5g22510 | MYB39 | –5.1 | |
| WRKY family | FvH4_3g11140 | Probable WRKY14 | +3.9 |
| FvH4_6g10510 | Probable WRKY33 | +3.6 | |
| FvH4_7g16150 | Probable WRKY46 | +3.2 | |
| FvH4_7g31050 | Probable WRKY41 | +2.8 | |
| bZIP family | FvH4_6g46000 | Basic leucine zipper 43 | +2.8 |
| FvH4_6g02940 | Light-inducible protein CPRF2 | +2.7 | |
| FvH4_3g24800 | ABA-INSENSITIVE 5-like protein 1 | –5.0 | |
| FvH4_6g22750 | HBP-1b (c38) | –5.0 | |
| FvH4_6g18510 | ABA-INSENSITIVE 5 | –5.8 | |
| FvH4_4g21800 | HY5-like | –6.9 | |
| NAM family | FvH4_7g18000 | NAC domain-containing protein 68-like | +6.2 |
| FvH4_6g23460 | NAC domain-containing protein 55-like | −2.7 |
RNA-seq was performed on EV-OE and FvMAPK3-OE fruits collected 25 DPA. Three biological replicates were performed, and expression changes were calculated as Log2(Fold-change); “–” indicates downregulated genes in FvMAPK3-OE fruits, “+” indicates upregulated genes in FvMAPK3-OE fruits. DEGs were selected by |Log2FC| ≥ 2.5 and P ≤ 0.05.
FvMAPK3 is a downstream component of low-temperature repression of anthocyanin accumulation
We previously showed that FvMAPK3 is activated by low temperatures in strawberry fruits (Figure 2, E and F). Flag-FvMAPK3 abundance also substantially increased in white stable OE of Flag-FvMAPK3 fruits after being subjected to 4°C for 60 min (Figure 5A). To investigate whether FvMAPK3 altered the sensitivity of strawberry fruits to low temperature, we characterized the effects of exposing white stage fruits to a 10°C treatment on anthocyanin accumulation in FvMAPK3-OE and FvMAPK3-cr fruits. The repression of anthocyanin accumulation at low temperatures appeared to be enhanced in FvMAPK3-OE fruits and attenuated in FvMAPK3-cr fruits (Figure 5, B and C), suggesting that FvMAPK3 is a downstream positive regulator of low-temperature-mediated repression of anthocyanin accumulation in strawberry fruits. In agreement with this observation, the expression of FvCHS1 and FvCHI was much lower in FvMAPK3-OE fruits transferred to 4°C, but not in FvMAPK3-cr fruits subjected to the same treatment (Figure 5, D and E). We hypothesized that besides the transcriptional regulation of their encoding genes, CHS and CHI may also be regulated by another mechanism in response to low temperature.
Figure 5.
FvMAPK3 is a downstream component of low-temperature-repressed anthocyanin accumulation. A, FvMAPK3 protein levels increase upon exposure of white FvMAPK3-OE fruits to 4°C for 60 min. B, Phenotypes of detached EV-OE and FvMAPK3-OE-L1 and -L4 strawberry fruits cultivated at 10°C for the indicated number of days. Bar, 0.5 cm. C, Phenotypes of attached EV-cr and FvMAPK3-cr-C15 and -C16 strawberry fruits cultivated at 10°C for the indicated number of days. Bar: 0.5 cm. D, Relative expression levels of reported regulators involved in anthocyanin biosynthesis in detached white strawberry fruits from EV-OE and FvMAPK3-OE-L1 and -L4 plants exposed to 4°C for 24 h. Values are means ± sem of three biological replicates. Statistical significance was determined by Student’s t test (*P < 0.05, **P < 0.01). E, Relative expression levels of reported regulators involved in anthocyanin biosynthesis in detached white strawberry fruits from EV-cr and FvMAPK3-cr-C15, and -C16 plants exposed to 4°C for 24 h. Values are means ± sem of three biological replicates. Statistical significance was determined by Student’s t test (*P < 0.05, **P < 0.01). F, ABA content in detached white strawberry fruits (F. vesca, cv Fragola di Bosco) cultivated at 25°C (control conditions) or exposed to 4°C for 12, 36, or 60 h. Values are means ± sem of three biological replicates. Statistical significance was determined by Student’s t test (*P < 0.05, **P < 0.01). G, Cold-related DEGs between strawberry fruits of FvMAPK3-OE-L4 and EV-OE plants. Log2 fold-change was used to evaluate the alteration in gene expression and direction between the two genotypes. H, FvMAPK3 interacts with FvICE1 in a Y2H assay.
Notably, the expression of the transcription factor genes FvMYB10 and FvRAP was lower in FvMAPK3-OE fruits but higher in FvMAPK3-cr fruits transferred to 4°C for 24 h (Figure 5, D and E). FvRAP1 was reported to regulate anthocyanin accumulation independently of FvMYB10 in strawberry fruits, suggesting that MAPK3 responds to low-temperature signals through both a MYB10-dependent and a MYB10-independent pathway at the transcriptional level (Figure 5, D and E; Gao et al., 2020). We also observed that FvRAV1 expression is higher in both FvMAPK3-OE and FvMAPK3-cr fruits exposed to 4°C for 24 h, suggesting that FvRAV1 may not be involved in FvMAPK3-mediated anthocyanin accumulation. Alternatively, FvRAV1 may function independently of FvMAPK3 in low-temperature signaling (Figure 5, D and E). In addition, the expression of FvRIF was not affected by 4°C exposure in either FvMAPK3-OE or FvMAPK3-cr fruits. The results suggested that although the expression of FvRIF is downregulated in FvMAPK3-OE fruits, it may not be involved in FvMAPK3-mediated repression of anthocyanin accumulation at low temperatures (Figures 4, G, 5, D and E).
Plants mainly respond to low-temperature stress by modulating endogenous ABA levels or by activating the ICE1–CBF pathway (Eremina et al., 2016). Because ABA was also shown to induce fruit ripening in strawberry (Jia et al., 2011), we measured the ABA contents of white stage strawberry (F. vesca, cv Fragola di Bosco) fruits cultivated at 25°C (control conditions) or exposed to 4°C for 12–60 h. However, we detected no significant differences in ABA contents among all fruits (Figure 5F). According to our RNA-seq data, both FvCBF1 and FvCBF3 were significantly upregulated in FvMAPK3-OE fruits (Figure 5G; Zhang et al., 2019). Co-expression analysis also indicated that FvMAPK3 is strongly co-expressed with the cold-responsive transcription factor genes FvICE1, FvCBF1, FvCBF3, and BASIC TRANSCRIPTION FACTOR3 (FvBTF3) (Supplemental Figure S4D). We also determined that FvMAPK3 interacts with FvICE1 (Figure 5H). However, how FvMAPK3 relays the low-temperature signaling needs to be explored further.
FvMAPK3 interacts with and phosphorylates FvCHS1 and FvMYB10
Since the specific output of MAPK cascades is determined by the identity of their substrates (Komis et al., 2018), we set out to identify proteins that interact with FvMAPK3 by performing immunoprecipitation followed by MS (IP-MS) using FvMAPK3-OE strawberry plants. We identified 619 proteins that appeared to bind to FvMAPK3 in vivo, including the UV receptor UVB-RESISTANCE8 (FvUVR8) and the low-temperature regulator FvBTF3 (Supplemental Data Set 2). Notably, FvCHS1 was among the list of putative FvMAPK3 interacting partners (Supplemental Figure S5A).
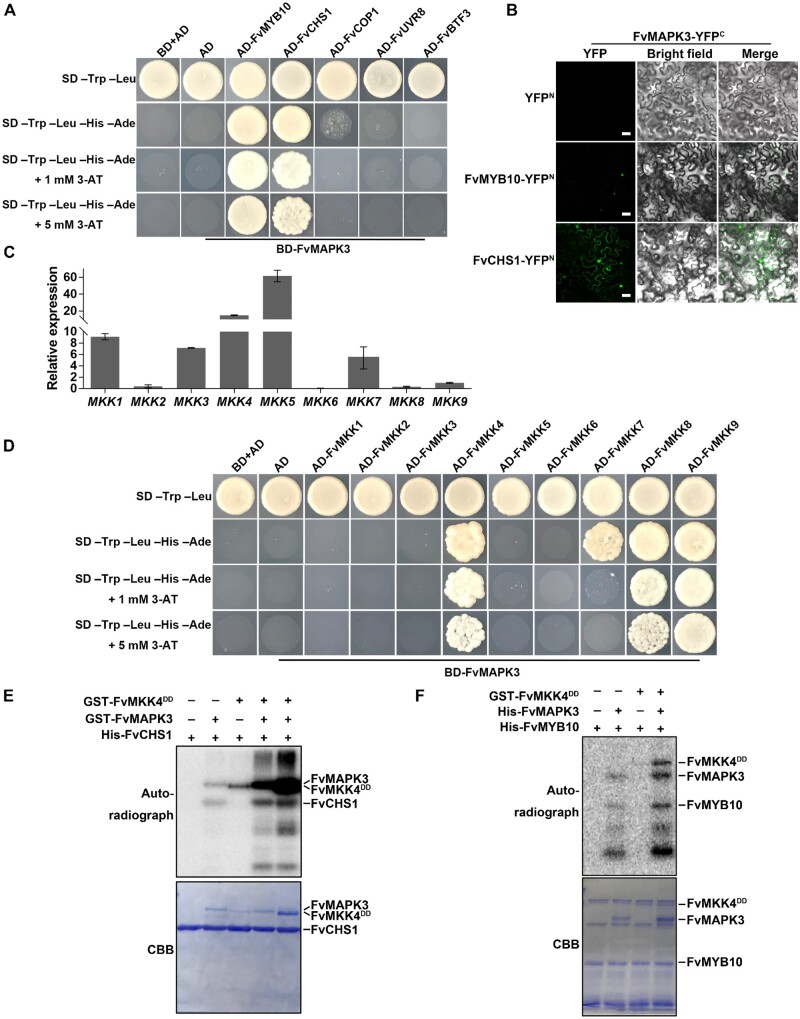
To verify the substrates of FvMAPK3, we tested the interaction between FvMAPK3 and FvUVR8, FvBTF3, FvCOP1, FvCHS1, and FvMYB10 in a Y2H assay. FvMAPK3 interacted with FvCHS1 and FvMYB10, but not with FvUVR8, FvBTF3, or FvCOP1 (Figure 6A). We validated the interaction of FvMAPK3 with FvCHS1 and FvMYB10 using a BiFC assay (Figure 6B). These results suggested that FvCHS1 and FvMYB10 are substrates for FvMAPK3.
Figure 6.
The FvMKK4–FvMAPK3 cascade phosphorylates FvCHS1 and FvMYB10. A, FvMAPK3 interacts with FvMYB10 and FvCHS1, as seen in a Y2H assay. B, Interaction analysis of FvMAPK3 with FvMYB10 and FvCHS1 by BiFC in N. benthamiana leaves. YFP signals were observed under 488-nm excitation. Bars: 50 µm. C, Relative expression levels among MKK genes in turning stage strawberry fruits, as determined by RT-qPCR. FvACTIN was used as the internal reference. Values are means ± sem of three biological replicates. D, FvMKK4, FvMKK7, FvMKK8 and FvMKK9 interact with FvMAPK3. cDNAs for the nine fruit-expressed FvMKK genes were cloned into pGADT7 and tested for interaction against FvMAPK3 cloned into pGBKT7. E, FvMAPK3 phosphorylates FvCHS1 in vitro. Recombinant GST-FvMKK4DD, GST-FvMAPK3, and His-FvCHS1 proteins were mixed in pairs for kinase activity assays. To test whether FvMKK4 has a dose-dependent effect on FvMAPK3 activation, 0.1 and 0.5 µg recombinant GST-FvMKK4DD proteins were used in Lanes 4 and 5, respectively. Phosphorylated FvCHS1 was visualized by autoradiography (top). Recombinant kinases and substrates were detected by CBB staining (bottom). F, FvMAPK3 phosphorylates FvMYB10 in vitro. Purified GST-FvMKK4DD, His-FvMAPK3, and His-FvMYB10 recombinant proteins were mixed in kinase reaction buffer and then separated by SDS–PAGE. The autoradiograph (top) and the CBB staining (bottom) of the proteins are shown.
In general, MAPKs need to be activated by specific upstream MKKs to exert their kinase activity and conduct their specific biological functions (Cobb and Goldsmith, 1995). The strawberry genome encodes nine MKK family members, with the expression of six FvMKKs being detectable in strawberry fruits (Figure 6C;Supplemental Figure S6). Of those, only FvMKK4, FvMKK7, FvMKK8, and FvMKK9 interacted with FvMAPK3 in yeast cells (Figure 6D). Relative FvMKK8 and FvMKK9 transcript levels were extremely low in turning stage fruits, while FvMKK4 was highly expressed at this stage (Figure 6C). Accordingly, we tested the activation capacity of FvMKK4 on FvMAPK3 using an in vitro kinase activity assay. Recombinant GST-FvMAPK3 showed basal autophosphorylation activity, but that of FvMKK4 rose significantly in a dose-dependent manner in the presence of FvMAPK3 (Figure 6E). Although recombinant GST-FvMAPK3 was able to phosphorylate FvMYB10 and FvCHS1 in vitro, adding recombinant FvMKK4 to the kinase reaction significantly enhanced FvMAPK3-mediated phosphorylation of FvCHS1 and FvMYB10 (Figure 6, E and F). These results suggested that FvMYB10 and FvCHS1 are output substrates of the FvMKK4–FvMAPK3 cascade in strawberry fruit.
The FvMKK4–FvMAPK3 cascade represses anthocyanin biosynthesis by enhancing FvKFB1-mediated degradation of FvCHS1 at low temperatures
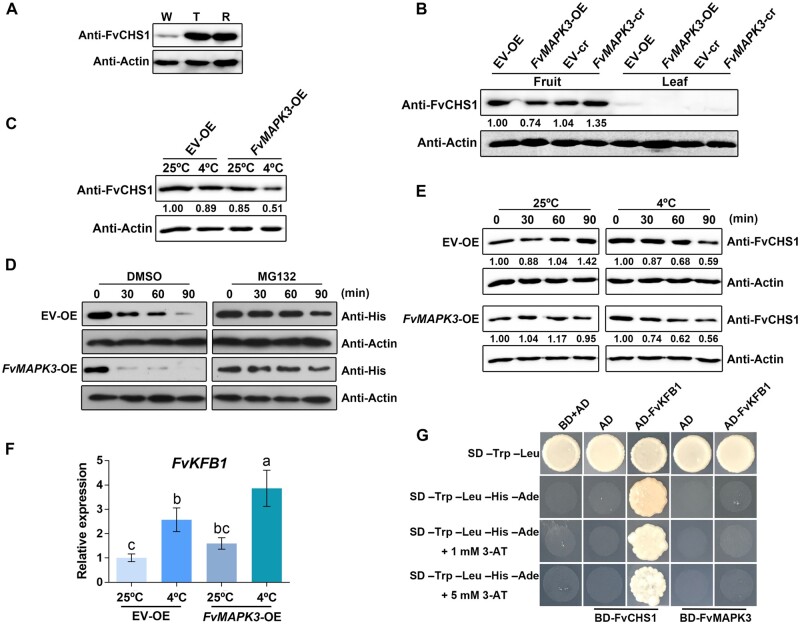
We identified five CHS genes in the strawberry genome (Supplemental Figure S5, B and C). FvCHS1 was the most highly expressed in fruits (Supplemental Figure S5D). We also measured FvCHS1 protein levels in leaves and fruits using an antibody we raised against FvCHS1 (Supplemental Figure S5; Supplemental Data Set 3). We detected FvCHS1 in strawberry fruits at the white fruit stage, followed by a sharp rise in protein abundance thereafter and until fruits were fully ripe (Figure 7A). In contrast, FvCHS1 protein was almost undetectable in leaves (Figure 7B), suggesting that FvCHS1 mainly participates in ripening-related anthocyanin biosynthesis and has a minor role in vegetative tissues. Since we had determined that FvCHS1 and FvMAPK3 physically interact in vitro and in vivo, we characterized FvCHS1 protein levels in FvMAPK3-OE and FvMAPK3-cr fruits: compared to fruits from their respective controls, FvMAPK3-OE fruits had lower FvCHS1 levels, whereas FvMAPK3-cr accumulated more FvCHS1 (Figure 7B). Since these experiments were performed at 25°C, we tested what effects, if any, transferring fruits to low temperature might have on FvCHS1 protein levels. The low temperature of 4°C reduced FvCHS1 protein levels, especially in FvMAPK3-OE fruits compared to their EV-OE control fruits, indicating that MAPK3 regulates low-temperature-mediated repression of anthocyanin accumulation by regulating FvCHS1 protein levels (Figure 7C).
Figure 7.
FvMAPK3 accelerates the degradation of FvCHS1 at low temperature. A, Immunoblot analysis of increasing FvCHS1 protein levels along with strawberry fruit ripening. W, white fruit; T, turning fruit; R, red fruit. FvCHS1 was detected with an anti-FvCHS1 antibody. Anti-actin was used as loading control. B, FvCHS1 protein levels in turning fruits and young leaves of EV-OE, FvMAPK3-OE, EV-cr, and FvMAPK3-cr plants. C, FvCHS1 protein levels diminish in white FvMAPK3-OE fruits compared to EV fruits and are more pronounced after treatment at 4°C for 30 min. D, Cell-free degradation assay of recombinant His-FvCHS1. Recombinant His-FvCHS1 protein was incubated with total proteins extracted from EV-OE or FvMAPK3-OE fruits at 37°C with 50 µM MG132 or DMSO only (carrier control). His-FvCHS1 protein was detected with anti-His antibody. E, Low-temperature-mediated degradation of FvCHS1 protein is accelerated in FvMAPK3-OE fruits. Total proteins from EV-OE and FvMAPK3-OE fruits were extracted for immunoblot analysis with anti-FvCHS1 antibody. Anti-actin was used as loading control. F, Relative FvKFB1 expression levels in EV-OE and FvMAPK3-OE fruits cultivated at 25°C (control conditions) or shifted to 4°C for 24 h. Values are means ± sem of three biological replicates. Statistical significance was determined by Tukey’s test; significant differences at the P < 0.05 level are indicated by different letters. G, FvKFB1 interacts with FvCHS1 in a Y2H assay.
Protein levels are determined by a balance between protein biosynthesis and protein degradation, the latter involving ubiquitination to mark proteins for degradation (Vierstra, 2012; Trujillo, 2018). We, therefore, investigated whether FvMAPK3 might affect FvCHS1 protein degradation in a cell-free system. To this end, we incubated recombinant His-FvCHS1 with protein extracts from EV-OE and FvMAPK3-OE white fruits. FvCHS1 degradation was significantly accelerated upon incubation with FvMAPK3-OE fruit extracts relative to EV-OE extracts (Figure 7D). In addition, the proteasome inhibitor MG132 blocked FvMAPK3-mediated degradation of FvCHS1, pointing to degradation of FvCHS1 via ubiquitination (Figure 7D). Interestingly, compared to the cell-free system, FvCHS1 protein was more stable in strawberry fruits at 25°C, but transferring fruits to low temperatures (4°C) rapidly initiated the degradation of FvCHS1, and even more so in FvMAPK3-OE fruits (Figure 7E), suggesting that FvMAPK3 may increase the degradation of FvCHS1 by sensing low-temperature stress.
It was recently reported that Arabidopsis CHS protein levels are regulated by KFBCHS-mediated ubiquitination and degradation (Zhang et al., 2017a). Here, we determined that the expression of FvKFB1, the strawberry homolog to Arabidopsis KFBCHS, rises in response to low temperature and in FvMAPK3-OE fruits (Figure 7F;Supplemental Figure S7). We then established that FvKFB1 interacts with FvCHS1, in agreement with the results from Arabidopsis, but not with FvMAPK3 (Figure 7G;Supplemental Figure S7). These results indicate that FvMAPK3 mediates low-temperature-mediated repression of anthocyanin accumulation by regulating the degradation of FvCHS1 in strawberry fruits.
Because FvSnRK2.6 phosphorylates FvMAPK3 and both proteins are involved in anthocyanin accumulation (Figure 2, C and D; Han et al., 2015), we examined whether FvCHS1 was a substrate for phosphorylation by FvSnRK2.6 or the FvSnRK2.6–FvMAPK3 module using an in vitro kinase activity assay. However, FvSnRK2.6 failed to interact with or phosphorylate FvCHS1; in addition, adding FvSnRK2.6 to a kinase reaction already containing FvMAPK3 had no significant effect on FvMAPK3-mediated phosphorylation of FvCHS1 (Supplemental Figure S8, A and B).
FvMAPK3 represses FvMYB10-mediated anthocyanin biosynthesis by decreasing its transcriptional activity at low temperatures
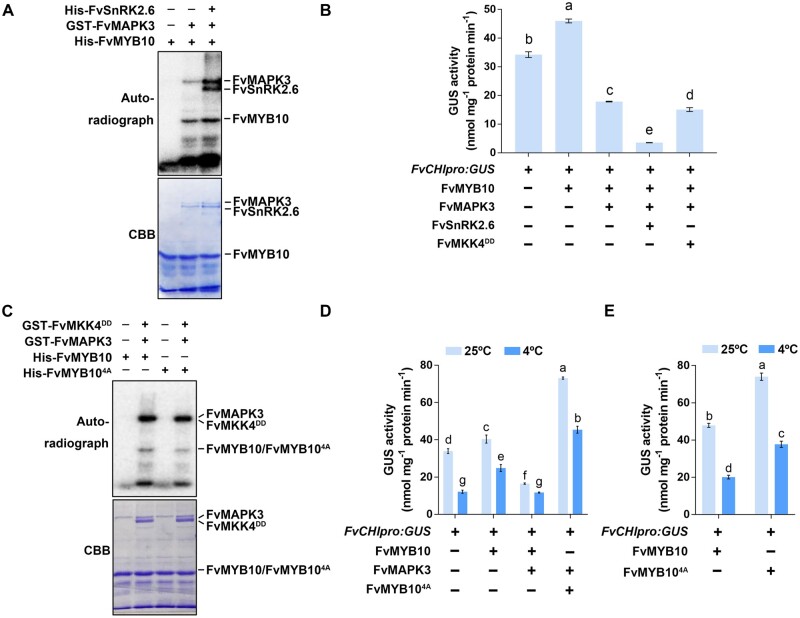
The FvMKK4–FvMAPK3 module phosphorylated FvMYB10, while FvSnRK2.6 itself phosphorylated FvMAPK3 (Figures 2, C and D and 6, F); we thus hypothesized that the FvSnRK2.6–FvMAPK3 module might target FvMYB10 for posttranslational modification. We, therefore, characterized the phosphorylation profile of FvMYB10 when incubated with recombinant FvSnRK2.6 and the FvSnRK2.6–FvMAPK3 module. We did not observe any evidence of direct phosphorylation of FvMYB10 by FvSnRK2.6 (Supplemental Figure S9A), but FvSnRK2.6 did enhance FvMAPK3-mediated FvMYB10 phosphorylation in vitro (Figure 8A).
Figure 8.
The FvSnRK2.6–FvMAPK3 module decreases the transcriptional activity of FvMYB10. A, FvSnRK2.6 increases the FvMAPK3-mediated phosphorylation of FvMYB10. Recombinant His-FvSnRK2.6, GST-FvMAPK3, and His-FvMYB10 proteins were mixed for in vitro kinase assays. Phosphorylated FvMYB10 was visualized by autoradiography (top). Recombinant FvSnRK2.6, FvMAPK3, and FvMYB10 proteins were detected by CBB staining (bottom). B, Detecting changes in FvMYB10 transcriptional activity. FvMKK4DD, FvSnRK2.6, FvMAPK3, and FvMYB10 were used as effector constructs and co-transformed with the FvCHIpro:GUS reporter. Statistically significant differences between samples were determined by Tukey’s test; significant differences at the P < 0.05 level are indicated by different letters. Values are means ± sem of three biological replicates. C, Phosphorylation analysis of FvMYB10 and its mutant form FvMYB104A. Purified recombinant GST-FvMKK4DD, GST-FvMAPK3, His-FvMYB10, and His-FvMYB104A proteins were mixed in kinase reaction buffer to detect the effects of the FvMKK4DD–FvMAPK3 module on the phosphorylation levels of FvMYB10 and FvMYB104A. The autoradiograph (top) and CBB staining (bottom) are shown. D, Detecting the effects of low temperature on FvMYB10 and FvMYB104A transcriptional activity. FvMAPK3, FvMYB10, and FvMYB104A were used as effector constructs and co-transformed with the FvCHIpro:GUS reporter. After 3 days, fruits were transferred to 4°C for 24 h or maintained at the control temperature of 25°C. Values are means ± sem of three biological replicates. Statistically significant differences between samples were determined by Tukey’s test; significant differences at the P < 0.05 level are indicated by different letters. E, The transcriptional activity of FvMYB104A is higher than that of FvMYB10. FvMYB10, FvMYB104A, and FvCHIpro:GUS were used as effector and reporter constructs co-transformed into big green strawberry fruits. After 3 days, fruits were transferred to 4°C for 24 h or maintained at the control temperature of 25°C. Values are means ± sem of three biological replicates, analyzed by Tukey’s test.
We then transiently co-expressed FvMAPK3 with its upstream kinase FvMKK4, FvSnRK2.6, and its downstream component FvMYB10 in strawberry fruits to determine whether these genes influenced FvCHI expression, based on a transcriptional reporter construct with the β-GLUCURONIDASE (GUS) reporter gene driven by the FvCHI promoter (Figure 8B). The transient expression of FvMYB10 alone significantly induced the FvCHIpro:GUS reporter, suggesting that FvMYB10 has high transcriptional activity in strawberry fruits. However, co-expressing FvMYB10 with FvMAPK3 resulted in a strong repression of FvCHIpro:GUS relative to FvMYB10 transient expression alone. Co-expressing either FvSnRK2.6 or FvMKK4 further enhanced the repression of FvMYB10 transcriptional activity imposed by FvMAPK3. Together, these results indicate that the FvSnRK2.6–FvMAPK3 and FvMKK4–FvMAPK3 signaling modules regulate FvMYB10 transcriptional activity through phosphorylation and thus regulate anthocyanin accumulation in strawberry fruits.
We sought to establish the phosphorylated residues in FvMYB10 based on conserved motifs that might be targeted by kinase and identified four putative sites (T-105, T-132, T-133, and S-163; Supplemental Figure S9B). We mutated all four sites to alanine (A) to obtain a phospho-dead mutant of FvMYB10, named FvMYB104A. We used FvMYB10 and FvMYB104A as substrates for in vitro kinase activity assays in the presence of recombinant FvMAPK3 and FvMKK4: The phosphorylation level of FvMYB104A was lower than that of FvMYB10, although not completely abolished (Figure 8C), indicating that these four residues are important for FvMAPK3-mediated phosphorylation. We also determined whether FvMAPK3-mediated phosphorylation regulated the degradation of FvMYB10 using a cell-free degradation assay. To this end, we incubated FvMYB10 and FvMYB104A with protein extracts from FvMAPK3-OE or EV-OE fruits; however, the degradation patterns seen for FvMYB10 and FvMYB104A were comparable and independent of the extracts used, suggesting that FvMAPK3 does not directly affect the degradation rate of FvMYB10 (Supplemental Figure S9C).
To examine whether the phosphorylation of FvMYB10 by FvMAPK3 affects the repression of anthocyanin accumulation at low temperature, we transiently co-expressed FvMYB10 and FvMYB104A with FvMAPK3 in strawberry fruits and measured the effects of low-temperature exposure on the FvCHIpro:GUS reporter used earlier as a measure of FvMYB10 and FvMYB104A transcriptional activity (Figure 8D). While low temperature and the transient expression of FvMAPK3 repressed the transcriptional activity of FvMYB10, this repression was largely eliminated when FvMYB104A was co-expressed. In addition, FvMYB104A exhibited a much higher transcriptional activity than FvMYB10 (Figure 8E). Notably, even when exposed to low temperatures, FvMYB104A showed strong activation of FvCHIpro:GUS expression. These results indicate that FvMYB104A has a higher transcriptional activity on the FvCHI promoter than FvMYB10. We propose that FvMYB104A offers a means to increase anthocyanin contents in strawberry fruits and may reduce poor fruit coloration caused by low temperature, making it a valuable candidate for strawberry breeding. However, we also observed that the basal GUS activity level generated by FvCHIpro:GUS was not significantly repressed by FvMAPK3–FvMYB10 co-transformation, as might have been expected (Figure 8D). We hypothesize that the effects of FvMAPK3 on FvCHI expression may be dependent on the existing FvMYB10 levels, at least in this experiment. Whether FvMAPK3 can regulate CHI/CHI at other levels of regulation and whether FvMAPK3 regulates the expression of other FvMYB10-downstream genes such as F3H and DFR via a MYB10-dependent way remain unclear.
Discussion
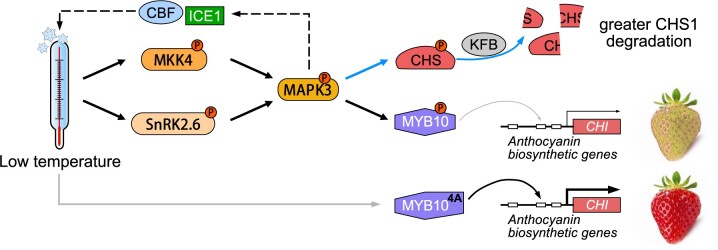
In this study, we revealed that FvMAPK3 is a regulator of strawberry fruit ripening and illustrated the regulatory pathways by which FvMAPK3 mediates poor coloration in strawberry fruit in response to low temperature (Figure 9). At low temperatures, FvMAPK3 expression is induced, followed by the activating phosphorylation of the encoded kinase. Activated FvMAPK3 then increases the sensitivity of strawberry fruits to low temperature, possibly by regulating the FvICE1–FvCBF1/FvCBF3 pathway, leading to the repression of anthocyanin accumulation in strawberry fruits. Activated FvMAPK3 represses anthocyanin accumulation in response to low temperature by two mechanisms. First, FvMAPK3 acts downstream of FvMKK4 and FvSnRK2.6 to phosphorylate FvMYB10, thereby decreasing its transcriptional activity. Second, FvMAPK3 phosphorylates FvCHS1 and enhances its FvKFB1-mediated degradation. In addition, a mutant version of FvMYB10 that is less susceptible to phosphorylation by FvMAPK3 (FvMYB104A) may have potential breeding value, enabling strawberry fruits to increase their anthocyanin contents under normal conditions and maintain fruit coloration even under low-temperature stress.
Figure 9.
MAPK3 is an important regulator of low-temperature response and ripening-related anthocyanin accumulation in strawberry fruits. When strawberry fruits are exposed to low-temperature stress, fruits perceive the low temperature possibly through the MAPK3-ICE1-CBFs signaling pathway, whereby MAPK3 is activated by SnRK2.6- and MKK4-mediated phosphorylation. The MKK4–MAPK3 module then catalyzes the phosphorylation of the rate-limiting enzyme CHS1 and upregulates the expression of the ubiquitination-related gene KFB1 to accelerate CHS1 degradation. At the same time, both the MKK4–MAPK3 and SnRK2.6–MAPK3 modules allow the phosphorylation of the transcription factor MYB10, the central activator of anthocyanin biosynthesis, leading to a reduction in its transcriptional activity. MAPK3, therefore, inhibits ripening-related anthocyanin accumulation by downregulating the transcriptional activity of MYB10 and decreasing CHS1 protein levels in strawberry fruits. Additionally, compared to MYB10, its phosphorylation-dead mutant MYB104A shows a reduced level of MAPK3-mediated phosphorylation, has higher transcriptional activity upon CHI and is not sensitive to low temperatures; hence, MYB104A may protect strawberry fruits from delayed coloration due to low temperatures. Dark arrows indicate activation, and thin gray arrow shows repression. Blue arrows indicate the pathways only mediated by the “FvMKK4-FvMAPK3” module, while the dark arrows represent the pathways regulated by both the “FvSnRK2.6-FvMAPK3” and “FvMKK4-FvMAPK3” modules. Dashed lines indicate the low-temperature sensing pathway. Thick gray arrow indicates that the low-temperature response is blocked.
MAPK cascades are important conserved signaling modules that regulate plant growth and responses to diverse abiotic and biotic stresses (Zhang et al., 2018). Although all members of MAPK cascades (FvMKKKs, FvMKKs, and FvMAPKs) have been identified in strawberry and their expression patterns to abiotic and biotic stress have been evaluated by RT-qPCR (Wei et al., 2017; Zhou et al., 2017a), their functions in strawberry and other fruits remain largely unknown (Chen et al., 2018; He et al., 2020; Zhang et al., 2020a).
We discovered here that FvMAPK3 is a negative regulator of anthocyanin accumulation in strawberry fruits (Figures 3 and 4) that acts downstream of FvMKK4 and FvSnRK2.6 as a repressor of FvCHS1 and FvMYB10 activities (Figures 2 and 5–8). MAPK3 is a well-known member of multiple MAPK cascades with many described biological functions (Chen et al., 2018; He et al., 2020; Zhang et al., 2020a). However, because MAPK3 regulates plant fertility and a double mutation of MPK3 (the Arabidopsis MAPK3) and the closely related MPK6 is embryo lethal, the function of MAPK3 in reproductive organs and especially in fruits is unknown (Chen et al., 2018; He et al., 2020; Zhang et al., 2020a). In this study, manipulating FvMAPK3 by both transient expression and stable transformation modulated anthocyanin accumulation in strawberry fruits (Figures 3 and 4). Notably, FvMAPK3-OE and FvMAPK3-cr plants bore normal fruits under favorable cultivation conditions. The T1 generation of FvMAPK3-OE and FvMAPK3-cr transgenic plants was viable, although the germination rate of FvMAPK3-cr seeds was lower than that of EV-cr seeds (Figure 4;Supplemental Figure S3). Although the relative expression level of FvMAPK3 was increased from the big green stage to the red fruit stage (Figure 3A), FvMAPK3 acts as a negative regulator of anthocyanin accumulation in fruits (Figures 3 and 4). This result suggests that FvMAPK3 is mainly involved in the regulation of fruit ripening at the posttranslational level. Additionally, the expression of ripening-related genes involved in fruit softening and aroma production and the key ripening regulator gene FvRIF was also affected in transgenic fruits (Figures 3, F and 4, G), indicating that FvMAPK3 may have more roles besides regulating anthocyanin accumulation in strawberry fruits.
MAPK proteins regulate various biological processes by phosphorylating different substrates. Thus, identifying MAPK substrates helps elucidate the biological functions of the underlying MAPK cascades (Zhang et al., 2016; Dóczi and Bögre, 2018; Wang et al., 2020). Previous work showed that low phosphate conditions activate the MAKK9–MAPK3/MAPK6 module to repress the transcription of anthocyanin biosynthetic genes and anthocyanin accumulation in Arabidopsis (Lei et al., 2014). However, the substrate of the MAKK9–MAPK3/MAPK6 module in response to low-phosphate-mediated repression of anthocyanin accumulation has yet to be identified. In this study, we established that the transcription factor FvMYB10 and the rate-limiting anthocyanin biosynthetic enzyme FvCHS1 are substrates of FvMAPK3 and both participate in the regulation of anthocyanin accumulation in strawberry fruits (Figure 6;Supplemental Figure S5; Supplemental Data Set 2).
FvMYB10 is a well-established activator of anthocyanin biosynthesis in diploid and octoploid strawberry fruits (Lin-Wang et al., 2014; Medina-Puche et al., 2014; Castillejo et al., 2020), and its homologs MYB75 in Arabidopsis (also named PRODUCTION OF ANTHOCYANIN PIGMENT1 [PAP1]), McMYB10 in crabapple (Malus crabapple), PavMYB10.1 in sweet cherry (Prunus avium L.), PcMYB10 in European pear (Pyrus communis), and SmMYB113 in eggplant (Solanum melongena L.) have also been reported to regulate anthocyanin biosynthesis, indicating that MYB10 is a powerful and conserved regulator of anthocyanin biosynthesis in plants (Telias et al., 2011; Wang et al.,2013b; Jin et al., 2016; Li et al., 2016, 2018; Zhou et al., 2020). In strawberry fruits, MYB10 regulates the expression of anthocyanin biosynthesis genes such as CHS1, CHI, DFR, and ANS individually or by forming an MYB–bHLH–WD repeat module with bHLH3/33 and TTG1, thereby regulating anthocyanin accumulation (Schaart et al., 2013; Wei et al., 2018). Compared to their transcriptional regulation, little is known about the posttranscriptional regulatory mechanisms of FvMYB10 and its homologs. In Arabidopsis, MYB75 is phosphorylated by MAPK4 to mediate high-light-induced anthocyanin accumulation, while MAPK3 and MAPK6 are not involved in this process (Li et al., 2016). However, MAPK3 and MAPK6 negatively regulated low-phosphate-mediated anthocyanin accumulation in Arabidopsis (Lei et al., 2014). These studies and our findings indicate that while the MAPK/MYB10 phosphorylation module may play a conserved role in the regulation of anthocyanin accumulation, the specific MAPK engaged in the module may be species dependent and respond to specific and distinct signals. Interestingly, both stable transformation and transient OE of FvMAPK3 significantly prevented anthocyanin accumulation in strawberry fruits (Figures 3 and 4), indicating that FvMAPK3 may have a more prominent role in regulating anthocyanin accumulation in fruits rather than in other tissues or that the FvMAPK3/FvMYB10 module can be regulated by ripening-related endogenous signals in strawberry fruit. We also noticed that the expression of FvMYB10 is extremely significantly repressed by low temperature (Figure 1E), suggesting that FvMYB10 transcript levels are also critical for the regulation of low-temperature-mediated anthocyanin accumulation. Given that FvMYB10 expression is suppressed in FvMAPK3-OE lines and increased in FvMAPK3-cr lines under both normal and low-temperature conditions (Figures 4 and 5), we postulate that how FvMAPK3 regulates FvMYB10 transcription is also important for regulating ripening and low-temperature sensing in strawberry fruits.
FvCHS1 is an important structural gene that controls strawberry fruit ripening (Hoffmann et al., 2006) and whose expression is regulated by the fruit ripening-related transcription factors FvMYB10, FvRAV1, FvRAP, and FvRIF (Lin-Wang et al., 2014; Medina-Puche et al., 2014; Gao et al., 2020; Zhang et al., 2020b; Martín-Pizarro et al., 2021). Because this gene encodes the rate-limiting enzyme in anthocyanin biosynthesis, the transcriptional regulation of CHS has been the focus of much work in different plants and organs (Hoffmann et al., 2006; Yuan et al., 2009; Hosokawa et al., 2013; Xu et al., 2017; Nakayama et al., 2019). Only recently has a posttranslational mode of regulation also been demonstrated for CHS, which involves KFBCHS-mediated degradation and ubiquitination in Arabidopsis (Zhang et al., 2017a). Our results show that the KFB/CHS module also functions in the regulation of anthocyanin accumulation in strawberry fruits and that FvMAPK3 acts upstream of the FvKFB1/FvCHS1 module by phosphorylating and enhancing the degradation of FvCHS1 and inducing FvKFB1 expression (Figures 6 and 7). Whether the MAPK3/CHS module functions are conserved in the context of anthocyanin accumulation should be explored further.
The accumulation of anthocyanin is strongly affected by environmental factors (Steyn et al., 2002; Zhang et al., 2010; Jaakola, 2013; Landi et al., 2015). Previous studies of low-temperature stress in strawberry showed that flavonoid compounds are closely associated with cold tolerance in strawberry seedlings and crowns. Moreover, CHS, F3H, and DFR were linked to cold tolerance in strawberry crowns (Koehler et al., 2012, 2015). Interestingly, low temperature reduced the antioxidant activity and anthocyanin and total phenolic contents extractable from strawberry fruits (Wang, 2006; Koehler et al., 2012). In agreement with these observations, we established that low temperatures prevent anthocyanin accumulation in attached and detached fruits over the entire fruit ripening process, greatly reducing the economic value of strawberry fruits (Figure 1). Previous studies have identified several regulators involved in low-temperature-mediated repression of anthocyanin accumulation, such as Arabidopsis LONG HYPOCOTYL5 (HY5), Arabidopsis CBF1, and apple (Malus domestica) MdMYB1, MdMYB23, MdMYB308L, and MdbHLH3 (Catalá et al., 2011; Xie et al., 2012; Zhou et al.,2017b; An et al., 2018, 2020). However, the underlying signal transduction mechanisms are unclear (Wang and Camp, 2000; Catalá et al., 2011; Jaakola, 2013; Koehler et al., 2015; Zhou et al., 2020).
In this study, we revealed that FvMAPK3 is activated at low temperatures (Figure 2), thus increasing FvKFB1-mediated degradation of FvCHS1 and decreasing the transcriptional activity of FvMYB10 (Figures 7 and 8). Besides FvMAPK3, FvMAPK4 and FvMAPK6 were also activated at low temperatures (Figure 2), raising the possibility that FvMAPK4 and FvMAPK6 might also contribute to low-temperature signaling during strawberry fruit ripening.
Combined with our transcriptional data and IP-MS results, we observed that FvMAPK3 is closely associated with several components of ABA signaling pathways (Supplemental Figure S4). However, low temperature had no effect on the ABA contents of strawberry fruits, and FvMAPK3 responded to low temperature, possibly by regulating the FvICE1/FvCBFs pathway (Figure 5). The exact nature of the relationship between FvMAPK3 and ABA signaling should be explored further in strawberry fruits. SnRK2.6/OST1 and MAPK3 were both reported to be involved in low-temperature signaling by phosphorylating the transcription factor ICE1 in Arabidopsis and rice (Ding et al., 2015; Li et al., 2017; Zhao et al., 2017; Zhang et al., 2017b), but the connection between SnRK2.6 and MAPK3 during low-temperature signaling is not clearly defined. We showed that FvSnRK2.6 acts as an enhancer of FvMAPK3-mediated repression of anthocyanin accumulation at low temperatures (Figure 8). Determining whether the SnRK2.6–MAPK3 module is functionally conserved in low-temperature signaling across plants may provide important insights into how plants respond to low-temperature stress.
Genome editing via CRISPR is an effective tool for gene functional analysis and molecular breeding (Chen et al., 2020; Gaston et al., 2020; Xing et al., 2020). To date, only genome-edited alleles of MAPK3 have been reported in tomato (Solanum lycopersicum). We generated knockout mutants in FvMAPK3 using diploid strawberry (F. vesca, cv Fragola di Bosco.), although the resulting FvMAPK3-cr strawberry lines underwent a ploidy change during tissue culture and are thus tetraploid (Supplemental Figure S3; Zhang et al., 2014). We also identified one line harboring the empty construct (EV-cr) that had undergone a similar doubling of its chromosome number as control (Supplemental Figure S3G). We obtained more tetraploid lines among all selected FvMAPK3-cr lines than among EV-cr lines, suggesting that FvMAPK3 might be involved in regulating strawberry ploidy. However, the underlying mechanism is not clear. Interestingly, most wild tetraploid resources are dioecious (Lei et al., 2008), but EV-cr and FvMAPK3-cr are both monoecious strawberry plants with normal fertility under favorable cultivation conditions. Several potential valuable traits observed in FvMAPK3-cr lines, such as robust stems, elongated fruiting branches, and oblate fruits, can be inherited from seed by the T1 generation (Supplemental Figure S3, D, E, and G). Whether the change in ploidy induced during transformation and tissue culture can be applied to strawberry breeding and the nature of the molecular mechanism responsible for ploidy levels should be studied further. Protein kinases have been proposed to act as rheostats in the tradeoff between growth and resistance to stress (Yang et al., 2019). The identification of specific phosphorylation sites in their substrates may provide valuable candidates to fine-tune breeding (Wang et al., 2018; Fu et al., 2021). Accordingly, we identified the phosphorylation sites in FvMYB10 that are associated with poor fruit coloration in response to low temperature. We propose that the phospho-dead mutant FvMYB104A may have potential breeding value, as it would allow strawberry fruits to increase their anthocyanin contents under normal conditions and maintain fruit coloration under low-temperature stress (Figures 8 and 9).
Materials and Methods
Plant materials and growth conditions
Octoploid strawberry F. × ananassa Duch. cv Benihoppe and diploid strawberry F. vesca, cv Fragola di Bosco were used in this study. Strawberry plants were grown in the greenhouse or in a growth chamber under a 12-h-light/12-h-dark photoperiod with a light intensity of 200–300 µmol m−2 s−1 (white fluorescent tubes, T5, 14 W) and 70% humidity, under day/night temperature cycles of 25°C/15°C.
Y2H assays
Y2H assays were performed with the GAL4-based Two-Hybrid System 3 according to the manufacturer’s instructions (Clontech, Mountain View, CA, USA). Full-length cDNAs for FvMAPK1-12, FvSnRK2.6, FvCHS1, FvMYB10, FvMKK1, FvMKK2, FvMKK3, FvMKK4, FvMKK5, FvMKK6, FvMKK7, FvMKK8, FvMKK9, FvKFB1, FvCOP1, FvUVR8, FvBTF3, and FvICE1 were cloned individually into pGADT7 or pGBKT7 vectors. Plasmids were co-transformed into yeast strain AH109 by the lithium acetate method and grown on minimal synthetic medium lacking Leu and Trp (–Leu –Trp), lacking Leu, Trp, His, and Ade (–Leu –Trp –His –Ade), or lacking Leu, Trp, His, and Ade and containing 3-AT (–Leu –Trp –His –Ade + 1 mM or 5 mM 3-AT). The primers used for cloning are listed in Supplemental Data Set 4.
BiFC assay
Full-length coding sequences (CDSs) for FvMAPK3, FvSnRK2.6, FvCHS1, and FvMYB10 were cloned into pSPYNE and pSPYCE vectors and transformed into Agrobacterium (Agrobacterium tumefaciens) strain GV3101. Agrobacterium cultures were resuspended in infiltration buffer (10-mM MES pH 5.6, 10-mM MgCl2, and 200-µM acetosyringone) to a final OD600 of 0.6. Pairs of Agrobacterium cultures harboring relevant constructs were co-infiltrated into 4- to 6-week-old N. benthamiana leaves to observe reconstitution of YFP signal with excitation at 488 nm using a confocal microscope (Olympus Fluoview FV1000) after 48–72 h of incubation, as described previously (Schütze et al., 2009). Primers used for cloning are listed in Supplemental Data Set 4.
Protein subcellular localization
The CDS of FvMAPK3 without stop codon was amplified by PCR, cloned into the pMDC83 vector, and transformed into Agrobacterium (strain GV3101). Agrobacteria cultures were resuspended in infiltration buffer to a final OD600 of 0.6 and infiltrated into epidermal cells of 4- to 6-week-old N. benthamiana leaves. After 48–72 h, the leaves’ epidermal cells were observed with a confocal laser scanning microscope (Olympus Fluoview FV1000). Primers used for cloning are listed in Supplemental Data Set 4.
In vitro pull-down assay
Recombinant GST-MAPK3, His-SnRK2.6, and glutathione-S-transferase (GST) were purified using glutathione Sepharose beads (GE Healthcare, Chicago, IL, USA) and Ni-NTA agarose (Novagen, Madison, WI, USA), as per the manufacturer’s manuals. GST or GST-FvMAPK3 bound to Glutathione Sepharose beads was incubated with His-FvSnRK2.6 at 4°C for 2 h. The beads were washed 5 times with phosphate-buffered saline buffer. The proteins were then eluted from beads and immunoblotted with Anti-His antibody.
Transient transformation of strawberry fruits
To generate the FvMAPK3 OE vector (FvMAPK3-OE), the full-length FvMAPK3 CDS was PCR amplified from strawberry first-strand cDNAs and recombined into the pH7WG2D vector (harboring the sequence for an N-terminal FLAG tag) using Gateway methods (Zhang et al., 2014). To construct the FvMAPK3-RNAi plasmid, two fragments of the FvMAPK3 CDS were inserted into vector pFGC5941 in opposite orientation on either side of the chalcone synthase A intron from Petunia hybrida (Supplemental Figure S10). The constructs and respective EVs were transformed into Agrobacterium strain EHA105 and grown at 28°C in Luria-Bertani (LB) medium. When the Agrobacterium cultures reached OD600 of 0.6–0.8, they were centrifuged at 3,000 g for 10 min at room temperature and resuspended in infiltration buffer (10-mM MES pH 5.6, 10-mM MgCl2, and 200-µM acetosyringone) and shaken for 2 h at room temperature. Big green octoploid fruits with consistent growth status were selected for transient transformation. The bacterial suspensions of EV-OE, FvMAPK3-OE, EV-RNAi, and FvMAPK3-RNAi were injected individually into fruits with a 1-mL syringe when the fruits became hygrophanous. After 9 days, injected fruits were harvested, and their seeds were removed, frozen in liquid nitrogen, and stored at –80°C for analysis of ripening-related parameters and gene expression (Wei et al., 2018). For each experiment, three biological repeats were performed, each consisting of 30 fruits.
Stable transformation of diploid strawberry
The FvMAPK3-OE vector was generated by cloning the full-length CDS for FvMAPK3 into the pH7WG2D vector. The CRISPR/Cas9 vector pYLCRISPR/Cas9 was used for genome editing of FvMAPK3 with two specific sgRNAs (Supplemental Data Set 4). The CRISPR vector was constructed based on previously described methods (Zeng et al., 2018). The resulting vectors (FvMAPK3-OE and FvMAPK3-cr) were then transformed into Agrobacterium strain EHA105 using diploid strawberry leaf disks as described (Oosumi et al., 2006). After selection of transformants with 2-mg/L hygromycin, all hygromycin-resistant strawberry seedlings were observed for eGFP fluorescence as independent confirmation, as the eGFP gene is harbored by the T-DNA of pH7WG2D. Relative transcript levels of target genes were also determined by RT-qPCR. Genomic DNA was extracted from hygromycin-resistant genome-edited strawberry seedlings for PCR analysis using specific primers and sequencing over the putative edited sites. Seeds of T0 transgenic strawberries were grown on half-strength Murashige and Skoog medium with 3-mg/L hygromycin to screen for homozygous mutants by PCR. The primers and sgRNA sequences are listed in Supplemental Data Set 4.
Recombinant protein production and purification
Full-length cDNAs for FvMAPK3, FvSnRK2.6, FvCHS1, and FvMYB10 were cloned into the pET30a, pGEX4T1, and pGEX6P1 vectors and introduced into Escherichia coli BL21(DE3) cells to produce recombinant His-FvMAPK3, GST-FvMAPK3, His-FvSnRK2.6, His-FvCHS1, and His-FvMYB10 proteins. The T227D/S233D (DD) point mutations were introduced into FvMKK4 (FvMKK4DD), while the T105A/T132A/T133A/S163A (4A) mutations were introduced into FvMYB10 (FvMYB104A) by site-directed mutagenesis. Recombinant proteins were produced and purified using Glutathione Sepharose beads (GE Healthcare) and Ni-NTA agarose (Novagen) according to the manufacturer’s instructions and as previously described (Zhao et al., 2017).
Protein extraction and immunoblot analysis
Total proteins from strawberry fruits and leaves were extracted in extraction buffer (phosphate buffer pH 7.8, 1-mM EDTA, 10% [v/v] glycerol, 0.5% [v/v] Triton X-100, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail, and 1× phosphatase inhibitor cocktail). Collected tissues were ground to powder in liquid nitrogen. Protein extraction buffer was added to the powder on ice and then vortexed to homogenize the samples. Homogenates were centrifuged at 13,000 g for 10 min at 4°C. The resulting supernatants were analyzed by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblots using goat anti-mouse IgG, HRP-conjugated (1:2,000, CWBIO, Jiangsu, China), anti-rabbit IgG, peroxidase antibody produced in goat (1:10,000; Sigma-Aldrich, St Louis, MO, USA), anti-His (1:1,000, CWBIO), anti-GST (1:1,000, CWBIO), anti-Flag (1:500; Abmart, Shanghai, China), anti-β-actin (1:1,000, CWBIO), anti-Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (1:500; Cell Signaling Technology, Danvers, MA, USA), and anti-FvCHS1 antibodies. The anti-FvCHS1 polyclonal antibody was produced by Beijing Protein Innovation using recombinant full-length His-FvCHS1.
In vitro phosphorylation assay
Recombinant His-FvMAPK3, GST-FvMAPK3, GST-FvMKK4DD, or His-FvSnRK2.6 proteins were incubated in reaction buffer (20-mM Tris–HCl pH 7.5, 10-mM MgCl2, 1-mM DTT, and 50-µM ATP) for 30 min at 30°C as previously described to activate His-FvMAPK3 or GST-FvMAPK3 proteins (Li et al., 2017). Recombinant His-FvCHS1, His-FvMYB10, and His-FvMYB104A proteins were then individually incubated with activated His-FvMAPK3 or GST-FvMAPK3 in kinase reaction buffer (20-mM Tris–HCl pH 7.5, 10-mM MgCl2, 1-mM DTT, 25-µM ATP, and 1-µCi [γ-32p] ATP) for 30 min at 30°C. The reactions were terminated by adding SDS loading buffer. Samples were then subjected to 10% SDS–PAGE to separate proteins. The gel was exposed to a phosphor imager screen for 12–24 h and visualized with a Typhoon 9410 imager.
Liquid chromatography tandem mass spectrometry assay
To identify the phosphorylation sites of FvMAPK3, purified recombinant GST-FvMAPK3 and His-FvSnRK2.6 proteins were incubated in reaction buffer (20-mM Tris–HCl pH 7.5, 10-mM MgCl2, 1-mM DTT, and 50-µM ATP) for 30 min at 30°C. The reaction was stopped by the addition of SDS loading buffer. After SDS–PAGE, the gel was stained with Coomassie brilliant blue (CBB), and the gel band containing GST-FvMAPK3 was subjected to tryptic digestion and then analyzed with a nanoLC-LTQ-Orbitrap XL (Thermo, San Jose, CA, USA).
Low-temperature treatments
The effects of low-temperature stress on fruit ripening and anthocyanin accumulation were analyzed using both detached fruits and attached fruits. Detached fruits collected at the big green stage, white stage, and turning stage were incubated at 4°C, 10°C, or 25°C under a light intensity of 50-µmol m−2 s−1 and 70% relative humidity in a growth chamber. Ten fruits at each stage were exposed to the same temperature treatment, and three biological repeats were performed in one individual experiment. Three individual experiments were performed. For attached fruits, strawberry plants carrying three fruits each were transferred to growth chambers set to 4°C, 10°C, or 25°C under a light intensity of 50-µmol m−2 s−1 and 70% relative humidity. Three biological replicates were performed at each temperature, with each biological replicate consisting of three plants. The phenotypes of detached and attached fruits were recorded every day.
To detect the expression changes of anthocyanin-related genes, detached white fruits were exposed to 25°C or 4°C for 24 h, frozen in liquid nitrogen, and stored at –80°C. The expression levels were determined by RT-qPCR. For each treatment, 10 fruits were pooled as one sample. Three biological repeats using 30 fruits were performed for one sample in one individual experiment.
Measurement of anthocyanin contents
For anthocyanin content analysis, 50-mg dried strawberry fruits were mixed with 2% formic acid–methanol (v/v) solution for 24 h at 4°C and centrifuged at 12,000 g for 5 min. The supernatant was filtered through a 0.22-µm filter for further analysis. Anthocyanin content was measured by high-performance liquid chromatography as previously described (Gu et al., 2019).
Co-immunoprecipitation/MS
To identify the proteins that bind to FvMAPK3 in vivo, we performed a co-immunoprecipitation followed by MS analysis on FvMAPK3-OE strawberry plants. Total proteins were extracted from control (EV-OE) and FvMAPK3-OE strawberry seedlings in extraction buffer (phosphate buffer pH 7.8, 1-mM EDTA, 10% [v/v] glycerol, 0.5% [v/v] Triton X-100, 1-mM DTT, 1-mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail, and 1× phosphatase inhibitor cocktail) and then incubated with anti-FLAG antibody (CWBIO) and protein A + G agarose (CWBIO, CW0349S). Beads with protein were washed 3–5 times using extraction buffer and eluted with SDS loading buffer. After SDS–PAGE, the gel was analyzed by LC–MS/MS.
Cell-free degradation assays
Cell-free degradation assays for FvMYB10 and FvCHS1 were performed as previously described with a few changes (Li et al., 2017). Total proteins of control (EV-OE) and FvMAPK3-OE fruits were extracted and incubated with recombinant His-MYB10, His-MYB104A, or His-FvCHS1 proteins in buffer (50-mM Tris–MES pH 8.0, 500-mM sucrose, 1-mM MgCl2, 10-mM EDTA, and 5-mM DTT). The mixture was divided into two equal volumes; to one, 50 µM MG132 was added to inhibit protein degradation, while the other samples received an equal amount of DMSO (MG132 carrier) as control. Both samples were incubated at 37°C, and a 30-µL aliquot was taken out every 30 min at the beginning of the incubation period (0 min) and until 90 min. His-MYB10, His-MYB104A, and His-FvCHS1 proteins were detected by immunoblot analysis using anti-His antibody as above.
In vivo degradation assays
White stage strawberry fruits from control (EV-OE) and FvMAPK3-OE plants were transferred from 25°C to 4°C for 30, 60, or 90 min, or maintained at 25°C. Total proteins of treated EV-OE and FvMAPK3-OE fruits were extracted in extraction buffer (phosphate buffer pH 7.8, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, 1 mM DTT, 1-mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail, and 1× phosphatase inhibitor cocktail). Proteins were separated on SDS–PAGE as above and examined by immunoblot analysis with anti-FvCHS1 antibody.
Transcriptome deep sequencing (RNA-seq) analysis
Total RNA was extracted from fruits of control (EV-OE) and FvMAPK3-OE plants at 25-day postanthesis using the E.Z.N.A. Total RNA Kit (Omega, Bienne, Switzerland). RNA integrity was assessed using the RNA Nano 6000 Assay Kit on a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Sequencing was performed on an Illumina Novaseq platform as 150-bp paired-end reads and mapped using the F. vesca subsp. vesca (assembly FraVesHawaii_1.0) reference genome. (https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/184/155/GCF_000184155.1_FraVesHawaii_1.0/). Genes with an adjusted P < 0.05 as determined by DESeq2 were considered differentially expressed (Love et al., 2014). Three independent biological replicates were analyzed in this experiment.
RT-qPCR analysis
Total RNA was extracted from strawberry fruits with the E.Z.N.A. Total RNA Kit (Omega) following the manufacturer’s instructions. First-strand cDNAs were synthesized from 1-µg total RNA with M-MLV reverse transcriptase (Promega, Madison, WI, USA). qPCR was performed on a Rotor Gene Q cycler (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and using FastSYBR Mix (Low ROX). FvACTIN was used as the internal reference. Relative expression was calculated using the 2−ΔΔCT method. Primer sequences used for RT-qPCR are listed in Supplemental Data Set 5.
GUS activity assay
Full-length cDNAs for FvMAPK3, FvSnRK2.6, FvMKK4DD, FvMYB10, and FvMYB104A were placed under the control of the cauliflower mosaic virus 35S promoter. The FvCHI promoter was cloned upstream of the GUS reporter gene in the pCAMBIA1301 vector to construct the reporter system. Agrobacteria cultures were resuspended in infiltration buffer (10-mM MES pH 5.6, 10-mM MgCl2, and 200-µM acetosyringone) to a final OD600 of 0.6 for each construct. Big green strawberry fruits were selected to inject the constructs, as previously described (Wei et al., 2018). After 3 days, fruits were transferred to 4°C for 24 h or maintained at the control temperature of 25°C, collected, and frozen in liquid nitrogen. GUS activity was determined as described (Wei et al., 2018). For each construct, three independent biological replicates were performed, each consisting of 10 transformed fruits. Primer sequences used for cloning are listed in Supplemental Data Set 4.
Determination of plant ploidy
Young strawberry leaves (2–4 cm2) were selected and thinly sliced into 2 mL of cell lysis buffer using a sharp blade. The mixture was filtered through an 80-µm filter membrane and centrifuged at 1,000 g and 4°C for 5 min to collect nuclei. Nuclear DNA was then labeled in propidium iodide solution for 20 min in the dark and loaded onto a FACSCalibur analyzer (BD Biosciences) for flow cytometry analysis. A strawberry plant with known ploidy was used as control.
Sequence analysis and phylogenetic analysis
Protein sequence alignments were performed using ClustalX version 2.1 with default parameters (Thompson et al., 1997). The phylogenetic trees were constructed via the neighbor-joining method in MEGA version 5.0 software with 1,000 bootstrap replicates. The protein sequences used to construct the phylogenetic trees are provided in Supplemental Files S1–S5.
Statistical analysis
Two-tailed Student’s t tests were used on two groups of data points in GraphPad Prism version 5.0. (*P < 0.05, **P < 0.01). When comparing multiple groups, analysis of variance (ANOVA) was performed using SPSS version 13.0. Summary statistics are given in Supplemental File S6.
Accession numbers
Sequence data from this article can be found in Supplemental Data sets 6, 7, and 8.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. FvSnRK2.6 interacts with FvMAPK3.
Supplemental Figure S2. Phylogenetic analysis of MAPKs in strawberry.
Supplemental Figure S3. Construction of FvMAPK3 OE and genome-edited transgenic diploid strawberry.
Supplemental Figure S4. Expression profiling of differentially expressed genes (DEGs) between EV-OE and FvMAPK3-OE fruits.
Supplemental Figure S5. FvCHS1 binds to FvMAPK3.
Supplemental Figure S6. Identification of MKK genes in strawberry.
Supplemental Figure S7. Identification of KFB genes in strawberry.
Supplemental Figure S8. Test of interaction and phosphorylation potential between FvSnRK2.6 and FvCHS1.
Supplemental Figure S9. Analysis of FvMYB10 phosphorylation sites and degradation pattern.
Supplemental Figure S10. Sequence used for the FvMAPK3-RNAi construct.
Supplemental Data Set 1. List of DEGs altered more than two-fold in FvMAPK3-OE fruits relative to EV-OE fruits.
Supplemental Data Set 2. Identification of specific FvMAPK3-binding proteins by co-immunoprecipitation-MS.
Supplemental Data Set 3. Identification of proteins in turning fruits for FvCHS1 antibody specification.
Supplemental Data Set 4. Sequences of the primers used in vector construction.
Supplemental Data Set 5. Primers used for RT-qPCR analysis.
Supplemental Data Set 6. Fragaria vesca ID and GenBank accession numbers mentioned in this work.
Supplemental Data Set 7. GenBank accession numbers of other species mentioned in this work.
Supplemental Data Set 8. The protein sequence of MAPK3 in diploid and octoploid strawberry in this work.
Supplemental Files S1–S5. Alignments used to generate phylogenetic trees.
Supplemental File S6. Summary of statistical tests.
Supplementary Material
Acknowledgments
We thank Prof. Wensuo Jia for his helpful suggestions and for providing tissue-culture and transformation facilities. We thank Dr Kathleen Farquharson for her professional suggestions on the writing.
Funding
This work was supported by the National Key Research and Development Program (2018YFD1000200), the National Natural Science Foundation of China (31772284 and 32072551), 111 Project (B17043) and the 2115 Talent Development Program of China Agricultural University.
Conflict of interest statement. None declared.
B.L. and W.M. designed the experiments and analyzed the data. W.M. performed most of the experiments. L.W., Y.H., Y.C., and M.S. made some of the constructs. Q.F., L.L., L.P.L., and K.Z. prepared reagents and detected the expression of some genes. B.L., W.M., and Z.H. wrote the article.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: libingbing@cau.edu.cn.
References
- Aharoni A, Ric De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JNM, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332 [DOI] [PubMed] [Google Scholar]
- Almeida JRM, D’Amico E, Preuss A, Carbone F, Ric de Vos CH, Deiml B, Mourgues F, Perrotta G, Fischer TC, Bovy AG, et al. (2007) Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria × ananassa). Arch Biochem Biophys 465: 61–71 [DOI] [PubMed] [Google Scholar]
- An JP, Li R, Qu FJ, You CX, Wang XF, Hao YJ (2018) R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J 96: 562–577 [DOI] [PubMed] [Google Scholar]
- An JP, Wang XF, Zhang XW, Xu HF, Bi SQ, You CX, Hao YJ (2020) An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol J 18: 337–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Waurich V, Wagner H, Ramos R, Oiza N, Muñoz P, Triviño JC, Caruana J, Liu Z, Cobo N, et al. (2020) Allelic variation of MYB10 is the major force controlling natural variation in skin and flesh color in strawberry (Fragaria spp.) fruit. Plant Cell 32: 3723–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci USA 108: 16475–16480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yang D, Zhang Y, Wu L, Zhang Y, Ye L, Pan C, He Y, Huang L, Ruan YL, et al. (2018) Evidence for a specific and critical role of mitogen-activated protein kinase 20 in uni-to-binucleate transition of microgametogenesis in tomato. New Phytologist 219: 176–194 [DOI] [PubMed] [Google Scholar]
- Chen Y, Mao W, Liu T, Feng Q, Li L, Li B (2020) Genome editing as a versatile tool to improve horticultural crop qualities. Hortic Plant J 6: 372–384 [Google Scholar]
- Cobb MH, Goldsmith EJ (1995) How MAP kinases are regulated. J Biol Chem 270: 14843–14846 [DOI] [PubMed] [Google Scholar]
- Davik J, Daugaard H, Svensson B (2000) Strawberry production in the Nordic countries. Adv Strawb Prod 19:13–18 [Google Scholar]
- Ding Y, Jia Y, Shi Y, Zhang X, Song C, Gong Z, Yang S (2018) OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J 37: e98228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32: 278–289 [DOI] [PubMed] [Google Scholar]
- Ding Y, Shi Y, Yang S (2019) Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytologist 222: 1690–1704 [DOI] [PubMed] [Google Scholar]
- Droillard MJ, Boudsocq M, Barbier-Brygoo H, Laurie`re C (2002) Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett 527: 43–50 [DOI] [PubMed] [Google Scholar]
- Dóczi R, Bögre L (2018) The quest for MAP kinase substrates: gaining momentum. Trend Plant Sci 23: 918–932 [DOI] [PubMed] [Google Scholar]
- Edger PP, VanBuren R, Colle M, Poorten TJ, Wai CM, Niederhuth CE, Alger EI, Ou S, Acharya CB, Wang J, et al. (2018) Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 7: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina M, Rozhon W, Poppenberger B (2016) Hormonal control of cold stress responses in plants. Cell Mol Life Sci 73: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM (1988) Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol 8: 1985–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Liu Y, Qin GWP, Zi H, Xu Z, Zhao X, Wang Y, Li Y, Yang S, Peng C, et al. (2021) The TOR–EIN2 axis mediates nuclear signalling to modulate plant growth. Nature 591: 288–292 [DOI] [PubMed] [Google Scholar]
- Gao Q, Luo H, Li Y, Liu Z, Kang C (2020) Genetic modulation of RAP alters fruit coloration in both wild and cultivated strawberry. Plant Biotechnol J 18: 1550–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston A, Osorio S, Denoyes B, Rothan C (2020) Applying the Solanaceae strategies to strawberry crop improvement. Trend Plant Sci 25: 130–140 [DOI] [PubMed] [Google Scholar]
- Gu Z, Men S, Zhu J, Hao Q, Tong N, Liu ZA, Zhang H, Shu Q, Wang L (2019) Chalcone synthase is ubiquitinated and degraded via interactions with a RING-H2 protein in petals of Paeonia ‘He Xie’. J Exp Bot 70: 4749–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Lu Z, Shan J, Ye W, Dong N, Lin H (2020) ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell 32: 2763–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Dang RH, Li JX, Jiang JZ, Zhang N, Jia MR, Wei LZ, Li ZQ, Li BB, Jia WS (2015) Sucrose nonfermenting1-related protein kinase2.6, an ortholog of open stomata1, is a negative regulator of strawberry fruit development and ripening. Plant Physiol 167: 915–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C, Caruana J, Schiksnis E, Liu Z (2016) Genome-scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci Rep 6: 29017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Giusti M (2010) Anthocyanins: natural colorants with health-promoting properties. Annu Rev Food Sci Technol 1: 163–187 [DOI] [PubMed] [Google Scholar]
- He X, Wang C, Wang H, Li L, Wang C (2020) The function of MAPK cascades in response to various stresses in horticultural plants. Front Plant Sci 11: 952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, Kalinowski G, Schwab W (2006) RNAi-induced silencing of gene expression in strawberry fruit (Fragaria × ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J 48: 818–826 [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Yamauchi T, Takahama M, Goto M, Mikano S, Yamaguchim Y, Tanaka Y, Ohno S, Koeda S, Doi M, et al. (2013) Phosphorus starvation induces post-transcriptional CHS gene silencing in Petunia corolla. Plant Cell Rep 32: 601–609 [DOI] [PubMed] [Google Scholar]
- Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18: 477–483 [DOI] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Wang Y, Sun M, Li B, Han Y, Zhao Y, Li X, Ding N, Li C, Ji W, et al. (2013) Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytologist 198: 453–465 [DOI] [PubMed] [Google Scholar]
- Jin W, Wang H, Li M, Wang J, Yang Y, Zhang X, Yan G, Zhang H, Liu J, Chen K (2016) The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol J 14: 2120–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z(2013) Genome-Scale transcriptomic insights into Early-Stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25: 1960–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL (2004) β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol 135: 1674–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler G, Rohloff J, Wilson RC, Kopka J, Erban A, Winge P, Bones AM, Davik J, Alsheikh MK, Randall SK (2015) Integrative “omic” analysis reveals distinctive cold responses in leaves and roots of strawberry, Fragaria × ananassa ‘Korona’. Front Plant Sci 6: 826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler G, Wilson RC, Goodpaster JV, Sonsteby A, Lai X, Witzmann FA, You JS, Rohloff J, Randall SK, Alsheikh M (2012) Proteomic study of low-temperature responses in strawberry cultivars (Fragaria × ananassa) that differ in cold tolerance. Plant Physiol 159: 1787–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G, Šamajová O, Ovečka M, Šamaj J (2018) Cell and developmental biology of plant mitogen-activated protein kinases. Annu Rev Plant Biol 69: 6.1–6.29 [DOI] [PubMed] [Google Scholar]
- Landi M, Tattini M, Gould KS (2015) Multiple functional roles of anthocyanins in plant-environment interactions. Environ Exp Bot 119: 4–17 [Google Scholar]
- Ledesma NA, Nakata M, Sugiyama N (2008) Effect of high temperature stress on the reproductive growth of strawberry cvs. ‘Nyoho’ and ‘Toyonoka’. Sci Hortic 116: 186–193 [Google Scholar]
- Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Dai H, Zhao M, Wu W (2008) Study on the tetraploid Fragaria species distributed in China. J Fruit Sci 25: 358–361 [Google Scholar]
- Lei L, Li Y, Wang Q, Xu J, Chen Y, Yang H, Ren D (2014) Activation of MKK9-MPK3/MPK6 enhances phosphate acquisition in Arabidopsis thaliana. New Phytologist 203: 1146–1160 [DOI] [PubMed] [Google Scholar]
- Li H, Ding Y, Shi Y, Zhang X, Zhang S, Gong Z, Yang S (2017) MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev Cell 43: 1–13 [DOI] [PubMed] [Google Scholar]
- Li KT, Zhang J, Kang YH, Chen MC, Song TT, Geng H, Tian J, Yao YC (2018) McMYB10 modulates the expression of a ubiquitin ligase, McCOP1 during leaf coloration in crabapple. Front Plant Sci 9: 704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang W, Gao J, Yin K, Wang R, Wang C, Petersen M, Mundy J, Qiu JL (2016) MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28: 2866–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K, McGhie TK, Wang M, Liu Y, Warren B, Storey R, Espley RV, Allan AC (2014) Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front Plant Sci 5: 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Chen J, Ren X, Yuan J, Han X, Mao L, Ying T, Luo Z (2018) One novel strawberry MADS-box transcription factor FaMADS1a acts as a negative regulator in fruit ripening. Sci Hortic 227: 124–131 [Google Scholar]
- Martín-Pizarro C, Vallarino JG, Osorio S, Meco V, Urrutia M, Pillet J, Casañal A, Merchante C, Amaya I, Willmitzer L, et al. (2021) The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell 33: 1574–1593 Doi:10.1093/plcell/koab070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Puche L, Cumplido-Laso G, Amil-Ruiz F, Hoffmann T, Ring L, Rodríguez-Franco A, Caballero JL, Schwab W, Munoz-Blanco J, Blanco-Portales R (2014) MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. J Exp Bot 65: 401–417 [DOI] [PubMed] [Google Scholar]
- Mirahmadi F, Hanafi QM, Mohammadi H (2012) Effect of low temperatures on physico-chemical properties of different strawberry cultivars. ARPN J Agric Biol Sci 7: 564–569 [Google Scholar]
- Nakayama T, Takahashi S, Waki T (2019) Formation of flavonoid metabolons: functional significance of protein-protein interactions and impact on flavonoid chemodiversity. Front Plant Sci 10: 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosumi T, Gruszewski HA, Blischak LA, Baxter AJ, Wadl PA, Shuman JL, Veilleux RE, Shulaev V (2006) High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta 223: 1219–1230 [DOI] [PubMed] [Google Scholar]
- Salvatierra A, Pimentel P, Moya-León MA, Herrera R (2013) Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry 90: 25–36 [DOI] [PubMed] [Google Scholar]
- Schaart JG, Dubos C, Fuente IRDL, Houwelingen AMML, Vos RCH, Jonker HH, Xu WJ, Routaboul JM, Lepiniec L, Bovy AG (2013) Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria×ananassa) fruits. New Phytol 197: 454–467 [DOI] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Plant Signal Trans Methods Protocol 479: 189–202 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Ryder CD, Cevik V, Hammond JP, Popovich A, King GJ, Vrebalov J, Giovannoni JJ, Manning K (2011) A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria × ananassa Duch.) fruit, a non-climacteric tissue. J Exp Bot 62: 1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155: 349–361 [DOI] [PubMed] [Google Scholar]
- Taghavi T, Siddiqui RK, Rutto L (2019) The effect of preharvest factors on fruit and nutritional quality in strawberry. Strawberry Pre-and Post-harvest Management Techniques for Higher Fruit Quality. BoD – Books on Demand, Norderstedt, Germany Doi:10.5772/intechopen.84619. [Google Scholar]
- Telias A, Lin-Wang K, Stevenson D, Cooney JM, Hellens RP, Allan AC, Hoover EE, Bradeen JM (2011) Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol 11: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur P, Kumar S, Malik JA, Berger JD, Nayyar H (2010) Cold stress effects on reproductive development in grain crops: an overview. Environ Exp Bot 67: 429–443 [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Ann Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M (2018) News from the PUB: plant U-box type E3 ubiquitin ligases. J Exp Bot 69: 371–384 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6: rs8. [DOI] [PubMed] [Google Scholar]
- Vallarino JG, Merchante C, Sánchez-Sevilla JF, de Luis Balaguer MA, Pott DM, Ariza MT, Casañal A, Posé D, Vioque A, Amaya I, et al. (2019) Characterizing the involvement of FaMADS9 in the regulation of strawberry fruit receptacle development. Plant Biotechnol J 18: 929–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallarino JG, Osorio S, Bombarely A, Casanal A, Cruz-Rus E, Sánchez-Sevilla JF, Amaya I, Giavalisco P, Fernie AR, Botella MA, et al. (2015) Central role of FaGAMYB in transition of strawberry receptacle from development to ripening. New Phytol 208: 482–496 [DOI] [PubMed] [Google Scholar]
- Vierstra RD (2012) The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol 160: 2–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang H, Yang Y, Li M, Zhang Y, Liu J, Dong J, Li J, Butelli E, Xue Z, et al. (2019) The control of red colour by a family of MYB transcription factors in octoploid strawberry (Fragaria × ananassa) fruits. Plant Biotechnol J 18:1169–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, He M, Yin J, Zhu X, Li Y, et al. (2018) A single transcription factor promotes both yield and immunity in rice. Science 361: 1026–1028 [DOI] [PubMed] [Google Scholar]
- Wang P, Hsu CC, Du Y, Zhu P, Zhao C, Fu X, Zhang C, Paez JS, Macho AP, Tao WA, et al. (2020) Mapping proteome-wide targets of protein kinases in plant stress responses. Proc Natl Acad Sci USA 117: 3270–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Andy Tao W, Zhu JK (2013a) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effector of abscisic acid action. Proc Natl Acad Sci USA 110: 11205–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SY (2006) Effect of pre-harvest conditions on antioxidant capacity in fruits. Acta Hortic 712: 299–305 [Google Scholar]
- Wang SY, Camp MJ (2000) Temperatures after bloom affect plant growth and fruit quality of strawberry. Sci Hortic 85: 183–199 [Google Scholar]
- Wang SY, Zheng W, Galletta GJ (2002) Cultural system affects fruit quality and antioxidant capacity in strawberries. J Agric Food Chem 50: 6534–6542 [DOI] [PubMed] [Google Scholar]
- Wang Z, Meng D, Wang A, Li T, Jiang S, Cong P, Li T (2013b) The methylation of the PcMYB10 promoter is associated with green-skinned sport in max red bartlett pear. Plant Physiol 162: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Mao W, Jia M, Xing S, Ali U, Zhao Y, Chen Y, Cao M, Dai Z, Zhang K, et al. (2018) FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J Exp Bot 69: 4805–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Chai Z, Xie Y, Gao K, Cui M, Jiang Y, Feng J (2017) Bioinformatics identification and transcript profile analysis of the mitogen-activated protein kinase gene family in the diploid woodland strawberry Fragaria vesca. PLoS One 12: e0178596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35: 1884–1897 [DOI] [PubMed] [Google Scholar]
- Xing S, Chen K, Zhu H, Zhang R, Zhang H, Li B, Gao C (2020) Fine-tuning sugar content in strawberry. Genome Biol 21: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Charles MT, Luo Z, Mimee B, Veronneau PY, Rolland D, Roussel D (2017) Preharvest ultraviolet C irradiation increased the level of polyphenol accumulation and flavonoid pathway gene expression in strawberry fruit. J Agric Food Chem 65: 9970–9979 [DOI] [PubMed] [Google Scholar]
- Yang G, Yu Z, Gao L, Zheng C (2019) SnRK2s at the crossroads of growth and stress responses. Trend Plant Sci 24: 672–676 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Chiu L, Li L (2009) Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta 230: 1141–1153 [DOI] [PubMed] [Google Scholar]
- Zeng DC, Ma XL, Xie XR, Zhu QL, Liu YG (2018) A protocol for CRISPR/Cas9-based multi-gene editing and sequence decoding of mutant sites in plants. Sci Sin Vitae 48: 783–794 [Google Scholar]
- Zhang G, Jia S, Yan Z, Wang Y, Zhao F, Sun Y (2020a) A strawberry mitogen-activated protein kinase gene, FaMAPK19, is involved in disease resistance against Botrytis cinerea. Sci Hortic 265: 109259 [Google Scholar]
- Zhang M, Su J, Zhang Y, Xu J, Zhang S (2018) Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr Opin Plant Biol 45: 1–10 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Folta KM, Davis TM (2014) Somatic embryogenesis, tetraploidy, and variant leaf morphology in transgenic diploid strawberry (Fragaria vesca subspecies vesca ‘Hawaii 4’). BMC Plant Biol 14: 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Chen S, Harmon AC (2016) Protein-protein interactions in plant mitogen-activated protein kinase cascades. J Exp Bot 67: 607–618 [DOI] [PubMed] [Google Scholar]
- Zhang X, Abrahan C, Colquhoun TA, Liu CJ (2017a) A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in Arabidopsis. Plant Cell 29: 1157–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, Lin Y, Luo Y, Wang X, Chen Q, Sun B, Wang Y, Li M, Tang H (2019) A transcriptomic analysis reveals diverse regulatory networks that respond to cold stress in strawberry (Fragaria×ananassa). Int J Genom 2019:7106092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zheng S, Liu Z, Wang L, Bi Y (2010) Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J Plant Physiol 168: 367–374 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li J, Li F, Liu H, Yang W, Chong K, Xu Y (2017b) OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev Cell 43: 731–743 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shi Y, Ma Y, Yang X, Yin X, Zhang Y, Xiao Y, Liu W, Li Y, Li S, et al. (2020b) The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol J 18: 2267–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Wang P, Si T, Hsu CC, Wang L, Zayed O, Yu Z, Zhu Y, Dong J, Tao WA, et al. (2017) MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev Cell 43: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Ren S, Han Y, Zhang Q, Qin L, Xing Y (2017a) Identification and analysis of Mitogen-Activated Protein Kinase (MAPK) Cascades in Fragaria vesca. Int J Mol Sci 18: 1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, He Y, Li J, Liu Y, Chen H (2020) CBFs function in anthocyanin biosynthesis by interacting with MYB113 in eggplant (Solanum melongena L.). Plant Cell Physiol 61: 416–426 [DOI] [PubMed] [Google Scholar]
- Zhou LJ, Li YY, Zhang CL, Xie XB, Zhao C, Hao YJ (2017b) The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple. Plant Cell Environ 40: 2068–2080 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.