Abstract
Cytosine methylation is a reversible epigenetic modification of DNA. In plants, removal of cytosine methylation is accomplished by the four members of the DEMETER (DME) family of 5-methylcytosine DNA glycosylases, named DME, DEMETER-LIKE2 (DML2), DML3, and REPRESSOR OF SILENCING1 (ROS1) in Arabidopsis thaliana. Demethylation by DME is critical for seed development, preventing experiments to determine the function of the entire gene family in somatic tissues by mutant analysis. Here, we bypassed the reproductive defects of dme mutants to create somatic quadruple homozygous mutants of the entire DME family. dme; ros1; dml2; and dml3 (drdd) leaves exhibit hypermethylated regions compared with wild-type leaves and rdd triple mutants, indicating functional redundancy among all four demethylases. Targets of demethylation include regions co-targeted by RNA-directed DNA methylation and, surprisingly, CG gene body methylation, indicating dynamic methylation at these less-understood sites. Additionally, many tissue-specific methylation differences are absent in drdd, suggesting a role for active demethylation in generating divergent epigenetic states across wild-type tissues. Furthermore, drdd plants display an early flowering phenotype, which involves 5′-hypermethylation and transcriptional down-regulation of FLOWERING LOCUS C. Active DNA demethylation is therefore required for proper methylation across somatic tissues and defines the epigenetic landscape of intergenic and coding regions.
Analysis of a mutant null for all four DNA demethylases in Arabidopsis vegetative tissues expands our understanding of the targets and function of DNA demethylation.
Introduction
Phenotypic stability depends on epigenetic stability. Cytosine DNA methylation is both stably inherited and dynamic. In many plant species, DNA methylation is concentrated in transposable elements and other repetitive sequences, transcriptionally silencing these genomic regions. DNA methylation also accumulates in gene coding regions, where it does not induce transcriptional silencing (Niederhuth et al., 2016; Bewick and Schmitz, 2017). Methylation patterns can be highly conserved over evolutionary time but also dynamic in specific developmental contexts, such as during reproduction (Gehring, 2019). These patterns are a result of methylation activity and demethylation activity, the coordination of which is required to maintain transgenerational epigenetic and phenotypic stability (Williams and Gehring, 2017). Methylation is established by de novo DNA methyltransferases, which are guided to their targets by small RNAs in a process termed RNA-directed DNA methylation (RdDM) (Matzke and Mosher, 2014). Symmetric DNA methylation is maintained by maintenance methyltransferase enzymes that copy patterns of DNA methylation after DNA replication.
Loss of DNA methylation can occur passively, when DNA methylation is not maintained after DNA replication, or by active removal by DNA demethylation pathways. Active DNA demethylation is initiated by the activity of a family of bifunctional HhH-GPD DNA glycosylases/lyases (Agius et al., 2006; Gehring et al., 2006; Morales-Ruiz et al., 2006; Penterman et al., 2007; Ortega-Galisteo et al., 2008). In Arabidopsis thaliana, there are four gene family members: DEMETER (DME), REPRESSOR OF SILENCING1 (ROS1), DEMETER-LIKE2 (DML2), and DML3 (Choi et al., 2002; Gong et al., 2002; Penterman et al., 2007; Ortega-Galisteo et al., 2008). These enzymes remove methylated cytosines through base excision repair (Roldán-Arjona et al., 2019). DME and ROS1 genes are present in plants from algae onward (Pei et al., 2019). The distribution of DML2 and DML3 is more limited, with DML3 present in monocots and dicots and DML2 present only in a subset of dicots (Pei et al., 2019).
Molecular and morphological phenotypes have been described for plants with mutations in one or more of the 5-mC DNA glycosylases in Arabidopsis and rice (Oryza sativa). Arabidopsis plants heterozygous for dme mutations have a visibly striking phenotype: 50% seed abortion (Choi et al., 2002). Seeds that inherit a mutant dme allele from the mother abort after several days of development and thus homozygous dme mutants have only rarely been recovered. In some Arabidopsis accessions, dme is also not fully transmitted through the male parent (Schoft et al., 2011). DME is expressed in the polar nuclei and the central cell (the female gamete that is the progenitor of the endosperm) before fertilization and is required to establish gene imprinting in the endosperm after fertilization (Choi et al., 2002; Gehring et al., 2006). In the central cell, DME demethylates specific loci (Park et al., 2016) and this hypomethylated state is transmitted to the endosperm after fertilization, such that the maternally inherited endosperm genome is hypomethylated compared with the paternally inherited endosperm genome (Gehring et al., 2006, 2009; Hsieh et al., 2009; Ibarra et al., 2012). DME-dependent endosperm hypomethylated sites are enriched for fragments of transposable elements that reside near genes (Gehring et al., 2009). DME is also active in the pollen vegetative cell and similar targets are hypomethylated in a DME-dependent manner in both the vegetative cell and endosperm (Calarco et al., 2012; Ibarra et al., 2012). Like Arabidopsis, rice central cells and vegetative cells are hypomethylated (Kim et al., 2019; Park et al., 2016). Mutations in a rice DME homolog (termed ROS1a) have a similar phenotype as Arabidopsis dme mutants––maternal null alleles disrupt endosperm development and mutant maternal or paternal alleles are only rarely transmitted to progeny (Ono et al., 2012). ROS1a is also responsible for DNA demethylation in the vegetative cell (Kim et al., 2019) and likely also in the central cell. Although Arabidopsis DME is mostly highly expressed in reproductive tissues, expression is also detected in vegetative tissues (Mathieu et al., 2007; Park et al., 2017; Schumann et al., 2019). The function of DME outside of reproductive tissues is less well understood.
Arabidopsis plants with mutations in any of the other three DNA glycosylases lack visibly dramatic phenotypes under standard growth conditions and all null mutants are viable singly and in combination (Gong et al., 2002; Penterman et al., 2007; Ortega-Galisteo et al., 2008). However, some phenotypes have been observed. Stomatal precursor-cell density is increased in ros1 leaves due to hypermethylation and transcriptional silencing of a negative regulator of precursor cells (Yamamuro et al., 2014). Additionally, ros1 and ros1; dml2; dml3 (rdd) triple mutants exhibit impaired tracheary element differentiation, resulting in a high frequency of protoxylem discontinuities (Lin et al., 2020). Plants with mutations in ros1 and rdd exhibit enhanced susceptibility to bacterial and fungal pathogens (Yu et al., 2013; Le et al., 2014; López Sánchez et al., 2016), which is associated with decreased expression of biotic stress genes and, in a handful of examined genes, promoter hypermethylation at TE sequences (Le et al., 2014; Halter et al., 2021). Genome-wide profiling of DNA methylation in ros1 plants and rdd plants indicates DNA hypermethylation at hundreds or thousands of intergenic regions and transposable elements that are also targeted by RdDM and are enriched near genes (Penterman et al., 2007; Lister et al., 2008; Tang et al., 2016). Most of the DNA methylation changes in rdd are not obviously associated with changes in gene transcription (Penterman et al., 2007; Lister et al., 2008). Thus, it is thought that one function of the DNA demethylation in vegetative tissues is to “clean up” after a robust DNA methylation system to keep genes free from methylation.
Although genetic approaches have proved fruitful in understanding the function of 5-mC DNA glycosylases, the inability to generate homozygous mutant dme plants has stymied full understanding of this gene family. RNAi was used to knockdown DME expression in vegetative tissues of the rdd triple mutant (Schumann et al., 2019). These plants are even more susceptible to a fungal pathogen and additional hypermethylation was shown at a few loci. These data suggest that vegetative tissue of rdd plants retain some demethylation activity from DME. Recently, Kim et al. (2021) isolated homozygous mutant dme-2 plants in the Landsberg erecta (Ler) background, which display root and shoot defects related to meristem alterations. To identify the full extent of demethylation activity, we created plants where dme was complemented only in the central cell, allowing us to examine methylation and transcription on a genome-wide scale in vegetative tissues null for all four 5-methylcytosine DNA glycosylases.
Results
Isolating quadruple demethylase mutants
In order to more fully understand the role of the DME family of 5-methylcytosine DNA glycosylases in Arabidopsis, we sought to isolate homozygous mutations in all four homologs in this family: DME, ROS1, DML2, and DML3. To rescue the dme seed abortion phenotype, we created a transgene in which the genomic coding sequence of DME was expressed under the central cell-specific promoter of AGL61 (Figure 1, A; Steffen et al., 2008). The transgene was transformed into the previously isolated rdd triple mutant in the Columbia-0 (Col-0) background (Penterman et al., 2007). Transgenic rdd plants were pollinated by heterozygous dme mutants to create heterozygous quadruple mutants (Figure 1, B). These heterozygotes were self-fertilized and seed abortion rates were quantified to test the ability of the pAGL61:DME transgene to complement the dme seed abortion phenotype. Whereas non-transgenic plants harboring the dme mutation exhibit 50% seed abortion, transgenic lines expressing pAGL61:DME exhibited minimal seed abortion, indistinguishable from rdd plants. Expression of DME in the central cell before fertilization is therefore sufficient to rescue the post-fertilization seed abortion phenotype.
Figure 1.
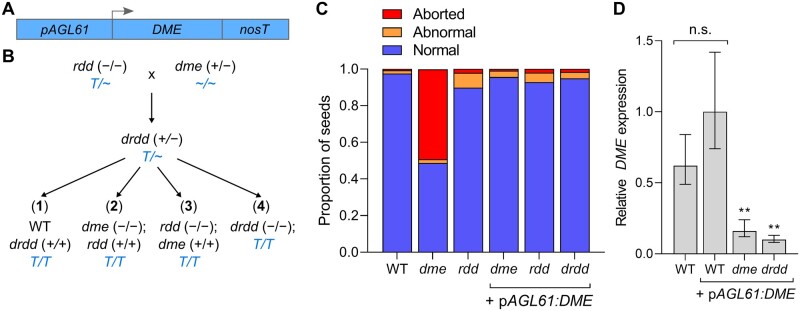
Generation of homozygous dme and quadruple drdd mutants. A, Schematic showing the construct designed to express DME specifically in central cells using the AGL61 promoter. B, Schematic showing the segregation of mutant genotypes. Transgenic rdd plants were pollinated by heterozygous (+/−) dme to generate heterozygous drdd F1 progeny. WT, dme, rdd, and drdd homozygotes were isolated from two subsequent generations. C, Proportion of aborted seeds in non-transgenic WT, dme, rdd plants, as well as dme, rdd, and drdd plants homozygous for the pAGL61:DME transgene. In total, 250–300 seeds were evaluated for each genotype. D, qRT-PCR measurement of DME expression in WT Col-0 plants and WT, dme, and drdd plants expressing pAGL61:DME. **P ≤ 0.001 (versus WT).
After self-fertilizing the heterozygous quadruple mutant, the following four genotypes were isolated over two subsequent generations: homozygous quadruple mutants (hereafter termed drdd), homozygous rdd mutants, homozygous dme mutants, and homozygous wild-type segregants (hereafter termed WT, and to serve as genetically closely related wild-type controls for subsequent experiments). All four of these genotypes were determined to be homozygous for a single transgene insertion and displayed seed abortion rates of <2%, similar to non-transgenic rdd (Figure 1, C).
To determine whether the AGL61 promoter was active outside of the central cell, we compared the expression of DME in leaves of wild-type Col-0 plants lacking the transgene to leaves of WT, dme, and drdd plants that were transgenic for pAGL61:DME (Figure 1, D). There was no significant difference in DME expression between Col-0 and WT, suggesting that pAGL61:DME is not significantly active in leaves. DME expression was 6.3- and 10-fold lower in dme and drdd, respectively (Figure 1, D), suggesting the AGL61 promoter can direct a low level of expression in leaf tissues. Thus, although perhaps not representing a complete loss-of-function of DME, this material is suitable to examine the consequences of substantially reduced DME activity in vegetative tissues.
DME is required for full demethylation in somatic tissue
To fully understand the function of DME outside of the central cell and the combined activity of all members of the DME family, paired DNA and RNA samples were collected from rosette leaf tissue of WT, dme, rdd, and drdd plants. Individual leaves from four separate biological replicate plants were sliced along the midvein, and DNA and RNA were extracted from each half, so that DNA methylation and gene expression could be analyzed from the same leaf.
Whole-genome bisulfite sequencing (BS-seq) was performed on DNA samples (Supplemental Dataset S1). In order to improve read depth for subsequent analyses, bisulfite-sequencing reads were combined for pairs of replicates to create two high-depth replicates for each genotype. Previously, the rdd triple mutant was constructed by back-crossing ros1 and dml2 T-DNA insertions originally from a Wassilewskija (Ws) background into a Col-0 background (Penterman et al., 2007). We therefore first analyzed the zygosity of Ws SNPs throughout the sequenced WT, dme, rdd, and drdd methylomes to determine the presence of chromosomal regions originating from Ws. Consistent with previous reports (Penterman et al., 2007), we observed regions of Ws homozygosity surrounding the ros1 and dml2 T-DNA insertions on chromosomes 2 and 3, respectively, in both rdd and drdd (Supplemental Figure S1). These regions were discarded in all methylation analyses, as strain-specific methylation differences between Col-0 and Ws could contribute to false positives in identifying differentially methylated regions (DMRs).
To assess the extent of methylation differences in dme, rdd, and drdd, differentially methylated dinucleotides (CG context) or individual cytosines (CH context) were identified between WT and each mutant genotype. Methylation differences were restricted to a minimum percentage difference of 35% for CG, 20% for CHG, and 15% for CHH. Differentially methylated CGs/Cs (DMCs) were then aggregated into windows, which were assigned a differential methylation score based on the quantity and proportion of DMCs (Williams and Gehring, 2017). This score was then used to identify DMRs (Figure 2, A). Using this approach, very few DMRs were identified between biological replicates of the same genotype (33 between WT replicates and 5 between drdd replicates) and similar numbers of differences were identified between WT and mutant genotypes, regardless of which pairwise comparison of biological replicates was performed (Supplemental Figure S2). In total, 8–13% of DMRs identified with one biological replicate were not differentially methylated in other replicate comparisons, suggesting little difference between biological replicates. To account for these differences between replicates and identify a final list of DMRs between genotypes, only CGs/Cs that were differentially methylated in both mutant replicates were included (see methods). As this approach required DMCs to have adequate read depth in both replicates in order to be counted, we estimate that our analysis has few false positives, but as a consequence likely offers an under-estimate of the true number of methylation differences among the genotypes.
Figure 2.
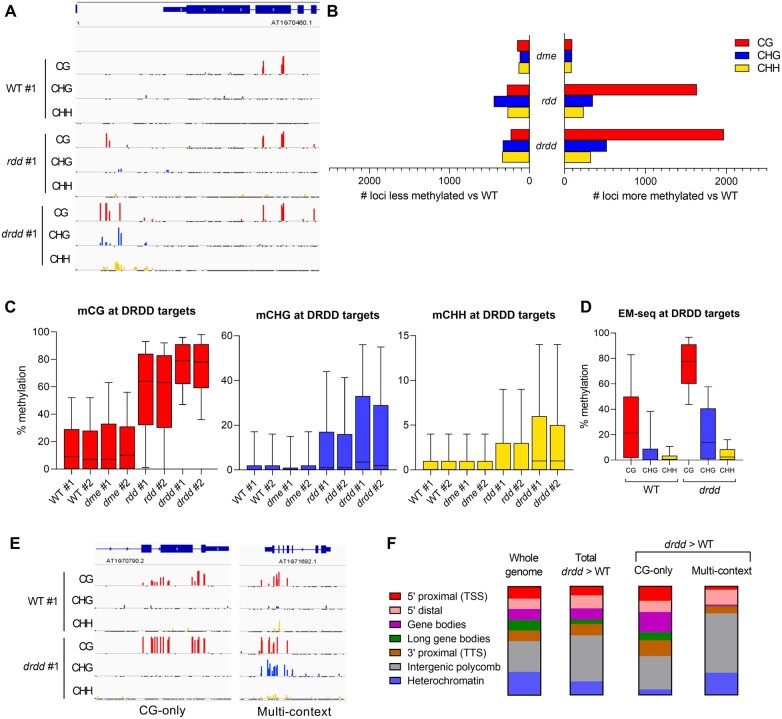
Exacerbated DNA hypermethylation in drdd mutants. A, Genome browser snapshot showing typical locus hypermethylated in drdd compared with WT and rdd. #1 denotes first of two biological replicates. B, Number of DMRs in dme, rdd, and drdd compared with WT. C, Methylation levels of WT, dme, rdd, and drdd at loci hypermethylated in drdd compared with WT (DRDD targets). Boxes denote the interquartile range, whiskers denote 10th and 90th percentiles. D, Methylation of WT and drdd at DRDD targets (identified by BS-seq) independently measured by EM-seq. E, Genome browser snapshots showing typical examples of a locus hypermethylated specifically in the CG context (CG-only) and a locus hypermethylated in all sequence contexts (multi-context). F, The distribution of loci hypermethylated in drdd compared with WT across different genomic chromatin states.
We identified 276 DMRs between WT and dme mutants (Figure 2, B). This suggests that DME does not have a large number of target loci in leaves that are independent of the other three demethylases. We observed 2,091 regions with increased methylation in rdd compared with WT. The quadruple mutant drdd exhibited even greater methylation differences compared with WT, with 1,265 loci more than rdd, including almost twice the number of regions with differential non-CG methylation. This implies that DME does function in somatic tissue, but redundantly with ROS1, DML2, and DML3. The set of loci hypermethylated in drdd compared with WT (2,601 regions after merging sequence contexts) therefore represents a set of genomic targets for the entire DME family, which we hereafter refer to as “DRDD target loci” (Supplemental Dataset S2).
To better understand the extent of DME activity at DRDD target loci, we quantified methylation levels at these regions in all four genotypes studied. Single mutant dme replicates had methylation levels similar to WT, suggesting that DME functions redundantly with the other demethylases in removing methylation at these targets (Figure 2, C). Whereas rdd triple mutants showed hypermethylation in all three contexts at DRDD target loci, this hypermethylation was much more pronounced in drdd (Figure 2, C). At the single-cytosine level, 72% of CGs within DRDD target loci were more highly methylated in drdd compared with rdd, as well as 53% of non-CG cytosines. These data suggest that in rdd plants, DME still actively removes methylation from the majority of DRDD target loci. To further verify the 2,601 DRDD target loci identified, we performed a single replicate of enzymatic methyl-sequencing (EM-seq; Feng et al., 2020) on WT and drdd leaf tissues, which confirmed the increased methylation of all 2,601 loci in drdd (Figure 2, D).
We next sought to better understand the genomic context of DRDD target loci, as well as their characteristic DNA methylation patterns. To do this, we identified the chromatin state of each DRDD target using a published dataset of chromatin states that incorporates information from 16 characteristics and histone modifications (Sequeira-Mendes et al., 2014). The overall set of DRDD targets was dispersed over a distribution of chromatin states highly similar to the entire Arabidopsis genome (Figure 2, F). DRDD target loci could be classified in either of two groups: “CG-only” loci, which lack non-CG methylation in both WT and drdd, and exhibit increased CG methylation in drdd, and “multi-context” loci, which exhibit hypermethylation in CG and either CHG and/or CHH contexts in drdd (Figure 2, E). While equally common (Supplemental Dataset S2), these two types of target loci occupied distinct chromatin states. CG-only DMRs were more commonly associated with gene bodies and the immediately adjacent 5′- and 3′-regions, whereas multi-context loci were more common in distal 5′-regions and other intergenic DNA associated with the repressive histone mark H3K27me3 (Figure 2, F).
Intergenic DNA enriched for H3K27me3 is typically adjacent to heterochromatin and colocalizes with many RdDM targets in euchromatin as well as the borders of heavily methylated TEs (Sequeira-Mendes et al., 2014). These differences in chromatin state distribution likely reflect the well-characterized differences between the distributions of gene body methylation (gbM), which is typically only on CG-dinucleotides, and the activities of the RdDM pathway, which is active at repetitive DNA and intergenic sequences within euchromatin and the borders of TEs (Stroud et al., 2014; Bewick and Schmitz, 2017). Gene bodies have not been previously reported as targets for active DNA demethylation. We found that two-thirds of CG-only DMRs hypermethylated in drdd were also more methylated in rdd than WT, but failed to meet cutoffs for differential methylation (Supplemental Figure S3). We note that these CG-only DMRs are unlikely to represent the trivial differences in CG methylation that can spontaneously arise between generations, termed spontaneous epimutations (Johannes and Schmitz, 2019); our approach for identifying DMRs was designed to avoid identifying large numbers of epimutations, as evidenced by the small number of significant CG methylation differences identified between WT and dme (<100 CG DMRs) and between WT replicates (33 CG DMRs), which shared the same grand-parental heritage as WT and drdd.
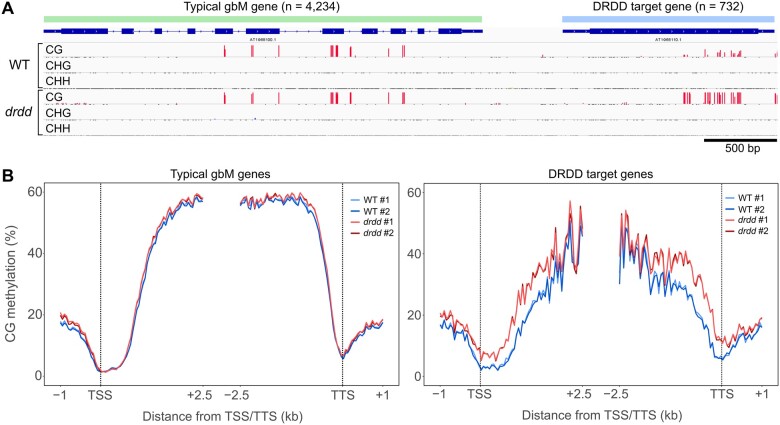
To further understand the role of active demethylation in targeting methylation within gene bodies, we sought to examine drdd methylation patterns within gene body methylated (gbM) genes (Zilberman et al., 2007; Bewick et al., 2016; Picard and Gehring, 2017). Across 4,234 Arabidopsis gbM genes identified by Bewick et al. (2016), we observed no differences in methylation between WT and drdd (Figure 3). We therefore sought to identify the genes directly overlapping CG-only hyperMRs, which we termed DRDD target genes. We identified 732 DRDD target genes, which minimally overlapped previously described gbM genes (175 out of 4,234). DRDD target genes display distinct characteristics in WT. Most notably, CGs were intermediately methylated, suggesting they are methylated in a subset of cells, and consequently display a lower average methylation level than typical gbM genes (Figure 3, B). Active demethylation by DRDD at these target genes appears to be biased toward the 5′- or 3′-ends of the coding sequence (Figure 3, B). We therefore propose that the gene bodies targeted by DRDD represent a new class of gbM genes that appear to be specifically targeted in a subset of cell and tissue types, raising the possibility that gbM may play a functional role in some contexts.
Figure 3.
Active demethylation of a subset of gene bodies. A, Genome browser snapshot showing typical gene body methylated (gbM) gene as identified by Bewick et al. (2016), as well as a DRDD target gene which exhibits reduced gbM in WT. B, Ends analysis summarizing the average CG methylation level across typical gbM genes and DRDD target genes in WT and drdd mutants. #1 and #2 refer to independent biological replicates.
Demethylation by DRDD establishes tissue-specific methylation states
DRDD target loci exhibit low but often non-zero levels of CG methylation in wild-type leaves and are by definition hypermethylated in drdd. This suggests that both methylation and demethylation processes occur at these sites in wild-type rosette leaves (Figures 2, C, 3, A). The low level of CG methylation at DRDD target loci in WT leaves is in contrast to most CG sites in the genome, which are either not methylated or fully methylated (Figure 3, A). We hypothesized that the balance between methylation and demethylation occurring at DRDD target loci could be altered in different tissues or over developmental time. To test this, we generated whole-genome bisulfite sequencing data for four tissues from individual wild-type Col-0 plants: rosette leaves (of the identical developmental stage as our prior leaf samples), cauline leaves, closed flower buds, and mature green embryos. Cauline leaves and flower buds were collected simultaneously. Two replicates were sequenced for each tissue type, with each set of tissues collected from one individual plant.
We identified hundreds of regions differentially methylated at CG dinucleotides among tissues and thousands of regions differentially methylated in CHG or CHH contexts (Supplemental Figure S4). Embryos exhibited 2,612 regions with increased CHH methylation when compared with rosette leaves and 2,189 regions when compared with cauline leaves, suggesting increased activity of the RdDM or CMT2 pathways at this developmental stage, consistent with previous reports documenting embryo hypermethylation (Bouyer et al., 2017; Kawakatsu et al., 2017; Narsai et al., 2017). In addition, we observed increased CHG methylation at 11,588 regions in embryos or flower buds compared with both rosette and cauline leaves. CHH and CHG methylation sites typically exhibit partial methylation, with only a fraction of sites methylated in whole-tissue bisulfite sequencing data, suggesting cellular heterogeneity. It is therefore possible that differences in the cell type composition of different tissues could cause large differences in the levels of non-CG methylation observed in whole tissues. Rosette and cauline leaves exhibited highly similar methylation profiles in our dataset, suggesting that cell-type composition is predictor of methylation patterns.
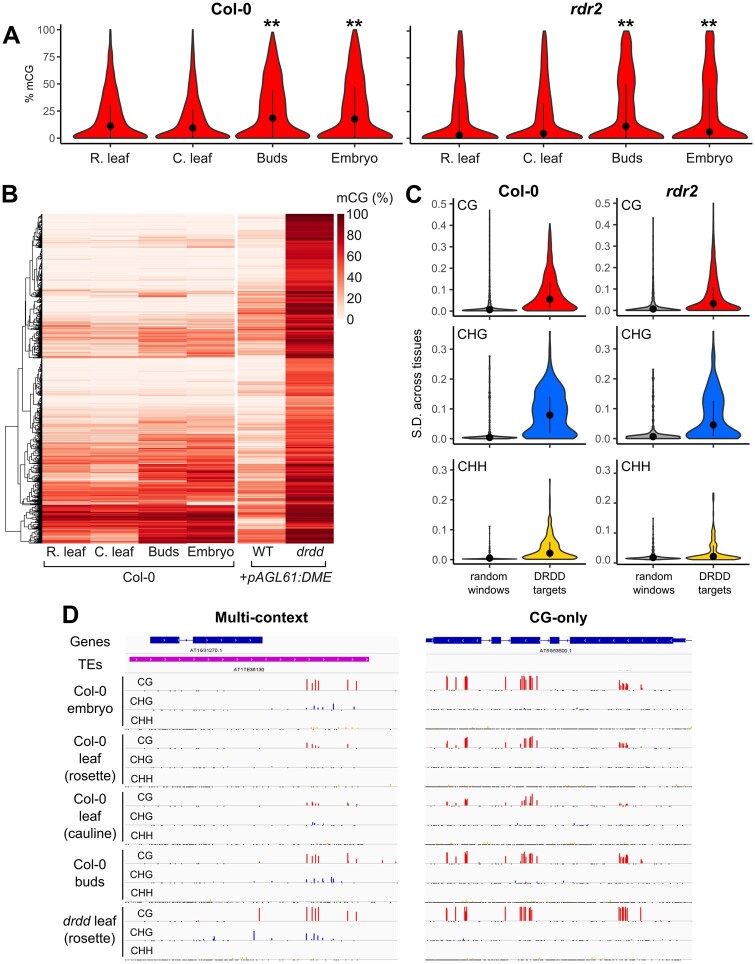
To evaluate the relationship between tissue-specific methylation differences and DRDD, we calculated the methylation levels of DRDD target loci in each tissue of our dataset, as well as a set of randomly selected 200 bp windows that had the characteristics necessary to be identified as a hypermethylated region (hyperMR) (see Methods). DRDD target loci exhibited increased methylation in flower buds and embryos compared with the leaf samples, most obviously at regions with increased CG methylation in drdd (Figure 4, A and B and Supplemental Figure S5). To control for genome-wide differences in the activity of the RdDM pathway between different tissue types (Bouyer et al., 2017), we also examined the methylation level of DRDD targets in all four tissues of an rdr2 mutant. RDR2 is involved in biogenesis of the small RNAs that direct DNA methylation. We found that many DRDD targets exhibited tissue-specific methylation differences in the absence of RdDM activity (Figure 4, A and Supplemental Figure S5). We also calculated the standard deviation in DNA methylation among tissues at DRDD targets and control regions in both WT and rdr2. The standard deviation of methylation levels between tissues was much higher at DRDD target loci than at randomly selected windows (Figure 4, C), suggesting that DRDD targets overlap with loci that exhibit developmentally dynamic epigenetic states, in many cases independent of RdDM activity. Lastly, >50% of regions identified to be CG hypomethylated in either rosette or cauline leaves compared with buds or embryos overlapped directly with DRDD target loci (Supplemental Figure S4). Thus, hundreds of loci exhibited high methylation levels in WT buds and embryos but reduced methylation in cauline and rosette leaves, as well as high methylation levels in drdd rosette leaves (Figure 4, B and D). We observed this DRDD-dependent demethylation in WT leaves at both CG-only and multi-context DRDD targets (Figure 4, D). The activity of DRDD at multi-context targets likely opposes the RdDM pathway, as these targets exhibited reduced variability between tissues in rdr2 mutants, whereas CG-only targets did not (Supplemental Figure S5).
Figure 4.
Demethylation by DRDD establishes tissue-specific methylation differences. A, CG methylation levels of four different tissues of WT and rdr2 mutants at 2,601 DRDD target loci. **P < 0.0001 (t-test, compared with rosette leaves). The black dots represent the median value and vertical lines denote the interquartile range. B, Heatmap showing methylation level of DRDD target loci across four different WT tissues. Methylation levels of rosette leaves from WT and drdd +pAGL61:DME are shown for comparison. C, Standard deviation of DNA methylation levels across tissues at DRDD target loci compared with random windows. D, Genome browser snapshots depicting typical examples of multicontext and CG-only DRDD targets in different WT tissues.
Demethylation by DRDD is associated with target gene regulation
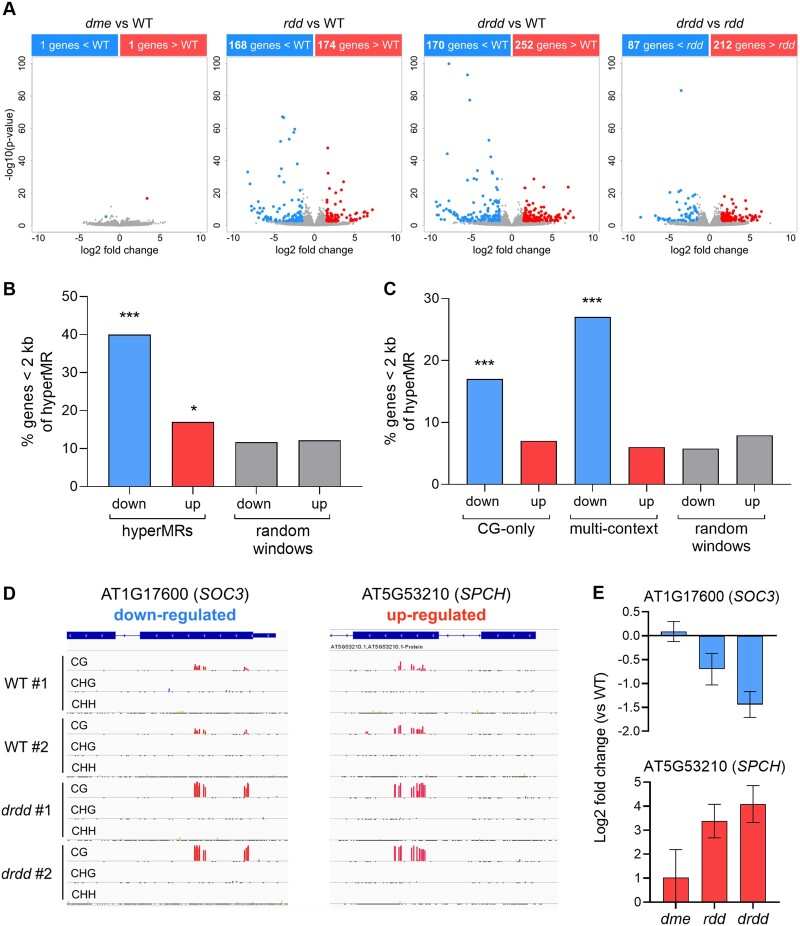
DNA methylation has variable impacts on gene expression. While many genes are not affected by changes to DNA methylation at proximal sequences, a subset of genes are strongly impacted by DNA methylation, often by transcriptional silencing and/or heterochromatin formation (Lister et al., 2008), although occasionally by promoting increased expression (Williams et al., 2015; Pignatta et al., 2018). To identify which genes might be impacted by the activity of DRDD demethylases, we performed RNA-seq on WT, dme, rdd, and drdd tissue isolated from the same leaf samples used for methylation profiling (Supplemental Dataset S1). WT and dme transcriptomes were almost identical (Figure 5, A), consistent with the minimal differences observed in methylation patterns between the two genotypes. We identified 168 downregulated and 174 upregulated genes in rdd compared with WT, and 170 downregulated and 252 upregulated genes in drdd compared with WT. Overall, the drdd transcriptome encompassed a larger dynamic range of expression changes relative to WT, suggesting that the most extreme expression changes were exacerbated in drdd compared with rdd (Figure 5, A, Supplemental Figure S6, and Supplemental Dataset S3).
Figure 5.
Demethylation by DRDD directly impacts expression of a subset of target genes. A, Volcano plots showing differentially expressed genes between dme, rdd, and drdd compared with WT, as well as drdd compared with rdd. B, Percentage of differentially expressed genes within 2 kb proximity to regions hypermethylated in drdd compared with WT. Proximity between differentially expressed genes and randomly selected genomic windows is shown as a negative control. C, Percentage of differentially expressed genes within 2 kb proximity of CG-only or multi-context DRDD target loci. D, Genome browser snapshots showing two examples of CG-only DRDD target loci that are also differentially expressed. E, Gene expression differences of example CG-only target genes in dme, rdd, and drdd compared with WT as determined by DESeq2. Error bars represent standard error between biological replicates. ***P < 0.0001; *P < 0.05 (hypergeometric test).
To examine whether these expression changes were linked to DNA methylation differences between drdd and WT, we calculated the distance between up and down-regulated genes and the closest DMR. Regions of increased DNA methylation (hyperMRs) were frequently adjacent to down-regulated genes, with 40% (45 out of 113 genes, P ≤ 0.0001 (Ws regions excluded)) of down-regulated genes residing within 2 kb of a hyperMR (Figure 5, B). This association is consistent with previous studies of ros1 and rdd mutants, which suggested that a small subset of methylation gains in these mutants caused transcriptional silencing of proximal genes (Lister et al., 2008; Yamamuro et al., 2014; Halter et al., 2021). We also observed proximity between hyperMRs and some up-regulated genes, with 17% of up-regulated genes residing <2 kb of a hyperMR (34 out of 205 genes, P = 0.03). This association is marginally enriched compared with the number of genes expected by chance, as determined by calculating the distance between hyperMRs and control groups of randomly selected 200 bp windows (Figure 5, B).
The proximity between a subset of hyperMRs and down-regulated genes was detectable for both multi-context (30 genes) and CG-only (19 genes) hyperMRs (Figure 5). An association between down-regulated genes and multi-context hyperMRs was expected, as methylation in multiple sequence contexts is a hallmark of the RdDM pathway, which typically functions in transcriptional silencing. The association of CG-only DMRs with decreased expression in drdd was more surprising, as CG gbM has mostly been associated with minimal impacts on gene regulation in plants (Bewick and Schmitz, 2017; Picard and Gehring, 2017). For example, the locus encoding the defense and cell death regulator SUPPRESSOR OF CHS1 (SOC3) is down-regulated in drdd mutants, and exhibits hypermethylation of CG dinucleotides within the first exon (Figure 5, D–E and Supplemental Figure S7). Conversely, we also observed that increased SPEECHLESS expression in drdd was associated with increased CG gbM (Figure 5, D–E).
To further understand the link between expression and gbM, we identified 13 genes that were differentially expressed between WT and drdd and directly overlap CG-only hyperMRs. We then analyzed their tissue-specific methylation and expression levels (as defined by Klepikova et al., 2016), to see if developmentally modulated methylation patterns could be associated with gene regulation. In all down-regulated genes, there was a strong inverse correlation between expression and methylation level across tissues (Supplemental Figure S8). Additionally, four out of six upregulated genes exhibited strong positive correlations between expression and methylation level, suggesting that CG gbM can be associated with up-regulation. DRDD may actively remove methylation from these coding regions to facilitate higher or lower expression, or the altered transcriptional output of these genes may induce a different epigenetic state in a DRDD-dependent manner (Figure 5, D and E).
To better understand if the gene expression differences between WT and drdd mutants are associated with any particular biological functions, we performed GO term analysis (Supplemental Figure S6). Genes with decreased expression in drdd were significantly associated with one GO term, immune response, which was also associated with genes that increased in expression (Supplemental Figure S6). Several defense-response genes down-regulated in drdd were previously identified as targets of ROS1 and are in transposon-rich rapidly evolving regions of chromosomes, which tend to be frequently targeted by the RdDM pathway (Le et al., 2014; Halter et al., 2021). We observed greater increases in the expression of epidermal development genes in drdd than in rdd (Supplemental Figure S6). GO terms for regulation of hormone levels, response to osmotic stress, and response to cold were also significantly enriched among up-regulated genes (Supplemental Figure S6).
DRDD enzymes remove methylation from the FLOWERING LOCUS C (FLC) locus and are required to delay flowering
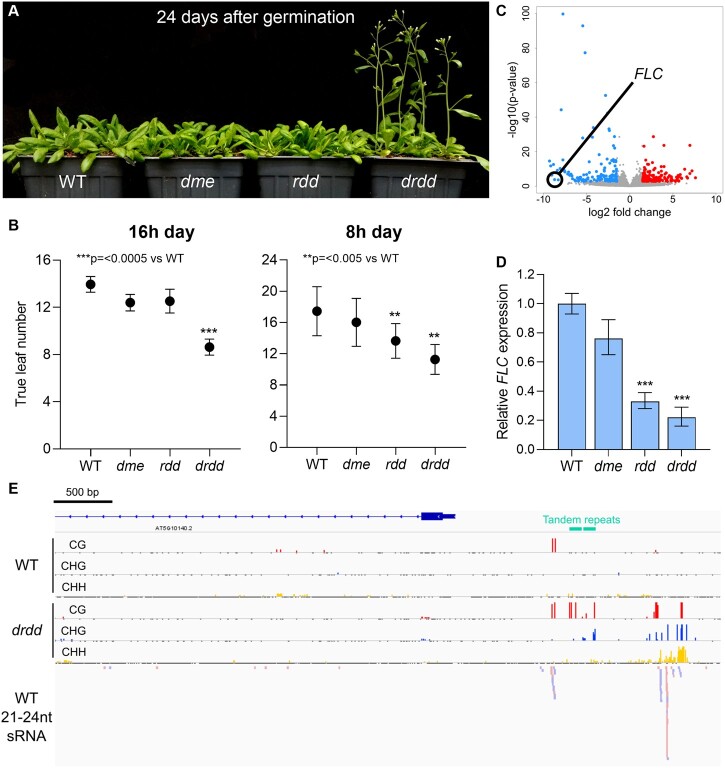
During long-day (16 h) growth conditions, we observed that drdd mutants were early flowering compared with WT, dme, and rdd plants, transitioning to flowering after establishing eight-nine true leaves (Figure 6, A, Supplemental Figure S9, and Supplemental Dataset S4). The early flowering phenotype observed in drdd is unlikely to be caused by the presence of Ws regions, as these regions are also present in rdd (Supplemental Figure S1). Under short day (8 h) growth conditions, we observed an early flowering phenotype both in drdd and rdd, which has not been noted in previous studies, although the effect was greater for drdd (Figure 6, B, Supplemental Figure S9, and Supplemental Dataset S4). We therefore hypothesized that active DNA demethylation in Arabidopsis might regulate flowering time, perhaps by altering the epigenetic state at genes important for regulating the transition to flowering. In our RNA-seq dataset, the flowering time regulator FLC had significantly reduced expression in comparisons of WT versus drdd and rdd versus drdd. In fact, FLC transcripts were not detected in drdd mutants (Figure 6, C). We then sought to verify the expression levels of FLC using reverse transcription-quantitative PCR (RT-qPCR). FLC expression was clearly reduced in both rdd and drdd mutant seedlings compared with WT, with a 3-fold and 5-fold lower relative transcript abundance, respectively (Figure 5, D). Complex epigenetic regulation of the FLC locus by histone modification pathways has been well-described (Wu et al., 2020), but whether it is connected to DNA methylation in cis remained unknown.
Figure 6.
DRDD enzymes delay flowering and remove DNA methylation proximal to FLOWERING LOCUS C. A, Flowering phenotype of long-day grown WT, dme, rdd, and drdd plants 24 days after germination. B, Flowering time of WT, dme, rdd, and drdd plants grown in long (16 h) and short (8 h) day conditions. Flowering time is measured by the number of true leaves present at the initial time of bolting. Error bars represent standard deviation between biological replicates. C, Differentially expressed genes in drdd compared with WT, with FLC highlighted among strongly down-regulated genes. D, Relative FLC expression in 7-day-old seedlings measured by quantitative real-time PCR. E, Genome browser snapshot of EM-seq methylation data and Col-0 embryo small RNA-seq data (Erdmann et al., 2017) at the intergenic region 5′ of FLC.
To assess the DNA methylation state of the FLC locus, we analyzed the DNA methylation profiles of WT, dme, rdd, and drdd plants in our methylation profiling data. We observed minimal DNA methylation in the transcribed region or 3′-region of FLC, but saw evidence of DNA methylation in all three sequence contexts in a region 800–2,139 bp 5′ of the transcriptional start site of the FLC coding region. To better define the methylation patterns of this region with greater depth, we performed enzymatic-methyl sequencing (EM-seq) on WT and drdd leaves. The EM-seq data clearly showed large increases in methylation in all sequence contexts across this region (Figure 6, E). We further verified this methylation change by performing bisulfite PCR on three separate fragments within this region (Supplemental Figures S10, S11). Across these bisulfite PCRs, we observed no methylation in WT or dme plants, but increased methylation in drdd, and to a lesser extent rdd, consistent with the observed flowering time phenotypes (Figure 5, B and E). Thus, DRDD enzymes target the region 5′ of FLC for demethylation. Hypermethylation of this region is correlated with reduced FLC expression, as well as earlier flowering time.
As this region is highly methylated in all three sequence contexts in drdd, we hypothesize that it is targeted by RdDM. Indeed, 23–24 nucleotide small RNAs homologous to this precise region, which could participate in RdDM, are present within multiple WT small RNA datasets (Lee et al., 2012; Jeong et al., 2013; Erdmann et al., 2017). Additionally, we also identified a pair of 100 bp tandem repeats with high sequence similarity within the DRDD target region. Tandem repeats have been shown to recruit the RdDM pathway upstream of other genes, including the flowering time regulator FWA (Chan et al., 2006; Henderson and Jacobsen, 2008). Thus, FLC is an exemplar of a locus that lacks DNA methylation in WT leaves due to active DNA demethylation, rather than due to lack of methylation targeting.
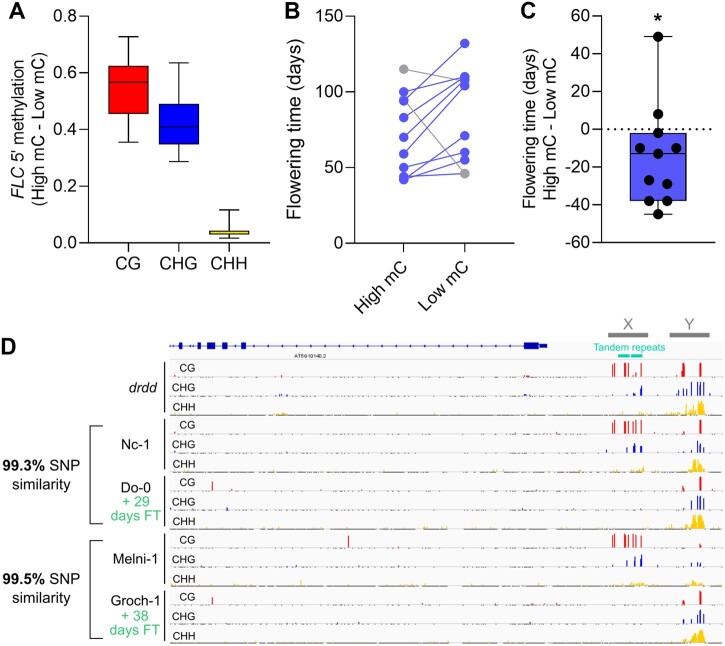
To further investigate the link between DNA methylation 5′ of FLC and flowering time, we examined methylation patterns of the FLC 5′ region across 653 ecotypes for which methylation coverage was available (Kawakatsu et al., 2016). We identified a short sequence upstream of the tandem repeats targeted by RdDM/DRDD that exhibited highly variable methylation levels across ecotypes, which we term the ecotype variable region (region Y, Figure 7, D). Conversely, we found that the tandem repeats were seldom methylated, exhibiting methylation levels comparable to drdd in only 14 ecotypes (region X, Figure 7, D). If DNA methylation of the FLC tandem repeats reduces flowering time as we hypothesize, then we would predict these 14 ecotypes to exhibit reduced flowering time compared with closely related ecotypes. For 11 out of 14 these ecotypes (for which both DNA methylation and flowering time data were available), we identified the most closely related ecotype that lacked methylation at the FLC tandem repeats, using the 1001 genomes project SNP identity tool (tools.1001genomes.org/strain_id/). In all cases, the genomic SNP identity of related ecotypes was >99%. This provided a dataset of genetically similar ecotype pairs with large differences in FLC methylation levels (Figure 7, A). While the 11 highly methylated ecotypes exhibited variable flowering times—likely reflecting a wide variation in geography, reproductive strategy, and developmental phenotypes—overall they exhibited a significantly reduced flowering time compared with closely related and lowly methylated strains (Figure 7, B and C). This data are therefore consistent with the hypothesis that high levels of methylation directed to the tandem repeats upstream of FLC is associated with a reduced flowering time, and may point to a role for DNA (de)methylation in modulating the flowering time of plants in the wild.
Figure 7.
Natural variation in FLC 5′ methylation and flowering time. A, The difference in DNA methylation levels at FLC tandem repeats between 11 pairs of closely related ecotypes exhibiting divergent DNA methylation 5′ of FLC. High mC and Low mC represent the ecotype groups with high or low FLC 5′ methylation levels, respectively. B, Plot showing flowering time of related pairs of high mC and low mC ecotypes. C, Distribution of flowering time difference across all ecotype pairs. *P < 0.05, paired t-test. D, Genome browser snapshots showing methylation patterns 5′ of FLC in drdd mutants and two example pairs of ecotypes. % SNP similarity refers to genomic SNP profiles (0 SNPs at FLC). FT, flowering time. X denotes the DRDD target region overlapping the tandem repeats, whereas Y denotes the ecotype variable region.
Discussion
In this study, we sought to elucidate the role of the DME family of 5-mC DNA glycosylases in somatic tissues of A. thaliana. To this end, we generated somatic homozygous mutants of dme as well as quadruple drdd mutants. dme mutants exhibited similar phenotypes, DNA methylation profiles, and transcriptional profiles to WT plants, suggesting that DME does not perform a distinct individual function within leaf tissue, with the caveat that our mutants are likely not a true dme null due to possible low-level expression of the pAGL61:DME transgene outside of the central cell. Nevertheless, when combined with mutations to the other three members of the DME-family to generate drdd mutants, a number of previously unknown targets and functions for active demethylation in somatic tissues were brought to light.
Active DNA demethylation targets a subset of gene bodies
Compared with WT, drdd mutants exhibited DNA methylation gains at 2,601 loci. We estimate the true number of DRDD targets to be even higher, as we employed stringent cutoffs for both read depth and the effect size of methylation changes. Approximately half of the DRDD targets gained methylation in all three sequence contexts, likely due to DNA methylation deposited by the RdDM pathway. These targets are similar in manner to previously described RDD targets—loci proximal to TEs or intergenic sequences that are targeted by RdDM, and exhibit an increase or “spreading” of DNA methylation in the absence of demethylase activity (Penterman et al., 2007; Lister et al., 2008; Tang et al., 2016). However, we also identified approximately 1,000 targets unlike those previously described in demethylation mutants, in which DNA methylation present in the CG-only context was removed. These regions are typically located within open chromatin, such as introns, exons, or untranslated regions and are referred to as gbM. However, the sites we describe here are distinct from typical gbM sites, in which CG methylation at individual sites is close to 100% (Picard and Gehring, 2017). In WT plants, the CG-only regions we identified exhibit intermediate methylation levels, in which CG dinucleotides are methylated at 5–50% (Figure 3). This low to intermediate methylation level implies that the CG sites are methylated in only a fraction of cells in the tissue. Although many of the CG-only target loci exhibit low methylation levels in WT leaves, they are more highly methylated in buds and embryos (Figure 4 and Supplemental Figure S5). Importantly, these loci are also highly methylated in drdd leaves, suggesting their methylation state is established by the active removal of methylation by DRDD enzymes in a subset of leaf cells. The function, if any, of gbM has been enigmatic. Globally, gbM is associated with moderately expressed genes (Zilberman et al., 2007), but some plant species have dispensed with this aspect of methylation patterning altogether (Bewick et al., 2016). Nevertheless, we observed 11% of down-regulated genes contained a CG-only DRDD target within their gene body, suggesting that methylation at these loci may indeed impact gene expression. These genes also exhibited a strong inverse correlation between CG methylation level and expression level across tissues (Supplemental Figure S8). Thus, we propose that active demethylation may act in a specific subset of cell types, cell cycle, or endoreduplication states in WT tissues, and that the intermediate methylation levels in WT plants are a consequence of heterogeneity in methylation state between cells. The absence of this heterogeneity within drdd mutants could lead to interesting future research on the establishment of methylation patterns within specific cell types.
The discovery of CG-only DRDD targets within gene bodies also raises questions about possible targeting mechanisms for DRDD demethylases. Previous work has proposed that active demethylation by ROS1 may be targeted by specific histone modifications associated with transcriptionally repressed intergenic sequences in Arabidopsis, such as H3K18ac and H3K27me3 (Qian et al., 2014; Tang et al., 2016). Our results are not fully consistent with this view, as we observed DRDD targets in transcriptionally active genes, and across a broad range of chromatin states (Figure 2, F) not dissimilar to the overall distribution of chromatin states in the Arabidopsis genome. It is possible that functional diversification between ROS1 and DME may contribute to such differences in their individual targeting.
Active DNA demethylation targets FLOWERING LOCUS C and is associated with reduced flowering time
FLC is a key regulator of the transition to flowering in Arabidopsis. The transcriptional silencing of FLC under prolonged exposure to cold mediates the vernalization response in some accessions via a Polycomb repressive complex switch (Costa and Dean, 2019). In addition, FLC also regulates the autonomous flowering pathway in early-flowering strains of Arabidopsis, such as Col-0 (Wu et al., 2020). FLC is expressed in the shoots of Col-0 plants in the initial weeks after germination, but expression decreases immediately prior to flowering (Pien et al., 2008; Choi et al., 2009). FLC has been established as a model locus for epigenetic and transcriptional control, and is beginning to be understood with a high degree of complexity (Wu et al., 2020). Despite extensive previous research, connections between FLC regulation and DNA methylation have been limited and indirect. While changes to FLC transcript abundance have been reported in some DNA methyltransferase and DNA methylation-binding mutants (Sheldon et al., 1999; Finnegan et al., 2005; Peng et al., 2006; Yaish et al., 2009), targeting of the FLC locus by either methylation or demethylation pathways has not previously been reported.
FLC is one of the most highly down-regulated genes identified by RNA-seq in drdd compared with WT. This expression change was consistent with the early flowering phenotype of drdd mutants in both long and short day photoperiods. We also observed a difference in methylation patterning proximal to the FLC locus that is correlated with the observed transcript abundance (Figure 6, E). It is possible that DNA methylation at this region could interfere with the binding of key regulators of FLC transcription. The FLC regulator FRIGIDA has been shown to form a super-complex with the H3K4 methyltransferase complex COMPASS-like at FLC (Li et al., 2018). Binding of this complex has been detected at a broad region encompassing 500 bp on either side of the FLC transcriptional start site. While this region does not directly overlap the sequences hypermethylated in rdd and drdd mutants, or 14 ecotypes that exhibit high methylation levels at this region (Figure 7), it is possible that demethylation of this region is required to establish the chromatin landscape necessary to facilitate binding of this super-complex.
As FLC appears to be targeted by DRDD, yet is completely unmethylated in most WT strains, an interesting question is whether this intergenic region is co-targeted by methylation and demethylation pathways in WT leaves, resulting in a constant battle between the addition and removal of DNA methylation. Active DNA demethylation at this locus may maintain epigenetic homeostasis (Williams and Gehring, 2020) by preventing the over-reach of methylation-targeting pathways, or methylation of FLC might be important for modulating expression at some point in the life cycle. Small RNAs with homology to the 5′-intergenic region of FLC (Lee et al., 2012; Jeong et al., 2013; Erdmann et al., 2017) could drive the activity of the RdDM pathway, only for methylated cytosines to be subsequently removed by DRDD. Furthermore, FLC is an imprinted gene in the endosperm of some Arabidopsis accessions (Pignatta et al., 2014), a phenomenon that has been observed at other loci with methylated 5′ regulatory intergenic regions. Understanding why dynamic methylation and demethylation pathways appear to be engaged in a tug-of-war at this locus will be an exciting avenue for future research.
In summary, we propose that active demethylation in somatic tissues by DRDD plays an important role in maintaining epigenetic states that can influence transcriptional activity. This activity appears to be important in protecting the chromatin landscape at FLC, a locus at which a number of dynamic epigenetic mechanisms converge. Similar to reproductive development, active demethylation by DRDD may also be developmentally regulated, and may act as a mechanism to establish divergent epigenetic states between cell and tissue types.
Materials and methods
Plant material
Triple homozygous mutant A. thaliana ros1-3; dml2-1; dml3-1 plants in the Col-0 background (Penterman et al., 2007) were transformed with pAGL61:DME via floral dipping (Clough and Bent, 1998). Twelve single-insertion transformants were selected and pollinated with dme-2 (Col-gl background) (Choi et al., 2002) heterozygote mutant pollen to generate F1 progeny heterozygous for all four DRDD demethylase genes. Two of these T1 lines exhibited a complete rescue of seed abortion. Quadruple heterozygous plants from one of these lines were self-fertilized, and over two subsequent generations of segregation the following genotypes were isolated, each homozygous for the pAGL61:DME transgene: dme, rdd, drdd, and DRDD WT segregants (to serve as a closely related WT control). The selfed progeny of the initial plant of each genotype was used for all subsequent experiments. To assess seed abortion, siliques were harvested after drying and seeds examined under a dissecting microscope. For tissue methylation profiling, WT plants were segregated from an rdr2-1 ros1-7 double heterozygote, as described in Williams and Gehring (2017). rdr2 plants were homozygous for the rdr2-1 allele.
pAGL61::DME transgene
The transgene to rescue DME expression in central cells was created by amplifying the promoter of AGAMOUS-LIKE 61 (AGL61), using the primers originally designed by Bemer et al. (2008), (F primer: CAACCGATTTGACAAATGCCCGAAACCGA, R primer: TTTTTGTATGGAGGGTTTTAGTTGCTTTTCT), the full genomic coding sequence (introns included) of DME (F primer: ATGAATTCGAGGGCTGATCCG, R primer: TTAGGTTTTGTTGTTCTTCAATTTGCTC), and cloning both fragments into pENTR-TOPO-D via Gibson assembly (overhang sequences are not included in the primers above). The assembled pAGL61:DME construct was then transferred to the binary vector pMDC99 (Curtis and Grossniklaus, 2003) using LR clonase.
Flowering time assay
Plants were sown such that every row of the flats contained one plant of each genotype (WT, dme, rdd, and drdd) with the order iterating by one with each successive row. Flats were grown in a Conviron CMP6050 Control System at 22°C and 50% relative humidity, with 16 h of 120 µmol light from Philips Master TL-D 58W/840 bulbs and 8 h of darkness per day. For short day experiments, the growth cycle was 8 h of light and 16 h of darkness. Starting at 2 weeks of age, all plants were visually inspected three times per week. The number of rosette leaves was recorded once a bolt was visible. Populations were compared using a one-way ANOVA with post hoc Tukey’s test (Supplemental Dataset S4). The long day flowering time assay was repeated three times independently and the short-day assay was repeated twice, using >30 plants of each genotype for each experiment.
Bisulfite sequencing
Four replicate samples of the fifth true leaf of 3-week-old WT, dme, rdd, and drdd individual plants were collected (16 plants total). Leaf samples were split along the midvein, with one half each used for DNA and RNA extractions. DNA was extracted using a CTAB protocol and bisulfite conversion was performed on 200 ng DNA using an Invitrogen MethylCode bisulfite conversion kit. Bisulfite-sequencing libraries were then generated using a QIAGEN Qiaseq Methyl Library Kit, with 11 cycles of amplification and each individual replicate separately indexed. Samples were then sequenced using an Illumina HiSeq 2500 rapid run using 100 × 100 bp paired-end protocol at the Whitehead Institute Genome Technology Core. All 16 samples were multiplexed equally in two separate lanes, to avoid batch effects in sequencing. To improve genomic coverage for downstream analyses, the reads for every two replicates were pooled, creating two high-depth biological replicates for each genotype.
To sequence different tissue types, two WT Col-0 replicates and a single rdr2 replicate were sampled throughout each individual plant’s development. The fifth true leaf was collected on day 21; the second cauline leaf and closed flower buds from the primary inflorescence were collected on day 35; and a pool of 30 mature green embryos were collected on day 50. DNA extraction, bisulfite conversion, library preparation, and sequencing were conducted as above, with the following modifications: (1) embryo samples yielded less than 200 ng of DNA, so reduced quantities were used for each replicate; (2) libraries were amplified with 6–10 cycles; and (3) samples were equally multiplexed across three lanes and sequenced using an Illumina HiSeq2500 standard run with 100 × 100 bp paired-end reads at the Whitehead Institute Genome Technology Core.
DNA methylation analysis for bisulfite-sequencing data
Prior to mapping, adapters were trimmed using Trim Galore (Babraham Bioinformatics), trimming 8 or 10 bp from the 5′-end of reads, and enforcing a 3′-end quality of >25%. Reads were mapped to the Araport11 genome using Bismarck 0.20.1 (Krueger and Andrews, 2011), allowing for 1 mismatch per read and removing PCR duplicates. Methylation values for each cytosine were calculated using the Bismark methylation extractor function. The efficiency of bisulfite conversion was verified by quantifying the percentage of methylation for reads mapped to the chloroplast. Bisulfite conversion rates were >99.7% across all samples. Before identifying DMRs, SNPs homologous to the Ws-2 genome were identified as described (Picard and Gehring, 2017), and their zygosity was plotted across each chromosome (Supplemental Figure S1). This is because the ros1 and dml2 T-DNA insertions in rdd were originally introgressed from a Ws background. In order to exclude genomic regions (and associated methylation differences) originating from the Ws ecotype, two regions were excluded from all subsequent methylation analyses: chromosome 2 (8,802,496–15,397,296 bp) and chromosome 3 (677,340–5,117,803 bp).
To identify DMRs, the genome was divided into 200 bp windows overlapping by 100 bp. Symmetrical cytosines within CG base pairs were combined to make a single averaged data point, as the two opposite-stranded cytosines within CG base pairs are not statistically independent. A “methylation score” was then calculated for each window based on the density of differentially methylated cytosines (DMCs) within each window (Williams and Gehring, 2017). Each hypermethylated cytosine/CG was assigned a score of +1 and each hypomethylated cytosine/CG was assigned a score of −1, with the added stringency that DMCs had to be present in both biological replicates for each sample. The minimum methylation difference for each cytosine context was as follows: CG—35%, CHG—20%, and CHH 15%. Cytosines with fewer than five reads coverage in each sample being compared were excluded from the analysis. For each window, the number of DMCs were then divided by the total number of cytosines with adequate coverage. A higher score is therefore assigned to windows in which hypermethylated DMCs are proximal to each other, whereas a lower score is assigned to windows in which hypomethylated DMCs are proximal to each other. Windows with a score greater than (not including) +2 were called hyperMRs, windows with a score less than (not including) −2 were called hypoMRs. DRDD targets were identified by combining all windows hypermethylated in drdd compared with WT in any sequence context and merging together using bedtools merge. DRDD targets were split into two categories based on their methylation profiles. CG-only targets were identified based on the absence of non-CG methylation (both CHG and CHH <5%) in drdd, whereas multi-context targets were defined by possessing methylation in both CG and at least one other sequence context (>5% CHG and/or CHH). The presence of DRDD target loci in each chromatin state of the genome was determined by intersecting DRDD targets with the chromatin states identified by Sequeira-Mendes et al. (2014). Chromatin states 4 and 5 were combined to represent “Intergenic polycomb” and chromatin states 8 and 9 were combined to represent “heterochromatin”.
Random control windows were selected from windows with five or more reads coverage for each cytosine and methylation levels low enough in WT to be detected as hypermethylated in drdd (e.g. <65% CG methylation, <80% CHG methylation, and <85% CHH methylation). Random control windows for comparison against multi-context DRDD targets were required to have <65% CG methylation and either <80% CHG methylation or <85% CHH methylation in WT.
Enzymatic methyl sequencing
Single biological replicates of the fifth true leaf of 3-week-old WT and drdd individuals were collected. DNA was extracted using a CTAB protocol and enzymatic methyl sequencing (EM-Seq) was carried out using 100 ng of input DNA. Input DNA was first sheared in 1× TE buffer using a Covaris S220 to an average fragment size of 200 bp (175W, 10% duty factor, 200 cycles/burst, 180 s). After shearing, a 2.5× bead cleanup was performed with a final elution in water. EM-seq was conducted using the NEBNext Enzymatic Methyl-Seq Kit according to the manufacturer’s directions; including the provided spike-in controls (methylated and unmethylated DNA). Six cycles of PCR amplification were performed. Samples were equally multiplexed across three lanes and sequenced using an Illumina HiSeq2500 standard run with 100 bp paired-end reads at the Whitehead Institute Genome Technology Core.
DNA methylation analysis for EM-Seq data
Adapter trimming and read mapping were performed as described above for the bisulfite sequencing data, except that spike-in DNA sequences were included in the reference genome. Methylation values for each cytosine were also calculated as above. The efficiency of enzymatic conversion was determined by quantifying the percent of methylation for reads mapping to the spike-in methylated and unmethylated DNA sequences.
RNA-seq
RNAs were isolated from 3-week-old leaf samples (explained above) using a QIAGEN RNeasy plant mini kit. In total, 400 ng total RNA was used to generate RNA-seq libraries using a QIAGEN QIAseq Stranded mRNA Select Kit, with 13 cycles of amplification. An additional round of purification using QIAseq beads was performed to remove adapter dimers. RNA-seq was performed on an Illumina HiSeq 2500 using a 50-bp single-end protocol at the Whitehead Institute Genome Technology Core. All samples were multiplexed equally in two separate lanes to avoid batch effects in sequencing.
Gene expression analysis
Prior to mapping, adapters were trimmed using Trim Galore (Babraham Bioinformatics), trimming 9 bp from the 5′-end of reads, and enforcing a 3′-end quality of >25%. Reads were mapped to the Araport11 genome using STAR, permitting 0.05 mismatches as a fraction of total read length and discarding reads that did not map uniquely (Dobin et al., 2013). Differentially expressed genes were identified by running htseq-count and DESeq2 (Love et al., 2014), ensuring a minimum of two-fold change in expression and a Benjamini–Hochberg corrected P-value <0.05. Proximity between differentially expressed genes and DMRs was calculated using BEDTools (closest). As a negative control, the proximity between differentially expressed genes and 10 sets of randomly selected 200 bp windows was calculated for each comparison. For example, proximity to 2,601 DRDD target DMRs was compared with proximity to 2,601 random windows for which hypermethylation could have been detected (e.g. <65% CG, <80% CHG, and <85% CHH methylation in WT). Genes mapping to chromosomal regions with homology to Ws were omitted from the differential expression and DMR proximity analyses shown in Figure 4. GO term analysis was performed using DAVID (Dennis et al., 2003).
Locus-specific bisulfite-PCR
DNA was extracted using a CTAB protocol, and bisulfite conversion was performed on 200 ng DNA using an Invitrogen MethylCode bisulfite conversion kit. Bisulfite PCR was performed using a hot-start Taq polymerase [either Ex-Taq HS (Clontech) or DreamTaq HS (Thermo Scientific)] with an annealing temperature of 50°C and an extension temperature of 68°C. PCR products were purified with a MinElute Gel Extraction Kit (QIAGEN) and cloned into the pJET1.2/blunt vector (CloneJET PCR Cloning Kit; Thermo Scientific). Individual colonies (15–25 per locus) were either PCR screened with pJET F and R primers, or plasmids were extracted with a QIAprep Spin Plasmid Miniprep Kit (QIAGEN). PCR screen amplicons or plasmids were sequenced by Sanger sequencing using the PJET1-2F universal primer. Sequences were checked manually for quality, and vector and primer sequences were removed with SnapGene (V5.2.4). Sequences were aligned with Clustal Omega (Sievers et al., 2011) and methylation state was analyzed using CYMATE (Hetzl et al., 2007). Primers for locus-specific bisulfite PCR are in Supplemental Table S1.
Reverse transcription-quantitative PCR
RNA was isolated from the 5th leaf of 21-day-old plants using TRIzol Reagent (Invitrogen) according to manufacturer’s instructions. Genomic DNA was removed by treatment with Amplification-grade DNase I (Invitrogen). cDNA was prepared from 500 to 750 ng RNA (standardized within each batch) with Superscript II Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions, with polyadenylated transcripts selected for through use of an oligo-dT primer. qPCR was performed on a StepONE Plus Real-Time PCR system with Fast SYBR-Green PCR master mix (Applied Biosystems). DME primers were F: CGAGGAAGGGCTGATTCCTTCAT R: TCCATGGCGAAAAACGTCTATCTC. FLC primer sequences were as previously described (Csorba et al., 2014): FLC_F: AGCCAAGAAGACCGAACTCA, and FLC_R: TTTGTCCAGCAGGTGACATC. Reactions were normalized to reference gene AT1G58050 (Czechowski et al., 2005). All qPCR reactions were performed with technical triplicates. Cycling conditions were as follows: 95°C for 20 s followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. Relative fold change in expression was determined using the ΔΔCt method (Livak and Schmittgen, 2001).
Accession numbers
All high-throughput sequencing data are available in NCBI GEO under accession GSE191307.
Supplemental material
Supplemental Figure S1. Zygosity of Ws SNPs in rdd and drdd mutants.
Supplemental Figure S2. Identification of hyperMRs across individual replicates.
Supplemental Figure S3. Methylation of individual CGs within hyperMRs of rdd and drdd mutants.
Supplemental Figure S4. DNA methylation differences between tissue types.
Supplemental Figure S5. Methylation levels of DRDD targets across tissues in WT and rdr2.
Supplemental Figure S6. Significantly enriched GO terms in drdd versus WT.
Supplemental Figure S7. Bisulfite PCR validation of CG-only DRDD targets.
Supplemental Figure S8. Correlation between expression and CG methylation across tissues.
Supplemental Figure S9. Additional flowering time assay replicates.
Supplemental Figure S10. Bisulfite PCR analysis of FLC in WT and demethylation mutants.
Supplemental Figure S11. Individual cytosine methylation data for FLC.
Supplemental Table S1. Primers for locus-specific bisulfite PCR.
Supplemental Dataset S1. Properties of sequencing libraries generated in this study.
Supplemental Dataset S2. List of DRDD target regions.
Supplemental Dataset S3. Differentially expressed genes identified by DESeq2.
Supplemental Dataset S4. ANOVA of flowering time data.
Supplementary Material
Acknowledgments
We are grateful to Enrico Calvanese for performing bisulfite PCR validation experiments and Maria José Aranzana for performing a flowering time assay replicate.
B.P.W. and M.G. conceived the study. B.P.W., L.L.B., and D.A.P. designed and performed experiments and data analysis. B.P.W. and M.G. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Mary Gehring (mgehring@wi.mit.edu).
Funding
Research in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award R01GM112851 to M.G.
Conflict of interest statement. None declared.
References
- Agius F, Kapoor A, Zhu J-K (2006) Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA 103: 11796–11801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Wolters-Arts M, Grossnkilaus U, Angenent GC (2008) The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell 20: 2088–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Ji L, Niederhuth CE, Willing EM, Hofmeister BT, Shi X, Wang L, Lu Z, Rohr NA, Hartwig B, . et al. (2016) On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci USA 113: 9111–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Schmitz RJ (2017) Gene body DNA methylation in plants. Curr Opin Plant Biol 36: 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Kramdi A, Kassam M, Heese M, Schnittger A, Roudier F, Colot V (2017) DNA methylation dynamics during early plant life. Genome Biol 18: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MTA, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijó JA, Becker JD. et al. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW-L, Zhang X, Bernatavichute YV, Jacobsen SE (2006) Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol 4: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Hyun Y, Kang M-J, In Yun H, Yun J-Y, Lister C, Dean C, Amasino RM, Noh B, Noh Y-S, et al. (2009) Resetting and regulation of flowering locus C expression during Arabidopsis reproductive development. Plant J Cell Mol Biol 57: 918–931 [DOI] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110: 33–42 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Costa S, Dean C (2019) Storing memories: The distinct phases of Polycomb-mediated silencing of Arabidopsis FLC. Biochem Soc Trans 47: 1187–1196 [DOI] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA 111: 16160–16165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: R60. [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann RM, Satyaki PRV, Klosinska M, Gehring M (2017) A small RNA pathway mediates allelic dosage in endosperm. Cell Rep 21: 3364–3372 [DOI] [PubMed] [Google Scholar]
- Feng S, Zhong Z, Wang M, Jacobsen SE (2020) Efficient and accurate determination of genome-wide DNA methylation patterns in Arabidopsis thaliana with enzymatic methyl sequencing. Epigenetics Chromatin 13: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA, Jaligot E, Sheldon CC, Peacock WJDennis ES (2005) The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant J 44: 420–432 [DOI] [PubMed] [Google Scholar]
- Gehring M (2019) Epigenetic dynamics during flowering plant reproduction: Evidence for reprogramming? New Phytol 224: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S (2009) Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh T-F, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL (2006) DEMETER DNA glycosylase establishes MEDEA Polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu J-K (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814 [DOI] [PubMed] [Google Scholar]
- Halter T, Wang J, Amesefe D, Lastrucci E, Charvin M, Singla Rastogi M, Navarro L (2021) The Arabidopsis active demethylase ROS1 cis-regulates defence genes by erasing DNA methylation at promoter-regulatory regions. eLife 10: e62994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE (2008) Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev 22: 1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Scheid OM (2007) CyMATE: A new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J 51: 526–536 [DOI] [PubMed] [Google Scholar]
- Hsieh T-F, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, . et al. (2012) Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D-H, Thatcher SR, Brown RSH, Zhai J, Park S, Rymarquis LA, Meyers BC, Green PJ (2013) Comprehensive investigation of microRNAs enhanced by analysis of sequence variants, expression patterns, ARGONAUTE loading, and target cleavage. Plant Physiol 162: 1225–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F, Schmitz RJ (2019) Spontaneous epimutations in plants. New Phytol 221: 1253–1259 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Huang SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, . et al. (2016) Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Nery JR, Castanon R, Ecker JR (2017) Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol 18: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Ono A, Scholten S, Kinoshita T, Zilberman D, Okamoto T, Fischer RL (2019) DNA demethylation by ROS1a in rice vegetative cells promotes methylation in sperm. Proc Natl Acad Sci USA 116: 9652–9657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Park J-S, Lee J, Lee KK, Park O-S, Choi H-S, Seo PJ, Cho H-T, Frost JM, Fischer RL, et al. (2021) The DME demethylase regulates sporophyte gene expression, cell proliferation, differentiation, and meristem resurrection. Proc Natl Acad Sci USA 118: e2026806118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88: 1058–1070 [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T-N, Schumann U, Smith NA, Tiwari S, Au PCK, Zhu Q-H, Taylor JM, Kazan K, Llewellyn DJ, Zhang R, et al. (2014) DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol 15: 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Gurazada SGR, Zhai J, Li S, Simon SA, Matzke MA, Chen X, Meyers BC (2012) RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics 7: 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jiang D, He Y (2018) FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat Plants 4: 836–846 [DOI] [PubMed] [Google Scholar]
- Lin W, Sun L, Huang R-Z, Liang W, Liu X, He H, Fukuda H, He X-Q, Qian W (2020) Active DNA demethylation regulates tracheary element differentiation in Arabidopsis. Sci Adv 6: eaaz2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ddCt method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- López Sánchez A, Stassen JHM, Furci L, Smith LM, Ton J (2016) The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J 88: 361–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Čaikovski M, Smathajitt C, Paszkowski J (2007) Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marín MI, Martínez-Macías MI, Ariza RR, Roldán-Arjona T (2006) DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA 103: 6853–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Gouil Q, Secco D, Srivastava A, Karpievitch YV, Liew LC, Lister R, Lewsey MG, Whelan J (2017) Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol 18: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth CE, Bewick AJ, Ji L, Alabady MS, Kim KD, Li Q, Rohr NA, Rambani A, Burke JM, Udall JA, . et al. (2016) Widespread natural variation of DNA methylation within angiosperms. Genome Biol 17: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Yamaguchi K, Fukada-Tanaka S, Terada R, Mitsui T, Iida S (2012) A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J 71: 564–574 [DOI] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldán-Arjona T (2008) Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol 67: 671–681 [DOI] [PubMed] [Google Scholar]
- Park J-S, Frost JM, Park K, Ohr H, Park GT, Kim S, Eom H, Lee I, Brooks JS, Fischer RL, et al. (2017) Control of DEMETER DNA demethylase gene transcription in male and female gamete companion cells in Arabidopsis thaliana. Proc Natl Acad Sci USA 114: 2078–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Kim MY, Vickers M, Park J-S, Hyun Y, Okamoto T, Zilberman D, Fischer RL, Feng X, Choi Y, et al. (2016) DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc Natl Acad Sci USA 113: 15138–15143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Zhang L, Li J, Shen C, Qiu P, Tu L, Zhang X, Wang M (2019) Tracing the origin and evolution history of methylation-related genes in plants. BMC Plant Biol 19: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Cui Y, Bi Y-M, Rothstein SJ (2006) AtMBD9: A protein with a methyl-CpG-binding domain regulates flowering time and shoot branching in Arabidopsis. Plant J 46: 282–296 [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL (2007) DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA 104: 6752–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard CL, Gehring M (2017) Proximal methylation features associated with nonrandom changes in gene body methylation. Genome Biol 18: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inzé D, Avramova Z, Dean C, Grossniklaus U (2008) ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D, Erdmann RM, Scheer E, Picard CL, Bell GW, Gehring M (2014) Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. eLife 3: e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D, Novitzky K, Satyaki PRV, Gehring M (2018) A variably imprinted epiallele impacts seed development. PLoS Genet 14: e1007469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Miki D, Lei M, Zhu X, Zhang H, Liu Y, Li Y, Lang Z, Wang J, Tang K, et al. (2014) Regulation of active DNA demethylation by an α-crystallin domain protein in Arabidopsis. Mol Cell 55: 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán-Arjona T, Ariza RR, Córdoba-Cañero D (2019) DNA base excision repair in plants: An unfolding story with familiar and novel characters. Front Plant Sci 10: 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft VK, Chumak N, Choi Y, Hannon M, Garcia-Aguilar M, Machlicova A, Slusarz L, Mosiolek M, Park J-S, Park GT, et al. (2011) Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci USA 108: 8042–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U, Lee JM, Smith NA, Zhong C, Zhu J-K, Dennis ES, Millar AA, Wang M-B (2019) DEMETER plays a role in DNA demethylation and disease response in somatic tissues of Arabidopsis. Epigenetics 14: 1074–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Aragüez I, Peiró R, Mendez-Giraldez R, Zhang X, Jacobsen SE, Bastolla U, Gutierrez C (2014) The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen JG, Kang I-H, Portereiko MF, Lloyd A, Drews GN (2008) AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol 148: 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Lang Z, Zhang H, Zhu J-K (2016) The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat Plants 2: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Gehring M (2020) Principles of epigenetic homeostasis shared between flowering plants and mammals. Trends Genet 36: 751–763 [DOI] [PubMed] [Google Scholar]
- Williams BP, Gehring M (2017) Stable transgenerational epigenetic inheritance requires a DNA methylation-sensing circuit. Nat Commun 8: 2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Pignatta D, Henikoff S, Gehring M (2015) Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet 11: e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Fang X, Zhu D, Dean C (2020) Autonomous pathway: FLOWERING LOCUS C repression through an antisense-mediated chromatin-silencing mechanism. Plant Physiol 182: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaish MWF, Peng M, Rothstein SJ (2009) AtMBD9 modulates Arabidopsis development through the dual epigenetic pathways of DNA methylation and histone acetylation. Plant J 59: 123–135 [DOI] [PubMed] [Google Scholar]
- Yamamuro C, Miki D, Zheng Z, Ma J, Wang J, Yang Z, Dong J, Zhu J-K (2014) Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat Commun 5: 4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Lepère G, Jay F, Wang J, Bapaume L, Wang Y, Abraham A-L, Penterman J, Fischer RL, Voinnet O, et al. (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA 110: 2389–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.