Abstract
The spatiotemporal development of somatic tissues of the anther lobe is necessary for successful fertile pollen production. This process is mediated by many transcription factors acting through complex, multi-layered networks. Here, our analysis of functional knockout mutants of interacting basic helix–loop–helix genes Ms23, Ms32, basic helix–loop–helix 122 (bHLH122), and bHLH51 in maize (Zea mays) established that male fertility requires all four genes, expressed sequentially in the tapetum (TP). Not only do they regulate each other, but also they encode proteins that form heterodimers that act collaboratively to guide many cellular processes at specific developmental stages. MS23 is confirmed to be the master factor, as the ms23 mutant showed the earliest developmental defect, cytologically visible in the TP, with the most drastic alterations in premeiotic gene expression observed in ms23 anthers. Notably, the male-sterile ms23, ms32, and bhlh122-1 mutants lack 24-nt phased secondary small interfering RNAs (phasiRNAs) and the precursor transcripts from the corresponding 24-PHAS loci, while the bhlh51-1 mutant has wild-type levels of both precursors and small RNA products. Multiple lines of evidence suggest that 24-nt phasiRNA biogenesis primarily occurs downstream of MS23 and MS32, both of which directly activate Dcl5 and are required for most 24-PHAS transcription, with bHLH122 playing a distinct role in 24-PHAS transcription.
Sequential tapetal bHLHs regulate protein-coding genes, including hundreds of transcription factor and known male sterility genes, and the 24-nt phasiRNA biogenesis pathway in maize anthers.
IN A NUTSHELL.
Background: Correct spatiotemporal development of somatic tissues in the anther is necessary for successful fertile pollen production. This development is mediated by many transcription factors acting through complex, multi-layered networks. MS23, MS32, bHLH122, and bHLH51 are sequentially involved in somatic development of the anther lobe based on studies in Arabidopsis, rice, and maize. Both the temporal order in which these four bHLH proteins function and their roles in anther development and 24-nt phased secondary small interfering RNA (phasiRNA) biogenesis are largely unclear. 24-nt phasiRNAs are highly abundant in maize anthers during early meiosis.
Question: Our goal is to clarify the hierarchy and roles of four maize bHLHs—MS23, MS32, bHLH122, and bHLH51—in tapetal cell differentiation and 24-nt phasiRNA biogenesis.
Findings: Functional analysis of knockout mutants in interacting basic-helix-loop-helix genes Ms23, Ms32, bHLH122, and bHLH51 established that male fertility requires all four genes, expressed sequentially in the tapetum. Not only do they regulate each other, but their encoded proteins also form heterodimers that act collaboratively to guide many cellular processes at specific developmental stages. MS23 is confirmed to be the master factor, whose mutation leads to the earliest developmental defect, which is cytologically visible in the tapetum; ms23 mutants exhibit the most drastic alterations in pre-meiotic anther gene expression. Mutations in the four bHLH genes result in misregulation of more than 800 downstream transcription factors. The male sterile ms23, ms32, and bhlh122-1 mutants lack 24-nt phasiRNAs and the precursor transcripts from the corresponding 24-PHAS loci. The bhlh51-1 mutant has wild-type levels of both precursors and small RNA products. Both MS23 and MS32 activate Dcl5, which encodes an endoribonuclease that cleaves 24-PHAS precursors, producing 24-nt phasiRNAs. MS23 and MS32 are also required for the transcription of most 24-PHAS loci, with additional and distinctive action by bHLH122 in regulating these loci.
Next steps: We plan to identify the direct targets of each bHLH in order to organize a more detailed transcription factor hierarchy in the context of anther development and 24-nt phasiRNA biogenesis. We would also like to apply this knowledge to improve crop yield.
Introduction
As maize (Zea mays) is one of the most important crops in the USA, much attention has focused on male-sterile mutants in this crop, motivated by their potential use in hybrid seed production (Wan et al., 2019). Within maize anthers, germinal cells are fated to undergo meiosis early in lobe ontogeny, yielding the haploid gametophytes that become pollen. Somatic anther lobe development involves a series of tightly orchestrated events to build the four cell types required for viable pollen biogenesis. This complex process involves the sequential deployment of diverse transcription factors (TFs) with key roles whose timing of action is defined by defects in loss-of-function mutants (Higginson et al., 2003; Yang et al., 2003; Schreiber et al., 2004; Millar and Gubler, 2005; Ito et al., 2007; Yang et al., 2007; Vernoud et al., 2009; Liu et al., 2010; Murmu et al., 2010; Li et al., 2011; Phan et al., 2012; Fernández Gómez and Wilson, 2014; Cai et al., 2015; Lou et al., 2018; Zhang et al., 2018; Su et al., 2019; Xiao et al., 2019). We previously reported that four basic helix–loop–helix (bHLH) TFs—MS23, MS32, bHLH122, and bHLH51—are sequentially involved in somatic development in anther lobes, particularly in supporting correct patterns of tapetal cell proliferation (Nan et al., 2017). Analysis of the classic male-sterile mutants ms23 and ms32 showed that MS23 and MS32 are required for anther fertility (Chaubal et al., 2000; Moon et al., 2013; Nan et al., 2017). Recent studies demonstrated that mutants with little or no function of the two other bHLHs show male sterility: a recently discovered ethyl methanesulfonate-derived mutant of bHLH51 (Liu et al., 2021) and CRISPR-Cas9 (the RNA guided Cas9 nuclease from the microbial clustered regularly interspaced short palindromic repeats)-mediated knockout mutants of both bHLH122 and bHLH51 all show male sterility (Jiang et al., 2021).
Orthologs of these four bHLH genes are required for normal anther development and fertility in rice (Oryza sativa) and Arabidopsis thaliana, demonstrating conserved aspects of anther development (Sorensen et al., 2003; Jung et al., 2005; Li et al., 2006; Zhang et al., 2006, 2008; Xu et al., 2010, 2014; Feng et al., 2012; Ji et al., 2013; Niu et al., 2013; Fu et al., 2014, 2020; Ko et al., 2014; Zhu et al., 2015; Cui et al., 2016; Liu et al., 2018; Ono et al., 2018; Zheng et al., 2020). As summarized in Table 1, Ms23 and bHLH122 are closely related maize paralogs, each with a corresponding ortholog in rice, that is, TIP2 and EAT1/DTD, respectively. Both TIP2 and EAT1/DTD are indispensable for fertile pollen production. In Arabidopsis, there are three homologous genes—bHLH010, bHLH089, and bHLH091—which are functionally redundant, because single mutants are fertile and only the triple mutant is completely sterile (Zhu et al., 2015). The ortholog of Ms32 is UDT1 in rice and DYT1 in Arabidopsis; bHLH51 is TDR in rice and AMS in Arabidopsis.
Table 1.
Orthologs of Ms23, Ms32, bHLH122, and bHLH51 in maize, rice, and Arabidopsis
| Gene Name | Ms23 | bHLH122* | Ms32 | bHLH51 |
|---|---|---|---|---|
| Gene ID | Zm00001d008174 | Zm00001d017724 | Zm00001d006564 | Zm00001d053895 |
| Chromosome | 8 | 2 | 5 | 4 |
| Rice ortholog (gene name) | Os01g293100 (TIP2) | Os04g0599300 (EAT1/DTD) | Os07g0549600 (UDT1) | Os02g0120500 (TDR) |
| Arabidopsis ortholog (gene name) |
|
At4g21330 (DYT1) | At2g16910 (AMS) | |
Numerous loci that generate phased secondary small-interfering RNAs (phasiRNAs) have been identified by extensive small RNA (sRNA) sequencing in plants. The reproductive phasiRNAs are enriched in anthers (Johnson et al., 2009; Song et al., 2012; Zhai et al., 2015; Xia et al., 2019). Canonical reproductive phasiRNA biogenesis is initiated by 22-nt miRNA-mediated cleavage of RNA polymerase II transcripts of two classes of PHAS loci, namely miR2118 for the 21-nt and miR2275 for the 24-nt classes. Next, the 3′-portions of cleaved transcripts are converted into double-stranded RNAs and precisely chopped by DICER-LIKE4 (DCL4) into 21-nt products, or by DCL5 into 24-nt phasiRNAs (Song et al., 2012; Zhai et al., 2015; Teng et al., 2020). The 21-nt phasiRNAs are highly abundant during initial cell fate acquisition in maize anthers (Zhai et al., 2015) and are important for male fertility in both maize (Zhai et al., 2015) and rice (Nonomura et al., 2007; Araki et al., 2020). Maize 24-nt phasiRNAs accumulate in conjunction with meiosis initiation, peak during meiosis, and persist afterward at lower levels (Zhai et al., 2015; Teng et al., 2020). Multiple lines of evidence indicate that a tapetal layer is required for 24-nt phasiRNA biogenesis, while defects in other lobe cell types are unimportant (Zhai et al., 2015; Nan et al., 2017; Ono et al., 2018; Teng et al., 2020).
Maize Ms23 and its rice ortholog were shown to be important for 24-nt phasiRNA biogenesis (Nan et al., 2017, Ono et al., 2018). Anthers of ms23 mutants have five instead of the normal four wall layers, without a fully differentiated TP or normal meiosis progression (Chaubal et al., 2000). The fourth and fifth anther wall layers, previously termed t1 and t2, never reach the binucleate state as a normal TP cell would (Chaubal et al., 2000; Nan et al., 2017). Dcl5 mRNA and 24-nt phasiRNAs are eliminated in ms23 (Zhai et al., 2015; Nan et al., 2017). Also, the abundances of miR2275 isoforms (the 22-nt microRNA trigger of 24-nt phasiRNA production) are altered but the microRNA persists in ms23, suggesting that MS23 is not a direct regulator of miR2275 transcription (Zhai et al., 2015). The rice EAT1 ortholog of maize bHLH122 was suggested to be a key regulator triggering 24-nt phasiRNA biogenesis in the anther TP (Ono et al., 2018). EAT1 protein accumulates specifically in the TP, when meiocytes are progressing from leptotene to diplotene, and promotes transcription of 101 24-PHAS loci and Dcl5 in rice (Ono et al., 2018).
In this study, to further clarify the roles of the bHLH122 and bHLH51 genes, we generated knockout mutants to observe cellular phenotypes and to perform RNA-seq and sRNA-seq analyses, together with analysis of the historic maize male-sterile mutants ms23 and ms32. Key questions addressed include interactions among the TFs and their co-regulated gene sets, and the requirement for these TFs to produce 24-nt phasiRNAs. A major goal was to identify anther processes affected sequentially by the absence of each bHLH and to better understand the complex development of tapetal cells. These cells are initially proliferative. They then cease mitosis and undergo sequential redifferentiation to perform distinctive roles that support premeiotic cells, including digesting excess callose (Wang et al., 2010); providing structural and nutrient support during the 6 days of meiosis (Nan et al., 2011); synthesizing and secreting pollen wall components deposited on haploid microspores as they develop into pollen grains (Wan et al., 2020); generating a sticky “landing pad” to attach haploid microspores to the locule wall (Tsou et al., 2015); and finally undergoing programmed cell death (PCD) to contribute final exine components and structural support to the locular wall (Varnier et al., 2009). Here, we sought to organize the bHLH cascade to define steps in TP differentiation and to understand their roles in 24-nt meiotic phasiRNA biogenesis.
Results
Mutant analysis demonstrates that bHLH51 and bHLH122 are required for male fertility
The genes responsible for the historic male-sterile mutants ms23 and ms32 have been cloned and described (Moon et al., 2013; Nan et al., 2017). We previously reported that bHLH122 and bHLH51 are transcribed later in anther development and are regulated by MS23 and MS32; hetero-dimers among these four bHLHs have also been documented, as well as bHLH51 homodimers (Nan et al., 2017). To further understand the functions of bHLH122 and bHLH51, we generated transgenic mutant lines using CRISPR–Cas9 technology (Char et al., 2017). As illustrated in Figure 1A, insertions or deletions were introduced near the guide RNA target sites (dotted lines) in the coding regions upstream of their bHLH domains. Lines with frameshift mutations resulting in early translation termination were selected for further analysis. All homozygous mutant lines examined were male sterile with no anther exertion and no viable pollen detected, indicating that both bHLH122 and bHLH51 are required for anther fertility (Figure 1, B and C), thus corroborating two recent reports (Jiang et al., 2021; Liu et al., 2021).
Figure 1.

DNA sequences and tassel fertility status of bHLH51 and bHLH122 mutants. A, Mutants were generated with the CRISPR–Cas9 system and sequenced to pinpoint alterations. Based on the annotated bHLH51 and bHLH122 genes from the published version 4 of the B73 maize genome, bHLH51 (purple) is ∼3-kb long with three major transcripts, while bHLH122 (green) is ∼3.5-kb long with one major transcript. Introns are shown as line segments, and exons are shown as colored boxes, with coding sequences with predicted bHLH domains shown in gray and untranslated regions shown in white. Triangles indicate the direction of transcription. Arrows point to mutated sites, as indicated by dotted lines. Sequences of two mutant lines per gene alongside the wild-type alleles are shown with the mutations highlighted in red (+1A in bhlh51-1, −CC deletion in bhlh51-2, +1T in bhlh122-1, and −1C in bhlh122-2). B, Mature tassel on a bhlh122-1 mutant plant without anther exertion at maturity compared to a bhlh122-1//bHLH122 heterozygous sibling plant. C, Mature tassel on a bhlh51-1 mutant plant lacks exerted anthers compared to a bhlh51-1//bHLH51 heterozygous sibling plant shedding pollen. Insets in (B) and (C) show pollen viability tests via I2-PI staining.
Using confocal microscopy, we evaluated propidium iodide-stained anthers representing 10 days of development and scored the timing and type of any somatic cell abnormalities. The progression of normal development is presented in Figure 2. By the 0.8 mm stage, a fertile maize anther is composed of four wall layers: external epidermis, endothecium, middle layer (ML), and TP, surrounding the central premeiotic cells. All four mutants appeared to reach the four-wall layer stage normally. In ms23 mutant anthers, extra periclinal divisions in the presumptive TP were noted one day later, at the ∼1-mm anther stage (Figure 2). At the 1.5-mm stage, tapetal cells in fertile anthers increase in thickness and contain dense cytoplasm, but these features were missing in ms23 anthers. Aberrant cells (highlighted in blue in Figure 2) appeared in both ms32 and bhlh122-1 anther walls by 1.5 mm. Anther wall organization was still normal in the bhlh51-1 mutant at this stage; however, the cytoplasm in bhlh51-1 TP looked less dense, and the TP did not expand in depth compared to fertile siblings. At the 2.0 mm stage, the ML in fertile anthers starts to compress, reducing in thickness, a prelude to degeneration, while the ML of all four mutant anthers retained their thickness. Aberrant cells also started to appear in bhlh51-1 anthers at 2.0 mm, and more aberrant cells were found in ms23, ms32, and bhlh122-1 anthers; the defining feature of ms23 anthers was five well-formed wall layers generated by an ectopic periclinal division of each pretapetal cell (Supplemental Figure S1). Both t1 and t2 layers shared similar features with a fertile TP when viewed in the XY plane (Supplemental Figure S1; Nan et al., 2017) but were unable to reach the binucleate state (Chaubal et al., 2000). Hence, we suspect that MS23 is actively involved in blocking cell plate formation after the nuclear divisions in these pretapetal cells. By 3.0 mm, the ML in fertile anthers is nearly absent, and the haploid microspores have attached to tapetal cells. None of the bHLH mutants in this study reached this developmental status. The presumptive tapetal cells in ms32 remain mononucleate (Chaubal et al., 2000).
Figure 2.

Progression of normal anther lobe development and defects in bHLH mutant anthers. Longitudinal confocal images of anther wall layers of ms23, ms32, bhlh122-1, and bhlh51-1 mutants compared to those of heterozygous fertile sibling plants from 1:1 segregating populations. Confocal images of anthers were collected from anthers of different lengths from 1.0 to 3.0 mm. Aberrant cells arising from ectopic periclinal divisions are colored in blue. Scale bar = 20 µm.
We conclude that the primary defects in all four mutants are in tapetal differentiation; transcripts of all four genes were predominantly detected in the tapetal layer of fertile anthers by RNA in situ hybridization (Supplemental Figure S2; Moon et al., 2013), supporting this conclusion. One key finding of our previous study is that bHLH122 transcript levels dropped drastically at the 2.0-mm anther stage, and no protein was detected in total soluble protein extracted from 1.0 or 2.0 mm anthers (Nan et al., 2017).
In summary, all four bHLH mutants have an aberrant TP, and we confirmed the temporal order of defects: the earliest defects are in m23, followed by ms32, bhlh122-1, and finally in bhlh51-1. We also generated double mutant lines, as shown in Supplemental Figure S3; both Ms23 and Ms32 were epistatic to both bHLH122 and bHLH51 while showing an additive phenotype in the double mutant of ms23 + ms32, as previously described (Nan et al., 2017). One unexpected observation was the sporadic presence of extra cell(s) in ms32 + bhlh122-1 and bhlh122-1 + bhlh51-1 as early as 1.0 mm. These ectopic cells were produced after the anther had formed four wall layers (0.8 mm) but before the TP initiated exine secretion at the end of meiosis (2.5-mm stage).
A global Y2H analysis identifies MS23 binding partners
In an attempt to assemble a comprehensive list of MS23 binding partners, we conducted a global yeast two-hybrid (Y2H) screening by next-generation sequencing (NGS) using a male inflorescence cDNA prey library screened against the full-length MS23 protein as the bait. A total of 19 genes that were enriched compared to both the cDNA library and empty vector controls were identified using two-fold change and false discovery rate (FDR) ≤ 0.05 as the cutoff (Supplemental Figure S4A; Supplemental Data Set S1). Among these, two bHLH proteins, that is, MS32 and bHLH51, were identified (Supplemental Data Set S1). We discovered the MS23 and MS32 interaction here using full-length Ms23 coding sequence as bait, whereas no strong interaction was detected previously using only the bHLH domains of MS23 and MS32 (Nan et al., 2017).
Altogether, as shown in Supplemental Figure S4B, MS23 interacted with MS32 and bHLH51, while bHLH122, the paralog of MS23, also interacted with both MS32 and bHLH51. Of these two bHLH proteins, only bHLH51 formed homodimers in the previous Y2H assay (Nan et al., 2017).
Dynamic gene expression patterns during anther development and dependence on bHLH TFs
To investigate the gene networks closely associated with each of the four bHLH TFs, we analyzed RNA-seq data from the anthers of each mutant and heterozygous fertile sibling controls in segregating families (Supplemental Data Set S2). Based on the timing of the cellular defects and the expression time course established previously (Nan et al., 2017), anthers of 1.0 (premeiosis), 1.5 (leptotene–zygotene), and 2.0 mm (pachytene) in length were selected for the study of ms23 and ms32; an extra stage at 3.0 mm (postmeiosis) was added for bhlh122 and bhlh51 (Nan et al., 2011; Nelms and Walbot, 2019). Correlation heatmaps of RNA-seq data between samples for the 5,000 genes with the highest variance in abundance between mutant and fertile samples (Figure 3) clearly demonstrate the reproducibility of the replicates. ms23 and ms32 were distinctive at all three stages (Figure 3A). In contrast, bhlh122-1 and bhlh51-1 were similar to each other and to their fertile controls at 1.0 mm, before there were cytological defects. However, divergence became evident at the intermediate stages, and the mutants were highly distinctive at 3.0 mm (Figure 3B).
Figure 3.

Correlation heatmaps of maize anther RNA-seq data. A, Correlation heatmap between ms23 (red) and ms32 (blue) replicates at the 1.0, 1.5, and 2.0 mm stages. Two or three biological replicates of RNA-seq libraries sequenced from staged, pooled anthers of each genotype were analyzed. The similarity analysis of the biological replicates (listed as R1, R2, and R3) was generated with the ComplexHeatmap R package based on the 5,000 most varying genes across all samples. The replicates and stages are listed across the top of the heatmaps, and the samples in the same order are present on the y-axis. B, Correlation heatmap between bhlh51-1 (purple) and bhlh122-1 (green) replicates at 1.0, 1.5, 2.0, and 3.0 mm.
A large deletion removed the entire Ms23 coding sequence in the ms23 allele (Nan et al., 2017); therefore, no Ms23 transcript was detectable in ms23 anthers (Figure 4A). Similarly, Ms32 transcript levels of only 13.9% (relative to wild-type) could be detected in sterile ms32 anthers at 1.5 mm (Figure 4A). bHLH122 and bHLH51 transcript levels peaked in 1.5 and 2.0 mm fertile anthers, respectively, while in ms23 and ms32 anthers at 1.0–2.0 mm, the transcript levels were drastically reduced (Figure 4A). These results suggest that the expression of bHLH122 and bHLH51 depends on Ms23 and Ms32. Surprisingly, Ms32 mRNA was 13.7 times more abundant in ms23 anthers at 2.0 mm compared to its fertile siblings (Figure 4A).
Figure 4.
Transcript abundances of bHLH genes in bHLH mutants. A, RNA-seq quantification of Ms23, Ms32, bHLH51, and bHLH122 transcripts in fertile ms32//Ms32 (striped circles), ms23 (solid circles), and ms32 (open circles) sterile anthers at the 1.0, 1.5, and 2.0 mm stages. ms32//Ms32 and ms32 were siblings from the same segregating family, and ms23 had a comparable genetic background. B, RNA-seq quantification of Ms23, Ms32, bHLH51, and bHLH122 transcripts millimeter in fertile (striped circles), bhlh122-1 (solid circles), and bhlh51-1 (open circles) sterile anthers at the 1.0, 1.5, 2.0, and 3.0 stages. Fertile, bhlh122-1, and bhlh51-1 were siblings from the same segregating family. Dot sizes represent total read numbers (CP20M) mapped to the genes of interest and were normalized together in (A) or (B) across the genotypes and time points. At least two replicates were used for each sample.
The transcript abundances of bHLH122 and bHLH51 CRISPR–Cas9 mutants were only moderately reduced, with 60% lower levels in bhlh122-1 anthers at 1.5 mm and 32% lower levels in bhlh51-1 anthers at 2.0 mm (Figure 4B). The predicted peptides contain frameshift errors and early termination; if translated, the proteins would lack the bHLH domains. These alleles are unable to sustain pollen fertility despite the presence of substantial transcript. Both the bHLH51 abundance in bhlh122-1 anthers and the bHLH122 abundance in bhlh51-1 anthers were unaltered (Figure 4B), suggesting that these genes are independent of each other. In bhlh51-1 anthers, the peak expression of Ms23 at 2.0 mm was no longer observed, while Ms32 transcript abundance was significantly higher at 2.0 mm (2.9 times) and 3.0 mm (2.2 times) compared to fertile anthers. The bimodal expression pattern of bHLH122 is consistent with the expression pattern of its rice ortholog EAT1 (Ono et al., 2018). The dual expression peaks of bHLH122 commonly observed in fertile anthers at 1.5 and 3.0 mm had disappeared in the bhlh51-1 background, with only a minor peak at 1.5 mm remaining (Figure 4B). A subset of these expression patterns were validated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR; Supplemental Figure S5; Supplemental Data Set S3; and Supplemental File S1). In summary, bHLH51 and bHLH122 appear to function downstream of MS23 and MS32, while bHLH51 and bHLH122 exhibit feedback regulation or have an indirect impact on Ms23 and Ms32 expression.
To test the proposed bHLH hierarchy, we transiently expressed bHLH-expressing constructs in maize leaf protoplast cells, followed by qRT-PCR assays (see “Materials and methods” for details on the experimental design). Ms23 mRNA expression is anther-specific (Nan et al., 2017). Ms23 transcripts were barely detectable in protoplasts transfected with 35S:GFP, with a significant increase in the protoplasts after transfection with 35S:Ms23 (Supplemental Figure S6A; Supplemental Data Set S4); this was also true for Ms32 (Supplemental Figure S6B). Expression of these individual constructs had no impact on the other bHLHs. As expected, bHLH122 mRNA abundance was elevated in protoplasts transfected with 35S:bHLH122, but not with Ms23+Ms32 (Supplemental Figure S6C), suggesting that an unknown additional factor is required if bHLH122 is directly regulated by MS23 + MS32. Surprisingly, bHLH51 mRNA abundance increased in protoplasts transfected not only with 35S:bHLH51 (as expected), but also with Ms23+Ms32 (3.9- to 13.6-fold increase), Ms32 + bHLH122 (44.0- to 342.5-fold), or Ms23 + Ms32+bHLH122 (24.8- to 183.3-fold), but not with Ms23 + bHLH122 (Supplemental Figure S6D; Supplemental Data Set S4; Supplemental File S1). These results confirm the notion that bHLH51 functions downstream of the other three bHLHs. Strong upregulation in the transient assay required MS32 and bHLH122; however, based on RNA-seq, bHLH51 abundance was not significantly altered in bhlh122 anthers (Figure 4;Supplemental Figure S5B). We conclude that the combination of MS23 + MS32 is sufficient to promote bHLH51 expression in anthers in the absence of bHLH122.
Differential expression trends over time, and affected processes that are shared and distinctive to each bHLH mutant
As shown in Venn tables (Figure 5), at 1.0, 1.5, and 2.0 mm, the majority of differentially expressed (DE) genes were found exclusively in ms23 (n = 2,674, 4,615, and 4,323, respectively), followed by a much smaller subset in ms32 (n = 688, 405, and 643, respectively). At 1.5 mm, a substantial subset of DE genes was shared by both ms23 and ms32 (n = 715). A second subset was shared among ms23, ms32, and bhlh122-1 (n = 243), demonstrating the cascade of bHLH122-dependent processes downstream of both MS23 and MS32. At 2.0 mm, more DE genes were found in both ms23 and ms32 (n = 984), while the second most abundant, shared subset was now present among ms23, ms32, and bhlh51-1 (n = 359). At 3.0 mm, a large cohort of 4,207 DE genes was found exclusively in the bhlh51-1 mutant, making them candidates for regulation by the bHLH51 homodimer (Nan et al., 2017). A smaller subset of 665 DE genes was possibly co-regulated by the bHLH122 and bHLH51 heterodimer.
Figure 5.

Counts of DE genes in four bHLH mutant anthers. Venn table of DE genes unique to one mutant or shared with one, two, or all three other mutants. Bar charts of gene counts in each category are displayed, with gene counts over 100 highlighted in bold text. Significance was determined with a minimum fold change of 2 with a P ≤ 0.05.
We then determined how the expression of other TF genes is affected in these mutants. Among the 2,947 TF genes surveyed, more than a third (n = 1,036) were significantly differentially regulated (Supplemental Data Set S5). Among these, 41 TF genes were downregulated in both ms23 and bhlh51-1, including 11 that were affected by AP2-EREBPs (Supplemental Data Set S5), indicating that many ethylene processes are present in their downstream cascade. Many male sterility genes were also DE in one or more of these bHLH mutants; for example, some classic male sterility genes, including Ms7, Ms8, Ms25, Ms26, Ms30, Ms33, and Ms44 were downregulated more than 20-fold at various stages (Supplemental Data Set S6), pointing to the essential roles of the bHLHs in choreographing downstream cascades. qRT-PCR further validated that Ms26 was significantly downregulated in ms23, ms32, and bhlh51-1 at 3.0 mm (Supplemental Figure S5).
Gene ontology analysis of the DE genes in the four mutants
In an attempt to pinpoint processes that are positively regulated by these bHLHs, we performed gene ontology (GO) analysis of the DE genes using version 2 of AgriGO (Tian et al., 2017). Lists of DE genes in individual mutants at different anther stages were uploaded and compared against the Complete GO database (Supplemental Data Set S7) and the Plant Slim GO database (Supplemental Figure S7). Results from both databases highlighted several GO terms tightly associated with processes crucial for anther development, including cell growth and cell differentiation, which are required to sustain cell proliferation and organ growth. Lipid synthesis, lipid transport, and lipid metabolism were also highlighted: these processes contribute to the biogenesis of wax and other lipid-derived materials in exine, which are secreted postmeiotically by tapetal cells into the locule to coat maturing microspores. Novel sets of GO terms were also discovered, including many associated with photosynthesis, including chlorophyll biosynthesis, responses to light, light reactions, and dark reaction. These photosynthesis-related terms were exclusively associated with downregulated genes in ms23 anthers at the 1.0, 1.5, and 2.0 mm stages (Supplemental Figure S8A; Supplemental Data Set S8), indicating that MS23, likely with other unknown factor(s), positively regulates photosynthetic processes. qRT-PCR also validated the expression pattern of the PsbS1 gene, encoding a putative chlorophyll A–B binding subunit in photosystem II (Supplemental Figure S4). Tapetal cells contain proplastids; only endothecial cells contain differentiated chloroplasts (Murphy et al., 2015), which are evident from 1.0 mm onwards. We suggest that TP may be required to modulate endothecial cell development, including chloroplast biogenesis and function, through cell-to-cell interactions.
Starting at 1.5 mm through 2.0 mm, genes enriched in another set of cell cycle-related GO terms, that is, cytokinesis and nuclear division, were upregulated only in ms23. At these stages, mitosis has slowed or ceased in most anther cell types (Kelliher and Walbot, 2011). However, in ms23, the pretapetal cells each undergo an ectopic periclinal division to form a bilayer, a major process because approximately one-third of the cells in a normal anther are tapetal by 2.0 mm (Kelliher and Walbot, 2011); the complete extra layer is a distinctive feature not observed in other plant species or other bHLH mutants. At 1.5 mm, genes enriched in several sets of GO terms related to sporopollenin biosynthesis associated with pollen exine formation were downregulated in all four mutants, particularly bhlh122-1 and bhlh51-1 (Supplemental Data Set S7). In both bhlh122-1 at 2.0 mm and bhlh51-1 at 3.0 mm, genes encoding negative regulators of PCD were upregulated. A recent study confirmed these two TFs regulate the senescence of tapetal cells by PCD (Liu et al., 2021). Studies in rice, Arabidopsis, and other plants have defined additional TFs that regulate this important process (Zhang et al., 2006; Zhu et al., 2008; Feng et al., 2012; Ma et al., 2012; Xu et al., 2014; Ferguson et al., 2017; Verma, 2019), including orthologs of these bHLHs (Li et al., 2006). At 3.0 mm, genes involved in major processes such as proteolysis, transport (ion, water, amino acids, hormones, etc.), and lipid biosynthesis, for example, Ms7, Ms20, Ms26, and chls1 (Supplemental Data Set S9), were predominantly downregulated in bhlh51-1 mutant anthers; these genes are predicted to contribute to the postmeiotic gene cascade associated with pollen biogenesis.
MS23, MS32, and bHLH122 are indispensable for 24-nt phasiRNA biogenesis
sRNAs are known to be highly enriched in developing maize anthers (Zhai et al., 2015). We used published and newly generated sRNA-seq data to investigate sRNA homeostasis in the bHLH mutant anthers. At 1.0 mm, there was no significant difference in 21- or 24-nt sRNA abundance in ms23 or ms32 anthers compared to the fertile control (Supplemental Figure S9A). In contrast, 24-nt sRNA abundance peaked at 1.5 and 2.0 mm in fertile anthers, whereas 24-nt sRNA levels decreased by 15%–45% at 1.5 and 2.0 mm in sterile ms23 and ms32 anthers versus fertile anthers (Supplemental Figure S9, B and C), suggesting roles for MS23 and MS32 in 24-nt sRNA biogenesis. Similarly, while 24-nt sRNA abundances were comparable at 1.0 and 3.0 mm (Supplemental Figure S9, D and G) among bhlh51-1, bhlh122-1, and the fertile controls, 24-nt sRNA levels decreased by 35% and 40% in bhlh122-1 sterile anthers at 1.5 and 2.0 mm, respectively (Supplemental Figure S9, E and F). In contrast, the abundance of 24-nt sRNAs in sterile bhlh51-1 anthers was similar to that of fertile anthers (Supplemental Figure S9, D–G).
In maize anthers, 45%–60% of 24-nt sRNAs are 24-nt phasiRNAs at 1.5–2.5 mm (Zhai et al., 2015). We, therefore, examined the expression of genes associated with sRNA biogenesis and processes in addition to 24-nt phasiRNA products. Similar to previous findings in ms23 (Zhai et al., 2015), the abundances of 24-PHAS precursors and Dcl5, as well as 24-nt phasiRNAs, were significantly reduced in ms32 anthers versus the fertile controls (Figure 6A). Interestingly, miR2275 family members, the triggers of 24-nt phasiRNA biogenesis (mainly miR2275a and miR2275b/c), persisted or accumulated in ms23 and ms32 anthers at 2.0 mm compared to the fertile controls, implying that miR2275 production is an earlier, distinctive event that is regulated separately from 24-phasiRNA production (Figure 6A;Supplemental Figure S10, A and B). Similar to ms23 and ms32, 24-PHAS precursors and 24-phasiRNAs were almost absent in bhlh122-1, while miR2275 and Dcl5 transcript abundances in bhlh122-1 were normal (Figure 6B;Supplemental Data Set S10). Considering that bHLH122 functions downstream of MS23 and MS32, this finding suggests that bHLH122 is specialized for regulating 24-PHAS transcription in maize and does not participate in ensuring normal levels of the microRNA precursor or Dcl5. Interestingly, abundances of 24-PHAS, Dcl5 mRNA, and 24-nt phasiRNAs are normal in bhlh51-1 anthers, except that miR2275 was more abundant at 1.0 mm (Figure 6B;Supplemental Figure S10C), suggesting that bHLH51 is not a direct regulator of any component of 24-nt phasiRNA biogenesis. The finding that Dcl5 was not affected in either bhlh122-1 or bhlh51-1 indicates that Dcl5 is regulated separately and probably slightly earlier than the 1.5-mm stage. Changes in the levels of 21-nt phasiRNA pathway components (miR2118, 21-PHAS mRNA, and 21-nt phasiRNAs) were minor in ms23 and ms32 (Supplemental Figure S11A). As expected, the levels of 21-nt TAS3-derived tasi-ARFs were normal in all four mutants (Supplemental Figure S11). Therefore, the regulatory contributions of MS23, MS32, and bHLH122 are centered on 24-nt phasiRNAs.
Figure 6.
Quantification of miR2275, 24-PHAS, Dcl5, and 24-nt phasiRNA in bHLH mutants. A, Quantification of miR2275, 24-PHAS mRNA, Dcl5 mRNA, and 24-nt phasiRNA in fertile ms32//Ms32 (striped circles), ms23 (solid circles), and ms32 (open circles) sterile anthers at the 1.0, 1.5, and 2.0 mm stages. ms32//Ms32 and ms32 were siblings from the same segregating family, and ms23 had a comparable genetic background. B, Quantification of miR2275, 24-PHAS mRNA, Dcl5 mRNA, and 24-nt phasiRNA in fertile (striped circles), bhlh122-1 (solid circles), and bhlh51-1 (open circles) sterile anthers at the 1.0, 1.5, 2.0, and 3.0 mm stages. Fertile, bhlh122-1, and bhlh51-1 were siblings from the same segregating family. Dot sizes represent total read numbers (CP20M) of genes or sRNAs and were normalized together under the same subscript line in (A) or (B) across the genotypes and time points. At least two replicates were used for each sample.
24-PHAS transcripts are differentially regulated in the mutants
Previously, 176 24-PHAS loci were identified in maize (Zhai et al., 2015). These loci were classified into five distinct groups based on RNA-seq abundances (Figure 7, A and B). Most 24-PHAS transcripts were absent at 1.0 mm and abundant at 1.5 mm in fertile ms32/Ms32, bhlh51-1, and Hi-II fertile control anthers, whereas these transcripts were absent in ms23, ms32, and bhlh122-1. Surprisingly, 10 moderately abundant 24-PHAS mRNAs in Group I were present at all stages, regardless of genotype (Figure 7, A and B; Supplemental Data Set S11), and were also present in seedlings (Supplemental Figure S12), suggesting that they are MS23, MS32, and bHLH122-independent. This group constitutes a new 24-nt phasiRNA class.
Figure 7.
Quantification of 176 24-PHAS mRNAs and 24-nt phasiRNAs in bHLH mutants. A, RNA-seq quantification of 176 24-PHAS mRNA abundances at 1.0, 1.5, and 2.0 mm in fertile ms32//Ms32, ms23, and ms32 sterile anthers. ms32//Ms32 and ms32 were siblings from the same segregating family, and ms23 had a comparable genetic background. B, RNA-seq quantification of 176 24-PHAS mRNA abundances at 1.0, 1.5, 2.0, and 3.0 mm in fertile, bhlh122-1, and bhlh51-1 sterile anthers. Fertile, bhlh122-1, and bhlh51-1 were siblings from the same segregating family. C, 24-nt phasiRNAs abundances quantification by sRNA-seq at 1.0, 1.5, and 2.0 mm in fertile ms32//Ms32, ms23, and ms32 sterile anthers. D, Quantification of 24-nt phasiRNA abundance by sRNA-seq at 1.0, 1.5, 2.0, and 3.0 mm in fertile plus bhlh122-1 and bhlh51-1 sterile anthers. In (A)–(D), individual loci were plotted along the y-axis, and were clustered into five groups (I, II, III, IV, and V) based on abundance pattern similarity. Replicates were contained within a box below the genotype and anther size label. At least two replicates were used for each sample.
The levels of precursors from 131 24-PHAS loci were significantly reduced in ms23, ms32, and bhlh122-1, while 35 others were either low abundance or inconsistently detected between genotypes (Supplemental Data Set S11). These findings suggest that MS23, MS32, and bHLH122 regulate the majority of 24-PHAS precursors. We further grouped these 131 downregulated 24-PHAS mRNAs based on their expression patterns (Figure 7, A and B; Supplemental Figure S12). In the major Group II (n = 96), almost no reads were detected in ms23, ms32, or bhlh122-1, suggesting they are exclusively promoted by MS23, MS32, and bHLH122. Group III (n = 20) was virtually absent in ms23 and ms32 but marginally detected in bhlh122-1 at 1.5 mm, suggesting their transcription depends mainly on MS23 and MS32 but only partially on bHLH122. In Groups IV and V (n = 15), transcription initiated as early as 1.0 mm, with the levels of Group IV (n = 8) members significantly reduced after 2.0 mm in Hi-II and bhlh51-1 and absent in bhlh122-1. Group V (n = 7) maintained minimal read levels in ms23, ms32, and bhlh122-1, as well as at 3.0 mm in Hi-II and bhlh51-1. These results suggest that these 24-PHAS mRNAs (n = 35) in Groups III, IV, and V are bHLH122-dependent but can also adopt other regulatory mechanisms to promote transcription.
To determine any positive impacts of these bHLHs on Dcl5 and 24PHAS_NO15, and Hen1 expression, we performed maize leaf protoplast transfection followed by qRT-PCR (see “Materials and methods” for details of the experimental design). These three sample genes were differentially downregulated in ms23, ms32, and bhlh122 (Figure 6;Supplemental Figure S5; Supplemental Data Set S10 and S11). While minimal Dcl5 transcripts were detected in protoplasts harboring 35S:GFP, Dcl5 was significantly induced (19.4- to 210.4-fold) in protoplasts co-transfected with 35S:Ms23, 35S:Ms32, and 35S:bHLH122 (Supplemental Figures S6, A–C and S13A; Supplemental Data Set S4; Supplemental File S1); interestingly, the induction was lower, while still statistically significant, with Ms23 + Ms32 (3.2- to 43.0-fold). Based on RNA-seq analysis, Dcl5 requires both MS23 and MS32, but its expression was not significantly altered in bhlh122 anthers (Figure 6; Supplemental Figure S5). The transient gene expression assays suggested that bHLH122 might boost Dcl5 expression in the presence of MS23 and/or MS32. Interestingly, 24PHAS_NO15 and Hen1 were not induced by any transfection combinations tested (Supplemental Figure S13, B and C), indicating that either they are not direct targets or an unknown additional factor is required.
The 24-nt phasiRNAs were mostly absent in ms23, ms32, and bhlh122-1 but their levels appeared normal in sterile bhlh51-1 anthers at 1.5 and 2.0 mm, matching their abundances in fertile anthers (Supplemental Figure S9, B, C, E, and F). It is worth noting that all 24-nt phasiRNAs were depleted in ms23 and ms32, despite the observation that the precursor transcript production of Groups I, IV, and V PHAS loci appears to be MS23- or MS32-independent (Figure 7, A and B; Supplemental Figure S12). The 24-nt phasiRNAs from a few bHLH122-independent loci from Group III persisted in bhlh122-1 anthers; this observation indicates that DCL5 is not only expressed but also functional in bhlh122-1 anthers at 1.5 mm (Figure 7B;Supplemental Figure S9).
Discussion
A bHLH cascade regulates 24-nt phasiRNA production in the maize TP
All four bHLH single mutants in this study are male sterile and exhibit aberrant tapetal cell differentiation. These TP-regulating bHLH factors are conserved in angiosperms and regulate essential pathways for anther development. All four maize bHLH genes have orthologs in rice and Arabidopsis (Nan et al., 2017), and loss-of-function mutants of these orthologs causes male sterility (Sorensen et al., 2003; Jung et al., 2005; Li et al., 2006; Zhang et al., 2006; Fu et al., 2020). In our maize study, anthers at 1.0, 1.5, 2.0, and 3.0 mm were collected, spanning the peak of 24-nt phasiRNA abundance (Zhai et al., 2015). Most 24-nt phasiRNAs were absent in ms23, ms32, and bhlh122-1 single mutants (Figures 6 and 7), suggesting that MS23, MS32, and bHLH122 are required for 24-nt phasiRNA biogenesis in maize. This is consistent with the finding that the rice orthologs of these genes regulate pollen production and 24-nt phasiRNA biogenesis (Ono et al., 2018). These finding point to both conserved and different regulatory mechanisms between maize and rice, particularly in their interactions as heterodimers and shared gene regulatory mechanisms related to 24-nt phasiRNA biogenesis by MS23, MS32, and bHLH122. Most 24-nt phasiRNAs and their precursors are bHLH122-dependent. Dcl5 expression, on the other hand, is directly activated by MS23 and MS32 but is not bHLH122-dependent based on our RNA-seq analysis and transient protoplast study (Supplemental Figure S13). In contrast, Dcl5 expression is thought to be activated directly by the bHLH122 ortholog in rice (Ono et al., 2018). Despite the conservation of bHLH TFs in Arabidopsis, 24-nt phasiRNAs are absent in the Brassicaceae and other (but not all) eudicot families (Xia et al., 2019). Furthermore, they are conditionally required for male fertility in maize based on the finding that dcl5 mutants grown at low temperature exhibit normal pollen development (Teng et al., 2020), confounding the elucidation of the precise roles of these sRNAs in tapetal development.
Both previous and current findings indicate that the mRNA and protein products of these four bHLHs in maize and rice exhibit rapid accumulation and rapid declines in abundance (Nan et al., 2017; Ono et al., 2018). Therefore, MS23, MS32, and bHLH122 can rapidly turn transcription of 24-PHAS and Dcl5 mRNAs on and off to define the temporal appearance of the 24-nt phasiRNA pathway in the developing TP. There is a transcriptional activation hierarchy of MS23 and MS32 that regulates bHLH122 mRNA transcription first; bHLH122 is then required for the transcription of the majority of 24-PHAS mRNAs. Such a multi-step transcriptional regulatory network strongly resembles a classic TF cascade.
A cascade of gene expression in a multistep pathway normally results in amplification of the initial signal, as observed in processes such as the formation of spatial polarity during body axis establishment or segmentation in vertebrates and arthropods (Peel et al., 2005). Our results suggest that a high-level mechanism of expression control was adopted for 24-nt phasiRNA biogenesis in maize TP, while it is still unknown when, or how, this mechanism emerged during the evolution of flowering plants. An outstanding question is how conserved the regulatory cascade is. Rice EAT1 shows the ability to promote Dcl5 mRNA transcription (Ono et al., 2018), while it seems that MS23 and MS32 promote the transcription of Dcl5 in maize, but bHLH122 possibly does not. Meiotic 24-nt phasiRNAs were recently shown to be conserved broadly across angiosperms, with absences in some clades (Xia et al., 2019), but little is known about the TFs that regulate their expression, except in maize and rice. Besides the conventional meiotic 24-phasiRNA cohorts, surprisingly, a novel group of 24-nt early, premeiotic phasiRNAs are highly abundant in wheat and barley anthers, but not in rice or maize, suggesting that a different set of regulators functions in premeiotic anthers of wheat and barley (Bélanger et al., 2020).
There are obvious parallels between plant reproductive phasiRNAs and piRNAs in animal spermatogenesis. Mouse A-MYB seems to be the activator of pachytene piRNA precursor mRNA transcription, which is responsible for 95% of piRNAs in the adult mouse (Mus musculus) testis (Li et al., 2013), while in fruit fly (Drosophila melanogaster), Moonshiner forms heterodimers with Rhi, anchoring to H3K9me marks at piRNA clusters in heterochromatic regions (Andersen et al., 2017). These may be intriguing cases of convergent evolution between plant phasiRNAs and animal piRNAs that function in reproductive tissues to ensure normal germinal cell development and functions. Future work could pinpoint underlying evolutionary mechanisms that resulted in key roles for sRNAs in male organ development in the two kingdoms.
The function of 24-nt phasiRNAs in the TP
Loss of 24-nt phasiRNAs in maize dcl5 mutants disrupts TP developmental progression and causes male sterility under optimal growth regimes (Teng et al., 2020), but the function and underlying mechanism are still unclear. The delayed anther development in sterile dcl5 anthers begins at 1.5 mm; nonetheless, meiosis is completed normally in sterile dcl5 anthers. Therefore, the germinal transcriptional program and steps in meiosis are cell autonomous, independent of 24-nt phasiRNAs. In addition, a permissive condition with slightly lower day and night temperatures can partially rescue male infertility (Teng et al., 2020). To date, multiple attempts have failed to identify mRNA targets of 24-nt phasiRNAs that would involve phasiRNAs in canonical posttranscriptional gene silencing. These efforts included extensive bioinformatic searches for complementarity of phasiRNAs to anther mRNAs, degradome analysis of anthers of maize inbred line W23, and differential gene expression analysis in sterile dcl5 anthers (Zhai et al., 2015; Patel et al., 2018; Teng et al., 2020). Also, no targets for 24-nt phasiRNAs could be found in isolated rice meiocytes (Jiang et al., 2020). There is, however, elevated DNA methylation in the CHH context at 24-PHAS loci in isolated maize meiocytes (Dukowic-Schulze et al., 2016) and in whole anthers in which approximately half the DNA is from tapetal cells (Zhang et al., 2021). The increased CHH DNA methylation is absent in dcl5 and ms23 anthers, and in fertile anthers, the degree of CHH modification reflects the abundance of individual 24-nt phasiRNAs arising at that locus. These observations suggest that this cis-DNA modification depends on 24-nt phasiRNAs (Zhang et al., 2021). The importance of this cis-regulation of the 24-PHAS loci by the product 24-nt phasiRNAs is unclear at this point.
The TP, the innermost layer of the anther wall, surrounds the developing pollen mother cells, meiocytes, and haploid microspores. Historically, TP development was divided into four stages: initiation, differentiation, maturation, and degeneration. Instead, we view tapetal cells as undergoing sequential redifferentiation to perform discrete roles during anther growth. Discussions of tapetal roles (Ariizumi and Toriyama, 2011; Kelliher and Walbot, 2011; Zheng et al., 2019) indicate that the TP produces and secretes enzymes to digest the callose wall as the pollen mother cell forms, provides structural and nutrient support to meiocytes, synthesizes exine components during meiosis, and transports them after tetrad dissolution, generating an attachment surface for gametophytes, ultimately exhausting its contents through PCD. Here, using RNA-seq data acquired from fertile and four male-sterile mutants, we provided important insights into the gene regulation that underpins the sequence of tapetal activities.
Are 24-nt phasiRNAs required for tapetal cell functions or for tapetal support of meiosis? Meiosis has been reported to abort early at prophase I in both ms23 and ms32 anthers (Chaubal et al., 2000), while it is completed in sterile dcl5 and eat1 anthers (Ono et al., 2018; Teng et al., 2020). Furthermore, aberrant tapetal cells appear at 1.0 mm in ms23, which is much earlier than the alterations in dcl5 (1.5 mm; Teng et al., 2020) or the appearance of 24-nt phasiRNAs, suggesting that early TP differentiation events are independent of 24-nt phasiRNAs. After the formation of the tapetal layer, a characteristic and unusual hallmark of tapetal cells is the acquisition of binucleate status. The fact that no TP binucleation is observed in either ms23 or ms32 anthers (Chaubal et al., 2000) and that TP binucleation numbers are reduced in sterile dcl5 anthers (Teng et al., 2020) suggests that 24-nt phasiRNAs may participate in this cellular differentiation process. Quantification of 24-PHAS mRNA and 24-nt phasiRNAs showed no difference in abundance in sterile bhlh51-1 anthers compared to its fertile counterpart, suggesting that bHLH51 is dispensable for 24-nt phasiRNAs biogenesis. Nonetheless, in bhlh51-1 anthers, Ms23 expression is downregulated, while Dcl5, bHLH122, and miR2275 are upregulated, which could affect 24-nt phasiRNAs indirectly. Furthermore, a group of chromatin-silencing genes is DE in bhlh51-1, suggesting that bHLH51 might regulate an active transcriptional gene silencing mechanism that involves 24-nt phasiRNAs.
A dynamic cascade under the collaborative control of MS23, MS32, bHLH122, and bHLH51
Our primary goal was to confirm the order of action of four bHLH proteins required for normal anther wall development in maize and to define processes controlled by each TF. Based on the cytological evidence and the differential gene expression patterns in each mutant, the order of essential actions we infer is MS23, MS32, bHLH122, then bHLH51, which reverses the position of the final two factors compared to a recent publication (Jiang et al., 2021). Mutants of the broadly detectable (https://qteller.maizegdb.org/bar_chart_B73v4.php?name=Zm00001d006564) Ms32 transcript do not show any phenotype except male sterility, indicating it is mainly required for normal tassel development, when it is highly expressed during early meiosis.
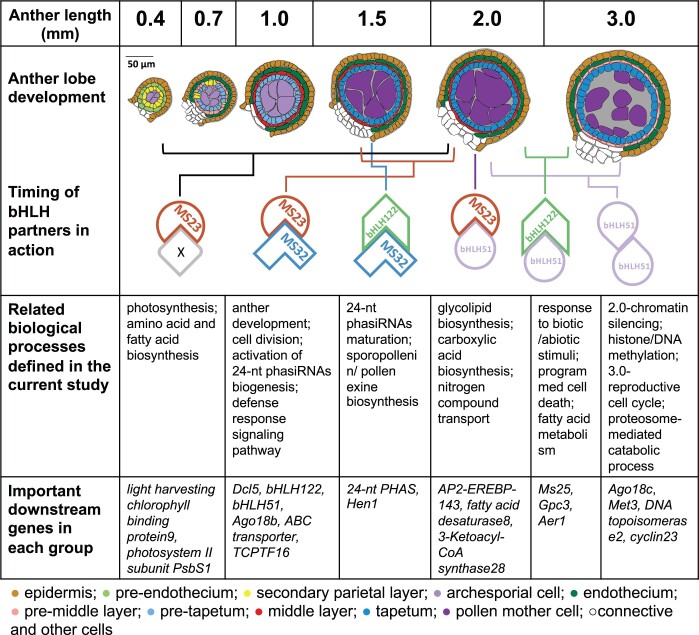
Unexpectedly, MS23, likely with an unknown factor “X” (Figure 8; Supplemental Figures S5 and S13D), is required to support photosynthetic processes, which occur in the endothecial cells, comprising an outer wall layer. We propose that an MS23-dependent signal(s) generated in the TP or its predecessor secondary parietal layer modulates endothecial development. Starting around 2.0 mm, the MS23 + bHLH51 heterodimer is required either directly or indirectly for the expression of many ethylene-associated TF genes and other processes, for example, fatty acid biosynthesis, lipid transport, and carbohydrate metabolism, that are essential for tapetal biosynthesis of exine and sporopollenin compounds and their subsequent secretion onto microspores. At 2.0 mm, bHLH51 is important for the expression of many genes associated with chromatin silencing via Pol II/RDR6- and methylation-mediated processes, which are distinctive from the Pol IV-mediated siRNAs regulated by the three other bHLHs (Supplemental Figure S8B; Matzke et al., 2015). At 3.0 mm, Ms23 is diminished, and bHLH122 is re-expressed; we suggest that there is a partner exchange and that bHLH122 + bHLH51 heterodimers and the bHLH51 homodimers are in charge of the final stages of tapetal development: secretion of the final tapetal cell contents to support pollen maturation and PCD of the layer.
Figure 8.
Proposed cascades of bHLH-associated pathways in maize anthers. Tracings of transverse confocal images of fertile anthers at various lengths are colored by cell type. Proposed partnerships and timing of actions are indicated, with important biological processes and downstream genes listed below each partner pair.
Our transient activation study in maize leaf protoplasts demonstrated that bHLH51 can be activated when either Ms23 + Ms32, Ms32 + bHLH122, or Ms23 + Ms32+bHLH122 are over-expressed (Supplemental Figure S6D). This demonstrates that bHLH51 functions downstream of the three other bHLHs and can be directly regulated by them. Our finding is also consistent with a recent report (Jiang et al., 2021) showing that the expression of bHLH122 together with Ms32 induced the expression of bHLH51. Studies in rice also showed that OsTIP2/OsbHLH142 (ortholog of MS23) is upstream of and directly binds to both OsEAT1 (ortholog of bHLH122) and OsTDR (ortholog of bHLH51), while OsTIP2/OsbHLH142 together with OsTDR can modulate OsEAT1 expression (Fu et al., 2014; Ko et al., 2014). Unexpectedly, bHLH122 does not appear to be a direct target of MS23 + MS32, although its expression is dependent on both, indicating that other unknown factors might be required for its expression in maize leaf protoplasts (Supplemental Figure S6C). Similar to what was proposed in rice (Fu et al., 2014), uncovering the timing of activity of these bHLH TFs in anthers could unveil another layer of regulatory complexity. Additionally, Dcl5 expression was induced when either Ms23 + Ms32 or Ms23 + Ms32 + bHLH122 were over-expressed (Supplemental Figure S13A), indicating that MS23 and MS32 directly regulate Dcl5 and that bHLH122 can further enhance this effect. This trio is also the key regulators of 24-PHAS production during the peak period of 24-nt phasiRNA biogenesis at 1.5 mm.
Collectively, discrete processes function downstream of particular bHLHs as they are deployed and modulated across developmental stages (Figure 8). These important insights will focus future research on discerning whether bHLH TFs act directly by binding to their target promoters or indirectly through their regulation of downstream TFs.
Materials and methods
Plant materials
Maize (Z. mays) plants were grown under greenhouse conditions (31°C/21°C, 14-h day/10-h night light cycle with supplemental LED and UV-A lamps providing ∼50% of summer sun conditions) except during the summer months (June through September), when they were grown in the field at Stanford, California, USA. Established 1:1 segregating lines of male-sterile to heterozygous ms23-ref and ms32-ref stocks (Nan et al., 2017) have been introgressed into the W23 background by several generations of backcrosses, selfing, and sibling crosses. These lines were used and referred to as ms23 and ms32 throughout this study; the heterozygous fertile samples in this background were used as controls.
The transgenic knockout lines for both bHLH122 and bHLH51 were generated in Bing Yang’s lab at the University of Missouri (Columbia) using CRISPR–Cas9 (the RNA-guided Cas9 nuclease from the microbial clustered regularly interspaced short palindromic repeats) technology with two guide RNAs, targeting the 5′-GGAATGGTCGGGGTACCAG-3′ and 5′-GCAGTGCAAGAACCTCGAGG-3′ regions of bHLH122 and bHLH51, respectively. The sequences of two complementary oligonucleotides were as follows: 5′-tgttGGAATGGTCGGGGTACCAG-3′//5′-aaacCTGGTACCCCGACCATTCC-3′ and 5′-gtgtGCAGTGCAAGAACCTCGAGG-3′//5′-aaacCCTCGAGGTTCTTGCACTGC-3′ (lowercase letters indicate cloning sequences). These sequences were cloned into a pGW-Cas9 vector as described previously (Char et al., 2017), and T-0 maize transgenic lines in the Hi-II background were obtained after transformation via Agrobacterium tumefaciens strain EHA101 at the Iowa State University Plant Transformation Facility. A total of 10 independent Basta-resistant lines were obtained, and mutations were further genotyped using PCR followed by sequencing. Selected lines were propagated and screened to generate 1:1 segregating lines that inherited only a single mutation in either bHLH122 or bHLH51 and lacked the original Cas9 transgene. Heterozygous sibling samples were used as controls for the studies related to this group.
Confocal microscopy
Anthers were collected under a dissecting microscope, measured with a micrometer with scales accurate from 0.01 µm to 5 mm (Fisher Scientific, Waltham, MA, USA), and stained with propidium iodide as described previously (Kelliher and Walbot, 2011). Images were collected on a Leica SP8 confocal microscope equipped with the LAS X acquisition software (Leica Microsystems Inc., Buffalo Grove, IL, USA) and processed with the ImageJ software package Fiji (Schindelin et al., 2012).
Global Y2H screening
An NGS-based, global Y2H screening for partners of MS23 in maize tassel was conducted (Next Interactions, Richmond, CA, USA). A male inflorescence cDNA library was generated with poly(A)-enriched RNA prepared from pooled total RNA (with RNA Integrity Number = 10) extracted from central spikelets, branch spikelets, and immature tassels collected from wild-type plants of the W23 inbred line (Supplemental Data Set S12). The resulting cDNA library was cloned into the pGAD yeast vector to generate the comprehensive prey library. The full-length Ms23 coding region was cloned into the pGBK yeast vector as the bait. The empty bait vector and the cDNA library (precloned) were two controls simultaneously run and analyzed to generate the list of significant interactions. A total of three replicates each were run, and positive clones were sequenced on the Illumina MiSeq System. Sequence read quality was verified using fastQC. The raw sequence reads were mapped to the genome using Subjunc aligner from Subread (Liao et al, 2019). The alignment bam files were compared against the Z. mays AGPv4 gene annotation GFF file, and raw counts for each gene were generated using the featureCounts tool from Subread. The raw count data were then processed to remove genes that were not expressed. Based on the average read count of all samples (4.5 million), a counts per million (CPM) cutoff of 1.00 corresponds to approximately 10 raw reads. If a gene had a CPM value >1.00 in at least two samples, it was included in the analysis. In this experiment, a total of 17,188 out of 39,324 annotated genes were detected and further analyzed. The raw count data of the expressed genes were transformed to log2CPM values, and statistics for differential expression analysis were computed using the R Limma package (Ritchie et al., 2015). The statistics values include log fold change (logFC), P-value, and FDR (or adjusted P-value corrected for multiple hypothesis testing with Benjamini–Hochberg procedure). The enrichment value for each gene was calculated as reads per kilobase per million. Interacting partners with MS23 were called based on their enrichment compared to both the cDNA and the pGBK empty vector controls using standard cutoff of LogFC ≥ 1 with FDR ≤ 0.05.
RNA in situ hybridization
Wild-type W23 tassels were fixed and embedded in paraffin following a standard protocol (Jackson et al., 1994). PCR-generated fragments with primer pairs as described in Supplemental Data Set S13 were transcribed using a DIG RNA Labeling Kit (T7) (Roche, http://www.roche-applied-science.com). Thin sections (7 μm) were hybridized to anti-sense probes as previously described (Zhang et al., 2014). Images contained anthers at around the 2 mm stage were selected.
RNA-seq and sRNA-seq library construction and sequencing
Staged anthers were collected directly into liquid nitrogen-chilled, 1.5-mL tubes and stored at −80°C until RNA extraction. Total RNA for sRNA-seq and RNA-seq libraries was isolated using the TRI Reagent (Sigma-Aldrich, St Louis, MO, USA). Total RNA quality was assessed by denaturing agarose gel electrophoresis and quantified using a Qubit RNA BR Assay Kit (ThermoFisher, Waltham, MA, USA). For library preparation, 20- to 30-nt RNAs were excised from a 15% polyacrylamide/urea gel, and ∼25 ng of sRNA was used for library construction with a NEBNext sRNA Library Prep Set for Illumina (New England Biolabs, Ipswich, MA, USA). For RNA-seq, 2 µg of total RNA was treated with DNase I (New England BioLabs) and cleaned with RNA Clean and Concentrator-5 (Zymo Research, Irvine, CA, USA). A NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England BioLabs) was used for library construction with 500 ng of treated RNA. Sequencing in single-end mode on an Illumina HiSeq 2500 or NextSeq (University of Delaware) yielded 51- and 75-bp reads for sRNA-seq and RNA-seq, respectively.
For sRNA-seq data, we first used Trimmomatic version 0.32 to remove the linker adaptor sequences (Bolger et al., 2014). The trimmed reads were then mapped to version 4 of the B73 maize genome using Bowtie (Kersey et al., 2016). Read counts were normalized to 10 or 20 million to allow for the direct comparison across libraries. phasiRNAs were designated based on a 24-nt length and mapping coordinates within the previously identified 176 24-PHAS loci, updated to version 4 of the B73 genome using the assembly converter tool followed by manual curation (Tello-Ruiz et al., 2018). If a 24-nt phasiRNA did not uniquely map to the genome, we divided the abundance equally to each location to which the read mapped, that is, a hits-normalized abundance. These hits-normalized abundances were then summed for each of the 176 loci to calculate the 24-nt phasiRNA abundances.
RNA-seq libraries were trimmed as above and mapped to version 4 of the B73 genome using TopHat version 2.0.12 (Trapnell et al., 2009). Gene counts based on the reference genome GFF annotation file were calculated using the featureCounts function in Rsubread. Gene counts were filtered to include only genes with at least 10 CPM in 2 or more samples and then normalized using the calcNormFactors function of the edge RR package (Robinson et al., 2010) with the “TMMwzp” method. Differential expression between genotypes (mutant versus fertile sib) was analyzed with the limma package in R using the lmFit and eBayes functions (Ritchie et al., 2015). Significant differential expression was considered a minimum fold change of 2 with a p-value ≤ 0.05. For differential expression analysis of the PHAS precursors, we were unable to assemble the transcripts that mapped to these regions due to the absence of annotation in any feature files. Instead, we conducted this analysis in parallel to the analysis of the sRNA data. We first normalized the mapped fragmented reads as counts per 10 or 20 million and applied the hits-normalized abundance approach described above. Using the annotated 24-PHAS loci (above), we then identified the summed abundances of RNA-seq transcript fragments that mapped to these regions to identify a representative abundance for each 24-PHAS precursor.
Replicate libraries were displayed together for construction of the heatmaps for 24-PHAS mRNA and 24-nt phasiRNAs, and we added 1 to all abundances to prevent log10(0) error while also giving the added benefit of removing negative log-transformed values. Heatmaps for miR2275 were generated for log-transformed average values of replicates.
Transient gene expression assay in maize leaf protoplasts
Maize protoplast isolation and transfection were performed using published protocols (Gomez-Cano et al., 2019). Briefly, B73 seeds were germinated and grown in the dark at 25°C for ∼12 days. Sections from the second leaves were cut and chopped into 0.5 mm strips with sharp razor blades. The leaf strips were digested in 10–20 mL freshly prepared enzyme solution, with 0.6 M mannitol, 20 mM KCl, 10 mM CaCl2, 1.5% Cellulase “ONOZUKA” RS (Yakult Japan), 0.4% Macerozyme R-10 (Yakult Japan), 0.1% BSA, and 20 mM MES pH5.7 in a Petri dish according to Gomez-Cano et al. (2019). The enzyme solution containing protoplasts was then filtered with a 35-µm strainer. A brief centrifugation collected the protoplasts, which were then resuspended in W5 solution (125 mM CaCl2, 154 mM NaCl, 5 mM KCl, and 2 mM MES pH 5.7). Protoplasts were counted using a hemocytometer under a light microscope or an automated cell counter (ThermoFisher). Finally, the protoplasts were pelleted again and resuspended in MMG solution (0.6 M mannitol, 15 mM MgCl2, and 4 mM MES pH 5.7) to a final density of 2.5 × 106 cells per milliliter at room temperature (RT).
The vectors containing the Ms23 and Ms32 coding sequences without stop codons were obtained from TFome (https://www.grassius.org/tfomecollection.php). A full-length cDNA clone of bHLH51 was obtained from the Arizona Genomics Institute (http://www.maizecdna.org). The bHLH122 coding sequence was cloned from maize anther mRNA. Each clone was sub-cloned by TOPO PCR cloning (ThermoFisher) into a pENTR vector and subsequently transferred to p35S-GWC-GFP or p35S-GFP-GWC to fuse in frame with N- or C-terminal GFP via LR Gateway recombination; p35S-GFP was used as a control. Eight sets of protoplasts transiently expressing single bHLH with either N- or C-terminal GFP driven by a constitutive 35S promoter (Ms23-GFP, GFP-Ms23, Ms32-GFP, GFP-Ms32, bHLH122-GFP, GFP-bHLH122, bHLH51-GFP, and GFP-bHLH51), and seven sets of protoplasts transiently expressing mixtures of circular plasmids mentioned above, containing two or three bHLHs (Ms23 + Ms32, Ms32 + bHLH122, Ms23 + bHLH51, bHLH122 + bHLH51, Ms23 + bHLH122, Ms32 + bHLH51, Ms23 + Ms32 + bHLH122), were compared to GFP without bHLH (35S:GFP). Three biological replicates (separate experiments) were performed for each set. Circular plasmids were prepared using a Midi Plasmid DNA Prep Kit (Zymo Research).
For each single plasmid transfection, 15 µg of plasmid DNA was used. For multi-bHLH transfection, a total of 15 µg of all plasmids combined was used, with equal quantities of each plasmid. Plasmid DNA in a total of 10 µL water was added to a 2 mL round-bottom tube and mixed with 100 µL of the prepared protoplasts in MMG solution. About 110 µL of PEG/Ca solution (40% PEG4000, 0.3 M mannitol, and 0.1 M CaCl2) was added to each tube, mixed thoroughly, and allowed to sit for 5 min at RT; the reaction was stopped by adding 0.44-mL W5 solution. The transfected protoplasts were collected by centrifugation, resuspended in 1-mL WI solution (0.6 M mannitol and 20 mM KCl, and 4 mM MES pH 5.7), and transferred to a 6-well culture plate precoated with 5% BSA. The culture plates were incubated in the dark at RT for 18–20 h. Transient transfection efficiency was assessed by determining the proportion of GFP-positive cells to total cells using a Zeiss Axiocam 512 with color CCD camera on a Zeiss Axio Zoom with green-light filters; the efficiency varied between 10% and 30%. Total protoplasts were collected with a brief spin in a 2 mL plastic tube, disrupted with glass beads (0.3–0.5 mm), and snap-frozen in liquid nitrogen in the tube.
Total RNA was isolated from protoplasts using TRI Reagent (Sigma-Aldrich, #T9424). Total RNA quality was assessed using an Agilent RNA 6000 Nano kit on a BioAnalyzer 2100, and quantified using a Qubit RNA BR Assay Kit (ThermoFisher). Total RNA was treated with DNase I (New England BioLabs) and purified using RNA Clean & Concentrator-5 Kits (Zymo Research). Reverse transcription was carried out with SuperScript™ III First-Strand Synthesis SuperMix (ThermoFisher). GoTaq qPCR master mix (Promega, Madison, WI, USA) was used on a BioRad C1000 thermocycler for quantitative PCR. Gene-specific primers (Supplemental Data Set S13) were designed to amplify 100–200 bp coding sequences, using Cyanase as the internal reference. Each qRT-PCR had at least two technical replicates, and relative gene expression data were calculated by performing the Pfaffl method (Pfaffl, 2001).
Statistical analysis
Relative gene expression data obtained by qRT-PCR were compared using one-way analysis of variance with genotypes as factors in anthers, or treatments as factors in transiently transformed maize protoplasts. For mutant anthers, means of the individual genotypes were compared with fertile anthers (ms32//Ms32 or Hi-II) using Dunnett’s post-hoc t test; in transiently transformed maize protoplasts, means of individual treatments were compared with 35S:GFP using Dunnett’s post-hoc t test. All statistical analyses were performed using either R (Liao et al., 2019) or data analysis tools in Excel. Results of all statistical analyses are presented in Supplemental File S1, along with original data in Supplemental Data S3 and S4. Individual data points within bar charts are presented in Supplemental Figures S5, S6, and Figure S13, and the bar charts were generated using Excel.
Accession numbers
Sequence data from this article can be found in the MaizeGDB under the following accession numbers: Ms23 (Zm00001d008174), Ms32 (Zm00001d006564), bHLH122 (Zm00001d017724), and bHLH51 (Zm00001d053895).
Requests for materials should be addressed to walbot@stanford.edu. Raw data were submitted to GEO under accession number GSE175915 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE175915] (including sRNA-seq as GSM5351110 to GSM5351153; and RNA-seq data as GSM5351154 to GSM5351197), and the processed data are available via our maize genome browser at https://mpss.danforthcenter.org. Other supporting data are available from the corresponding author upon request.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Confocal imaging of ms23 and fertile anthers at the 2.0 mm stage.
Supplemental Figure S2. Transcript detection in transverse sections of anthers.
Supplemental Figure S3. Confocal imaging of anther wall layers in selected double mutants.
Supplemental Figure S4. Global Y2H screening of interacting partners of MS23.
Supplemental Figure S5 . qRT-PCR validation data.
Supplemental Figure S6. qRT-PCR analysis of bHLH transcripts in transiently transformed maize protoplasts.
Supplemental Figure S7. GO analysis (http://systemsbiology.cau.edu.cn/agriGOv2/) of significant downregulated genes.
Supplemental Figure S8. Hierarchical graphs generated with the singular enrichment analysis algorithm provided by AgriGO version 2 (http://systemsbiology.cau.edu.cn/agriGOv2/).
Supplemental Figure S9. Total sRNA size distribution in bHLH mutants.
Supplemental Figure S10. Quantification of miR2275 abundance in bHLH mutants.
Supplemental Figure S11. Quantification of miR2118, 21-PHAS, 21-nt phasiRNA, and TAS3-derived tasi-arf in bHLH mutants.
Supplemental Figure S12. Maize genome browser data for representative 24-PHAS loci.
Supplemental Figure S13. qRT-PCR analysis of bHLH downstream transcripts in transiently transfected maize protoplasts.
Supplemental Data Set S1. List of potential interacting partners of full-length MS23.
Supplemental Data Set S2. Summary of RNA-seq and sRNA-seq libraries.
Supplemental Data Set S3. Relative gene expression levels detected by qRT-PCR.
Supplemental Data Set S4. qRT-PCR analysis of genes of interests in transfected maize protoplasts.
Supplemental Data Set S5. Transcript abundance (TP20M) of TF genes DE in one or more of the bHLH mutants at one or more anther stage.
Supplemental Data Set S6. DE genic male sterile genes in the four bHLH mutants examined in this study.
Supplemental Data Set S7. GO analysis of significantly downregulated genes in mutant anthers compared to anthers of the fertile sibling.
Supplemental Data Set S8. Transcript abundance (TP20M) of photosynthesis-associated genes (GO:0015979) that are exclusively downregulated in ms23 anthers.
Supplemental Data Set S9. Selected genes downregulated in bhlh51-1 at 3.0 mm under the GO term lipid biosynthetic process (GO:0008610).
Supplemental Data Set S10. Transcript abundance (TP20M) of sRNA-associated genes in bHLH mutant and fertile anthers.
Supplemental Data Set S11. Groups of 24-PHAS based on abundance patterns in bHLH mutants (CP20M).
Supplemental Data Set S12. Pooled RNA samples used for male inflorescence cDNA prey library construction for Y2H analysis.
Supplemental Data Set S13. Primer sequences used in this study.
Supplemental File S1. Analysis of variance tables.
Supplementary Material
Acknowledgments
The bhlh51 and bhlh122 CRISPR mutants were generously generated by Si Nian Char in Professor Bing Yang’s Lab at Iowa State University (now at University of Missouri–Columbia) and were key to initiating the overall study. We thank Joanna Friesner for her assistance with editing. We thank Mayumi Nakano for assistance with data handling. We thank Professor Erich Grotewold for kindly providing plasmids p35S-GFP, p35S-GFP-GWC, and p35S-GWC-GFP.
Funding
This work was supported by US National Science Foundation Plant Genome Research Program (NSF-PGRP) awards 1649424 and 1754097.
Conflict of interest statement. None declared.
G.L.N., B.C.M., and V.W. conceived of the project; G.L.N., C.T., and L.O. performed and interpreted experiments; J.F. and C.T. analyzed sequencing data; G.L.N. and C.T. wrote the manuscript with editing by V.W., B.C.M, and with input from all co-authors.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Virginia Walbot (walbot@stanford.edu) and Blake C. Meyers (bmeyers@danforthcenter.org).
References
- Andersen P, Tirian L, Vunjak M, Brennecke J (2017) A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 549: 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki S, Le NT, Koizumi K, Villar-Briones A, Nonomura KI, Endo M, Inoue H, Saze H, Komiya R (2020) miR2118-dependent U-rich phasiRNA production in rice anther wall development. Nat Commun 11:3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62:437–460 [DOI] [PubMed] [Google Scholar]
- Bélanger S, Pokhrel S, Czymmek KJ, Meyers BC (2020) Pre-meiotic, 24-nt reproductive phasiRNAs are abundant in anthers of wheat and barley but not rice and maize. Plant Physiol 184:1407–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CF, Zhu J, Lou Y, Guo ZL, Xiong SX, Wang K, Yang ZN (2015) The functional analysis of OsTDF1 reveals a conserved genetic pathway for tapetal development between rice and Arabidopsis. Sci Bull 60:1073–1082 [Google Scholar]
- Char SN, Neelakandan AK, Nahampun H, Frame B, Main M, Spalding MH, Becraft PW, Meyers BC, Walbot V, Wang K, et al. (2017) An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotech J 15:257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubal R, Zanella C, Trimnell MR, Fox TW, Albertsen MC, Bedinger P (2000) Two male-sterile mutants of Zea mays (Poaceae) with an extra cell division in the anther wall. Am J Bot 87:1193–1201 [PubMed] [Google Scholar]
- Cui J, You C, Zhu E, Huang Q, Ma H, Chang F (2016) Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28:1078–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukowic-Schulze S, Sundararajan A, Ramaraj T, Kianian S, Pawlowski WP, Mudge J, Chen C (2016) Novel meiotic miRNAs and indications for a role of phasiRNAs in meiosis. Front Plant Sci 7:762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Lu D, Ma X, Peng Y, Sun Y, Ning G, Ma H (2012) Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J 72:612–624 [DOI] [PubMed] [Google Scholar]
- Ferguson AC, Pearce S, Band LR, Yang C, Ferjentsikova I, King J, Yuan Z, Zhang D, Wilson ZA (2017) Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol 213:778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández Gómez J, Wilson ZA (2014) A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotech J 12:765–777 [DOI] [PubMed] [Google Scholar]
- Fu Y, Li M, Zhang S, Yang Q, Zhu E, You C, Qi J, Ma H, Chang F (2020) Analyses of functional conservation and divergence reveal requirement of bHLH010/089/091 for pollen development at elevated temperature in Arabidopsis. J Genet Genom 47:477–492 [DOI] [PubMed] [Google Scholar]
- Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W (2014) The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26: 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cano L, Yang F, Grotewold E (2019) Isolation and efficient maize protoplast transformation. BIO-PROTOCOL 101: e3346 [Google Scholar]
- Higginson T, Li SF, Parish RW (2003) AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J 35:177–192 [DOI] [PubMed] [Google Scholar]
- Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, Shinozaki K (2007) Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19:3549–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Ji C, Li H, Chen L, Xie M, Wang F, Chen Y, Liu YG (2013) A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol Plant 6: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Jiang P, Lian B, Liu C, Fu Z, Shen Y, Cheng Z, Qi Y (2020) 21-nt phasiRNAs direct target mRNA cleavage in rice male germ cells. Nat Commun 11:5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, An X, Li Z, Yan T, Zhu T, Xie K, Liu S, Hou Q, Zhao L, Wu S, et al. (2021) CRISPR/Cas9-based discovery of maize transcription factors regulating male sterility and their functional conservation in plants. Plant Biotech J 19:1769–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan GL, Walbot V, Sundaresan V, Vance V, Bowman LH (2009) Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res 19:1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17:2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Walbot V (2011) Emergence and patterning of the five cell types of the Zea mays anther locule. Dev Biol 350:32–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Selva D, Christensen M, Davis P, Falin LJ, Grabmueller C, et al. (2016) Ensembl Genomes 2016: more genomes, more complexity. Nucl Acids Res 44:D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SS, Li MJ, Ku MSB, Ho YC, Lin YJ, Chuang MH, Hsing HX, Lien YC, Yang HT, Chang HC. et al. (2014) The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 26:2486–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson ZA, Zhang D (2011) PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol 156: 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice Tapetum Degeneration Retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Moore MJ, Zamore PD (2013) Defining piRNA primary transcripts. Cell Cycle 12: 1657–1658 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2019) The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucl Acids Res 47:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Bao W, Liang W, Yin J, Zhang D (2010) Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development. J Inter Plant Biol 52:670–678 [DOI] [PubMed] [Google Scholar]
- Liu X, Yue Y, Gu Z, Huang Q, Pan Z, Zhao Z, Zheng M, Zhang Z, Li C, Cao M (2021) Fine mapping and candidate gene analysis for a novel male-sterile mutant Ms40 in maize. Plant Cell Rep 40:1957–1970 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhao Z, Wei G, Zhang P, Lan H, Zhang S, Li C, Cao M (2018) Characterization of the ZmbHLH122 transcription factor and its potential collaborators in maize male reproduction. Plant Growth Regul 85:113–122 [Google Scholar]
- Lou Y, Zhou HS, Han Y, Zeng QY, Zhu J, Yang ZN (2018) Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana. New Phytol 217:378–391 [DOI] [PubMed] [Google Scholar]
- Ma X., Feng B, Ma H (2012) AMS-dependent and independent regulation of anther transcriptome and comparison with those affected by other Arabidopsis anther genes. BMC Plant Biol 12:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJM (2015) RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 66:243–267 [DOI] [PubMed] [Google Scholar]
- Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17:705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Skibbe D, Timofejeva L, Wang CJR, Kelliher T, Kremling K, Walbot V, Cande WZ (2013) Regulation of cell divisions and differentiation by MALE STERILITY32 is required for anther development in maize. Plant J 76:592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmu J, Bush MJ, DeLong C, Li S, Xu M, Khan M, Malcolmson C, Fobert PR, Zachgo S, Hepworth SR (2010) Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol 154:1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KM, Egger RL, Walbot V (2015) Chloroplasts in anther endothecium of Zea mays (Poaceae). Am J Bot 102:1931–1937 [DOI] [PubMed] [Google Scholar]
- Nan GL, Ronceret A, Wang RC, Fernandes JF, Cande WZ, Walbot V (2011) Global transcriptome analysis of two ameiotic1 alleles in maize anthers: defining steps in meiotic entry and progression through prophase I. BMC Plant Biol 11:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan GL, Zhai J, Arikit S, Morrow D, Fernandes J, Mai L, Nguyen N, Meyers BC, Walbot V (2017) MS23, a master basic helix-loop-helix factor, regulates the specification and development of the tapetum in maize. Development 144:163–172 [DOI] [PubMed] [Google Scholar]
- Nelms B, Walbot V (2019) Defining the developmental program leading to meiosis in maize. Science 364: 52–56 [DOI] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun 4:1445. [DOI] [PubMed] [Google Scholar]
- Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N (2007) A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Liu H, Tsuda K, Fukai E, Tanaka K, Sasaki T, Nonomura KI (2018) EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLoS Genet 14:e1007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P, Mathioni S, Kakrana A, Shatkay H, Blake C., Meyers BC (2018) Reproductive phasiRNAs in grasses are compositionally distinct from other classes of small RNAs. New Phytol 220: 851–864 [DOI] [PubMed] [Google Scholar]
- Peel AD, Chipman AD, Akam M (2005) Arthropod segmentation: beyond the Drosophila paradigm. Nat Rev Genet 6:905–916 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucl Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HA, Li SF, Parish RW (2012) MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol Biol 78:171–183 [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl Acids Res 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber DN, Bantin J, Dresselhaus T (2004) The MADS box transcription factor ZmMADS2 is required for anther and pollen maturation in maize and accumulates in apoptotic bodies during anther dehiscence. Plant Physiol 134:1069–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Li P, Zhai J, Zhou M, Ma L, Liu B, Jeong DH, Nakano M, Cao S, Liu C, et al. (2012) Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J 69:462–474 [DOI] [PubMed] [Google Scholar]
- Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33:413–423 [DOI] [PubMed] [Google Scholar]
- Su Y, Liu J, Liang W, Dou Y, Fu R, Li W, Feng C, Gao C, Zhang D, Kang Z, et al. (2019) Wheat AGAMOUS LIKE 6 transcription factors function in stamen development by regulating the expression of Ta APETALA3. Development 146:dev177527. [DOI] [PubMed] [Google Scholar]
- Tello-Ruiz MK, Naithani S, Stein J, Gupta P, Campbell M, Olson A, Wei S, Preece J, Geniza MJ, Jiao Y, et al. (2018) Gramene 2018: unifying comparative genomics and pathway resources for plant research. Nucl Acids Res 46:D1181–D1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng C, Zhang H, Hammond R, Huang K, Meyers BC, Walbot V (2020) Dicer-like 5 deficiency confers temperature-sensitive male sterility in maize. Nat Commun 11:2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucl Acids Res 45:W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou CH, Cheng PC, Tseng CM, Yen HJ, Fu YL, You TR, Walden DB (2015) Anther development of maize (Zea mays) and longstamen rice (Oryza longistaminata) revealed by cryo-SEM, with foci on locular dehydration and pollen arrangement. Plant Reprod 28:47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnier AL, Jacquard C, Clément C (2009) Programmed cell death and microspore embryogenesis. InTouraev A, Forster BP, Jain SM, eds, Advances in Haploid Production in Higher Plants, Springer, Dordrecht, the Netherlands, pp 147–154. [Google Scholar]
- Verma N (2019) Transcriptional regulation of anther development in Arabidopsis. Gene 689: 202–209 [DOI] [PubMed] [Google Scholar]
- Vernoud V, Laigle G, Rozier F, Meeley RB, Perez P, Rogowsky PM (2009) The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J 59:883–894 [DOI] [PubMed] [Google Scholar]
- Wan X, Wu S, Li Z, Dong Z, An X, Ma B, Tian Y, Li J (2019) Maize genic male-sterility genes and their applications in hybrid breeding: progress and perspectives. Mol Plant 12: 321–342 [DOI] [PubMed] [Google Scholar]
- Wan X, Wu S, Li Z, An X, Tian Y (2020) Lipid metabolism: critical roles in male fertility and other aspects of reproductive development in plants. Mol Plant 13:955–983. [DOI] [PubMed] [Google Scholar]
- Wang D, Oses-Prieto JA, Li KH, Fernandes JF, Burlingame AL, Walbot V (2010) The male sterile 8 mutation of maize disrupts the temporal progression of the transcriptome and results in the mis-regulation of metabolic functions. Plant J 63:939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Chen C, Pokhrel S, Ma W, Huang K, Patel P, Wang F, Xu J, Liu Z, Li J, et al. (2019) 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat Commun 10:627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, You S, Kong W, Tang Q, Bai W, Cai Y, Zheng H, Wang C, Jiang L, Wang C, et al. (2019) A GARP transcription factor anther dehiscence defected 1 (OsADD1) regulates rice anther dehiscence. Plant Mol Biol 101:403–414 [DOI] [PubMed] [Google Scholar]
- Xu J, Ding Z, Vizcay-Barrena G, Shi J, Liang W, Yuan Z, Werck-Reichhart D, Schreiber L, Wilson ZA, Zhang D (2014) ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell 26:1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang DB, Wilson ZA (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22:91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Makaroff CA, Ma H (2003) The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15:1281–1295 [PMC free article] [PubMed] [Google Scholar]
- Yang C, Xu Z, Song J, Conner K, Vizcay Barrena G, Wilson ZA. ( 2007) Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19:534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Zhang H, Arikit S, Huang K, Nan GL, Walbot V, Meyers BC. ( 2015) Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci USA 112:3146–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wu S, An X, Xie K, Dong Z, Zhou Y, Xu L, Fang W, Liu S, Liu S, et al. (2018) Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotech J 16: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DS, Liang WQ, Yuan Z, Li N, Shi J, Wang J, Liu YM, Yu WJ, Zhang DB (2008) Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol Plant 1: 599–610 [DOI] [PubMed] [Google Scholar]
- Zhang H, Egger RL, Kelliher T, Morrow D, Fernandes J, Nan GL, Walbot V (2014) Transcriptomes and proteomes define gene expression progression in pre-meiotic maize anthers. Genes Genomes Genetics 3:993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Ma X, Wang C, Li Q, Meyers BC, Springer NM, Walbot V (2021) CHH DNA methylation increases at 24‐PHAS loci depend on 24‐nt phasiRNAs in maize meiotic anthers. New Phytol 229:2984–2997 [DOI] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133:3085–3095 [DOI] [PubMed] [Google Scholar]
- Zheng S, Li J, Ma L, Wang H, Zhou H, Ni E, Jiang D, Liu Z, Zhuang C (2019) OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc Natl Acad Sci USA 116: 7549–7558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, He L, Liu Y, Mao Y, Wang C, Zhao B, Li Y, He H, Guo S, Zhang L, et al. (2020) A study of male fertility control in Medicago truncatula uncovers an evolutionarily conserved recruitment of two tapetal bHLH subfamilies in plant sexual reproduction. New Phytol 228:1115–1133 [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008) Defective in Tapetal Development and Function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55:266–277 [DOI] [PubMed] [Google Scholar]
- Zhu E, You C, Wang S, Cui J, Niu B, Wang Y, Qi J, Ma H, Chang F (2015) The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J 83:976–990 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.