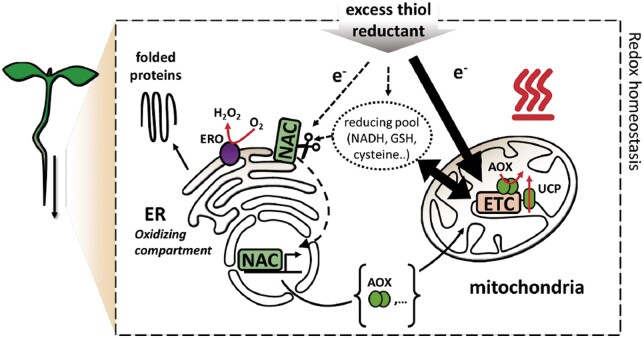
Maintaining the redox balance of each cell compartment is critical for cellular processes including protein maturation, signaling, and metabolism. The endoplasmic reticulum (ER) redox status is of paramount importance for proper folding of proteins in the secretory pathway. While antioxidant networks that buffer oxidative stress are well-studied, far less is known about reductive stress, which can be equally disruptive. In the cellular redox landscape, the mitochondrion is central because its electron transfer chain (mETC) constitutes a high-capacity electron sink. Some components of the mETC, such as the alternative oxidase AOX and uncoupling proteins, allow the mitochondrion to dissipate reducing power with little to no ATP generation. AOX1a in mitochondria is vital for redox balance in crosstalk with chloroplasts under light and drought stress (Giraud et al., 2008), but its functions in non-photosynthetic tissues have received less attention. In this issue of the Plant cell, Philippe Fuchs and colleagues (Fuchs et al., 2022) reveal a role of enhanced mitochondrial respiration in preserving ER function during reductive stress in Arabidopsis thaliana (see Figure).
Figure.
Hypothetical model for how the plant root copes with excess thiol reductant. Crosstalk between mitochondria and the ER allows the activation of a NAC transcription factor that enhances the mETC capacity to alleviate thiol-mediated reductive stress. This maintains an oxidized state in the ER, preventing protein misfolding. Adapted from Fuchs et al. (2022), Figure 8.
The authors observed that aox1a mutants exhibited greater impairment of root growth than the wild type upon treatment with dithiothreitol (DTT), a reducing reagent, pointing to a role for AOX1a in tolerance to thiol-based reductive stress. The authors hypothesized that the role of AOX1a might lie in its ability to increase mETC capacity while bypassing proton pumping. Uncoupling proteins also enable an increase in mETC activity by dissipating the proton gradient, and an uncoupling protein1 (ucp1) mutant showed similar hypersensitivity in root growth to DTT treatment as aox1a, reinforcing the role of enhanced mETC activity in counteracting reductive stress.
Fuchs et al. used fluorescent protein-based redox sensor (roGFP) variants to measure the redox potential of glutathione, the major thiol metabolite and the main DTT target. The ER was found to be the most oxidizing compartment and highly responsive to DTT treatment, whereas the cytosol, peroxisomes, and mitochondrial matrix maintained highly reducing glutathione pools. In addition, DTT treatment was associated with induced transcription of Unfold Protein Response (UPR) markers. This result indicates that DTT treatment affects oxidative protein folding in the ER. The main source of oxidative power is ER oxidoreductins (EROs) and ero mutants have lowered endogenous ER oxidation capacity. The authors showed that ero lines fail to grow after DTT treatment, reinforcing the hypothesis that DTT induces an ER reductive stress in roots. In aox1a and ucp1 lines, transcription of UPR markers was highly induced upon DTT treatment, similar to what was observed in ero mutant lines. This result suggests that ER acclimation to reductive stress depends on mitochondrial respiration. The root growth phenotype of an ero aox1a double mutant upon DTT treatment was more pronounced than that of either single mutant, further demonstrating the function of mitochondrial respiration in alleviating ER reductive stress.
To account for the observations, the authors hypothesized that the mETC can use DTT as an electron donor, thereby overloading the respiratory capacity. Indeed, the use of a genetically encoded biosensor showed that cytosolic NAD, the major mETC donor, became reduced during DTT treatment and that AOX1a absence exacerbated this response. Thanks to a genetically encoded pH sensor (cpYFP) and rhodamine 123, purified mitochondria were shown to build a proton gradient and a membrane potential when incubated with DTT. Using the same method, isolated mitochondria were found to use reduced glutathione, cysteine, and N-acetyl-cysteine, validating that the thiol group can be an electron donor for the mETC. Although exact mechanism for transfer of electron from thiol to mETC remains unclear, the rates observed were metabolically relevant, pointing to a considerable mitochondrial capacity for thiol detoxification.
AOX1a is upregulated upon H2O2 and antimycin A treatment and its induction is mediated by the ER-localized NAC domain containing protein 17 (ANAC017), a transcription factor that is released from the ER upon oxidative stress (Ng et al., 2013). Fuchs et al. showed that AOX1a is also upregulated upon DTT treatment in roots. Some anac017 mutant lines showed a similar root phenotype as the aox1a mutants under DTT and failed to fully induce AOX1a expression. This result suggests that ANAC017 retrograde signaling is involved in the reductive stress response to mediate AOX1a induction, boosting respiratory capacity.
Overall, this work leads to an interesting mechanistic model for redox balance and crosstalk between organelles as a reductive stress response (see Figure). Mitochondrion–ER redox crosstalk has been the focus of recent studies in yeast and mammalian cells (Hijazi et al., 2020) suggesting that underlying principles are conserved beyond plants.
References
- Fuchs P, Bohle F, Lichtenauer S, Ugalde JM, Feitosa Araujo E, Mansuroglu B, Ruberti C, Wagner S, Müller-Schüssele SJ, Meyer AJ, et al. (2022) Reductive stress triggers ANAC017-mediated retrograde signaling to safeguard the endoplasmic reticulum by boosting mitochondrial respiratory capacity. Plant Cell 34: 1375--1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Ho LH, Clifton R, Carroll A, Estavillo G, Tan YF, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al. (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijazi I, Knupp J, Chang A (2020) Retrograde signaling mediates an adaptive survival response to endoplasmic reticulum stress in Saccharomyces cerevisiae. J Cell Sci 133: jcs241539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, et al. (2013) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]