Abstract
The target of an antibody plays a significant role in the success of antibody-based therapeutics and diagnostics, and vaccine development. This importance is focused on the target binding site—epitope, where epitope selection as a part of design thinking beyond traditional antigen selection using whole cell or whole protein immunization can positively impact success. With purified recombinant protein production and peptide synthesis to display limited/selected epitopes, intrinsic factors that can affect the functioning of resulting antibodies can be more easily selected for. Many of these factors stem from the location of the epitope that can impact accessibility of the antibody to the epitope at a cellular or molecular level, direct inhibition of target antigen activity, conservation of function despite escape mutations, and even noncompetitive inhibition sites. By incorporating novel computational methods for predicting antigen changes to model-informed drug discovery and development, superior vaccines and antibody-based therapeutics or diagnostics can be easily designed to mitigate failures. With detailed examples, this review highlights the new opportunities, factors, and methods of predicting antigenic changes for consideration in sagacious epitope selection.
Keywords: antigen selection, epitope selection, antibody targeting, epitope accessibility, antibody engineering, protein engineering, drug targeting, model-informed drug discovery and development
Statement of Significance: Advances in protein engineering and antibody development have allowed focus on the target antigen for antibody-based design thinking to maximize the success of antibody development. Based on Model-Informed Drug Discovery and Development, considerations of epitope factors such as accessibility and locality allow for better epitope selection and interventions.
INTRODUCTION
Antibodies and their fragments are increasingly important in diagnostics and therapeutics development as evidenced in the ongoing COVID-19 pandemic [1, 2]. An already expensive process, diagnostics can fail owing to escape mutations on the epitope that compromise primer based kits [2–4] or diagnostic antibody binding [5], even with sagacious rational antibody design and engineering [6]. On therapeutics, antigenic epitope changes leading to escape mutations can contribute to drug failures. Thereby, to improve success, the Model Informed Drug Discovery and Development (MID3) [7, 8] has been in pilot by the U.S. Food and Drug Administration since 2018 [9] to support drug development [8, 10].
The ability to select the right single antigen for diagnostics, therapeutics, and to an extent that for vaccines targeting (e.g. choosing only the Spike over a whole virus), was augmented through recombinant technology, where purified target antigens could be produced and either injected into animals or used with in vitro antibody display methods e.g. phage display for antibody selection. The same technology also supported the targeting of specific epitopes on the antigen, where having antibodies specific to an epitope in a diagnostic kit can improve selectivity and specificity. This specificity is useful when differentiating between similar antigens e.g. between reverse transcriptase (RT) of viruses such as human immunodeficiency virus (HIV) and hepatitis B virus (HBV) or the RNA-dependent RNA polymerase (RdRp) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from Middle East Respiratory Syndrome (MERS), influenza, and severe acute respiratory syndrome (SARS). Yet, being too specific in diagnostics, as opposed for vaccinations, can also result in false negatives when the target epitope on the antigen mutates beyond antibody recognition [11].
With advances in peptide technology, short/stapled peptides can also be used without necessarily going to the deoxyribonucleic acid (DNA) level for recombinant expression and the commonly used whole cells or antigens [12, 13]. Recent developments in cyclic peptides and peptide vaccines further allow for immunization against specific conformational structures in epitopes instead of whole antigens through mimotopes [14] as B cell peptides [15]. Such methods of greater selectivity can support the development of therapeutics to reduce off-target effects, although some level of lower specificity could be of value for vaccines, and to an extent, diagnostics to target variants.
The selection of epitopes when incorporated into antibody design thinking is thus a paradigm shift from ‘chance-dependent’ antibody development to a more rational and purposeful approach. Given the dependence on the intended application in guiding toward higher specificity or to cater to minor changes, there are two categories [16] of (1) linear/continuous: defined as a stretch of amino acids sequences; (2) conformational/discontinuous: defined as sequence distal residues in close proximity through protein folding, with the latter conformational type more prevalent as B-cell epitopes.
About 96% of monoclonal antibody therapeutic candidates fail to make it to the market [17], costing close to tens of millions of dollars [18] for each failure, augmenting the MID3 approach that includes epitope prediction. Epitope prediction, traditionally based on amino acid physicochemical properties such as hydrophobicity, flexibility, solvent accessibility, and antigenicity [19–22], has seen augmentation by machine learning methods to show promise for cancer [23] and even hybrid experimental-computational approaches [24] involving deep neural network for major histocompatibility complex binding [25] and attention-based long short-term memory networks [26].
EPITOPES FACTORS
Accessible epitopes (cellular)
For epitope selection, particularly for therapeutics, accessibility of the epitope by the antibodies is perhaps the first and foremost consideration. In maximizing success, extracellular targets are typically picked for therapeutic antibody candidates with the exception of ‘intrabodies’ (where target cells produce the antibodies against intracellular antigens within itself [27]). Apart from intrabodies, whole antibodies against intracellular targets often have limited penetration of membranes (knowledge known to frequent flow cytometry users) requiring intracellular delivery methods [28, 29] not always feasible for therapeutics. Although unknown in prevalence, there are instances where intracellular oncogenic markers can be targeted because of externalization e.g. PRL3 [30]. Despite the cellular penetration handicap, antibodies have intrinsic advantages for targets unsuitable for small molecules [29] due to specificity requirement. Thus, personalized assessment of targeting intracellular proteins on a case-by-case basis.
With antibody intracellular delivery hurdles yet to be overcome, most current therapeutics and diagnostic whole antibody targets are extracellular antigens. There are many factors within extracellular targets such as posttranslational modification which can underlie the suitability and recognition of the epitopes. In one example, glycosylation has been shown to impede antibody recognition in both the ongoing COVID-19 pandemic [31] and HIV [32] when the modification occurred at epitope sites. Thus, even with good epitope bioinformatics-calculated prediction scores or good experimental results from bacterial produced non-glycosylated proteins (with rare exceptions), care must be taken for the effects of posttranslational modification on occluding or interfering with antibody recognition.
Epitope occlusion, in the form of cryptic epitope or cryptotopes [33], has been reported in numerous viruses such as the Norovirus [34], Influenza [35], Ebola [36], and HIV (Fig. 1A), where apart from posttranslational modifications methods, HIV utilized the hypervariable regions in the Env to occlude gp41 epitopes [37] from immune detection. On self-antigens, such occlusion of inducible cryptic epitopes plays a role in reducing autoimmunity [38], but in some cases can be exploited for differentiating disease states when the cryptotopes are exposed during pathogenesis e.g. prion disease [39]. In the unlikely situation where targeting the viral receptor is not easily achieved, blocking the viral target (host cell receptor) from viral spike binding can be performed, as in the case for SARS-CoV-2 [40, 41] and poliovirus [42]. However, care is needed to avoid affecting host cell activity by the antibody through over activation (e.g. by the mimicking of receptor stimulation and inducing of altered signaling, as well as the triggering of uncontrolled microthrombosis, cell lysis and neutrophil activation [43]) or preventing activation (e.g. inhibiting hyaluronan clearance by liver cells [44]).
Figure 1.

Accessibility of epitopes. (A) Targeting cryptotopes. (B) Accessibility of epitope to differing antibody isotypes. The location of the epitope on the target antigen can affect its accessibility by antibodies that are conjugated or multimeric in nature due to steric hindrances. This is evident in internal epitopes that would not be accessible to multimeric IgMs but accessible to monomeric Igs or antibody fragments. (C) Steric hindrances for multivalent binding by antibodies due to the location of the epitope on the target antigen. Created with BioRender.com.
Some antigens have the potential to bind antibodies at nonconventional complementarity determining regions by inducing binding pockets or the formation of stretches/patches on antibodies. This phenomenon was observed for nonconventional immunogenic molecules such as nickel [45] binding to trastuzumab and pertuzumab that can possibly underlie the disease pathogenesis of nickel type-I allergy. The other example of induced binding is in the molecular dynamics simulation of Trastuzumab binding to Her2, inducing a cryptotope that facilitated pertuzumab binding [46]. Although the synergism between the two clinical therapeutics (pertuzumab and trastuzumab) was shown experimentally to be due to the different epitopes on the same antigen without evidence of an induced pertuzumab epitope [47], the possibility of occluded epitopes ought to be sagaciously considered depending on the desired application and utility.
Accessible epitopes (molecular): Lessons from immunoglobulin M (IgM) for multi-specific antibodies
Apart from access at the intra/extracellular level and occluded/induced epitopes, obvious steric hindrances at the molecular level to access epitopes can impact the efficacy of the antibody (Fig. 1B). Although the earlier mentioned example [46, 47] showed synergistic binding of trastuzumab and pertuzumab simultaneously to their different epitopes on Her2, steric hindrances resulting from multiple whole or conjugated or multimeric antibodies are known to arise in flow cytometry [48].
When made into multimeric IgMs for multiple antigen binding, trastuzumab IgM could not have full occupancy of its Fab regions due to steric hindrances binding to multiple Her2. This was, however, not in the case for pertuzumab IgM [49], which showed higher avidity effects [50]. The sheer size of multimeric antibodies can be advantageous in agglutination for immune clearance and result in a better therapeutic or diagnostic than the traditional IgG, as evidenced in a nasal delivery for SARS-CoV-2 [51]. Yet, this advantage requires further studies for possible steric hindrance (Fig. 1C). With IgM being typically used in hemagglutination assays [52–55], one need to consider these steric effects on the accuracy of such IgM-based assays.
For this reason, the checklist for selecting any epitope for diagnostic or therapeutic application needs to account for molecular accessibility, especially when utilizing larger antibodies (whether conjugated, multimeric, or whole). It should be noted that the textbook primary antibody response is typically IgM and steric hindrances may underlie why IgMs tend to have lower affinities for the antigen.
Such steric hindrances also apply to the development of bi-specific antibodies requiring additional optimization [56] such as protein flexibility in accommodating antibodies as reflected recently in the Her2-Her3 extracellular dimer dynamics in the Her2-Her3-Nrg1β complex, which modulated receptor activity and accommodated trastuzumab and pertuzumab binding in vitro [57]. Nonetheless, the potential limitation of multi-specific antibodies by steric hindrances, especially those intended to engage whole cells [58] (the promises of bispecific T-cell engagers (BiTEs) for oncology [59]), should include accessibility as a major consideration. It is in this area that perhaps a more flexible antibody hinge at the constant heavy 1 domain of antibodies may alleviate some structural constraints as evidenced in immunoglobulin A [60] and other isotypes [50].
Conservation of epitopes: Lessons from viruses and omalizumab-immunoglobulin e
Since escape mutations in the epitope result in antibody recognition failure, one key criterion of epitope selection is its conservation and this occurs in at least two levels within the antigen: (1) in the presence of mutations and (2) conservation within the family to allow broad-spectrum targeting (Fig. 2). The earlier level of mutations within the species or viral type being important is exemplified in the recent COVID-19 pandemic, where SARS-CoV2 spike mutations led to decreased effectiveness of the early vaccines to novel variants [5]. On the intent for broad-spectrum protection, other viral vaccines such as that against the human papillomavirus found to induce cross-neutralizing antibodies [61] to its close relations are potential exploitation areas.
Figure 2.

Conservation of epitopes across highly mutable target antigens (top) would reduce the chance of escape mutations even as the pathogen accumulates mutations in the target antigen to prolong the effectiveness of the antibody in either detecting or neutralizing the target. Similarly, targeting conserved epitopes that are conserved in similar proteins (e.g. RT) across viruses or pathogens can allow broad-spectrum detection or therapeutic intervention (bottom), improving its market use, while also useful against emerging viruses utilizing replication enzymes e.g. RT or RdRp of the same protein family. Created with BioRender.com.
Likely due to faster escape mutations in microbial pathogens, there are more monoclonal antibody therapeutics used clinically against cancer than infectious diseases [62], where those targeting the latter tend to be polyclonal [63], perhaps in an attempt to cover more epitopes to mitigate escape. Nonetheless, in the search for conserved regions in pathogens, large sequence databases can provide some insights (such as HBVdb [64], Los Alamos Hepatitis C Virus [65], hemorrhagic fever virus [66], HIV [67, 68–69], Global Initiative on Sharing All Influenza Data [70], and Nextstrain [71]), but bearing in mind the constraints of extracellular targets for both neutralization of viruses for therapeutics and detection in diagnostics that do not require preprocessing to release internal intracellular contents. It should be noted that broad-spectrum targets of such nature are expectedly limited due to viral tropism.
With some foresight, overcoming cellular level accessibility issues e.g. intrabodies or novel delivery methods to target intracellular targets, conserved viral epitopes can expand to viral enzymes [72] for better search of potential broad-spectrum antivirals. This can be performed by searching for substrate analogues sites that indicate the presence of a functional domain [73, 74] that would be more conserved due to the preservation of enzyme functions.
In antibodies, the conserved target of omalizumab (Xolair®) to immunoglobulin E (IgE) fragment crystallizable (Fc) enabled it to be effective against type I allergies of varying allergen-specific IgEs [75]. Since the constant (C) region of antibodies is generally more conserved, omalizumab was sagaciously raised against the C-region, overlapping with the FcεRIα binding site. Although the existence of allotypes in the Cε-regions [76] can be an issue, other possible effects recently reported include the allosteric communication from the variable (V) region that could influence FcεRIα interaction [77]. Despite overlapping with the binding site of FcεRI, the V-region distal allosteric effects had negligible effects on omalizumab binding. This makes the omalizumab binding site a sagacious epitope that displayed the following benefits: (1) at a conserved region across various pathogenic factors (different allergen-specific IgEs); (2) at the allosterically insulated regions that are unaffected by the changes at hypervariable regions; and (3) relatively immutable.
Predicting new mutated epitopes: Lessons from viruses
The need for conserved epitopes in the face of mutations is evidenced by the challenge to existing COVID-19 vaccines by new variants [78]. In the lack of conserved epitopes, the alternative option is to preemptively predict possible escape mutations in the epitopes and design interventions that would remain effective against them. This is not only easily performed through recombinant or peptide methods discussed above, but even easier to implement as vaccines with the clinical use of mRNA vaccines [79].
Despite the lack of their reported incorporation for therapeutic or diagnostic development, the use of prediction methods in vaccines has been in place for a while with the example of the successful poliovirus RdRp in vitro platform [80] and similar attempts for HFMD causative enterovirus (EV) A71 [81]. These methods of predicting new mutations exhibit great promise despite being in vitro and can benefit from in silico augmentation involving network analysis [82], robust statistical model building [83], time series [84], stacking models [85], and random signature analysis [86, 87]. Nonetheless, it should be noted that EVs and lentiviruses like HIV mutate through recombination, whereas influenza utilizes the method of assortment (in the Orthomyxovirus family), requiring further research to get to a unified computational method capable of dealing with RNA viruses of different mutation methodologies. For reassortment viruses, it is important to consider that in silico simulations of the reassortment would be more efficient and safer to generate given the need of coinfection which can be constrained experimentally by biosafety and bioethics concerns in possible gain-of-function experiments. Regardless of the methodology approach, key features of tropism changes or polymerase activity in respiratory viruses, such as that mentioned for H5N8 [88] would be areas of importance to focus in species jumping and pandemic potential.
Experimentally, there is already notable progress in predicting mutations through the understanding of molecular biology and the innate biases of polymerases involved in replication (Fig. 3). Although such predictions are often deemed stochastic and unpredictable [89], recent evidence has shown that certain propensities in the genetic code [90] can confer a degree of predictability. It is with more knowledge about the target biology that renders mutations in the given target less stochastic and more predictable. This is demonstrated recently where even without immune or drug selection pressures, HIV RT displayed clear mutational biases in specific locations in HIV genes within a mimic of a single replication cycle [91]. Given that a high percentage of the generated mutations were found in patients, the rise of drug-resistant mutations in the HIV genes was not entirely stochastic nor required intense immune or drug selection pressures to emerge. As the ‘Godzilla’ of fast mutating viruses, HIV studies showed that the cross-resistance for protease inhibitors [92] and RT inhibitors [93] could be investigated by network analysis. Such analysis has showed how mutations can confer cross-resistance beyond one inhibitor. Thereby, in the vein of MID3, structural modeling studies can reveal the possible effects of emerging mutations with respect to drug resistance given the constraints of the mutations to the functional fitness of the target. It is also in this area that therapeutics which augments lethal mutagenesis for error catastrophe [94, 95] could be an alternative strategy for undruggable targets.
Figure 3.
Schematic of mutation prediction of RNA viruses that can be generated in vitro (adapted from Yeo et al. [91]) to determine innate viral polymerase mutation rates, hotspots, and biases for the incorporation into computational methods for better prediction. Such a platform would not only provide insights to the viral polymerases for augmentation toward lethal mutagenesis, but also improve preparedness to identify conserved regions and identify potential mutations of concern that can lead to escape to guide drug and diagnostic kit development. Created with BioRender.com.
Enabling better direct inhibition: Lessons from Her2 and IgM
Although alternative strategies exist in noncompetitive non-nucleoside RT inhibitors or non-nucleoside reverse transcriptase inhibitors (NNRTIs) [96] and the above-mentioned lethal-mutagenesis-based [97], the direct inhibition of targets is the most common therapeutic approach. This approach puts the focus of epitopes at the active/functional sites as key targets, to which the consideration of the antibody therapeutics over small molecules can augment the success given that larger molecules may inhibit binding better.
From the example of steric hindrances of trastuzumab and pertuzumab IgM to Her2, pertuzumab IgM inhibited cell proliferation better than pertuzumab IgG1 [49]. This was interesting because pertuzumab IgM being larger/multimeric, may have prevented Her2 homodimerization/activation (necessary for its oncogenic effects [98]) better than its IgG1 counterpart based on the stoichiometry of binding sites. This is especially so given that the experiments were performed in vitro without effector immune cell effects and that unlike the trastuzumab binding site [99], the pertuzumab binding site directly inhibited homodimerization [100].
Through steric hindrances by larger IgM molecules, the selection of epitopes directly at/near the active or dimerization sites allows for direct therapeutic inhibition as opposed to mere immune tagging for immune effector cells. When coupled with the use of multimeric or conjugated antibodies, better direct effects can be achieved (Fig. 4A). Such an approach is relevant not only in oncology where over expressed biomarkers lead to proliferation, but also in infectious diseases to neutralize virus attachment and entry [101]. With further understanding of viral spike proteins, epitopes that facilitate direct inhibition of potentially deadly viral pathological effects ought to be considered. Given the recent evidence showing the SARS-CoV-2 spike to also possess superantigenic characteristics that can cause hyperinflammation leading to multisystem inflammatory syndrome in children and cytokine storms in adult COVID-19 [102], such regions can be targeted through epitope selection of the region for mitigating the clinical morbidity, providing a much needed paradigm shift in treating viral infections [103] with pathogenesis roots in superantigenic activity. Although antibodies may already bind to such superantigenic motifs sites, further in-depth consideration of such recognition sites [104] will facilitate epitope selection for direct viral pathogenicity blocking in addition to that of attachment and entry.
Figure 4.
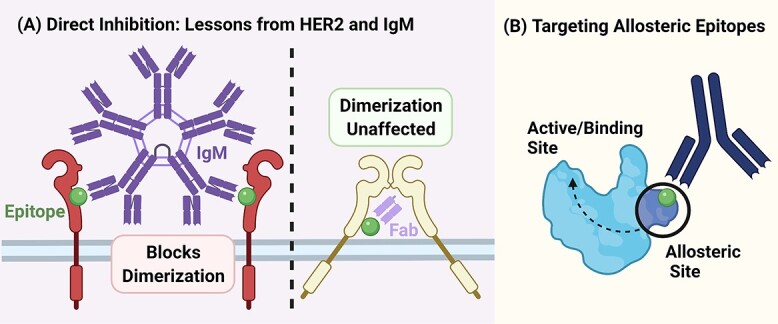
Choosing the epitopes for their potential functionality to (A) directly inhibit dimerization in the case of Pertuzumab IgM inhibiting Her2 dimerization better than its IgG counterpart due to its larger size [49]. In such cases, an antibody fragment such as a Fab alone may not necessarily provide sufficient steric hindrance to block dimerization. In the event where direct blocking may not be suitable, the targeting of (B) allosteric epitopes to influence active or functional sites can also be explored to increase target areas in the same target antigen. Created with BioRender.com.
Evident from oncology and HIV virology requiring multi-pronged interventions, where in the former, there is combinatorial synergistic use of trastuzumab and pertuzumab [47, 105], and in the latter, combinatorial antiretroviral therapy [106], there is still much to consider in additional epitopes.
The addition of more targetable sites within the same antigen especially in the absence of other ideal epitopes for direct intervention can open the way for allosteric epitopes. With clear lessons in the use of NNRTI and recent evidence of a non-enzymatic subunit being potentially druggable in HIV [107], distal epitopes can be used to influence the enzyme/target’s active sites by noncompetitive antibodies (Fig. 4B). Allosteric sites may not necessarily elicit major shape changes [108] and are thus often modulators than direct activators/inhibitors (see examples: IgE [77], FcεRI [109], IgA [60], microbial targets [110]). Such dampening of activity in oncology or virology can be what is precisely required at times to permit the necessary gain by the immune system. Furthermore, these allosteric sites in microbial and viral proteins could drive function-crippling mutations through drug resistance mechanisms at the drug binding site such as the non-cleavage site mutations contributing to drug resistance-associated protein fitness compensation in HIV-1 Gag [111, 112]. These sites could also be epitopes for intrabodies to prevent drug resistance or reduce protein fitness and be leveraged upon to dissociate already bound complexes by increasing dissociation (e.g. in the case of IgE-FcERI [109]).
With additional areas to target apart from active/functional sites, it may be easier to find common allosteric sites to develop broad-spectrum targeting antibodies. Such widening of the search field may overcome constraints present in extracellular localization. Although the allosteric analysis is currently most effectively performed computationally, it is important to be as inclusive of the entire protein structure/model as holistically possible to study the distal effects in a counterintuitive manner when compared with raising antibodies based on the reductionist approach in experiments of using purified recombinant proteins, regions, or peptides. Although new databases such as the Post Translational Modification Structural Database [113, 114] now including annotated posttranslational modification, the majority of crystal/nuclear magnetic resonance structures in structural databases without posttranslational modifications can result in incongruency of findings, especially when using cell lines with varying glycosylation patterns [115].
Applications, solutions, and conclusion—Holistic epitope selection
Sagacity in target and epitope can maximize the success of vaccine, therapeutics, and diagnostic development. Apart from deep understanding of the target biology, there are numerous factors ranging from accessibility at the cellular and molecular level, the inducibility, occlusion, conservation at family and species levels, influence by distal allosteric sites. Given the diversity of these factors, there will inevitably be difficulties for direct comparisons and implementations, especially when they should be considered holistically. Coupling such efforts with sagacious antibody development [6] involving swapping of elements [116], expensive developmental processes can be made more cost and time efficient.
ACKNOWLEDGEMENTS
Figures were created with BioRender.com. The authors would like to thank Zealyn Shi-Lin Heng for proofreading the manuscript.
Contributor Information
Samuel Ken-En Gan, Antibody & Product Development Lab, EDDC-BII, Agency for Science, Technology and Research (A*STAR), Singapore 138672, Singapore; APD SKEG Pte Ltd, Singapore 439444, Singapore.
Ser-Xian Phua, Antibody & Product Development Lab, EDDC-BII, Agency for Science, Technology and Research (A*STAR), Singapore 138672, Singapore.
Joshua Yi Yeo, Antibody & Product Development Lab, EDDC-BII, Agency for Science, Technology and Research (A*STAR), Singapore 138672, Singapore.
AUTHOR CONTRIBUTIONS
Conceptualization, S.K.-E.G.; Visualization, J.Y.Y. and S.-X.P.; Supervision, S.K.-E.G.; Writing—original draft, S.K.-E.G.; Writing—review & editing: S.K.-E.G., S.-X.P. and J.Y.Y.
FUNDING STATEMENT
The understanding that contributed to the insights were funded by Joint Council Office, Agency for Science, Technology, and Research, Singapore under Grant number JCO1334i00050. The staff are funded by the National Research Foundation (NRF) Singapore grant to Experimental Drug Development Centre (EDDC).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
DATA AVAILABILITY STATEMENT
No new data were generated or analysed in support of this research.
ETHICS AND CONSENT STATEMENT
Not applicable.
ANIMAL RESEARCH STATEMENT
Not applicable.
REFERENCES
- 1. El Jaddaoui, I, Allali, M, Raoui, S et al. A review on current diagnostic techniques for COVID-19. Expert Rev Mol Diagn 2021; 21: 141–60. [DOI] [PubMed] [Google Scholar]
- 2. Alpdagtas, S, Ilhan, E, Uysal, E et al. Evaluation of current diagnostic methods for COVID-19. APL Bioeng 2020; 4: 041506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. U.S. Food and Drug Administration . Genetic variants of SARS-CoV-2 may lead to false negative results with molecular tests for detection of SARS-CoV-2 - letter to clinical laboratory staff and health care providers. 2021. [https://www.fda.gov/medical-devices/letters-health-care-providers/genetic-variants-sars-cov-2-may-lead-false-negative-results-molecular-tests-detection-sars-cov-2].
- 4. U.S. Food and Drug Administration . SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. 2021. [https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests]
- 5. Eguia, RT, Crawford, KHD, Stevens-Ayers, T et al. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog 2021; 17: e1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ling, W-L, Lua, W-H, Gan, SK-E. Sagacity in antibody humanization for therapeutics, diagnostics and research purposes: considerations of antibody elements and their roles. Antibody Therapeutics 2020; 3: 71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krishnaswami, S, Austin, D, Della Pasqua, O et al. MID3: mission impossible or model-informed drug discovery and development? Point-counterpoint discussions on key challenges. Clin Pharmacol Ther 2020; 107: 762–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marshall, S, Madabushi, R, Manolis, E et al. Model-informed drug discovery and development: current industry good practice and regulatory expectations and future perspectives. CPT Pharmacometrics Syst. Pharmacol 2019; 8: 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. U.S. Food and Drug Administration . Model-Informed Drug Development Pilot Program. 2021. [https://www.fda.gov/drugs/development-resources/model-informed-drug-development-pilot-program]
- 10. Sou, T, Soukarieh, F, Williams, P et al. Model-informed drug discovery and development in pulmonary delivery: biopharmaceutical Pharmacometric Modeling for formulation evaluation of pulmonary suspensions. ACS Omega 2020; 5: 25733–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ascoli, CA. Could mutations of SARS-CoV-2 suppress diagnostic detection? Nat Biotechnol 2021; 39: 274–5. [DOI] [PubMed] [Google Scholar]
- 12. Meuleman, TJ, Dunlop, JI, Owsianka, AM et al. Immobilization by surface conjugation of cyclic peptides for effective mimicry of the HCV-envelope E2 protein as a strategy toward synthetic vaccines. Bioconjug Chem 2018; 29: 1091–101. [DOI] [PubMed] [Google Scholar]
- 13. Li, W, Joshi, MD, Singhania, S et al. Peptide vaccine: progress and challenges. Vaccine 2014; 2: 515–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geysen, HM, Rodda, SJ, Mason, TJ. A priori delineation of a peptide which mimics a discontinuous antigenic determinant. Mol Immunol 1986; 23: 709–15. [DOI] [PubMed] [Google Scholar]
- 15. Tobias, J, Battin, C, De Sousa Linhares, A et al. A new strategy toward B cell-based cancer vaccines by active immunization with Mimotopes of immune checkpoint inhibitors. Front Immunol 2020; 11: 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanchez-Trincado, JL, Gomez-Perosanz, M, Reche, PA. Fundamentals and methods for T- and B-cell epitope prediction. J Immunol Res 2017; 2017: 2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hingorani, AD, Kuan, V, Finan, C et al. Improving the odds of drug development success through human genomics: modeling study. Sci Rep 2019; 9: 18911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grignolo, A, Pretorius, S. Phase III trial failures: costly, but preventable. Appl Clin Trials 2016; 25: 36–42. [Google Scholar]
- 19. Kolaskar, AS, Tongaonkar, PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett 1990; 276: 172–4. [DOI] [PubMed] [Google Scholar]
- 20. Emini, EA, Hughes, JV, Perlow, DS et al. Induction of hepatitis a virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol 1985; 55: 836–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karplus, PA, Schulz, GE. Prediction of chain flexibility in proteins. Naturwissenschaften 1985; 72: 212–3. [Google Scholar]
- 22. Parker, JMR, Guo, D, Hodges, RS. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and x-ray-derived accessible sites. Biochemistry 1986; 25: 5425–32. [DOI] [PubMed] [Google Scholar]
- 23. Smith, CC, Chai, S, Washington, AR et al. Machine-learning prediction of tumor immunogenicity in the selection of therapeutic epitopes. Cancer Immunol Res 2019; 7: 1591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Denisova, GF, Denisov, DA, Bramson, JL. Applying bioinformatics for antibody epitope prediction using affinity-selected mimotopes relevance for vaccine design. Immunome Res 2010; 6: S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Phloyphisut, P, Pornputtapong, N, Sriswasdi, S et al. MHCSeqNet: a deep neural network model for universal MHC binding prediction. BMC Bioinformatics 2019; 20: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Noumi, T, Inoue, S, Fujita, H et al. Epitope prediction of antigen protein using attention-based LSTM network. J Inform Process 2021; 29: 321–7. [Google Scholar]
- 27. Trenevska, I, Li, D, Banham, AH. Therapeutic antibodies against intracellular tumor antigens. Front Immunol 2017; 8: 1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singh, K, Ejaz, W, Dutta, K et al. Antibody delivery for intracellular targets: emergent therapeutic potential. Bioconjug Chem 2019; 30: 1028–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Slastnikova, TA, Ulasov, AV, Rosenkranz, AA et al. Targeted intracellular delivery of antibodies: the state of the art. Front Pharmacol 2018; 9: 1208.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thura, M, Al-Aidaroos, AQ, Gupta, A et al. PRL3-zumab as an immunotherapy to inhibit tumors expressing PRL3 oncoprotein. Nat Commun 2019; 10: 2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wintjens, R, Bifani, AM, Bifani, P. Impact of glycan cloud on the B-cell epitope prediction of SARS-CoV-2 spike protein. npj Vaccines 2020; 5: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davis, D, Stephens, DM, Willers, C et al. Glycosylation governs the binding of Antipeptide antibodies to regions of hypervariable amino acid sequence within recombinant gp120 of human immunodeficiency virus type 1. J Gen Virol 1990; 71: 2889–98. [DOI] [PubMed] [Google Scholar]
- 33. Rieder, FJJ, Biebl, J, Kastner, M-T et al. Microbial Cryptotopes are prominent targets of B-cell immunity. Sci Rep 2016; 6: 31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindesmith, LC, Mallory, ML, Debbink, K et al. Conformational occlusion of blockade antibody epitopes, a novel mechanism of GII.4 human norovirus immune evasion. mSphere 2018; 3: e00518–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adachi, Y, Tonouchi, K, Nithichanon, A et al. Exposure of an occluded hemagglutinin epitope drives selection of a class of cross-protective influenza antibodies. Nat Commun 2019; 10: 3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Francica, JR, Varela-Rohena, A, Medvec, A et al. Steric shielding of surface epitopes and impaired immune recognition induced by the Ebola virus glycoprotein. PLoS Pathog 2010; 6: e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moscoso, CG, Xing, L, Hui, J et al. Trimeric HIV Env provides epitope occlusion mediated by hypervariable loops. Sci Rep 2014; 4: 7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Warnock, MG, Goodacre, JA. Cryptic T-cell epitopes and their role in the pathogenesis of autoimmune diseases. Rheumatology 1997; 36: 1144–50. [DOI] [PubMed] [Google Scholar]
- 39. Kang, H-E, Bian, J, Kane, SJ et al. Incomplete glycosylation during prion infection unmasks a prion protein epitope that facilitates prion detection and strain discrimination. J Biol Chem 2020; 295: 10420–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Starr, TN, Czudnochowski, N, Liu, Z et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 2021; 597: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tortorici, MA, Czudnochowski, N, Starr, TN et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 2021; 597: 103–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Minor, PD, Pipkin, PA, Hockley, D et al. Monoclonal antibodies which block cellular receptors of poliovirus. Virus Res 1984; 1: 203–12. [DOI] [PubMed] [Google Scholar]
- 43. Ludwig, RJ, Vanhoorelbeke, K, Leypoldt, F et al. Mechanisms of autoantibody-induced pathology. Front Immunol 2017; 8: 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weigel, JA, Raymond, RC, McGary, C et al. A blocking antibody to the Hyaluronan receptor for endocytosis (HARE) inhibits Hyaluronan clearance by perfused liver *. J Biol Chem 2003; 278: 9808–12. [DOI] [PubMed] [Google Scholar]
- 45. Su, CT-T, Lua, W-H, Poh, J-J et al. Molecular insights of nickel binding to therapeutic antibodies as a possible new antibody superantigen. Front Immunol 2021; 12: 676048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fuentes, G, Scaltriti, M, Baselga, J et al. Synergy between trastuzumab and pertuzumab for human epidermal growth factor 2 (Her2) from colocalization: an in silicobased mechanism. Breast Cancer Res 2011; 13: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lua, W-H, Gan, SK-E, Lane, DP et al. A search for synergy in the binding kinetics of Trastuzumab and Pertuzumab whole and F(ab) to Her2. Npj. Breast Cancer 2015; 1: 15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matos, DM. Steric hindrance: a practical (and frequently forgotten) problem in flow cytometry. Cytometry B Clin Cytom 2021; 100: 397–401. [DOI] [PubMed] [Google Scholar]
- 49. Samsudin, F, Yeo, JY, Gan, SK-E et al. Not all therapeutic antibody isotypes are equal: the case of IgM versus IgG in Pertuzumab and Trastuzumab. Chem Sci 2020; 11: 2843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lua, W-H, Ling, W-L, Yeo, JY et al. The effects of antibody engineering CH and CL in Trastuzumab and Pertuzumab recombinant models: impact on antibody production and antigen-binding. Sci Rep 2018; 8: 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yerabham, A, Ho, M. A novel IgM intranasal intervention against SARS-CoV-2. Antibody Ther 2021; 4: 171–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Duffy, KT, Wharton, PJ, Johnson, JD et al. Assessment of immunoglobulin-M immunosorbent agglutination assay (ISAGA) for detecting toxoplasma specific IgM. J Clin Pathol 1989; 42: 1291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cambiaso, CL, Galanti, LM, Leautaud, P et al. Latex agglutination assay of human immunoglobulin M antitoxoplasma antibodies which uses enzymatically treated antigen-coated particles. J Clin Microbiol 1992; 30: 882–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ryu, W-S. In: Ryu, W-S (ed). Molecular Virology of Human Pathogenic Viruses. Boston: Academic Press, 2017, 47–62 [Google Scholar]
- 55. Payne, S. In: Payne, S (ed). Viruses: From Understanding to Investigation. London: Academic Press, 2017, 37–52 [Google Scholar]
- 56. Mazor, Y, Hansen, A, Yang, C et al. Insights into the molecular basis of a bispecific antibody's target selectivity. MAbs 2015; 7: 461–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Diwanji, D, Trenker, R, Thaker, TM et al. Structures of the HER2–HER3–NRG1β complex reveal a dynamic dimer interface. Nature 2021; 600: 339–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Haber, L, Olson, K, Kelly, MP et al. Generation of T-cell-redirecting bispecific antibodies with differentiated profiles of cytokine release and biodistribution by CD3 affinity tuning. Sci Rep 2021; 11: 14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Einsele, H, Borghaei, H, Orlowski, RZ et al. The BiTE (bispecific T-cell engager) platform: development and future potential of a targeted immuno-oncology therapy across tumor types. Cancer 2020; 126: 3192–201. [DOI] [PubMed] [Google Scholar]
- 60. Su, CT-T, Lua, W-H, Ling, W-L et al. Allosteric effects between the antibody constant and variable regions: a study of IgA fc mutations on antigen binding. Antibodies 2018; 7: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kemp, TJ, Hildesheim, A, Safaeian, M et al. HPV16/18 L1 VLP vaccine induces cross-neutralizing antibodies that may mediate cross-protection. Vaccine 2011; 29: 2011–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lu, R-M, Hwang, Y-C, Liu, IJ et al. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci 2020; 27: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mullard, A. FDA approves antibody cocktail for Ebola virus. Nat Rev Drug Discov 2020; 19: 827. [DOI] [PubMed] [Google Scholar]
- 64. Hayer, J, Jadeau, F, Deléage, G et al. HBVdb: a knowledge database for hepatitis B virus. Nucleic Acids Res 2012; 41: D566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kuiken, C, Yusim, K, Boykin, L et al. The Los Alamos hepatitis C sequence database. Bioinformatics 2004; 21: 379–84. [DOI] [PubMed] [Google Scholar]
- 66. Kuiken, C, Thurmond, J, Dimitrijevic, M et al. The LANL hemorrhagic fever virus database, a new platform for analyzing biothreat viruses. Nucleic Acids Res 2011; 40: D587–D592.ss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Los Alamos National Laboratory . HIV Sequence Database, 2021. [http://www.hiv.lanl.gov/]
- 68. Kuiken, C, Korber, B, Shafer, RW. HIV sequence databases. AIDS Rev 2003; 5: 52–61. [PMC free article] [PubMed] [Google Scholar]
- 69. Kantor, R, Machekano, R, Gonzales, MJ et al. Human immunodeficiency virus reverse transcriptase and protease sequence database: an expanded data model integrating natural language text and sequence analysis programs. Nucleic Acids Res 2001; 29: 296–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shu, Y, McCauley, J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance 2017; 22: 30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hadfield, J, Megill, C, Bell, SM et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34: 4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bottoms, CA, White, TA, Tanner, JJ. Exploring structurally conserved solvent sites in protein families. Proteins 2006; 64: 404–21. [DOI] [PubMed] [Google Scholar]
- 73. Madu, IG, Roth, SL, Belouzard, S et al. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol 2009; 83: 7411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sundaravaradan, V, Hahn, T, Ahmad, N. Conservation of functional domains and limited heterogeneity of HIV-1 reverse transcriptase gene following vertical transmission. Retrovirology 2005; 2: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Okayama, Y, Matsumoto, H, Odajima, H et al. Roles of omalizumab in various allergic diseases. Allergol Int 2020; 69: 167–77. [DOI] [PubMed] [Google Scholar]
- 76. van Loghem, E, Aalberse, RC, Matsumoto, H. A genetic marker of human IgE heavy chains, Em(1)1. Vox Sang 1984; 46: 195–206. [DOI] [PubMed] [Google Scholar]
- 77. Lua, W-H, Su, CT-T, Yeo, JY et al. Role of the IgE variable heavy chain in FcεRIα and superantigen binding in allergy and immunotherapy. J Allergy Clin Immunol 2019; 144: 514–523.e515. [DOI] [PubMed] [Google Scholar]
- 78. Wang, R, Chen, J, Gao, K et al. Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics 2021; 113: 2158–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pardi, N, Hogan, MJ, Porter, FW et al. mRNA vaccines — a new era in vaccinology. Nat Rev Drug Discov 2018; 17: 261–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Weeks, SA, Lee, CA, Zhao, Y et al. A polymerase mechanism-based strategy for viral attenuation and vaccine development. J Biol Chem 2012; 287: 31618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen, T-C, Chang, H-Y, Lin, P-F et al. Novel antiviral agent DTriP-22 targets RNA-dependent RNA polymerase of enterovirus 71. Antimicrob Agents Chemother 2009; 53: 2740–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ozturk, K, Dow, M, Carlin, DE et al. The emerging potential for network analysis to inform precision cancer medicine. J Mol Biol 2018; 430: 2875–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Özcan ŞİmŞek, NÖ, ÖzgÜr, A, GÜrgen, F. Statistical representation models for mutation information within genomic data. BMC Bioinformatics 2019; 20: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yin, R, Luusua, E, Dabrowski, J et al. Tempel: time-series mutation prediction of influenza a viruses via attention-based recurrent neural networks. Bioinformatics 2020; 36: 2697–704. [DOI] [PubMed] [Google Scholar]
- 85. Yin, R, Tran, VH, Zhou, X et al. Predicting antigenic variants of H1N1 influenza virus based on epidemics and pandemics using a stacking model. PLoS One 2018; 13: e0207777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Goh, WWB, Wong, L. Why breast cancer signatures are no better than random signatures explained. Drug Discov Today 2018; 23: 1818–23. [DOI] [PubMed] [Google Scholar]
- 87. Omichessan, H, Severi, G, Perduca, V. Computational tools to detect signatures of mutational processes in DNA from tumours: a review and empirical comparison of performance. PLoS One 2019; 14: e0221235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yeo, JY, Gan, SK-E. Peering into avian influenza a(H5N8) for a framework towards pandemic preparedness. Viruses 2021; 13: 2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gold, B. Somatic mutations in cancer: stochastic versus predictable. Mutat Res Genet Toxicol Environ Mutagen 2017; 814: 37–46. [DOI] [PubMed] [Google Scholar]
- 90. Chan, K-F, Koukouravas, S, Yeo, JY et al. Probability of change in life: amino acid changes in single nucleotide substitutions. Biosystems 2020; 193-194: 104135. [DOI] [PubMed] [Google Scholar]
- 91. Yeo, JY, Koh, DW-S, Yap, P et al. Spontaneous mutations in HIV-1 gag, protease, RT p66 in the first replication cycle and how they appear: insights from an in vitro assay on mutation rates and types. Int J Mol Sci 2021; 22: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Su, CT-T, Ling, W-L, Lua, W-H et al. Structural analyses of 2015-updated drug-resistant mutations in HIV-1 protease: an implication of protease inhibitor cross-resistance. BMC Bioinformatics 2016; 17: 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chiang, RZ-H, Gan, SK-E, Su, CT-T. A computational study for rational HIV-1 non-nucleoside reverse transcriptase inhibitor selection and the discovery of novel allosteric pockets for inhibitor design. Biosci Rep 2018; 38: BSR20171113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Summers, J, Litwin, S. Examining the theory of error catastrophe. J Virol 2006; 80: 20–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bull, JJ, Sanjuán, R, Wilke, CO. Theory of lethal mutagenesis for viruses. J Virol 2007; 81: 2930–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Usach, I, Melis, V, Peris, J-E. Non-nucleoside reverse transcriptase inhibitors: a review on pharmacokinetics, pharmacodynamics, safety and tolerability. J Int AIDS Soc 2013; 16: 18567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Perales, C, Domingo, E. In: Domingo, E, Schuster, P (eds). Quasispecies: From Theory to Experimental Systems. Cham: Springer International Publishing, 2016, 323–39 [Google Scholar]
- 98. Brennan, PJ, Kumogai, T, Berezov, A et al. HER2/Neu: mechanisms of dimerization/oligomerization. Oncogene 2000; 19: 6093–101. [DOI] [PubMed] [Google Scholar]
- 99. Cho, H-S, Mason, K, Ramyar, KX et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin fab. Nature 2003; 421: 756–60. [DOI] [PubMed] [Google Scholar]
- 100. Franklin, MC, Carey, KD, Vajdos, FF et al. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 2004; 5: 317–28. [DOI] [PubMed] [Google Scholar]
- 101. Corti, D, Lanzavecchia, A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol 2013; 31: 705–42. [DOI] [PubMed] [Google Scholar]
- 102. Cheng, MH, Zhang, S, Porritt, RA et al. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc Natl Acad Sci U S A 2020; 117: 25254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Scaglioni, V, Soriano, ER. Are superantigens the cause of cytokine storm and viral sepsis in severe COVID-19? Observations and hypothesis. Scand J Immunol 2020; 92: e12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Deacy, AM, Gan, SK-E, Derrick, J. Superantigen recognition and interactions: functions, mechanisms and applications (provisionally accepted). Front Immunol 2021; 12: 731845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Maly, JJ, Macrae, ER. Pertuzumab in combination with Trastuzumab and chemotherapy in the treatment of HER2-positive metastatic breast cancer: safety, efficacy, and progression free survival. Breast Cancer 2014; 8: BCBCR.S9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bhatti, AB, Usman, M, Kandi, V. Current scenario of HIV/AIDS, treatment options, and major challenges with compliance to antiretroviral therapy. Cureus 2016; 8: e515–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Chan, K-F, Su, CT-T, Krah, A et al. An alternative HIV-1 non-nucleoside reverse transcriptase inhibition mechanism: targeting the p51 subunit. Molecules 2020; 25: 5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Tsai, C-J, del Sol, A, Nussinov, R. Allostery: absence of a change in shape does not imply that Allostery is not at play. J Mol Biol 2008; 378: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Phua, S-X, Chan, K-F, Su, CT-T et al. Perspective: the promises of a holistic view of proteins—impact on antibody engineering and drug discovery. Biosci Rep 2019; 39: BSR20181958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Blaise, P, Clevenbergh, P, Vaira, D et al. HIV resistance to antiretroviral drugs: mechanisms, genotypic and phenotypic resistance testing in clinical practice. Acta Clin Belg 2002; 57: 191–201. [DOI] [PubMed] [Google Scholar]
- 111. Su, CT-T, Kwoh, C-K, Verma, CS et al. Modeling the full length HIV-1 gag polyprotein reveals the role of its p6 subunit in viral maturation and the effect of non-cleavage site mutations in protease drug resistance. J Biomol Struct Dyn 2018; 36: 4366–77. [DOI] [PubMed] [Google Scholar]
- 112. Samsudin, F, Gan, SK-E, Bond, PJ. The impact of gag non-cleavage site mutations on HIV-1 viral fitness from integrative modelling and simulations. Comput Struct Biotechnol J 2021; 19: 330–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Arakhamia, T, Lee, CE, Carlomagno, Y et al. Posttranslational modifications mediate the structural diversity of Tauopathy strains. Cell 2020; 180: 633–644.e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Craveur, P, Rebehmed, J, de Brevern, AG. PTM-SD: a database of structurally resolved and annotated posttranslational modifications in proteins. Database 2014; 2014: bau041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Böhm, E, Seyfried, BK, Dockal, M et al. Differences in N-glycosylation of recombinant human coagulation factor VII derived from BHK, CHO, and HEK293 cells. BMC Biotechnol 2015; 15: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ling, W-L, Lua, W-H, Poh, J-J et al. Effect of VH–VL families in Pertuzumab and Trastuzumab recombinant production, Her2 and FcγIIA binding. Front Immunol 2018; 9: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.



