Abstract
The ampC and ampR genes of Enterobacter cloacae GN7471 were cloned into pMW218 to yield pKU403. Four mutant plasmids derived from pKU403 (pKU404, pKU405, pKU406, and pKU407) were isolated in an AmpD mutant of Escherichia coli ML4953 by selection with ceftazidime or aztreonam. The β-lactamase activities expressed by pKU404, pKU405, pKU406, and pKU407 were about 450, 75, 160, and 160 times higher, respectively, than that expressed by the original plasmid, pKU403. These mutant plasmids all carried point mutations in the ampR gene. In pKU404 and pKU405, Asp-135 was changed to Asn and Val, respectively. In both pKU406 and pKU407, Arg-86 was changed to Cys. The ease of selection of AmpR mutations at a frequency of about 10−6 in this study strongly suggests that derepressed strains, such as AmpD or AmpR mutants, could frequently emerge in the clinical setting.
Chromosomal class C β-lactamase is an inducible enzyme produced by Enterobacter cloacae and many other gram-negative bacilli (4, 14, 17, 29, 31, 38). The AmpD, AmpG, and AmpR proteins are reported to be involved in the induction of class C β-lactamase (32, 33, 35).
AmpD is a novel N-acetylmuramyl-l-alanine amidase that participates in the intercellular recycling of peptidoglycan fragments (11, 15). AmpD degrades cytoplasmic 1,6-anhydro-N-acetylmuramyl-tripeptide (1,6-anhMurNAc-tripeptide) to release the tripeptide l-Ala-d-Glu-meso-diaminopimelic acid (meso-DAP) for direct utilization in the construction of new peptidoglycans (15, 16). An ampD mutation that results in β-lactamase expression even in the absence of a β-lactamase inducer coincides with the accumulation of 1,6-anhMurNAc-tripeptide (15). Inactivation of AmpD leads to semiconstitutive or hyperinducible overproduction of AmpC in Citrobacter freundii and E. cloacae (8, 19, 21). On the other hand, AmpD mutants with increased levels of β-lactamase expression show one of three phenotypes (hyperinducible, derepressed, and partially derepressed), which are associated with different mutations or which may depend on environmental regulation of unknown genes (40). AmpG is a transmembrane protein involved in the permease for an N-acetylglucosaminyl (GluNAc)-1,6-anhMurNAc-tripeptide (15, 25). Dietz et al. (7) have reported that AmpG primarily affects aD-pentapeptide (disaccharide-pentapeptide; GluNAc-1,6-anhMurNAc-l-Ala-d-Glu-meso-DAP-d-Ala-d-Ala), a periplasmic muropeptide that is converted into the cytoplasmic signaling molecule for β-lactamase induction, aM-pentapeptide (monosaccharide-pentapeptide; L1,6-anhMurNAc-l-Ala-d-Glu-meso-DAP-d-Ala-d-Ala) (7). Without ampG, neither induction nor high-level expression of β-lactamase is possible (20). AmpR acts as a transcriptional activator by binding to a DNA region immediately upstream of the ampC promoter (2, 12, 24). In the absence of a β-lactam inducer, AmpR represses the synthesis of β-lactamase by 2.5-fold, whereas expression is induced 10- to 200-fold in the presence of a β-lactam inducer (22, 23). On the other hand, many clinical isolates of the family Enterobacteriaceae show high-level production of class C β-lactamase even without induction.
In the present study, we selected mutant strains by culture with an expanded-spectrum cephalosporin and a monobactam and examined the genetic background of ampC and ampR mutations that conferred high levels of resistance to β-lactam antibiotics, as well as compared the enzyme activity with that of the parental strain. The possible mechanisms by which these mutant strains had a strong response to an expanded-spectrum cephalosporin and a monobactam are discussed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. pACYC184 and pMW218 are vector plasmids that confer resistance to tetracycline-chloramphenicol and kanamycin, respectively, and were purchased from Nippon Gene (Tokyo, Japan) (5). pMW218 was derived from pSC101 (3).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. cloacae | ||
| GN7471 | Clinical isolate from Japan, resistance to cephaloridine and cefotiam | 30 |
| KU3261 | Clinical isolate from Japan | This study |
| KU3262 | Clinical isolate from Japan | This study |
| KU3263 | Clinical isolate from Japan | This study |
| ATCC 13047 | Purchased from ATCC; type strain, RF4738 | |
| E. coli | ||
| ML4947 | F−galK2 galT22 hsdR hsdM lacY1 metB1 relA supE44 RIFr | 13 |
| ML4953 | F−ampD9 galK2 galT22 hsdR hsdM lacY1 metB1 relA supE44 RIFr | ampD9 is deb9 (27) |
| Plasmids | ||
| pMS161 | 8-kb EcoRI fragment containing ampC and ampR from GN7471 cloned into pACYC184 | This study |
| pKU403 | 6-kb SalI fragment containing ampC and ampR from pMS161 cloned into pMW218 | This study |
| pKU404 | Mutant from pKU403 selected with aztreonam | This study |
| pKU405 | Mutant from pKU403 selected with ceftazidime | This study |
| pKU406 | Mutant from pKU403 selected with aztreonam | This study |
| pKU407 | Mutant from pKU403 selected with ceftazidime | This study |
| pKU408 | Prepared by deleting 1.7-kb SphI fragment containing ampR from pKU403 | This study |
| pKU409 | Prepared by deleting 1.7-kb SphI fragment containing ampR from pKU404 | This study |
| pKU410 | Prepared by deleting 1.7-kb SphI fragment containing ampR from pKU405 | This study |
| pKU414 | Prepared by deleting 1.7-kb SphI fragment containing ampR from pKU406 | This study |
| pKU411 | Prepared by deleting 4-kb BamHI-ScaI fragment containing ampC from pKU403 | This study |
| pKU412 | Prepared by deleting 4-kb BamHI-ScaI fragment containing ampC from pKU404 | This study |
| pKU413 | Prepared by deleting 4-kb BamHI-ScaI fragment containing ampC from pKU405 | This study |
| pKU415 | Prepared by deleting 4-kb BamHI-ScaI fragment containing ampC from pKU406 | This study |
| pACYC184 | Cloning vector, purchased from Nippon Gene (Tokyo, Japan); CPr TCr | 5 |
| pMW218 | Cloning vector, purchased from Nippon Gene (Tokyo, Japan); KMr | 3 |
RIF, rifampin; CP, chloramphenicol; TC, tetracyclin; KM, kanamycin; r, resistance; ATCC, American Type Culture Collection.
Antibiotics.
Reference samples of various antibiotics of known potency were kindly supplied in powder form by the respective manufacturers, as follows: ampicillin, Meiji Seika (Tokyo, Japan); cephaloridine, Shionogi (Osaka, Japan); cefotaxime, Nippon Hoechst Marion Roussel (Tokyo, Japan); cefotiam, Takeda Chemical Industries (Osaka, Japan); ceftazidime, Nippon Glaxo (Tokyo, Japan); aztreonam, Eisai (Tokyo, Japan); latamoxef, Shionogi; cefpodoxime, Sankyo (Tokyo, Japan); imipenem, Banyu Pharmaceutical (Tokyo, Japan); cefepime, Bristol-Myers Squibb K. K. (Tokyo, Japan); and kanamycin, Meiji Seika.
Determination of antibiotic sensitivity.
The MICs of the antibiotics were determined by the agar dilution method. Briefly, an overnight culture in Muller-Hinton broth (Nissui, Tokyo, Japan) was diluted to about 5 × 107 CFU/ml and was inoculated onto agar plates containing various concentrations of the test antibiotic by using an inoculating device which applied spots of bacterial suspensions containing 5 × 104 CFU.
Transformation of Escherichia coli.
Plasmid DNAs were isolated and were used to transform E. coli ML4947 (AmpD wild type) and ML4953 (AmpD mutant), as well as E. cloacae ATCC 13047 and clinical isolates of E. cloacae, by electroporation (6, 34, 37).
Cloning of ampC and ampR genes.
Genomic DNA was purified by the procedure of Marmur (28). Plasmid DNA was purified by extracting plasmid DNA by the small-scale alkaline method (37). Restriction enzymes and T4 DNA ligase were purchased from Takara shuzo (Kyoto, Japan) and Nippon Gene, respectively. The plasmid size was calculated from the sizes of the fragments obtained by cleaving the plasmid with restriction enzymes and by using λ phage DNA cleaved with HindIII as a molecular marker. PCR primers were obtained from Amersham Pharmacia Biotech (Tokyo, Japan). PCR was carried out according to the instructions with the GeneAmp PCR reagent kit (Perkin-Elmer Cetus, Emeryville, Calif.). All PCRs were performed on a Perkin-Elmer Cetus DNA thermal cycler (model 480) (34, 36).
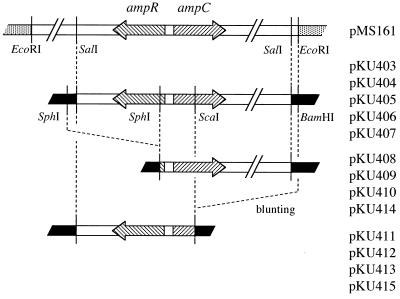
The genomic DNA from E. cloacae GN7471 was digested with EcoRI. The digested genomic DNAs were shotgun cloned into EcoRI-digested plasmid pACYC184 and were used to transform E. coli ML4947. Transformants with a plasmid carrying the E. cloacae genomic 8-kb fragment (containing ampC and ampR) were selected for increased resistance to cephaloridine. This hybrid plasmid (12 kb) was designated pMS161 (Fig. 1). pMS161 was digested with SalI and was ligated into the SalI site of pMW218. The resulting plasmid was used to transform E. coli ML4947. Transformants harboring this plasmid, which had a 6-kb SalI fragment carrying ampC and ampR, were selected with kanamycin by using the plasmid marker. This hybrid plasmid (9.9 kb) was renamed pKU403.
FIG. 1.
Cloning strategy for E. cloacae GN7471 ampC and ampR. pMS161 was used to clone an 8-kb EcoRI fragment containing ampC and ampR from E. cloacae GN7471 into pACYC184. pKU403 was constructed by cloning a SalI fragment of pMS161 into pMW218. pKU404, pKU405, pKU406, and pKU407 were mutated plasmids derived from pKU403. pKU408, pKU409, pKU410, and pKU414 were self-ligated at the SphI site of pKU403, pKU404, pKU405, and pKU406, respectively. pKU411, pKU412, pKU413, and pKU415 were also self-ligated at the BamHI-ScaI site of pKU403, pKU404, pKU405, and pKU406, respectively. Symbols: ░⃞, pACYC184; ■, pMW218.
To construct plasmids containing only the ampC gene, pKU403, pKU404, pKU405, or pKU406 was digested with SphI and was then self-ligated. These plasmids were used to transform E. coli ML4947, and plasmids with deletion of the 1.7-kb SphI fragment containing ampR were identified from the result of the plasmid DNA size obtained after digestion with SalI (8.7 kb) and by PCR. PCR primers AR3 and AR4, directed against the sequence for an ampR gene (GenBank accession no. AB016612), amplified a fragment of 743 bp. Forward primer AR3 (5′-CCGCCAGACACCTCAGTTTT-3′) is located between nucleotides 127 and 146 on the sequence, while reverse primer AR4 (5′-GTAACTCCCCAGGTCAATCC-3′) is located between nucleotides 869 and 850. The plasmids derived from pKU403, pKU404, pKU405, and pKU406 were named pKU408, pKU409, pKU410, and pKU414, respectively (Fig. 1). Similarly, to construct plasmids containing only the ampR gene, pKU403, pKU404, pKU405, and pKU406 were digested with BamHI-ScaI and were blunt ended at the BamHI site. The blunt end of the BamHI site was obtained with a DNA blunting kit (Takara shuzo) (37). After ligation, these plasmids were used to transform E. coli ML4947. Plasmids with deletion of the 4-kb BamHI-ScaI fragment containing ampC, which therefore carried only ampR, were identified from the plasmid DNA size obtained after digestion with SalI (5.9 kb) and by PCR. The PCR primers AC1 and AC2, directed against the sequence for an ampC gene (GenBank accession no. AB016611), amplified a fragment of 333 bp. Forward primer AC2 (5′-TTATCAGGGTCAGCCGCACT-3′) is located between nucleotides 262 and 281 on the sequence, while reverse primer AC1 (5′-GGTTTCCACTGCGGTTGCCA-3′) is located between nucleotides 594 and 575. The plasmids derived from pKU403, pKU404, pKU405, and pKU406 were named pKU411, pKU412, pKU413, and pKU415, respectively (Fig. 1).
Assay of β-lactamase activity.
β-Lactamase activity was detected as described previously (34). Briefly, imipenem (a carbapenem), a β-lactamase inducer, was added to mid-logarithmic-phase cultures and the cells were incubated for another 2 h. Imipenem was added at several concentrations (1/4× the MIC of imipenem) so that the cell protein concentration was not less than 75% compared with that for the controls. Cell lysis was negligible under these conditions, allowing enzyme activity to be assessed. The cells were harvested by centrifugation (1,700 × g, 10 min), resuspended in 3 ml of 50 mM potassium phosphate buffer (pH 7.0), and sonicated. After centrifugation at 14,000 × g for 10 min at 4°C, the β-lactamase activity and the protein concentration in the extract were measured and were compared between cultures. One unit of β-lactamase activity was defined as the amount of β-lactamase that hydrolyzed 1 μmol of cephalothin in 1 min at 30°C.
Isolation of ceftazidime- or aztreonam-resistant mutants.
Mutants with elevated levels of resistance to ceftazidime or aztreonam were obtained by plating about 109 CFU/ml of washed late-logarithmic-phase ML4953/pKU403 grown in L broth on agar plates containing ceftazidime or aztreonam at 4× to 16× the MIC.
DNA sequencing.
Analysis of the ampC and ampR sequences of pKU403 was performed as described by Sanger et al. (39). The DNA sequences of the ampR genes carried by pKU404, pKU405, pKU406, and pKU407 were determined with an ALFred DNA sequencer (Amersham Pharmacia Biotech) and the Thermo Sequenase fluorescence-labeled primer sequencing kit (Amersham Pharmacia Biotech). Sequencing primers were obtained from Amersham Pharmacia Biotech. The sequencing primers for the ampR gene, CY5AP4, CY5AR1 CY5AR2, and CY5AR3, were designed from the sequence of ampR (GenBank accession no. AB016612). Forward primers CY5AR1 (5′-CCCAGGAGAAGCTAAAAGTGG-3′) and CY5AR3 (5′-GATGGTCTTTGATTCGTCCGTG-3′) are located at nucleotides 352 to 372 and nucleotides 722 to 743, respectively, on the sequence, while reverse primers CY5AP4 (5′-TGCGTAAAACTGAGGTGTCTGGCG-3′) and CY5AR2 (5′-TAGGAGCGCAGCAGGGTAAACT-3′) are located at nucleotides 151 to 128 and 652 to 631, respectively.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the accession nos. AB016611 (ampC) and AB016612 (ampR).
RESULTS
Base sequences of ampC and ampR from E. cloacae GN7471.
The 8-kb DNA fragment from an EcoRI digest of E. cloacae GN7471 containing the ampC and ampR genes was introduced into the EcoRI site of pACYC184. The resulting plasmid, designated pMS161, was digested with SalI and was ligated into pMW218 at the SalI site, and the plasmid thus obtained was named pKU403. In this case, the DNA fragment was 6 kb in length. Next, the base sequence of pKU403 was determined. The degree of identity of ampC between E. cloacae GN7471 and E. cloacae MHN1 was 81.8%, while that between E. cloacae GN7471 and E. cloacae P99 was 81.8%. However, the degree of identity of ampC between E. cloacae P99 and MHN1 was 98.3% (10). In contrast, the degree of identity of ampR between DNA derived from E. cloacae GN7471 and that derived from E. cloacae MHN1 was 99.3% (12).
Isolation of mutants and MICs.
For isolation of mutants and determination of MICs, E. coli ML4953, which carried an AmpD mutant background, was used in order to avoid selecting only an AmpD mutant. Mutant strains were isolated from ML4953/pKU403 by selection with ceftazidime or aztreonam. When selection was performed with ceftazidime at 4× to 8× the MIC (2 to 4 μg/ml), mutants were obtained at a frequency of 6.2 × 10−6 to 2.0 × 10−6. With selection at 16× the MIC (8 μg/ml), mutants were obtained at a frequency of 3.4 × 10−8. When selection was performed with aztreonam at 4× to 8× the MIC (2 to 4 μg/ml), mutants were obtained at a frequency of 6.4 × 10−6 to 1.4 × 10−6, while the frequency of occurrence of mutants was 3.8 × 10−9 at 16× the MIC (8 μg/ml).
Among the 60 mutants thus obtained, a group of strains for which ceftazidime and aztreonam MICs were similarly high was chosen. Four strains were selected from this group at random. After DNA extraction and transformation of E. coli ML4953 (AmpD mutant), all four of these strains were confirmed to carry mutant plasmids, because a high-level cephalosporin resistance phenotype was transferred to ML4953. The mutant plasmids derived from selection with aztreonam at 8× the MIC were named pKU404 and pKU406, and those derived from selection with ceftazidime at 8× the MIC were named pKU405 and pKU407. All four plasmids were used to transform E. coli ML4947 (AmpD wild type) and ML4953 (AmpD mutant), and the MICs for each transformant were determined (Table 2).
TABLE 2.
MICs for E. coli mutants
| Host | Plasmid | Statusa | MIC
(μg/ml)b
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABPC | CER | CTM | LMOX | CTX | CAZ | CPDX | CFPM | AZT | IPM | |||
| ML4947 (AmpD wild type) | pKU403 | Wild type | 16 | 128 | 2 | 0.125 | 0.125 | 0.25 | 1 | 0.03 | 0.06 | 0.5 |
| pKU404 | H-mut | >128 | >128 | >128 | 8 | 32 | 64 | >128 | 0.25 | 32 | 1 | |
| pKU405 | L-mut | >128 | >128 | >128 | 4 | 16 | 32 | >128 | 0.125 | 16 | 2 | |
| pKU406 | I-mut | >128 | >128 | >128 | 2 | 8 | 8 | 128 | 0.06 | 4 | 2 | |
| pKU407 | I-mut | >128 | >128 | >128 | 2 | 8 | 8 | 128 | 0.06 | 4 | 2 | |
| pKU408 | pKU403 ΔampR | 8 | 16 | 2 | 0.125 | 0.125 | 0.25 | 2 | 0.03 | 0.125 | 0.5 | |
| pKU409 | pKU404 ΔampR | 16 | 16 | 4 | 0.125 | 0.125 | 0.25 | 2 | 0.03 | 0.125 | 0.5 | |
| pKU410 | pKU405 ΔampR | 8 | 8 | 2 | 0.125 | 0.125 | 0.25 | 2 | 0.03 | 0.06 | 0.25 | |
| pKU414 | pKU406 ΔampR | 8 | 16 | 4 | 0.125 | 0.125 | 0.25 | 2 | 0.03 | 0.125 | 0.25 | |
| pKU411 | pKU403 ΔampR | 8 | 2 | 0.125 | 0.125 | 0.06 | 0.125 | 0.5 | 0.03 | 0.06 | 0.25 | |
| pKU412 | pKU404 ΔampC | 4 | 2 | 0.25 | 0.125 | 0.06 | 0.125 | 0.5 | 0.03 | 0.06 | 0.25 | |
| pKU413 | pKU405 ΔampC | 8 | 4 | 0.5 | 0.125 | 0.125 | 0.125 | 1 | 0.03 | 0.06 | 0.25 | |
| pKU415 | pKU406 ΔampC | 8 | 4 | 0.25 | 0.125 | 0.125 | 0.125 | 0.5 | 0.03 | 0.06 | 0.25 | |
| pMW218 | Vector | 8 | 4 | 0.25 | 0.06 | 0.06 | 0.125 | 0.5 | 0.03 | 0.06 | 0.25 | |
| 8 | 2 | 0.25 | 0.125 | 0.06 | 0.125 | 0.5 | 0.03 | 0.06 | 0.25 | |||
| ML4953 (AmpD mutant) | pKU403 | Wild type | >128 | >128 | 128 | 2 | 0.5 | 0.5 | 32 | 0.03 | 0.25 | 1 |
| pKU404 | H-mut | >128 | >128 | >128 | 8 | 32 | 32 | >128 | 0.25 | 16 | 1 | |
| pKU405 | L-mut | >128 | >128 | >128 | 4 | 16 | 16 | >128 | 0.125 | 8 | 1 | |
| pKU406 | I-mut | >128 | >128 | >128 | 4 | 8 | 8 | >128 | 0.06 | 8 | 1 | |
| pKU407 | I-mut | >128 | >128 | >128 | 4 | 8 | 8 | >128 | 0.06 | 8 | 2 | |
| pKU408 | pKU403 ΔampR | 8 | 8 | 1 | 0.125 | 0.125 | 0.125 | 2 | 0.03 | 0.06 | 1 | |
| pKU409 | pKU404 ΔampR | 8 | 16 | 1 | 0.125 | 0.125 | 0.125 | 2 | 0.03 | 0.125 | 1 | |
| pKU410 | pKU405 ΔampR | 8 | 8 | 0.5 | 0.125 | 0.125 | 0.25 | 2 | 0.03 | 0.125 | 1 | |
| pKU414 | pKU406 ΔampR | 8 | 8 | 1 | 0.125 | 0.125 | 0.25 | 2 | 0.03 | 0.06 | 1 | |
| pKU411 | pKU403 ΔampC | 8 | 4 | 0.25 | 0.125 | 0.06 | 0.125 | 1 | 0.03 | 0.06 | 1 | |
| pKU412 | pKU404 ΔampC | 8 | 8 | 0.5 | 0.125 | 0.06 | 0.125 | 1 | 0.03 | 0.06 | 1 | |
| pKU413 | pKU405 ΔampC | 8 | 8 | 0.25 | 0.125 | 0.06 | 0.125 | 1 | 0.03 | 0.03 | 1 | |
| pKU415 | pKU406 ΔampC | 8 | 4 | 0.5 | 0.125 | 0.06 | 0.125 | 1 | 0.03 | 0.03 | 1 | |
| pMW218 | Vector | 4 | 4 | 0.25 | 0.125 | 0.06 | 0.125 | 1 | 0.03 | 0.06 | 1 | |
| 4 | 4 | 0.25 | 0.125 | 0.06 | 0.25 | 1 | 0.03 | 0.06 | 1 | |||
L-, I-, and H-mut, mutant plasmids with low, intermediate, and high levels of β-lactamase activity, respectively.
ABPC, ampicillin; CER, cephaloridine; CTM, cefotiam; LMOX, latamoxef; CTX, cefotaxime; CAZ, ceftazidime; CPDX, cefpodoxime; CFPM, cefepime; AZT, aztreonam; IPM, imipenem.
When E. coli ML4947 (AmpD wild type) was used as the host, the MIC of ampicillin for transformants carrying pKU404, pKU405, pKU406, or pKU407 was increased 16-fold or more compared with that for ML4947 carrying pKU403. Also, the MICs of cephems (cefotiam, latamoxef, ceftazidime, and cefotaxime) and aztreonam were increased 16-fold or more compared with those for ML4947 carrying pKU403. In addition, the MIC of cefepime increased twofold or more, and that of imipenem rose two- to fourfold. The MICs obtained when E. coli ML4953 (AmpD mutant) was the host were similar to those obtained when E. coli ML4947 was the host, and the MICs of ampicillin, cefotiam, latamoxef, and imipenem did not vary greatly.
On the other hand, the MICs for transformants carrying ΔampC or ΔampR plasmids were almost the same as the MICs for the parental strains.
β-Lactamase activities of mutants.
The β-lactamase activities encoded by the plasmids are shown in Table 3. When E. coli ML4947 (AmpD wild type) was the host cell, the β-lactamase activities encoded by pKU404, pKU405, pKU406, and pKU407 were about 470, 75, 160, and 180 times higher, respectively, than the activity encoded by the original plasmid (pKU403). However, the activity of the β-lactamase encoded by pKU403 increased about 50-fold when it was induced by imipenem, whereas the β-lactamase activity encoded by pKU405, pKU406, and pKU407 rose only three- to fivefold. The β-lactamase activity encoded by pKU404 was not induced by imipenem.
TABLE 3.
β-Lactamase activity
| Host | Plasmid | Statusb | β-Lactamase
activity (U/mg of protein)c
|
|
|---|---|---|---|---|
| Not induced | Induceda | |||
| ML4947 (AmpD wild type) | pKU403 | Wild type | 0.04 ± 0.01 | 1.94 ± 0.84 |
| pKU404 | H-mut | 18.86 ± 3.10 | 25.94 ± 5.53 | |
| pKU405 | L-mut | 2.99 ± 1.09 | 8.26 ± 0.68 | |
| pKU406 | I-mut | 6.34 ± 1.01 | 25.95 ± 4.01 | |
| pKU407 | I-mut | 7.10 ± 0.60 | 30.95 ± 7.66 | |
| pKU408 | pKU403 ΔampR | 0.14 ± 0.02 | 0.12 ± 0.01 | |
| pKU409 | pKU404 ΔampR | 0.13 ± 0.01 | 0.12 ± 0.01 | |
| pKU410 | pKU405 ΔampR | 0.13 ± 0.01 | 0.10 ± 0.01 | |
| pKU414 | pKU406 ΔampR | 0.13 ± 0.01 | 0.12 ± 0.01 | |
| pKU411 | pKU403 ΔampC | 0.01 | 0.01 | |
| pKU412 | pKU404 ΔampC | 0.01 | 0.01 | |
| pKU413 | pKU405 ΔampC | 0.01 | 0.01 | |
| pKU415 | pKU406 ΔampC | 0.01 | 0.01 | |
| pMW218 | Vector | 0.01 | 0.01 | |
| 0.01 | 0.01 | |||
| ML4953 (AmpD mutant) | pKU403 | Wild type | 4.89 ± 1.28 | 37.68 ± 9.53 |
| pKU404 | H-mut | 43.77 ± 5.88 | 41.54 ± 13.82 | |
| pKU405 | L-mut | 27.72 ± 5.11 | 48.48 ± 7.52 | |
| pKU406 | I-mut | 35.06 ± 4.46 | 41.39 ± 5.21 | |
| pKU407 | I-mut | 31.99 ± 2.97 | 38.28 ± 9.74 | |
| pKU408 | pKU403 ΔampR | 0.11 ± 0.00 | 0.09 ± 0.01 | |
| pKU409 | pKU404 ΔampR | 0.09 ± 0.01 | 0.06 ± 0.01 | |
| pKU410 | pKU405 ΔampR | 0.11 ± 0.02 | 0.08 ± 0.01 | |
| pKU414 | pKU406 ΔampR | 0.11 ± 0.01 | 0.07 ± 0.01 | |
| pKU411 | pKU403 ΔampC | 0.01 | 0.01 | |
| pKU412 | pKU404 ΔampC | 0.02 | 0.02 | |
| pKU413 | pKU405 ΔampC | 0.02 | 0.02 | |
| pKU415 | pKU406 ΔampC | 0.01 | 0.01 | |
| pMW218 | Vector | 0.01 | 0.01 | |
| 0.02 | 0.01 | |||
One to 4× the MIC of imipenem was used for induction of β-lactamase.
L-, I-, and H-mut, mutant plasmids with low, intermediate, and high levels of β-lactamase activity, respectively.
Values are means ± standard deviations of three independent experiments. Standard deviations for pKU411, pKU412, pKU413, pKU415, pMW218, and the host were <±0.001. One unit of β-lactamase activity was defined as the amount of β-lactamase that hydrolyzed 1 μmol of cephalothin in 1 min at 30°C.
When E. coli ML4953 (AmpD mutant) was used as the host, the β-lactamase activities encoded by pKU403, pKU404, pKU405, pKU406, and pKU407 were much higher compared with those when E. coli ML4947 was the host. Induction with imipenem resulted in an eightfold increase for pKU403 and a twofold increase for pKU405. However, no increase in activity was observed for pKU404, pKU406, or pKU407. These results indicated that pKU406 and pKU407 encoded similar levels of β-lactamase activity and had activities intermediate between those of pKU404, which encoded a high level of enzyme activity, and pKU405, which encoded a low level of enzyme activity. Hence, pKU404, pKU405, and pKU406 (which encoded different levels of enzyme activity) were used in subsequent experiments.
The specific enzyme activities encoded by ΔampR plasmids (pKU408, pKU409, pKU410, and pKU414) in E. coli ML4947 (AmpD wild type) and ML4953 (AmpD mutant) were 0.06 to 0.16 U/mg of protein and were two to four times higher than the β-lactamase activity encoded by ML4953/pKU403 but were markedly lower than the activities encoded by pKU404, pKU405, and pKU406. Similarly, since ΔampC plasmids (pKU411, pKU412, pKU413, and pKU415) lacked the structural gene for β-lactamase, their enzyme activities were always less than 0.02 U/mg of protein and did not differ from the activities of the host cells.
Amino acid sequence of AmpR.
Figure 2 shows the AmpR amino acid sequences encoded by pKU403, pKU404, pKU405, and pKU406 derived from E. cloacae GN7471, as well as those from the AmpR form of E. cloacae MHN1 and C. freundii OS60 (13, 25). G-538 in the base sequence of pKU403 was converted to A in pKU404, resulting in the replacement of Asp-135 by Asn. A-539 in the base sequence of pKU403 was converted to T in pKU405, and Asp-135 was replaced by Val. C-256 in the base sequence of pKU403 was converted to T in both pKU406 and pKU407, with Arg-86 being replaced by Cys.
FIG. 2.
Comparison of deduced amino acid sequences of AmpR from pKU403, pKU404, pKU405, pKU406, E. cloacae MHN1 (12), and C. freundii OS60 (24). Identical amino acids in all five sequences are marked with an asterisk. Nonidentical amino acids compared to those in pKU403 are underlined.
Effect of mutant AmpR on chromosomal β-lactamase.
In the experiment described above no difference in enzyme activity was found among ΔampR plasmids, while the β-lactamase activities encoded by pKU404, pKU405, and pKU406 were significantly higher than the activity of the pKU403-encoded β-lactamase. Therefore, the high level of enzyme activity encoded by the plasmids isolated in the present study appeared to be due to a mutation of ampR. To confirm this, ΔampC plasmids (pKU411, pKU412, pKU413, and pKU415) were used to transform E. cloacae ATCC 13047 as well as clinical isolates of E. cloacae (KU3261, KU3262, and KU3263), and the effects of mutations in AmpR were examined. Table 4 shows that the β-lactamase activities of the pKU411 transformants were almost the same as those of the host strains, whereas the activities of the pKU412, pKU413, and pKU415 transformants were 20 to 350 times, 10 to 130 times, and 15 to 250 times higher, respectively.
TABLE 4.
Effects of AmpR mutants against production of class C β-lactamase in E. cloacae ATCC 13047 and three clinical isolates
| Plasmid | β-Lactamase activity (U/mg
of protein)a
|
|||
|---|---|---|---|---|
| KU3261 | KU3262 | KU3263 | ATCC 13047 | |
| <0.02 | 0.06 | <0.02 | 0.07 | |
| pKU411 | 0.02 | 0.03 | <0.02 | 0.06 |
| pKU412 | 6.96 | 6.40 | 1.72 | 1.38 |
| pKU413 | 2.56 | 2.01 | 0.34 | 0.78 |
| pKU415 | 4.82 | 4.34 | 0.93 | 1.07 |
One unit of β-lactamase activity was defined as the amount of β-lactamase that hydrolyzed 1 μmol of cephalothin in 1 min at 30°C.
DISCUSSION
The frequency of occurrence of mutants that stably express derepressed class C β-lactamase in subpopulations of resistant organisms and the widespread use of β-lactams in the hospital environment have resulted in the emergence of clinically important endemic bacterial resistance (9). The differences between individual inducible strains that cause infection remain unclear, but these organisms appear to carry mutations in either AmpD or AmpR.
Many gram-negative bacilli (e.g., Enterobacter spp., C. freundii, Pseudomonas aeruginosa, and Serratia marcescens) produce chromosomal class C β-lactamases. The ampC gene used in the present study was derived from a clinical isolate, E. cloacae GN7471, and showed about 80% identity with those reported in E. cloacae P99 and E. cloacae MHN1 (10). However, microbiological comparison of E. cloacae GN7471 and E. cloacae P99 has shown that they belong to the same species (18). That is, among bacterial strains assigned to the same species by microbiological methods, classification into close relatives may be possible when identification is done at the gene level.
The degree of identity of ampR between DNA derived from E. cloacae GN7471 and that derived from E. cloacae MHN1 was 99.3%. In contrast, the degree of identity of AmpR between DNA derived from E. cloacae GN7471 and C. freundii OS60 was only 73.0%. However, the AmpR amino acid sequences of Arg-86, Gly-102, and Asp-135 were conserved between E. cloacae GN7471, E. cloacae MHN1, and C. freundii OS60 (Fig. 2).
As shown in Table 2, MICs appeared to be inconsistent with β-lactamase activity (Table 3). In E. coli ML4947 (AmpD wild type), the β-lactamase activity of pKU404 was sixfold higher than that of pKU405. In the case of E. coli ML4953 (AmpD mutant), the enzyme activity of pKU404 was only 1.6-fold higher than that of pKU405. On the contrary, the β-lactamase activities encoded by ΔampR plasmids (pKU408, pKU409, pKU410, and pKU414) in E. coli were markedly lower than the activities encoded by pKU404, pKU405, and pKU406. This result maybe indicates that β-lactamase was induced on a plate with drug and that the mutations in pKU404 and pKU405 are located in different sites of AmpR.
Two of the mutant plasmids obtained in the present study, pKU404 and pKU405, had single-base mutations of ampR, resulting in mutation of Asp-135. pKU406 and pKU407 also had only a one-base mutation, with a consequent change in Arg-86. Bartowsky and colleagues (1, 33) have reported on the variability of AmpR from C. freundii, and they isolated AmpR with alterations of Ser-35, Tyr-264, Gly-102, and Asp-135 by using nitrosoguanidine mutagenesis and site-directed mutagenesis. In our study, a change of wild-type Asp-135 to Asn (pKU404) or Val (pKU405) resulted in 470-fold and 75-fold increases in basal levels of β-lactamase expression, respectively, while a change of wild-type Arg-86 to Cys (pKU406 and pKU407) resulted in 160-fold and 180-fold increases, respectively (Table 3).
As for the mutations of Asp-135 (pKU404 and pKU405) and Arg-86 (pKU406 and pKU407), these amino acids also appear to be important for ampC activation. A change of either the 86th or the 135th amino acid of AmpR affected the ampC promoter. In other words, these mutants were considerably more active than wild-type AmpR as transcriptional activators for the ampC promoter. These high levels of expression of β-lactamase were shown in the presence or absence of a β-lactam inducer and in the AmpD wild type (ML4947) or AmpD mutant (ML4953).
On the other hand, for about 3% of clinical isolates the cefotaxime and ceftazidime MICs were less than 0.125 μg/ml, and we could not detect any class C β-lactamase in clinical isolates of E. cloacae (data not shown). In this study, we selected three isolates of E. cloacae (KU3261, KU3262, and KU3263) with β-lactamase activities of 0.02, 0.03, and <0.02 U/mg of protein, respectively. As shown in Table 4, the activities of pKU412, pKU413, and pKU415 transformants (ΔampR plasmids) were 20 to 350 times, 10 to 130 times, and 15 to 250 times higher, respectively.
In the present study, since mutant plasmids were selected by using AmpD mutant strains as the host cells, the resulting mutants may also have had mutations at sites other than ampD. On the other hand, the frequency of selection of AmpD mutants was about 10−5 in another study (27). The frequency of occurrence of stably derepressed class C β-lactamase mutations in a bacterial population can be as high as 10−5 (26). The existence of such mutants has serious clinical implications with regard to the generation of AmpC-producing strains during selective therapy with broad-spectrum β-lactams. These strains appear to carry a mutation of either AmpD or AmpR. The selection of AmpR mutants at a frequency of 10−6 or less strongly suggests that frequent generation of derepressed mutant strains, such as AmpD mutants, might occur first in the clinical setting, followed by selection of AmpR mutants. In the real situation both ampR and ampD are chromosomal single-copy genes. In these experiments, ampR is on a multicopy plasmid. This clearly affects the mutation frequency. Hence, it is much more likely that clinical E. cloacae isolates resistant to β-lactamase contain ampD mutations than ampR mutations, since any harmful event to the AmpD basically increases the level of resistance, whereas specific ampR mutations are needed to create an AmpR that works as an activator even in the absence of a muropeptide inducer. The potentially interesting aspect of this study is that an ampD mutation may perhaps be followed by an ampR mutation, creating further resistance. The problem is that it is not known if a single-copy version of the ampR mutants studied here actually will increase the level of β-lactamase expression in an ampD knockout mutant of E. cloacae. It is not known whether replacement of a single-copy version of the mutated ampR gene by wild-type ampR on the chromosome of E. cloacae will actually increase the level of resistance in AmpD wild-type strains.
ACKNOWLEDGMENTS
We thank Y. Ohya for excellent technical assistance.
This work was supported by grant 09670296 (to M.I.) from the Japanese Ministry of Education and by a grant for diagnosis of antibiotic resistance from the Japanese Ministry of Health and Welfare.
REFERENCES
- 1.Bartowsky E, Normark S. Purification and mutant analysis of Citrobacter freundiiAmpR, the regulator for chromosomal AmpC β-lactamase. Mol Microbiol. 1991;5:1715–1725. doi: 10.1111/j.1365-2958.1991.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 2.Bartowsky E, Normark S. Interactions of wild-type and mutant AmpR of Citrobacter freundiiwith target DNA. Mol Microbiol. 1993;10:555–565. doi: 10.1111/j.1365-2958.1993.tb00927.x. [DOI] [PubMed] [Google Scholar]
- 3.Bernardi A, Bernardi F. Complete sequence of pSC101. Nucleic Acids Res. 1984;12:9415–9426. doi: 10.1093/nar/12.24.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen S N, Chang A C, Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coliby R-factor DNA. Proc Natl Acad Sci USA. 1972;69:2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietz H, Pfeifle D, Wiedemann B. The signal molecule for β-lactamase induction in Enterobacter cloacaeis the anhydromuramyl-pentapeptide. Antimicrob Agents Chemother. 1997;41:2113–2120. doi: 10.1128/aac.41.10.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrhardt A F, Sanders C C, Romero J R, Leser J S. Sequencing and analysis of four new Enterobacter ampDalleles. Antimicrob Agents Chemother. 1996;40:1953–1956. doi: 10.1128/aac.40.8.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung-Tomc J C, Gradelski E, Huczko E, Dougherty T J, Kessler R E, Bonner D P. Differences in the resistant variants of Enterobacter cloacaeselected by extended-spectrum cephalosporins. Antimicrob Agents Chemother. 1996;40:1289–1293. doi: 10.1128/aac.40.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galleni M, Lindberg F, Normark S, Cole S, Honore N, Joris B, Frere J-M. Sequence and comparative analysis of three Enterobacter cloacae ampCβ-lactamase genes and their products. Biochem J. 1988;250:753–760. doi: 10.1042/bj2500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holtje J V, Kopp U, Ursinus A, Wiedemann B. The negative regulator of β-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol Lett. 1994;122:159–164. doi: 10.1111/j.1574-6968.1994.tb07159.x. [DOI] [PubMed] [Google Scholar]
- 12.Honore N, Nicolas M H, Cole S T. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 1986;5:3709–3714. doi: 10.1002/j.1460-2075.1986.tb04704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue M, Itoh J, Mitsuhashi S. pMS76, a plasmid capable of amplification by treatment with chloramphenicol. Plasmid. 1983;9:86–97. doi: 10.1016/0147-619x(83)90033-1. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs C, Frere J M, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in gram-negative bacteria. Cell. 1997;88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs C, Huang L, Bartowsky E, Normark S, Park J T. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 1994;13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs C, Joris B, Jamin M, Klarsov K, Beeumen J V, Mengin-Lecreulx D, Heijenoort J V, Park J T, Normark S, Frere J M. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol Microbiol. 1995;15:553–559. doi: 10.1111/j.1365-2958.1995.tb02268.x. [DOI] [PubMed] [Google Scholar]
- 17.Jones R N, Baquero F, Privitera G, Inoue M, Wiedemann B. Inducible β-lactamase-mediated resistance to third-generation cephalosporins. Clin Microbiol Infect. 1997;3:7–20. [Google Scholar]
- 18.Joris B, Dusart J, Frere J-M, Beeumen J V, Emanuel E L, Petursson S, Gagnon J, Waley S G. The active site of the P99 β-lactamase from Enterobacter cloacae. Biochem J. 1984;223:271–274. doi: 10.1042/bj2230271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopp U, Wiedemann B, Lindquist S, Normark S. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob Agents Chemother. 1993;37:224–228. doi: 10.1128/aac.37.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korfmann G, Sanders C C. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob Agents Chemother. 1989;33:1946–1951. doi: 10.1128/aac.33.11.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindberg F, Lindquist S, Normark S. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundiiβ-lactamase. J Bacteriol. 1987;169:1923–1928. doi: 10.1128/jb.169.5.1923-1928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindberg F, Normark S. Common mechanism of ampC β-lactamase induction in enterobacteria: regulation of the cloned Enterobacter cloacaeP99 β-lactamase gene. J Bacteriol. 1987;169:758–763. doi: 10.1128/jb.169.2.758-763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindberg F, Westman L, Normark S. Regulatory components in Citrobacter freundii ampCβ-lactamase induction. Proc Natl Acad Sci USA. 1985;82:4620–4624. doi: 10.1073/pnas.82.14.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindquist S, Lindberg F, Normark S. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampCβ-lactamase gene. J Bacteriol. 1989;171:3746–3753. doi: 10.1128/jb.171.7.3746-3753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindquist S, Weston-Hafer K, Schmidt H, Pul C, Korfmann G, Erickson J, Sanders C, Martin H H, Normark S. AmpG, a signal transducer in chromosomal β-lactamase induction. Mol Microbiol. 1993;9:703–715. doi: 10.1111/j.1365-2958.1993.tb01731.x. [DOI] [PubMed] [Google Scholar]
- 26.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maejima T, Ohya Y, Mitsuhashi S, Inoue M. Cloning and expression of the gene(s) for chromosome-mediated β-lactamase production of Proteus vulgaris in Escherichia coli. Plasmid. 1987;18:120–126. doi: 10.1016/0147-619x(87)90039-4. [DOI] [PubMed] [Google Scholar]
- 28.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 29.Medeiros A A. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin Infect Dis. 1997;24:19–45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 30.Minami S, Inoue M, Mitsuhashi S. Purification and properties of a cephalosporinase from Enterobacter cloacae. Antimicrob Agents Chemother. 1980;18:853–857. doi: 10.1128/aac.18.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minami S, Yotsuji A, Inoue M, Mitsuhashi S. Induction of β-lactamase by various β-lactam antibiotics in Enterobacter cloacae. Antimicrob Agents Chemother. 1980;18:382–385. doi: 10.1128/aac.18.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Normark S. β-Lactamase induction in gram-negative bacteria is intimately linked to peptidoglycan recycling. Microb Drug Resist. 1995;1:111–114. doi: 10.1089/mdr.1995.1.111. [DOI] [PubMed] [Google Scholar]
- 33.Normark S, Bartowsky E, Erickson J, Jacobs C, Lindberg F, Lindquist S, Hafer K W, Wikstrom M. Mechanisms of chromosomal β-lactamase induction in gram-negative bacteria. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell walls. Amsterdam, The Netherlands: Elsevier Science B. V.; 1994. pp. 485–503. [Google Scholar]
- 34.Okamoto R, Okubo T, Inoue M. Detection of genes regulating β-lactamase production in Enterococcus faecalis and Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:2550–2554. doi: 10.1128/aac.40.11.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J T. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol. 1995;17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 36.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Sanders C C, Sanders W E. Emergence of resistance to cefamandole: possible role of cefoxitin-inducible β-lactamases. Antimicrob Agents Chemother. 1979;15:792–797. doi: 10.1128/aac.15.6.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stapleton P, Shannon K, Phillips I. DNA sequence differences of ampD mutants of Citrobacter freundii. Antimicrob Agents Chemother. 1995;39:2494–2498. doi: 10.1128/aac.39.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]