Abstract
Background and aim:
Epigenetic modifications exhibit promising evidence in etiology and prognosis of important diseases such as inflammatory bowel diseases (IBD). In addition to complex factors involved in IBD, a trend toward better prognosis have been reported in older ages of disease onset. Gastrointestinal mucous layer is one of the important components which is disturbed in the disease course. Integrity of this layer is maintained with an anti-inflammatory factor called trefoil factors (TFF). We investigated the methylation status of TFF1 gene in IBD patients alongside with correlation of its alteration level with age of disease onset.
Methods:
We analyzed the promoter methylation status of TFF1 gene, using the real-time quantitative multiplex methylation specific PCR (QM-MSP). DNA was extracted from colorectal biopsies of 15 Crohn disease cases and 15 healthy controls. Correlation analysis was performed between unmethylated DNA level and age through Pearson correlation coefficient (PPC) test and simple linear regression models.
Results:
Our data didn’t provide significant positive correlation of age and TFF1 hypomethylation in Crohn patients (r = .518, p = .058).
Conclusions:
In conclusion, our case-control study didn’t show significant alteration in TFF1 methylation status in CD patients. (www.actabiomedica.it)
Keywords: Inflammatory bowel disease, Crohn disease, Epigenetics, Methylation, Aging
Introduction
Inflammatory bowel diseases (IBD), comprised of Crohn’s Disease (CD) and Ulcerative Colitis (UC), are a group of chronic inflammatory diseases which significantly affect the patients’ quality of life. UC and CD differ on the site and extension of the inflammation. CD can occur from mouth to anus and cause a severe inflammation which may extend to the entire thickness of bowel wall. However, UC is usually restricted to the innermost layer of the colon and rectum. Although the exact mechanisms underlying the occurrence and prognosis of both diseases demand further studies, several factors are reported to be involved such as genetic, immune responses, intestinal microbiota and interaction of genome with environmental factors without alteration in genome sequence, regarded as “epigenetic” factors (1, 2). Epigenetic modifications of promoter can occur through cytosine methylation in specific cytosine guanine rich sites called CpG islands. Several genes may undergo aberrant methylation in promoter site such as hypo- or hyper methylation which may lead to gene over- or under expression, respectively (2). Aging may also alter the methylation state of specific genes through various mechanisms (3). On the other hand, age of the IBD onset have been reported to influence disease severity and prognosis. For instance, Crohn disease is exacerbated in early life onsets (4). Among various factors perturbed in this disease, mucin layer and its associated factors seem intriguing in disease propagation and prognosis. Mucin layer provides a protective barrier between epithelial layer and gut microbiota (5). Mucin is co-secreted with small peptides called trefoil factors (TFF) which participate in mucosa protection and repairing through increasing mucin layer viscosity and elasticity. Therefore, these factors are considered to exert their protective role during the course of gut inflammatory diseases such as IBD (6). Trefoil factors (TFF) are basically expressed in gastrointestinal tract epithelial cells and their genes are clustered on chromosome 21q22.3. They play a role in epithelial defense and repairing (7). There are 3 subtypes of trefoil factor family (TFF) among which TFF1 seems more intriguing to be observed in IBD. Normally, TFF1 is expressed in foveolar cells of superficial epithelium in stomach and maintains its normal structure and functions (8, 9). Clinical studies on IBD demonstrated that gene expression of all three TFFs were altered at the site of mucosal damage in IBD (10). This alteration is reported to be the response of epithelial cells to inflammatory cytokines, as their innate protection. Due to the anti-inflammatory roles of TFF1, it’s considered as a protective and curative factor in IBD patients (11).Variations in expression of TFF can be associated with diverse environmental factors acting through Epigenetics. For instance, TFF1 promotor methylation variations have been reported in retinoblastoma cell lines and prostate cancer (12, 13). We hypothesized that epigenetic control of TFF expression through methylation of CpG Islands may have a role in IBD patients according to their age, as it plays an important role in IBD prognosis and severity. Therefore, a correlation analysis between TFF methylation and aging in CD patients is reported.
Methods
Tissue samples
Colorectal samples were obtained from men and women undergoing colonoscopic mucosal resection at the gastroenterology clinics of Kasra and Laleh hospitals in Tehran, Iran, between May 2014 and July 2015. Study population consisted of 15 CD patients (8 females, 7 males), and 14 age-matched healthy controls (8 females, 6 males) undergoing routine colonoscopy checkup. Healthy controls didn’t have any history of IBD or other gastrointestinal abnormalities, approved by histopathological examination. All patients were informed of the use of their specimen and written consent was obtained from all participants. Tissue collection and protocol of this study were approved by the Ethics Committee of Tehran University of Medical Sciences, Tehran, Iran (96-02-154-34947).
DNA preparation and bisulfite conversion
High Pure PCR Template Preparation Kit (Roche) was applied to isolate genomic DNA of subjects’ colonic mucosa. Bisulfite modification of genomic DNA was done by MethylEdge™ Bisulfite Conversion System (Promega, Madison, WI) through treatment with sodium bisulfite. During the conversion process, all cytosine residues are converted to uracil whereas 5-methylcytosine (5mC) remains unmodified. Bisulfite-modified DNA specimens were aliquoted and stored at -20°C.
Methylation Analysis
Real-time quantitative multiplex methylation specific PCR (QM-MSP) based on SYBR green (Lo, Watanabe et al. 2009) was applied to evaluate the methylation status of CpG sites across the promoter region of TFF1 gene. QM-MSP required two sequential steps of PCR reactions performed with a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). The first pre-amplification PCR reaction (the multiplex step) was carried out with MethySYBR primers including external forward primer (EXT-F; 5’-AGTTTAGGTTTAGACGGAATGG-3’) and external reverse primer (EXT-R; 5’-TCTCCTCCAACCTAACCTTAAT-3’). The primer set were designed to enable the simultaneous amplification of many discrete target alleles in a single reaction. PCR reaction was performed in a volume of 25 µl containing 1 µl of converted genomic DNA following this protocol:
Denaturation at 95°C for 5 min
30 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s
Final extension at 72°C for 5 min.
In the second round of PCR, the specific methylated target is quantified from multiplex step products using both nested methylation- independent and methylation-specific primer sets (Li, Deuring et al. 2012)including nested methylation-specific forward (FM; CGGAATGGGTTTTATGAGTT-3’) and reverse primer (RM; 5’- CCGCCAAAATAAATACTATACTCA-3’). The bisulfite treated DNA was PCR amplified in a 10 μl reaction volume containing 5 µl SYBR® Green Master Mix, 0.25 µl of each of the methylated primers, 3.5 µl DDW and 1 μl of bisulfite-treated DNA. Cycling conditions were:
1 min at 95°C
30 cycles of 30 s at 94°C
1 min at 60°C
30 s at 72°C subsequently followed by 5 min at 72°C
CpG island prediction and primer blasting were performed using MethBlast tool. The methylation profile of the promoter CpG islands were defined based on UCSC database.
No untreated template controls were included in each run as negative controls. Fully converted methylated human plasmid DNA was used as a positive control for MSP in each run to serve as the 100% methylated reference for calculating the relative methylation percentages of DNA samples.
The ΔΔCq method was used to calculate the ratio of unmethylated versus total amplifiable bisulfite-treated DNA. The cycle of quantification (Cq) for the reaction between methylation-specific primers (MSP) and bisulfite-specific primers (BSP) was also obtained (Schmittgen and Livak 2008, Husseiny, Kuroda et al. 2012). Using a reference sample for standardization, this indicates the relative difference between the template of interest and a control template:
ΔΔCq= ΔCq sample – ΔCq plasmid
ΔCq sample= Cq MCP – Cq BSP
ΔCq plasmid = Cq MCP – Cq BSP
All cycle threshold (CT) values were obtained in the exponential phase and normalized by subtraction of the CT value. After normalization with the expression of PCR products amplified by external nested primers as internal control (BSP), The fold change in target gene samples was calculated using the 2-ΔΔCT method:
ΔCT= CT target gene - CT BSP products
ΔΔCT=ΔCT (samples) - ΔCT (controls)
Unmethylated DNA level= 2-ΔΔCT
Statistical Analysis
Unmethylated DNA levels were compared between the three study groups using one-way analysis of variance. The Pearson correlation coefficient (PPC) test was used to assess the correlation between age and hypomethylation. Furthermore, simple linear regression models were constructed for prediction of unmethylated DNA levels based on age. Based on these models, measurement outliers were determined as data points with a significantly extreme studentized residual value at the level of alpha = 0.05. Thus, one patient was identified as outliers in the CD study groups (Figure 2). All statistical analysis was performed using R version 3.5.1.
Figure 2.
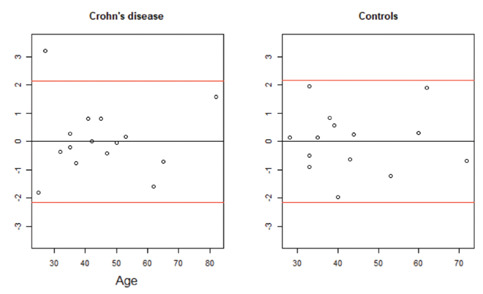
The red lines show the statistical significance level at alpha = 0.05 (±2.145 for CD [df = 14]; ±2.160 for Controls [df = 13]).
Figure 1.
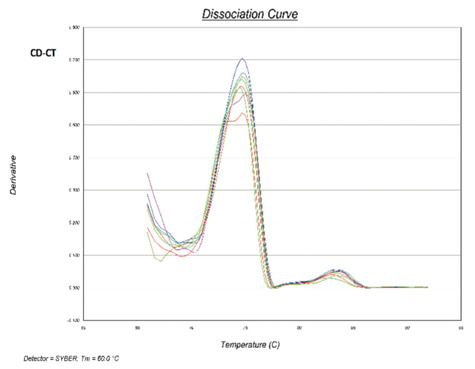
After bisulfite conversion, methylated DNA (with cytosine) exhibits higher melting temperature than unmethylated DNA (containing thymine). As seen in both CD patients, the higher peak of the melting curve is allocated to unmethylated DNA.
Results
Methylation state of the TFF1 gene in IBD patients
DNA methylation in TFF1 promotor was assessed in 15 CD patients and 14 healthy controls. A T-Test did not show a significant difference in hypomethylation levels between the two study groups, p = .0.062 (Figure 3).
Figure 3.
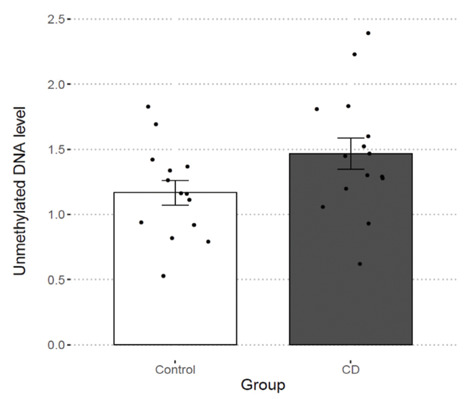
DNA methylation in TFF1 promotor in 15 CD patients and 14 healthy controls
Correlation of TFF1 promotor unmethylation and age in CD
We also studied the relationship between age and unmethylated DNA levels. Pearson correlation coefficients were calculated both including and excluding outliers and are presented in table 1. The correlation in CD patients is barely not significant, possibly requiring more data points to reach p < .05 (r = .518, p = .058). No significant correlation was observed in controls which strengthens our hypothesis on IBD-associated alteration on promotor methylation state, not the aging-associated epigenetic regulations.
Table 1.
Correlation between age and hypomethylation
| n | Including outliers | Excluding outliers | |||
|---|---|---|---|---|---|
| r | p-value | r | p-value | ||
| Crohn’s disease | 15 | .230 | .410 | .518 | .058 |
| Control | 14 | .237 | .413 |
Furthermore, simple linear regression models were constructed to predict hypomethylation levels based on age. the model constructed for CD patients is, once again, borderline significant (F(1,12) = 4.392, p = .058), with an R2 of 0.268. However, there does not seem to be any relationship between hypomethylation and age in healthy individuals (Figure 4).
Figure 4.
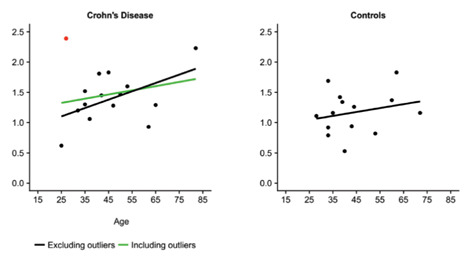
The red data points are the outliers. Simple linear regression models are shown both including (in) and excluding (ex) outliers. The formulas for the lines are as follows: UC: (in) .832 + .007 (age), R2 = .085, F(1,13) = 1.203, p = .293; (ex) .427 + .016 (age), R2 = .507, F(1,11) = 11.33, p = .006. CD: (in) 1.151 + .007 (age), R2 =.053, F(1,13) = .725, p = .410; (ex) .756 + .014 (age), R2 = .268, F(1,12) = 4.392, p = .058. Controls: .884 + .006 (age), R2 = .056, F(1,12) = .717, p = .414.
Discussion
It’s been reported that presentation of the IBD becomes prominently different as the age of the disease onset increases. Older patients tend to have milder and more stable disease. However, initial lesions of young adults and pediatrics are prone to further extension. To a lesser degree, our results supported this correlation in CD patients which is consistent with studies indicating less extensive disease in elderly patients (4). This alteration in promotor methylation may be due to the endogenous epigenetic tools of cells or the accumulative effects of lifestyle factors like smoking, breathing toxins, drugs, etc. The mechanisms involved in this protective epigenetic regulation of TFF1 in elder patients is currently unknown but further studies on cytokine profile analysis in young and elderly patients may also reveal a mechanism for TFF1 hypomethylation in old age. There are reports that TNFɑ upregulates TFF1 in gastric cells (16, 17). Interestingly, factors involved in IBD initiation and propagation such as IL1β and IL6, are reported to downregulate TFF1 transcription (18). In order to investigate the role of TFF1 downregulation in the disease initiation, further studies can analyze TFF1 promotor methylation level before and after the disease incidence in the form of cohort studies. TFF1 has also demonstrated tumor suppressing effects in several studies. For instance, TFF1 downregulation through promotor hypermethylation is reported in esophageal squamous cell carcinoma and gastric cancer (19, 20). Therefore, many studies support the clinical application of TFF1 in IBD and even cancers; however, it still requires further studies to find the optimum dose and delivery methods (11). In conclusion, our case-control study didn’t show significant alteration in TFF1 methylation status in CD patients.
Acknowledgements:
Research reported in this publication was supported by Elite Researcher Grant Committee under award number [982804] from the National Institute for Medical Research Development (NIMAD), Tehran, Iran.
Conflict of Interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article
References
- Ko JK, Auyeung KK. Inflammatory bowel disease: etiology, pathogenesis and current therapy. Current pharmaceutical design. 2014;20(7):1082–96. doi: 10.2174/13816128113199990416. [DOI] [PubMed] [Google Scholar]
- Yi JM, Kim TO. Epigenetic Alterations in Inflammatory Bowel Disease and Cancer. Intestinal Research. 2015;13(2):112–21. doi: 10.5217/ir.2015.13.2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Pfeifer GP. Aging and DNA methylation. BMC Biol. 2015;13:7. doi: 10.1186/s12915-015-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duricova D, Burisch J, Jess T, Gower-Rousseau C, Lakatos PL, EpiCom E. Age-related differences in presentation and course of inflammatory bowel disease: an update on the population-based literature. J Crohns Colitis. 2014;8(11):1351–61. doi: 10.1016/j.crohns.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Boltin D, Perets TT, Vilkin A, Niv Y. Mucin function in inflammatory bowel disease: an update. Journal of clinical gastroenterology. 2013;47(2):106–11. doi: 10.1097/MCG.0b013e3182688e73. [DOI] [PubMed] [Google Scholar]
- Thim L, Madsen F, Poulsen SS. Effect of trefoil factors on the viscoelastic properties of mucus gels. Eur J Clin Invest. 2002;32(7):519–27. doi: 10.1046/j.1365-2362.2002.01014.x. [DOI] [PubMed] [Google Scholar]
- Buda A, Jepson MA, Pignatelli M. Regulatory function of trefoil peptides (TFF) on intestinal cell junctional complexes. Cell Commun Adhes. 2012;19(5-6):63–8. doi: 10.3109/15419061.2012.748326. [DOI] [PubMed] [Google Scholar]
- Song J-Y, Kim B-W, Lee A-W, Lee K-Y, Chung I-S, Lee B-I, et al. Expression of MUC5AC and Trefoil Peptide 1 (TFF1) in the Subtypes of Intestinal Metaplasia. Clinical Endoscopy. 2012;45(2):151–4. doi: 10.5946/ce.2012.45.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobler L, Mejias-Luque R, Garrido M, Pera M, Badia-Garrido E, de Bolos C. Activation of the NF-kB pathway downregulates TFF-1 in gastric carcinogenesis. Virchows Arch. 2013;463(4):497–507. doi: 10.1007/s00428-013-1469-2. [DOI] [PubMed] [Google Scholar]
- Wright NA, Poulsom R, Stamp G, Van Norden S, Sarraf C, Elia G, et al. Trefoil peptide gene expression in gastrointestinal epithelial cells in inflammatory bowel disease. Scandinavian journal of gastroenterology Supplement. 1992;193:76–82. doi: 10.3109/00365529209096010. [DOI] [PubMed] [Google Scholar]
- Aamann L, Vestergaard EM, Gronbaek H. Trefoil factors in inflammatory bowel disease. World J Gastroenterol. 2014;20(12):3223–30. doi: 10.3748/wjg.v20.i12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippeit C, Busch M, Dunker N. Epigenetic control of trefoil factor family (TFF) peptide expression in human retinoblastoma cell lines. Cell Physiol Biochem. 2014;34(3):1001–14. doi: 10.1159/000366316. [DOI] [PubMed] [Google Scholar]
- Vestergaard EM, Nexo E, Torring N, Borre M, Orntoft TF, Sorensen KD. Promoter hypomethylation and upregulation of trefoil factors in prostate cancer. Int J Cancer. 2010;127(8):1857–65. doi: 10.1002/ijc.25209. [DOI] [PubMed] [Google Scholar]
- Longman RJ, Poulsom R, Corfield AP, Warren BF, Wright NA, Thomas MG. Alterations in the composition of the supramucosal defense barrier in relation to disease severity of ulcerative colitis. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 2006;54(12):1335–48. doi: 10.1369/jhc.5A6904.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low D, Mizoguchi A, Mizoguchi E. DNA methylation in inflammatory bowel disease and beyond. World J Gastroenterol. 2013;19(32):5238–49. doi: 10.3748/wjg.v19.i32.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike T, Shimada T, Fujii Y, Chen G, Tabei K, Namatame T, et al. Up-regulation of TFF1 (pS2) expression by TNF-alpha in gastric epithelial cells. J Gastroenterol Hepatol. 2007;22(6):936–42. doi: 10.1111/j.1440-1746.2007.04861.x. [DOI] [PubMed] [Google Scholar]
- Muzes G, Molnar B, Tulassay Z, Sipos F. Changes of the cytokine profile in inflammatory bowel diseases. World J Gastroenterol. 2012;18(41):5848–61. doi: 10.3748/wjg.v18.i41.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossinger V, Kayademir T, Blin N, Gott P. Down-regulation of TFF expression in gastrointestinal cell lines by cytokines and nuclear factors. Cell Physiol Biochem. 2002;12(4):197–206. doi: 10.1159/000066279. [DOI] [PubMed] [Google Scholar]
- Gonzaga IM, Soares Lima SC, Nicolau MC, Nicolau-Neto P, da Costa NM, de Almeida Simao T, et al. TFF1 hypermethylation and decreased expression in esophageal squamous cell carcinoma and histologically normal tumor surrounding esophageal cells. Clin Epigenetics. 2017;9:130. doi: 10.1186/s13148-017-0429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Ortiz CI, Lopez-Vidal Y, Arredondo-Hernandez LJR, Castillo-Rojas G. Genetic Alterations in Gastric Cancer Associated with Helicobacter pylori Infection. Front Med (Lausanne) 2017;4:47. doi: 10.3389/fmed.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]


