Abstract
Despite significant studies on the COVID-19 pandemic, scientists around the world are still battling to find a definitive therapy against the ongoing severe global health crisis. In this study, advanced computational approaches have been employed to identify bioactive food constituents as potential SARS-CoV-2 PLpro inhibitors-modulators. As a validated antiviral drug target, PLpro has gained tremendous attention for therapeutics developments. Therefore, targeting the multifunctional SARS-CoV-2 PLpro protein, ∼1039 bioactive dietary compounds have been screened extensively through novel techniques like negative image-based (NIB) screening and molecular docking approaches. In particular, the three different models of NIB screening have been generated and used to re-score the dietary compounds based on the negative image which is created by reversing the shape and electrostatics features of PLpro protein's ligand-binding cavity. Further, 100 ns molecular dynamics simulation has been performed and MM-GBSA based binding free energies have been estimated for the final proposed four dietary compounds (PC000550, PC000361, PC000558, and PC000573) as potential inhibitors/modulators of SARS-CoV-2 PLpro protein. Employed computational study outcome also has been compared with respect to the earlier experimentally investigated compound GRL0617 against SARS-CoV-2 PLpro protein, which suggests much greater interaction potential in terms of binding affinity and other energetic contributions for the proposed dietary compounds. Hence, the present study suggests that proposed dietary compounds can be suitable chemical entities for modulating the activity of PLpro protein or can be further utilized for optimizing or screening of novel chemical surrogates, however also needs experimental evaluation for entry in clinical studies for better assessment.
Keywords: SARS-CoV-2 PLpro, Food compounds, Virtual screening, Molecular docking, Molecular dynamics, MM-GBSA
Graphical abstract
1. Introduction
Since the first outbreak reported somewhere in Wuhan city in Hubei province of China in December 2019, the Coronavirus Disease 2019 (COVID-19) has been one of the worst pandemics in history, infecting a large number of individuals and posing a great concern to human health and public safety [1,2]. The World Health Organization (WHO) declared the COVID-19 emergence as epidemic and pandemic, respectively, on the January 30 and March 11, 2020, immediately after evaluating the contagious nature of COVID-19 illnesses that stance a significant worldwide health catastrophe [3,4]. As of February 28, 2022, there have been 434,154,739 confirmed COVID-19 cases and 5,944,342 deaths reported globally, according to the World Health Organization's “Weekly Operational Update on COVID-19″, published on March 1, 2022. Despite the rapid development of COVID-19 vaccines by different industries like Pfizer/BioNtech, Moderna, and Johnson & Johnson and mass administration of those vaccines for human immunization, however, transmission prevention of this highly virulent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants is greatly limited. Specifically, due to the re-emergence of various new SARS-CoV-2 variants of concerns (VOC) like Delta and Omicron, etc., the severity of this COVID-19 infection is still active and also rapidly spreading in some parts of the world, including India, South Africa, Brazil, the United States of America, and few countries in the European Region. Certainly, the vaccine's effectiveness will be a great concern against those continuously re-emerging variants of SARS-CoV-2. Therefore, for the last two years or more, the COVID-19 has wreaked havoc ravage to the world. However, to date, there is no active oral chemotherapeutic treatment measures have been developed to fight against this pandemic. In view of the ongoing active contagion of SARS-CoV-2 in its stupendous scale and annihilative impact on mankind, there is an extreme need for the development of effective antivirals with some novel mode of function to preserve the lives of COVID-19 victims.
Among several appealing antiviral drug targets of SARS-CoV-2, spike glycoprotein and two other proteases namely papain-like protease (PLpro) and main protease or 3-chymotrypsin-like protease (Mpro/3CLpro) represent to be very important and most studied drug target for COVID-19 drug therapeutics development [[5], [6], [7], [8], [9]]. In the present study, an exhaustive in-silico analysis of inhibiting or modulating the SARS-CoV-2 PLpro protein has been extensively employed, particularly due to the multifunctional biological functions of SARS-CoV-2 PLpro protein. Beyond the ability to process the viral polyproteins, PLpro of coronaviridae family is also responsible for cutting the three different N-terminal regions on ORF1a to produce three nsp1, nsp2 and nsp3, which are critically involved in host modulation and viral replication process [10,11]. Moreover, due to some degree of similarity in the structural organization with other proteases families including SARS-CoV-1 [12], targeting the SARS-CoV-2 PLpro protein can promptly dictate some possible therapeutic relevance for managing the COVID-19 pandemic situation. Involvement of SARS-CoV-2 PLpro protein in the processing of viral polyproteins and cleaving proteinaceous post-translational modifications on host proteins has already been established [12,13]. It has been also reported that the specificity of cleavage relies on recognition of tetrapeptidyl LXGG motif or pattern which primarily juxtaposed between viral proteins nsp1 and nsp2, nsp2 and nsp3, and nsp3 and nsp4 [14]. The yielded product is caused by means of hydrolysis of the peptide bond on the carboxyl side of glycine at the P1 position [15]. Moreover, additional proteolytic activities of SARS-CoV PLpro has also been reported earlier like removing K48-crosslinked ubiquitin (Ub) and Ub-like (Ubl) protein interferon-induced gene 15 (ISG15) from cellular proteins [7,13], which plays an important function in the regulation of host innate immune responses to viral infection [13]. The structural organization of SARS-CoV PLpro demonstrated that the active site of PLpro is made up of catalytic triad amino acid residues: Cys111–His272–Asp286 [9]. Particularly, the secondary structure of the SARS-CoV-2 PLpro protein stood like ‘thumb-palm-fingers’ architecture [16]. The thumb sub-domain of SARS-CoV-2 PLpro is comprised of six α-helices and a small β-hairpin. On the other hand, the fingers sub-domain is composed of six β-strands and two α-helices. The finger sub-domain of SARS-CoV-2 PLpro is most complex and also includes a zinc-binding site which is critically important for maintaining structural integrity and protease activity [16]. Based on earlier studies, the catalytic triad of CoVs' PLpro was located at the interface between the thumb and palm sub-domains [17,18]. Particularly, the proteolytic actions of COV's PLpro is carried out in part by the catalytic cysteine-protease in which amino acid residue Cys111 acts as a nucleophile, residue His272 acts as a general acid/base, and another catalytic residue Asp286 is linked to the histidine residue to help it align and deprotonate Cys111 [19]. Therefore, targeting the catalytic pocket is much more efficient for the development of selective SARS-CoV-2 candidate therapeutic compounds because it could allosterically inhibit the active site by triggering loop closure. Antiviral drug-candidate like chemical entities will be identified using the SARS-CoV-2 PLpro structure-guided technique therefore may be useful in not only decreasing CoV's replication but also in minimizing the dysregulation of signaling cascades in CoV's-infected cells.
In the last two years and so, in order to expedite the drug-discovery process against COVID-19 therapeutic development, scientific organizations across the globe have adopted multiple research projects including computational aspects, and henceforth, employed several advanced chemometric methodologies to dig out small chemical entities as inhibitors of the SARS-CoV-2 PLpro protein [[20], [21], [22], [23], [24], [25], [26], [27], [28], [29]]. However, little research has been conducted on active dietary constituent-based modulators or inhibitors identification against SARS-CoV-2 PLpro protein, which has yet to be discovered on a large scale. There is a number of evidence that suggests that bioactive food chemicals or food metabolites can interact with living cells or proteins or other drug entities and cause changes in physiological activities [[30], [31], [32]]. Moreover, bioactive food chemicals have either harmful or beneficial effects on human health and help to prevent or manage many diseases like metabolic disorders, obesity, type 2 diabetes, cancer, cardiovascular, and neurological problems [30,[33], [34], [35], [36], [37], [38], [39], [40], [41]]. According to many scientific studies and epidemiological data, intake of bioactive natural foodstuffs, such as vegetables and fruits, is associated with increased potential health benefits, including a lowering the risk of several chronic diseases such as cancer and anti-inflammatory diseases [36,42,43]. Numerous commonly known bioactive compounds found in different sources of foodstuffs, viz. ascorbic acid, anthocyanins, caffeine, catechins, curcumin, ellagic acid, salicylic acid, sulforaphane, quercetin, gallic acid, polyphenols, oleuropein, epigallocatechin, capsaicin, and other food substances may directly contribute toward a broad range of beneficial health effects [35,36,44,45]. However, understanding the molecular interaction mechanisms of such food components in disease-modifying implications has stayed a mystery. Moreover, based on biological assays in the different human cell and tissue samples, earlier several studies also demonstrated the significant role of bioactive dietary components in selective gene expression and large-scale epigenetic alteration, mostly by influencing phosphorylation and post-translational modification events [44,[46], [47], [48], [49]]. Therefore, in this present study, it has been aimed to identify a few potential plant-derived food constituents or food metabolites that can suppress or impede the biological activity of the SARS-CoV-2 PLpro enzyme by using various advanced computational methods. Particularly, exhaustive computational screening approaches including negative image-based (NIB) screening against dietary food compounds or food metabolites, molecular docking, and molecular dynamics (MD) simulations, and further MM-GBSA method-based binding free energy evaluation of SARS-CoV-2 PLpro protein-food compound complexes has been implemented. In particular, advanced computational techniques like NIB-based model generation was utilized to achieve better insight of ligand's binding site framework, and thereby specifically detect the neutral filler atoms, and negatively and positively charged cavity points based on negative image [50,51]. Further, the generated NIB models were considered for similarity score based screening of dietary compounds. The employed NIB methodology has been successfully implemented in improving docking performance and validated as an efficient screening tool in many novel compound discovery projects [[51], [52], [53]]. Employing such an advanced level of screening protocol the current study has identified four compounds viz. PC000550, PC000361, PC000558, and PC000573, which also exhibited to have a significant intermolecular binding affinity toward the studied SARS-CoV-2 PLpro, and hence can act as potential inhibitors/modulators for SARS-CoV-2 PLpro protein.
2. Materials and methods
2.1. Selection and preparation of food compounds and SARS-CoV-2 PLpro protein
A total of 1203 bioactive food chemicals or metabolites were retrieved from the FoodComEx database (available at https://foodcomex.org/). The entire chemical library of the FoodComEx (Food Compound Exchange) database consists of compounds or metabolites that may be directly found in various food items or can be prepared from food items. Specifically, metabolites of food compounds are usually produced by human, mammalian metabolism, or gut microbiota. Compounds are present in the FoodComEx database belong to the diverse classes of organic compounds such as pheniramine, pteridines, benzothiazoles, retinoids, estrogens, polyphenols, phenylpropanoids, quinic acids, alkaloids, and polyketides, etc. All compounds were downloaded in two-dimensional (2D) molecular structural data format (.sdf) and further converted into three-dimensional (3D) format. Particularly, using the Discovery Studio, initially, all molecules were checked for redundancies and removing bad valency compounds, and then converted into a three-dimensional (3D) organization. After careful curation of the collected compounds dataset, 1039 food compounds were found to be correct with their structural organizations and therefore considered in subsequent modeling analyses. In order to prepare each ligand compound, hydrogen atoms were added to each compound. In addition, using the OpenBabel - an open-source software tool [54], the entire compound dataset was converted into ‘.pdbqt’ format. The ‘.pdbqt’ is an essential format required in AutoDock Vina (ADV) tool [55], kept untouched till further use. Precisely, the ‘.pdbqt’ format representation is quite similar to the Protein Data Bank (PDB) format but its includes partial charges and AutoDock4 atom types. In addition, following the same protocol of ligand preparation and file format conversion, another compound GRL0617 was also prepared that was bound to the SARS-CoV-2 PLpro protein (PDB ID: 7CJM), considered as a standard or control compound in the present study for better evaluation and comparison perspective.
2.2. Preparation of SARS-CoV-2 PLpro protein and grid configuration file generation
On the other hand, the SARS-CoV-2 PLpro crystal structure with PDB ID: 7CJM [56], was retrieved from the PDB repository [57]. In order to prepare the compounds and protein structure, an appropriate number of polar hydrogen atoms, Gasteiger charges were added/adjusted to the compounds and SARS-CoV-2 PLpro crystal structure using AutoDock Tools (ADT) [58]. All water (H2O) molecules and other small molecules or hetero atoms were deleted from the selected crystal structure of SARS-CoV-2 PLpro protein. Finally, all atoms in the crystal structure were assigned to the AD4 type and saved in .pdbqt file format. For docking grid configuration file generation, initially, the coordinates information of the SARS-CoV-2 PLpro catalytic triad residues Cys111/His272/Asp286 [16] were collected from the published article, which positioned at the interface between the thumb and palm subdomains of the respective PLpro protein. However, it was found that the compound GRL0617 bound with SARS-CoV-2 PLpro protein interacted slightly apart from the exact site of catalytic triad residues, and hence, coordinates information of residues at very adjacent to the catalytic triads, and as well as residues (Ser262-His272) around the bound ligand compound GRL0617 were considered for docking configuration file generation. Precisely, in the present study, grid coordinates information were not used to entirely or exactly confine all of the catalytic triads, but their close vicinity residues were considered. Therefore, along the X-, Y-, and Z-axes, the binding site's center coordinates were chosen as 6.09, 21.00, and −6.90, respectively for the generation of grid box coordinates for molecular docking execution, specifically residues around the bound co-crystallized non-covalent inhibitor compound GRL0617, which might also be considered as allosteric site for the studied protein. Moreover, during docking, the grid dimensions size were set to 60 × 46 × 60 Å along with the X-, Y-, and Z-axes, respectively.
2.3. Negative image-based (NIB) model generation and ShaEP screening of food compounds
Three different negative image-based (NIB) models were generated using the program PANTHER [59]. Particularly, the SARS-CoV-2 PLro protein inhibitor compound GRL0617, or TTT (residue 401), that is bound at the active site in the PDB ID: 7CJM, was considered to define the cavity center during the NIB model creation, with PANTHER. For the execution of PANTHER, the box radius was defined as 10 Å. Three different NIB models were created based on assigning different parameters like keeping the bound standard GRL0617 inhibitor at the centroid, defining the ligand distance limit of 1.2 Å, and body-centered cubic packing settings. During NIB models generation, the same ligand binding site's center coordinates as 6.09, 21.00, and −6.90, along the X, Y, and Z axes, were specified. The NIB model generation focused solely on the shape/electrostatics complementarity between the ligand poses and the receptor/protein's ligand-binding site. Precisely, in the NIB model building, a negative image was generated by capsizing the shape/electrostatics or charge features of a ligand-binding cavity. Further, the resulting all three NIB model was used by ShaEP [60] - a similarity comparison algorithm for screening of all dietary compounds at the 3D structures. The ShaEP calculates the total similarity score within the range 0–1 by comparing the shape and electrostatic potential (ESP) of the cavity-based NIB model with the attached ligand molecule [52]. During ligand screening, by default, the shape and ESP are given equal weight i.e. 50:50 wt, in the total scoring by default, and this usually works well. In the present study, the ShaEP execution was carried out with multiple conformers setting for all three NIB models that were generated using PANTHER. After the execution of ShaEP program, screening of all dietary compounds was performed based on the shape and electrostatic similarity score, and therefore, an user defined arbitrary value of ‘shape_similarity’ score ≥0.6 was consider as threshold for further screening.
2.4. Molecular docking using AutoDock Vina, AutoDock tools and HADDOCK
The dietary compounds or food metabolites obtained from the NIB model based screening in the previous step was further subjected to the molecular docking execution using the ADV following standard protocol [55,61]. The grid coordinate information saved in the configuration file (mentioned in section 2.2), was used for the execution of molecular docking of all NIB model screened dietary compounds with SARS-CoV-2 PLpro protein. The widely accepted docking program ADV was installed on a Linux-based operating system used to execute the docking procedure for all compounds including the standard compound GRL0617. After successful docking execution, the binding affinity score of the standard compound GRL0617 was checked and considered as a threshold score for further screening of the dietary compounds. The binding affinity score of the standard compound GRL0617 was considered as the threshold and used for further screening criteria. The entire docking protocol execution was performed following the default settings. More details of the employed ADV protocol can be found in our previous studies [[62], [63], [64]]. In particular, molecular docking of all NIB model based screened compounds were allowed for docking execution with a maximum of nine binding modes to generate for each ligand compound. The exhaustiveness of search value was set to eight, as default. After successful execution of ADV protocol, each output generated .pdbqt files were separated using the vina_split tool for investigating the binding mode of each conformer and their corresponding binding affinity score was also checked. Moreover, two other docking programs HADDOCK and ADT were also used to perform molecular docking to re-check the conformational sampling and to get their optimal docking poses for the compounds. Following the previously described protocol [65,66], execution of the docking through HADDOCK [67] was carried out at the bound site of standard compound GRL0617. In HADDOCK, for each compound, a minimum of 193 structures, in 2–4 clusters were generated with 94–99.0% of water refined models. On the other hand, using the AutoGrid module of ADT [58], and considering the same grid center coordinates used in ADV, the atomic grid file was generated for SARS-CoV-2 PLpro. Following the default parameters setting with the Lamarckian genetic algorithm, the ‘AutoDock’ run module execution was performed for generating the ligand poses.
2.5. Molecular dynamic simulations study
The MD simulation is a fascinating approach that used a molecular force field to simulate the macro-molecular system by following Newton's equation of motion [68]. The dynamic simulation approach is widely used to explore the behavior of the molecules before testing it in the experimental laboratory. Herein, all proposed dietary compounds and standard GRL0617 bound with SARS-CoV-2 PLpro protein were taken into consideration for a 100ns MD simulation under a microcanonical ensemble. The entire MD simulation run was carried out in Amber20 software package installed in a CentOS operating system having the configuration of 10th Generation Intel Core i9-10885H and NVIDIA® GeForce RTXTM 2070. Each food compound bound with PLpro protein complex was submerged into the TIP3P water model inside the truncated octahedron box [69]. The truncated octahedron box was considered for system preparation and a 10 Å distance was maintained from the wall of the box. The protein and all dietary compounds topology files were generated using the ff14SB and GAFF2 force fields, respectively [70,71]. Moreover, a required number of Na+ and Cl− were adjusted to the entire system. The ionic strength of the system was set to 0.1 M to maintain the physiological pH in the execution of the simulation. The graphical processor unit (GPU) enabled module of Amber20 (pmemd.cuda) was used to perform the simulation [72]. The entire SARS-CoV-2 PLpro protein-dietary compound system's temperature was maintained at 300 K using a Langevin thermostat during simulation. With the help of the Monte Carlo barostat, the collision frequency was adjusted to 2 ps−1 at 1 atm. The covalent bond associated with hydrogens was controlled with the help of the SHAKE algorithm. The threshold of 8 Å was used to address the short-range electrostatic interactions and the particle mesh Ewald technique was implemented to maintain the long-range electrostatic interactions. To equally distribute the water and ions throughout the system, each system was equilibrated for a 10 ns time period through NVT and NPT ensembles. Followed by the equilibration, the MD simulation was performed for each complex. For exploration of the dynamic nature of PLpro and dietary compound in each system, a number of statistical parameters included PLpro backbone RMSD, ligand RMSD, root-mean square fluctuation (RMSF), radius of gyration (RoG), solvent accessible surface area (SASA), and, number of hydrogen bonds between SARS-CoV-2 PLpro protein and ligand compound were calculated from the MD simulation trajectories.
2.6. MM-GBSA based binding free energy and per-residue decomposition energy estimation
Moreover, the binding free energy (ΔG) of any small molecule bound with any respective target is one of the crucial parameters to determine the interaction affinity towards the respective target. It is illustrated that the binding free energy obtained from multiple frames such as MD simulation trajectory is considered to be more authentic and reliable in comparison to the binding energy calculated through any conventional forcefield approach or in molecular docking based approach. Therefore, in the current study, using the MD simulation trajectory of each complex, the binding free energy of the proposed four dietary compounds including the standard GRL0617 was estimated through the molecular mechanics-generalized born surface area (MM-GBSA) [[73], [74], [75]] method. The calculation was performed on accounting for the last 10000 MD trajectory frames i.e. approximate last 20 ns MD simulation run span for each protein-ligand system considered for binding free energy estimation. The binding free energy of each molecule was calculated through the following mathematic equations.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
The ΔG bind is can be obtained (equation (1)) by the subtraction of added free energy of the receptor (ΔG rec) and ligand (ΔG lig) from the free energy of complex (ΔG com). From the expression (2) it can be seen that the ΔG bind is the difference between two terms, enthalpy (ΔH) and entropy (TΔS). The enthalpy term can be obtained from GBSA and the entropy can be achieved from the normal mode analysis (NAM) and interaction entropy (IE) methods. The ΔH is implied by molecular mechanical energy (ΔEMM) and solvation-free energy (ΔEsol). The ΔEMM is the combination of intra-molecular (ΔEint), electrostatic (ΔEele) and the van der Waals interaction (ΔEvdw) energies. Further, the free energy of solvation (ΔGsol) can be embodied by the addition of polar (ΔGpol) and non-polar (ΔGnpol) energies. Finally, it is worth indicating that the modified Generalized Born (GB) [76] was applied to get the ΔGpol and ΔGnpol that were achieved from the LCPO algorithm [77] which is based on SASA.
The energetic contribution of individual key amino acid residue which was found to participate in several intermolecular binding interactions with the proposed compounds was assessed for MM-GBSA based per-residue energy decomposition approach using trajectories of last 20 ns. Specifically, some essentially important key amino acid residues (such as Asp164, Pro247, Pro248, Tyr268, Gln269, Cys270, His272, Tyr273, Asp286, and Asp302) found to interact in molecular docking study with the final proposed molecules were considered to calculate the per-residue decomposition energy [78,79].
2.7. Entropy estimation of the protein-ligand complexes
Following the normal-mode analysis (NMA) [79,80] the interaction entropy was calculated from MD simulated trajectories for all proposed and standard compounds-SARS-CoV-2 PLpro protein complexes. Due to the high computational cost of NMA, the last 50 trajectory frames were considered for the entropy calculation of each complex. All entropy terms were calculated through harmonic approximation with NMA. Three different entropy components like translational, rotational and vibrational entropic energy terms were calculated for the entire protein-ligand complex, and separately for individual ligand and protein, and in order to get the final entropic terms, their differences were calculated from entropic terms of the entire protein-ligand complex.
2.8. In silico ADMET analysis of proposed compounds
All identified screened inhibitor/modulator compounds were further evaluated for absorption, distribution, metabolism, excretion, and toxicity (ADMET) profile analysis. Using two highly recognized web-based application tools, namely, SwissADME [81] and pkCSM [82] were used for assessing the Lipinski's rule of five (RoF) [83] and other important ADMET properties include human intestinal absorption, water solubility, hERG I/hERG II inhibitor, skin sensibility, and mutagenicity. In order to support the clinical use of all proposed compounds, using these two tools, different medicinal chemistry and other drug-likeness properties were measured, specifically for assessing the ability of proposed compounds for being the drug-like characteristics.
3. Results and discussion
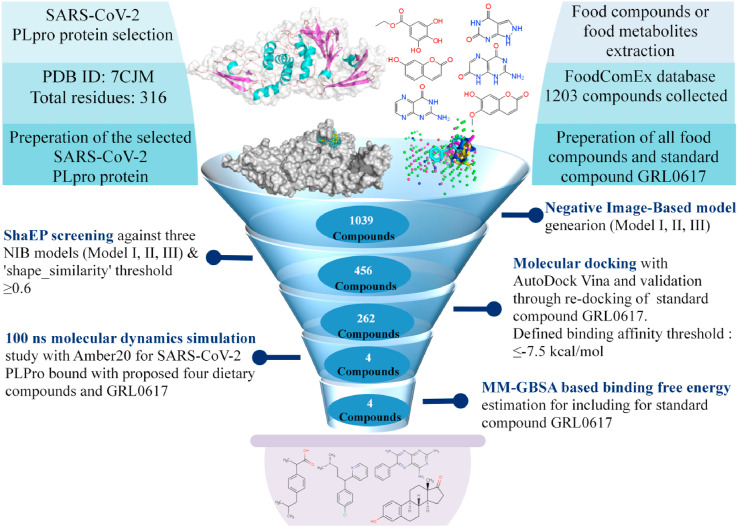
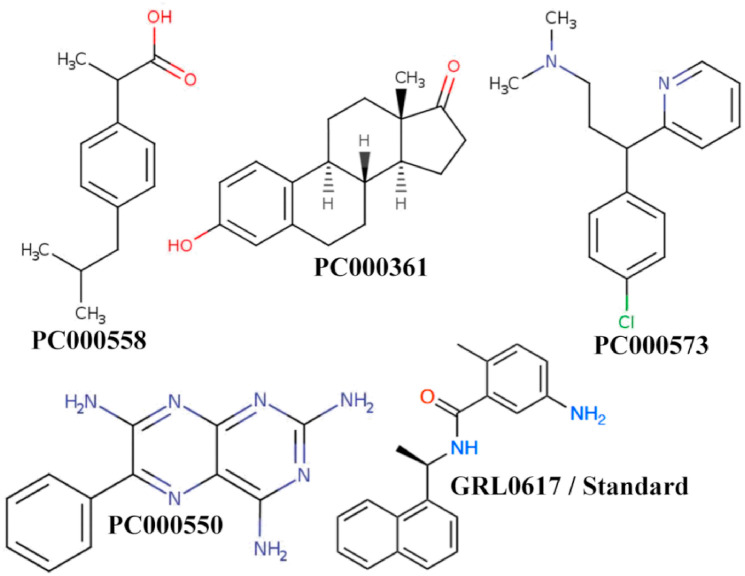
Structure-based virtual screening is a promising computational drug discovery technique for identifying possible lead-like compounds for a certain bio-macromolecular protein target. Usually, a chemical database contains a huge number of chemical entities, computational methods or algorithms can be used to swiftly examine them for possible interactions. In the same vein, a set of over 1000 food compounds or metabolites was extracted from the FoodComEx database and then screened against the SARS-CoV-2 PLpro protein using a variety of computational approaches. Primarily, in the current study, three NIB models were generated and screened by ‘shape_similarity’ score and followed by molecular docking and classical MD simulations analyses were employed to dig out some prominent food chemical entities which may create stable interactions formation with SARS-CoV-2 PLpro protein at the active site or close proximity residues and so can modulate or impede the biological function. The illustration of the entire workflow of employed study analyses is given in Fig. 1 . In detail, employing such exhaustive screening protocol which is integrated with an advanced and novel technology like NIB screening, finally, four potential food compounds or food metabolites (PC000550, PC000361, PC000558, and PC000573) were identified. The 2D chemical representation of all identified compounds is depicted in Fig. 2 . Specifically, all identified proposed compounds PC000550, PC000361, PC000558, and PC000573 are commonly known as Triamterene, Estrone, Ibuprofen, and Chlorpheniramine, respectively. Interestingly, compound PC000558 which is alternatively known as Ibuprofen is an aromatic homomonocyclic compound widely marketed as a nonsteroidal anti-inflammatory drug (NSAID) and used for relief of symptoms of arthritis, primary dysmenorrhoea, and fever. The compound PC000558 belongs to the class of organic compounds known as phenylpropanoid acids which are mostly used in the food industry to preserve and maintain the original aroma quality of frozen foods. Although this compound is not a direct food compound however such class of metabolite can be prepared from cinnamic acid (found in cinnamon bark and its products) by hydrogenation.
Fig. 1.
Schematic workflow for identification of SARS-CoV-2 PLpro inhibitors/modulators based on NIB screening, molecular docking and dynamics simulation analyses.
Fig. 2.
Two-dimensional (2D) structural representation of final proposed dietary inhibitors/modulators of SARS-CoV-2 PLpro protein.
3.1. Analysis of NIB models and ShaEP based screening
Three NIB models (Model I, II and III) or negative images were generated using the same coordinates of the SARS-CoV-2 PLpro protein bound with standard GRL0617, however, opting for different options like restraining ligand distance and specifying box radius dimension. Such conditional implementation resulted in three different NIB models, hence, focusing on comparing their ranking based on all dietary compounds close to the center of the SARS-CoV-2 PLpro protein's ligand-binding site. The obtained three NIB models (Model I, II and III) for the standard compound GRL0617 bound with SARS-CoV-2 PLpro protein is depicted in Fig. 3 A. It was observed that all three cavity-based NIB models generated were able to fit different cavity parts of the SARS-CoV-2 PLpro protein. On the other hand, three NIB models of the proposed four dietary compounds are also depicted in Fig. 3B. Interestingly, it was also noticed that all identified dietary compounds profoundly occupy the major part of all three generated NIB models.
Fig. 3.
Generated cavity-based NIB models (Model I, II, and III) shape/electrostatics (transparent surface with charge potential) for the SARS-CoV-2 PLpro protein bound with standard compound GRL0617 (A). The fitness state of all proposed dietary compounds into the generated three NIB models (Model I, II, and III) for the SARS-CoV-2 PLpro protein (B). The red and blue dots in each NIB model indicate the negative and positive cavity dots constituting the negative image. Green, cyan and magenta dots are neutral cavity dots in NIB model I, II, and III, respectively constituting the negative image. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Further, in order to calculate the shape and electrostatics similarity score of each dietary compound, internally it was calculated and compared directly against each generated NIB model (Model I, II, and III). A stringent screening protocol for 1039 dietary compounds was followed based on ShaEP executed ‘shape_similarity’ score. A user-defined threshold value of ≥ 0.6 ‘shape_similarity’ score was applied to all three generated NIB models. Implementation of such criterion resulted in sieving out of 1, 163, 292 dietary compounds for Model I, II and III, respectively. It was found that the ‘shape_similartiy’ score extends from 0.02 to 0.61, 0.04 to 0.76, 0.60 to 0.80 for NIB Model I, II, and III, respectively.
3.2. Molecular docking based interaction profile analyses of identified food compounds
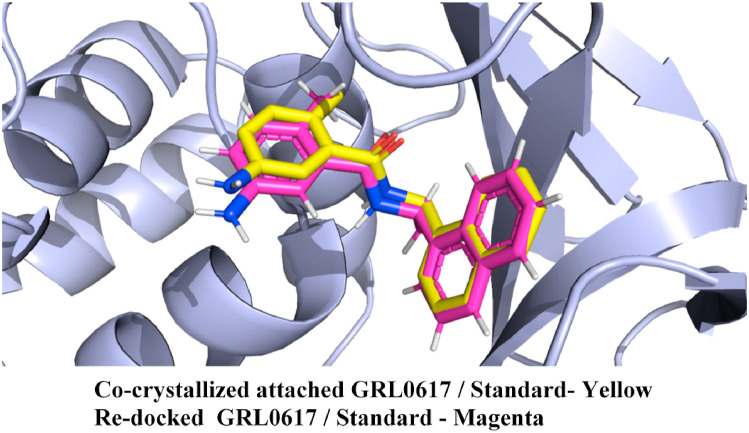
The intermolecular interaction profile of the final proposed four dietary compounds and the standard compound GRL0617 were extensively analyzed using the protein-ligand interaction profiler (PLIP) program [84]. Initially, the employed docking protocol was validated in the present study by checking the binding interactions and orientation of the standard inhibitor compound GRL0617 which were re-docked at the SARS-CoV-2 PLpro cavity and superimposed against the originally bound co-crystal pose of GRL0617. RMSD value was found to be 0.91 Å for the superimposed structural poses of originally bound co-crystal and re-docked standard compound GRL0617, and their superimposed orientation is depicted in Fig. 4 . It was indeed interesting to witness the re-docking analysis which was successfully able to reproduce almost identical binding orientation for the standard compound GRL0617 at the active site region of SARS-CoV-2 PLpro protein, and therefore implying true effectiveness of the employed docking protocol.
Fig. 4.
Superimposed orientation of the co-crystallized and re-docked standard compound GRL0617 with SARS-CoV-2 PLpro protein reveals almost identical binding orientation with a low RMSD value of 0.91 Å, suggesting successful validation of the employed docking protocol.
Detailed intermolecular binding interaction profiles of all proposed four dietary compounds bound with SARS-CoV-2 PLpro protein are given in Fig. 5 . In molecular docking analyses, the binding affinity scores were found to be −7.9, −8.2, −8.6, −8.4 and −8.1 kcal/mol for the proposed food compounds PC000550, PC000361, PC000558, PC000573, and GRL0617, respectively. Intermolecular interaction analyses disclose that all dietary food compounds were able to form a number of energetically important and biologically relevant interaction profiles, which included hydrogen bonds (HB), hydrophobic contacts and pi(π)-stacking, π-cation, and salt bridge interactions, etc. All intermolecular bonds formation were found to be within the energetically favorable and acceptable bond distances. Detailed intermolecular interaction analysis of GRL0617 revealed the participation of HB interactions with amino acid residues Asp164, Tyr268, Gln269, and Tyr273 of SARS-CoV-2 PLpro. A few amino acid residues of SARS-CoV-2 PLpro protein such as Pro247, Pro248, Tyr264, Tyr268, and Gln269 were found to interact with GRL0617 through hydrophobic contacts. Amino acid residue Tyr268 were also found to be involved in the creation of the π-stacking interaction with the standard compound GRL0617. In-depth analysis of molecular docking study of the proposed dietary compound PC000550 as potential PLPro inhibitor/modulator and its two amine groups attached in the terminal heterocyclic ring were showed the formation of two HB interactions with residue Tyr273. The phenyl ring of PC000550 was found to be crucial to establish one of each hydrophobic and π-stacking contact with another important aromatic amino acid residue Tyr268 of PLpro protein. In addition, the pyrimidine ring and one amine group of compound PC000550 were also formed salt-bridge interaction with Asp164 and Asp302 of SARS-CoV-2 PLpro protein, respectively. The molecular docking predicated intermolecular interaction profile of another proposed dietary compound PC000361 was found to form two types of intermolecular interactions, such as HB interaction and hydrophobic contacts. The presence of hydroxyl group in PC000361 was revealed as critical to connect with residues Arg166, Tyr273 and Asp302 through HB interactions. Moreover, all the cyclic rings of PC000361 were participated to establish hydrophobic contacts with Pro247, Pro248, Tyr264, Tyr268, and Thr301. Interestingly, the compound PC000361 revealed nearly similar types of hydrophobic contacts profile as of GRL0617, and hence possibly suggesting similar types of interaction mechanisms for modulating the action SARS-CoV-2 PLpro protein. The molecular docking-based intermolecular interaction analysis of compound PC000558 revealed HB interaction formation between two amino acid residues (Asp164 and Tyr273) and the terminal oxygen atom (hydroxyl oxygen) present in dietary compound PC000558. Beyond the above mentioned binding interaction for PC000558, five hydrophobic contacts were also found which were mostly facilitated with the phenyl ring and alkyl group of the small molecule, and, three amino acid residues Pro248, Tyr264 and Tyr268 of SARS-CoV-2 PLpro protein. As illustrated in Fig. 5, PC000573 showed multiple intermolecular interactions profile such as HB, hydrophobic, π-stacking and salt bridge interactions with SARS-CoV-2 PLpro protein. In particular, HB interaction was formed with residue Gln269 and pyridine of PC000573. Chlorobenzene of PC000573 was seen to form hydrophobic contacts with Pro248, Tyr264 and Tyr268. The pyridine ring of PC000573 was also found to be crucial to form hydrophobic interactions with Asl164 and Gln269. Dimethylamine of PC000573 was seen to form two salt bridges with residues Asp164 and Asp302 of SARS-CoV-2 PLpro protein. Beyond the above, Tyr268 of SARS-CoV-2 PLpro protein was revealed as an important amino acid to interact with two cyclic rings of PC000573 through π-stacking. Interestingly, it was observed that all dietary compounds docked and interacted nicely following a “two-winged” binding pattern at the active site cavity of SARS-CoV-2 PLpro protein. Overall, the binding mode and intermolecular interactions profile of all the four dietary chemical entities disclosed some critically interacting amino acid residues involvement at the active sub-sites of SARS-CoV-2 PLpro protein, and such intermolecular interactions associated with the palm sub-domain might possibly lead to a stable or controlled inhibition mechanism for SARS-CoV-2 PLpro protein. Surprisingly, most of the binding interactions were dominated in the region of residues Asp164, Pro248, Tyr264, Tyr268, and Gln269 of SARS-CoV-2 PLpro protein. Interestingly, most of the intermolecular binding interactions were shaped at very close proximity region of the catalytic triad residues or the interface of the thumb and finger subdomain, in the active site of the narrow deep two-winged space of SARS-CoV-2 PLpro protein.
Fig. 5.
Molecular binding interactions of proposed food compounds bound with SARS-CoV-2 PLpro protein obtained in molecular docking analyses.
It is indeed interesting to note that all four dietary compounds perfectly occupied the biologically relevant substrate-binding region of the SARS-CoV-2 PLpro protein. In Fig. 3, the surface view presentation of the SARS-CoV-2 PLpro protein is displayed which shows the different binding orientations of four proposed dietary molecular compounds (in stick representation) under this study including the standard compound GRL0617 suggesting tight binding affinity at the active site cavity. Such tight binding interaction affinity certainly might dictate some degree of improved therapeutic properties for SARS-CoV-2 PLpro protein and might also assist as the rationale for developing or identifying or optimizing much better chemical entities for suppressing the activity of SARS-CoV-2 PLpro protein. As illustrated in Fig. 6 , all dietary compounds docked and fit well into the P3/P4 region of the substrate-binding pocket of SARS-CoV-2 PLpro protein with a ‘two-winged’ pattern. Certainly, such binding orientation concurrently occupies the P3 and P4 pockets to block substrate access.
Fig. 6.
Molecular binding mode or orientation in 3D view of all proposed food compounds including the standard compound GRL0617 (displayed in the stick) inside the active site cavity of SARS-CoV-2 PLpro protein (displayed in surface view presentation).
Moreover, the ADT and HADDOCK-based molecular docking interaction analyses also corroborated with the study finding of ADV. Specifically, the conformational and binding orientation of the proposed four compounds has been found to be quite similar to the ADV. The HADDOCK docking results have been given as supplementary data (Table S1 and Fig. S1 in supplementary file) for all four proposed compounds including the standard compound GRL0617. In accord with ADV results, ADT results (Table S2 and Fig. S1 in supplementary file) also show strong intermolecular potentiality for all proposed compounds. From Fig. S1, it has been noticed that conformational orientations for all compounds obtained through two docking methods (ADT and HADDOCK) reveal very low RMSD value as < 2 Å, for three compounds (PC000550, PC000558, and PC000361). However, only for the compound PC000573, a little bit higher superimposed RMSD value has been observed. Such observation certainly indicates that conformational orientation or binding organization obtained in ADV and other tools for all proposed compounds are highly likely accurate and accepted for detailed conformational and interaction analyses.
3.3. Molecular dynamics simulation
The dynamic nature and interaction stability of SARS-CoV-2 PLpro protein bound with all identified potential dietary compounds including the standard inhibitor GRL0617 were explored using an all-atoms 100 ns MD simulation. The best lowest conformer docking pose of each food compound attached with PLpro protein was exposed to conventional MD simulations study. The MD simulation trajectories were explored to govern various trajectories analyzing parameters such as protein backbone RMSD, RMSF, RoG and SASA, and binding free energy using the MM-GBSA based methods. Values of each trajectory analyzing parameters in terms of maximum, minimum and average values for RMSD of both the PLpro backbone and ligand atoms, RMSF, RoG, intermolecular hydrogen bond interaction frequencies, and SASA profiles are given in Table 1 .
Table 1.
Average, maximum and minimum SARS-COV2- PLpro backbone RMSD, RMSF, RoG, and SASA values obtained for protein backbone and proposed dietary compounds during 100 ns MD simulation.
| Compounds | PC000550 | PC000361 | PC000558 | PC000573 | GRL0617 | |
|---|---|---|---|---|---|---|
| Backbone RMSD (Å) | Min. | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Max. | 3.575 | 3.289 | 3.256 | 3.676 | 2.797 | |
| Average | 2.079 | 1.991 | 1.972 | 2.010 | 1.643 | |
| RMSF (Å) | Min. | 1.424 | 2.028 | 1.501 | 2.061 | 1.886 |
| Max. | 12.674 | 22.779 | 19.824 | 23.330 | 16.664 | |
| Average | 5.560 | 11.097 | 8.230 | 9.315 | 8.242 | |
| Ligand RMSD (Å) | Min. | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Max. | 0.617 | 0.736 | 1.510 | 1.010 | 1.795 | |
| Average | 0.260 | 0.281 | 0.735 | 0.556 | 0.453 | |
| RoG (Å) | Min. | 23.229 | 23.153 | 23.228 | 23.326 | 23.114 |
| Max. | 24.496 | 24.383 | 24.471 | 24.576 | 24.304 | |
| Average | 23.809 | 23.694 | 23.808 | 23.907 | 23.732 | |
| SASA (Å2) | Min. | 23.229 | 23.153 | 23.228 | 23.326 | 23.114 |
| Max. | 24.496 | 24.383 | 24.471 | 24.576 | 24.304 | |
| Average | 23.809 | 23.694 | 23.808 | 23.907 | 23.732 |
3.3.1. RMSD analyses of the SARS-CoV-2 PLpro bound with identified proposed food compounds
The PLpro backbone RMSD of each frame bound with proposed molecules and GRL0617 were calculated and it is given in Fig. 7 . It can be seen that the PLpro backbone complex with each of the small molecules was maintained stability from the beginning to the end of the simulation. No noticeable deviation of the backbone was found when bound with proposed PLpro ligands. The average PLpro backbone RMSD was found to be 2.079, 1.991, 1.972, 2.010 and 1.643 Å when bound with PC000550, PC000361, PC000558, PC000573, and GRL0617, respectively. It is also interesting to note that the maximum RMSD among all frames was seen as 3.676 Å which substantiated that not a single frame was found extreme deviation during the MD simulation. Low average and consistent variation of RMSD sufficiently corroborated the stability of each complex in dynamic states.
Fig. 7.
RMSD values of SARS-CoV-2 PLpro protein backbone bound with proposed food compound PC000550, PC000361, PC000558, PC000573, and standard compound GRL0617.
Likewise, the RMSDs of protein backbone atoms, the deviation of individual ligands inside the PLpro active site was assessed through the calculation of ligand RMSD. For each ligand, the RMSD of the individual frame was calculated from the MD simulation trajectories and are given in Fig. 8 . Among proposed PLpro inhibitor/modulator compounds, except few frames of PC000558, all other frames of each proposed molecule were found to have consistency in RMSD values throughout the simulation phase. Close structural observation suggests that compound PC000558 is consisting of a single phenyl ring attached with two flexible chains at both terminals which might be the reason for such a different deviation in the RMSD values for a little number of frames. Moreover, the presence of greater number of rotatable bonds in the compound PC000558 also might be the plausible cause of introducing such fluctuation during simulation. Particularly, the rotatable bonds in both terminals can create some conformational swing that is probably exerted upon the induced-fit effect during simulation. Indeed, such conformational switching might have some effect on maintaining the arrangement of intermolecular interactions with SARS-CoV-2 PLpro protein. Similar to the compound PC000558, a number of frames of GRL0617 after about 50ns were shown higher deviation compared to other consistent frames. On close observation of the standard molecule GRL0617, it can be seen that presence of flexible and rotatable long chain might be the reason behind the higher deviation in dynamic states. From Table 1, the difference between average and maximum RMSD of each molecule was calculate as 0.357, 0.455, 0.775, 0.454 and 1.342 Å for PC000550, PC000361, PC000558, PC000573 and GRL0617, respectively. The above low value for each molecule undoubtedly explained the compactness of the molecules in MD simulation.
Fig. 8.
Ligand RMSD proflile for compounds PC000550, PC000361, PC000558, PC000573, and standard compound GRL0617 bound with SARS-CoV-2 PLpro protein.
3.3.2. RMSF analyses of the SARS-CoV-2 PLpro bound with identified proposed food compounds
The fluctuation of each of the amino acids of PLpro can also explain its role to hold the protein-ligand complex in dynamic states. Hence, the fluctuation of individual PLpro amino acids was calculated and it is given in Fig. 9 . It can be seen that the pattern of fluctuation of RMSF of each amino acid bound with proposed molecules and standard was almost similar. Moreover, not a single amino acid was found to fluctuate differently or abnormally during the entire simulation time. It is important to mention that the RMSF of terminal amino acids were found higher due free-floating of these regions. The average RMSF also can give an idea about the amino acid consistency in the MD simulation. From Table 1, the average RMSF of PC000550, PC000361, PC000558, PC000573 and GRL0617 was found to be 5.560, 11.097, 8.230, 9.315 and 8.242 Å respectively. It was noticed that amino acid residues approximately extending from 16 to 27, 34 to 48, 183 to 195, and 216 to 236 deviated on higher magnitudes for a few of identified food compounds in comparison to other compounds. The loop regions spanning form amino acid residues ∼185–200 and ∼220–230, which are positioned away from the binding pocket, are likely to be the cause of these significant fluctuations. Furthermore, no proposed dietary compounds were identified to form any kind of molecular binding contact at those locations of the investigated protein, hence such RMSF variations in these amino acid residues were expected. Not only did the current study uncover such an intriguing finding, but similar changes in residue levels in that specific location of the SARS-CoV-2 PLpro protein have also been described in a number of previous investigations [[85], [86], [87], [88], [89]]. From above average value and discussion along with Fig. 9, it can be concluded that in the dynamic states all amino acids of PLpro were remained consistent and held the molecules tightly.
Fig. 9.
RMSF profile of SARS-CoV-2 PLpro backbone bound with proposed food compounds PC000550, PC000361, PC000558, PC000573, and standard compound GRL0617.
3.3.3. RoG analyses of the SARS-CoV-2 PLpro bounds with identified proposed food compounds
In the MD simulation, the rigidity and compactness of any macromolecule bound with a small molecule are crucial to explain the stability of the complex. The RoG is a statistical parameter obtained from the MD simulation trajectories that can illustrate the rigidity and compactness of the molecules in the dynamic states. Moreover, the folding and unfolding of the protein are substantiated by low and high deviation of RoG value respectively. For each frame of PLpro bound with proposed and standard molecules, the RoG was calculated and it is given in Fig. 10 . Each of the complexes was seen as extremely compact and tight during the MD simulation. Not a single system was seen to have a higher deviation that corroborated the folding of the protein in the dynamic states. The difference between the highest and lowest RoG value will give an idea about the overall deviation of the parameter. The above deviation was found to be 1.267, 1.230, 1.243, 1.250 and 1.190 Å for PC000550, PC000361, PC000558, PC000573 and GRL0617 respectively. It is interesting to note that for all complexes the difference between the highest and lowest RoG is close to each other clearly explained the low and steady deviation of the system.
Fig. 10.
RoG profile of SARS-CoV-2 PLpro backbone bound with proposed food compounds PC000550, PC000361, PC000558, PC000573, and standard compound GRL0617.
3.3.4. H-bond interactions profile of the SARS-CoV-2 PLpro bound with identified proposed food compounds during MD simulation
Hydrogen bonding between protein and ligand is an essential and important factor to hold the molecules inside the active site cavity. In the MD simulation, the conformation of both protein and ligands are altered which leads to break down the existing and re-generation of new HB bond. To explore the distribution of the total number of HBs that exist in each frame, the number of HB was calculated from the MD simulation trajectories and it is given in Fig. 11 . Herein, the MD simulation trajectory-based H-bond distribution profile displayed in Fig. 11 was derived using CPPTRAJ that was considered the H-bond distance cut-off ≤ 3 Å. From Fig. 11, it can be seen that most of the frames for each complex either retained the existing HBs or re-formed it. It is important to note that most of the frames failed to form any HB with compound PC000558. In detail, all the proposed molecules were successful to keep or re-form HBs with the ligand-binding amino acids of PLpro and retained inside the projected binding site or cavity.
Fig. 11.
Distributions of H-bond interaction profile, measured within the distance length of ≤3 Å for all proposed food compounds PC000550, PC000361, PC000558, PC000573, and standard compound GRL0617 bound with SARS-CoV-2 PLpro protein during 100 ns MD simulation.
3.3.5. SASA analyses of the SARS-CoV-2 PLpro bound with food compounds
The MD simulation trajectories of each complex were used to calculate the SASA for the assessment of volume alteration during the simulation and it is given in Fig. 12 . A small variation of SASA was observed for each of the complexes in the MD simulation. In particular, the lower and higher SASA profiles were found for PLpro bound with GRL0617 and PC000537, respectively. All remaining complexes were seen to have almost similar SASA profiles. Hence, an almost similar SASA profile of all complexes was certainly explained that not much volume changes happened in dynamic states and complexes were remained compact. Moreover, the acquired constant SASA profile for all dietary compounds bound with SARS-CoV-2 PLpro protein also signifies folding of protein stably without any interferences from surrounding solvent exposers and/or any specific hydrophobic interactions that might possibly occur in a dynamic state.
Fig. 12.
PLpro backbone SASA profile bound with PC000550, PC000361, PC000558, PC000573, and standard compound GRL0617.
3.4. Superiority of the present study outcomes in comparison with previously reported studies with relation to the importance of binding interactions and stability of the identified dietary compounds with SARS-CoV-2 PLpro protein
The novelty of the present study relies on the implementation of NIB screening against food compounds or metabolites which is directed to the identification of some potential chemical entities as SARS-CoV-2 PLpro protein inhibitors/modulators. Several studies have previously demonstrated interesting findings, specifically, the importance of numerous intermolecular interactions (like H-bond, hydrophobic, π-stacking, π-cation, salt bridge, etc.) between the SARS-CoV-2 PLpro protein and different small molecular entities reported employing various experimental techniques, including in silico approaches such as molecular docking and MD simulations. Surprisingly, in the current investigation also, several numbers of intermolecular interaction profiles were identified which are very similar and/or comparable with many previously reported studies, particularly, noting the interaction profile at the active site region of SARS-CoV-2 PLpro protein residues and identified bioactive dietary compounds. S. Rajpoot et al. [90], conducted a virtual screening on some naturally occurring drugs, such as Fluoxetine, Benzenebutyric acid, Naloxone, and Acetylsalicylic acid, showed binding affinity scores of −7.5, −6.5, −7.8, and −5.3 kcal/mol, respectively, in docking with SARS-CoV-2 PLpro protein. Several numbers of common residues (such as Pro248, Tyr264, Gly266, Tyr268, Gln269, and Tyr273) participated in the formation of intermolecular interactions identified in the present study were also found with the reported naturally occurring drugs. The above mentioned study also demonstrated the dynamic nature of two drugs Fluoxetine and Naloxone which stated conformational stability of the protein-ligand complexes at a very lower RMSD value observed through 50 ns MD simulation analyses [90]. In the present study also, relatively similar or better RMSD profiles were found for all the studied dietary compounds. Another virtual screening study revealed that Desacetylgedunin, a potential bioactive phyto-compound derived from Azadirachta Indica (Neem) exhibited a docking energy score of −7.3 kcal/mol with SARS-CoV-2 PLpro protein. However, the said study reported relatively large fluctuations in RMSD values (∼after 40 ns) studied in MD simulation analyses, suggesting less structural integrity for the studied SARS-CoV-2 PLpro protein-ligand complex. This could be owing to the fact that other than the active site or different amino acid residues of the PLpro protein were involved in different interaction formations with Desacetylgedunin [91]. A high-throughput drug screening study against SARS-CoV-2 PLpro protein conducted by Y. Xu et al. [10], identified two inhibitor compounds chloroxine and Tanshinone IIA sulfonate sodium which showed docking obtained binding affinity score of −5.9 and −8.6 kcal/mol, respectively. Particularly, the said study revealed that residues, namely Tyr268 and Arg166 of SARS-CoV-2 PLpro, involved in π-π stacking and cation-π interaction formation with Tanshinone IIA sulfonate, and compound chloroxine, exhibited intermolecular binding interaction with residue Arg65. Interestingly, their MD simulation study revealed various intermolecular interaction associations with some critical amino acid residues such as Tyr264, Tyr273, Tyr268, and Gln269, also observed in the present study for the proposed dietary compounds [10]. Another structure-activity relationship study highlighted some amino acid residues viz. Leu162, Asp164, Tyr268, and Gln269 as important residues to interact with some structural analogues of potential inhibitor compound GRL0617 (analogues as Jun9–53-2, Jun9–72-2, and Jun9–75-4) identified as potent inhibitor compounds for SARS-CoV-2 PLpro protein [92]. In relation to the same potential inhibitor compound GRL0617, many other studies also demonstrated the importance of residues close proximity catalytic site residues like Pro247, Pro248, Tyr264, Tyr268, Gln269 for establishing different types of intermolecular binding interactions (such H-bonds, hydrophobic, and π-π stacking and cation-π interaction, etc.) with GRL0617 [[93], [94], [95]], however, yielded higher binding affinity score −6.91 kcal/mol in molecular docking [95], in comparison to the present study findings.
In a previously reported article, virtual screening of only 176 phytochemicals from five West African antiviral culinary herbs identified the phytochemical compound Vernonioside A4 as the prominent inhibitor molecule against SARS-CoV-2 PLpro protein with a docking score of −7.2 kcal/mol [96], revealed intermolecular interaction (H-bond) with some important amino acid residues such as Pro248, Gln250, and Try268 of the PLpro protein. Moreover, the same study also demonstrated a maximum RMSD value ∼2.5 Å during a 100 ns MD simulation study for the PLpro-Vernonioside A4 complex. In our previous study [97], the intention of finding some potential bioactive food constituents as inhibitor-modulator of SARS-CoV-2 PLpro protein, efficiently identified some food compounds (FDB001395, FDB029219, FDB030757, and FDB031079) from FooDB exhibited to interact with many common residues like Asp164, Pro247, Pro248, Tyr264, Tyr268 and Gln269 of SARS-CoV-2, as found for other dietary compounds proposed in the current study. Very interestingly, like our previous finding on 100 ns MD simulation analyses of SARS-CoV-2 PLpro protein and proposed compound complexes showed similar pattern of trajectory analyzing parameters, in the present study also exhibited no such observable fluctuations in RMSD, RoG, and RMSF profiles. In the similar fashion, the current study attributed similar type of binding interaction pattern for the majority of the identified food compounds in molecular docking and simulation analyses. Overall, outcomes of the current study findings substantiated with various parameters like intermolecular binding interactions, structural integrity and stability of the SARS-CoV-2 PLpro protein bound with identified bioactive dietary compounds were found to be highly reliable in comparison to other study findings, and therefore, may have direct the way of actual mechanism for either modulating or inhibiting the normal biological action of the SARS-CoV-2 PLpro protein.
3.5. MM-GBSA based binding free energy estimation for protein-ligand complexes and per residue decomposition energy analysis
In order to explore the binding affinity of each PLpro proposed molecule and GRL0617 from the MD simulation trajectories, the binding free energy (ΔGbind) through the MM-GBSA approach was estimated. Two important components of ΔGbind such as electrostatic and van der Waals' energies were also calculated. The ΔGbind along with all components are given in Table 2 . It can be seen that ΔGbind of PC000550, PC000361, PC000558, PC000573 and GRL0617 was found to be −27.650, −31.637, −25.948, −27.447 and −26.254 kcal/mol, respectively. It is important to note that except PC000558, all other molecules were shown to have a higher binding affinity towards PLpro in comparison to GRL0617. PC000558 was found to have comparable binding free energy with GRL0617. Moreover, both electrostatic and van der Waals’ were also shown a high negative value that substantiated that both components significantly contributed to the ΔGbind. Therefore, it can be concluded that all the selected promising molecules possess a strong affinity towards PLpro and might be effective inhibitors for PLpro.
Table 2.
Average binding free energy of PC000550, PC000361, PC000558, PC000573 and standard compound GRL0617 calculated from the MD simulation trajectories.
| Food compounds | Different energy terms (in kcal/mol) |
|||
|---|---|---|---|---|
| aElec. (dSEM) | bvdW (dSEM) | ΔGbind | cStd. Dev. | |
| PC000550 | −44.221 (0.206) | −29.449 (0.051) | −27.650 | ±2.382 |
| PC000361 | −60.267 (0.222) | −30.439 (0.093) | −31.637 | ±3.795 |
| PC000558 | −36.310 (0.336) | −26.866 (0.147) | −25.948 | ±3.229 |
| PC000573 | −40.343 (0.226) | −32.822 (0.064) | −27.447 | ±2.434 |
| GRL0617 | −28.638 (0.279) | −34.127 (0.050) | −26.254 | ±3.065 |
Electrostatic.
Van der Waal's.
Standard deviation.
Standard error of Mean.
To understand the energy contribution of key amino acids found in binding interactions analysis was further considered for the calculation of per residue decomposition energy through the MM-GBSA method. As similar to the MM-GBSA based ΔGbind calculation, the last 20 ns MD simulation trajectories were used to calculate the per residue decomposition energy of Asp164, Pro247, Pro248, Tyr268, Gln 269, Cys270, His272, Tyr273, Asp286 and Asp302. The per-residue decomposition energy of the above mentioned residues is given in Table 3 and Fig. 13 .
Table 3.
The per-residue decomposition energy (kcal/mol) for important key amino acid residues of SARS-CoV-2 PLpro protein bound with proposed compounds.
| Key amino acid residues | per-residues interaction energies (kcal/mol) |
||||
|---|---|---|---|---|---|
| PC000550 | PC000361 | PC000558 | PC000573 | GRL0617 | |
| Asp164 | −1.676 | −0.141 | −1.706 | −2.239 | −1.521 |
| Pro247 | −0.811 | −1.318 | −0.413 | −0.064 | −1.093 |
| Pro248 | −0.928 | −2.505 | −1.923 | −1.958 | −1.884 |
| Tyr268 | −2.257 | −1.669 | −2.107 | −1.151 | −1.568 |
| Gln269 | −0.071 | −0.059 | −0.235 | −1.932 | −1.042 |
| Cys270 | −0.013 | −0.011 | −0.132 | −0.599 | −0.037 |
| His272 | −0.018 | −0.035 | −0.252 | −0.313 | −0.098 |
| Tyr273 | −1.904 | −2.017 | −2.301 | −0.060 | −1.024 |
| Asp286 | −0.503 | −0.003 | −0.002 | −0.002 | −0.002 |
| Asp302 | −1.853 | −1.886 | −0.328 | −0.346 | −0.748 |
Fig. 13.
Per-residue decomposition energy of some key amino acid residues of PLpro protein showed significant energy contribution for forming the stable complexes with identified food compounds including standard compound GRL0617(A); per-residue decomposition energy of all amino acid residues of SARS-CoV-2 PLpro protein interacted with all four identified food compounds including standard compound GRL0617 (B).
From Table 3 and Fig. 13A, it can be seen that amino acids bind to the respective ligand were shown to have high negative decomposition energy in the MD simulation. In particular, for some key residues Asp164, Tyr268, Tyr273 and Asp302 were found to be interacted with the compound PC000550 either through HB or hydrophobic interactions. In Table 3, the decomposition energy of almost all the above mentioned amino acid residues was found to have high negative energy values in comparison to others. A number of amino acids included Pro247, Pro248, Tyr268, Tyr273 and Asp302 were found to have less than −1.00 kcal/mol decomposition energy when bound with PC000361 also corroborated through the binding interactions with the molecular docking study. PC000558 was seen to interact with residues Asp164, Pro248, Tyr264, Tyr268 and Tyr273 in the molecular docking study that substantiated through high negative decomposition energies. Moreover, MM-GBSA based free decomposition energy of entire residue-ligand pairs has been displayed in Fig. 13B. It can be observed that except key ligand-binding amino acids and a few vicinity amino residues, all were found to have no significant contribution in the ligand-binding interactions during the MD simulation. Most of the key residues like Pro247, His272, Tyr273, Asp286 and Asp302, other amino residues in Table 3 were found to show strong energetic affinity toward the compound PC000573. The above observation was also substantiated with the binding interactions between PC000573 and key amino residues found in the docking study. The decomposition energy of amino acids interacting with the standard compound GRL0617 was also shown to have high negative energetic values for some key residues in comparison to the non-interacting amino acids. Hence, the above observation and discussion undoubtedly suggested that all final proposed molecules were strongly bound with the key amino residues of PLpro.
3.6. Entropy analyses of the protein-ligand complexes
In the present study, following the NMA protocol in harmonic approximation, total entropy (ΔS) contribution was evaluated for all protein-ligand complexes including the standard compound GRL0617. Different entropy terms generated through the NMA for all compound-protein complexes using the 50 trajectory frames from the last 20ns of MD simulation are given in Table 4 . It was found that translational entropy contribution for all compounds were concentrating around −12.000 kcal/mol. On the other hand, rotational energy term were found to be −9.870, −9.847, −9.468, −10.205, and −10.150 kcal/mol for compounds PC000550, PC000361, PC000558, PC000573, and GRL0617, respectively, Total entropy contribution (ΔS) in terms of differences (i.e. Complex - Receptor - Ligand), was found to be −17.482, −19.633, −9.490, −13.912, and −10.713 kcal/mol for compounds PC000550, PC000361, PC000558, PC000573, and GRL0617, respectively. Indeed, it was interesting to notice that the total entropic contribution of most of the proposed compounds (PC000550, PC000361, and PC000573) exhibited much better energy contribution, in comparison to the standard compound GRL0617. Although from a limited snapshot of 50 MD frames, the estimated entropy terms given in Table 4, it can be suggested that with relatively low energy entropy scores of all proposed compounds the binding interaction stability will not be affected in dynamic conditions.
Table 4.
Estimated total entropy (ΔS) and other different entropic energy terms for all proposed and standard compounds bound with SARS-CoV-2.
| Compounds | Different entropy terms (in kcal/mol) |
||||
|---|---|---|---|---|---|
| Translational | Rotational | Vibrational | ΔS | aStd. Dev. | |
| PC000550 | −12.635 | −9.870 | 5.0236 | −17.482 | ±3.3673 |
| PC000361 | −12.638 | −9.847 | 2.853 | −19.633 | ±3.150 |
| PC000558 | −12.395 | −9.468 | 12.372 | −9.490 | ±3.872 |
| PC000573 | −12.661 | −10.205 | 8.954 | −13.912 | ±1.702 |
| GRL0617 | −12.763 | −10.150 | 12.000 | −10.713 | ±2.336 |
Standard deviation.
3.7. Post MD simulations based on conformational arrangement and intermolecular binding interactions analyses
Sometimes it may be observed that molecular docking obtained intermolecular interaction may lose the initial interaction integrity during the MD simulation run, or even some new intermolecular interactions may also arise due to various forces governed to the protein-ligand system. Such as, the intermolecular binding interactions established during the MD simulations might have highlighted preferential binding orientation of the protein-ligand complexes. Analysis of protein-ligand interactions based on MD simulation data was therefore thoroughly investigated to understand the sustainability of the key interactions revealed in the MD simulation study. The intermolecular binding interaction profiles obtained from the last frame, specifically at 100 ns MD simulation for each protein-ligand complex was extracted and displayed in Fig. 14 . It should be noted that the numbers of intermolecular contacts may differ depending on the H-bond distance cut-off opted in different programs (such as CPPTRAJ (≤3 Å) and PLIP), especially for the H-bond distribution profile seen in MD simulation trajectories analysis (Fig. 11). Herein, in post-MD simulation analyses, all appeared H-bond interaction distances have been found within the range of 1.63–3.29 Å, whereas hydrophobic distances measured within the 2.86 to 3.92 Å for all compounds. In particular, three different types of intermolecular interactions (such as HB, hydrophobic and salt bridges) were observed for compound PC000550 with SARS-CoV-2 PLpro protein. Three amino acid residues Pro248, Tyr264, and Tyr268 established hydrophobic contacts with compound PC000550. Side chain atom of Tyr273 residue of SARS-CoV-2 PLpro protein formed H-bond interaction with PC000550. Another H-bond interaction was also found between the residue Thr301 and compound PC000550. Salt bridge interaction was observed with residue Asp302. It was truly interesting to observe that with a slight shift in binding orientation most of the intermolecular interactions for compound PC000550, sustained its initial binding interaction with the same (Tyr268, Tyr273, and Asp302) or very close proximity catalytic site residues (Pro248 and Tyr264) of SARS-CoV-2 PLpro protein. MD simulation based interaction profile of compound PC000361 revealed very similar types of intermolecular binding interactions as obtained in the molecular docking study. Particularly, amino acid residues Pro248, Tyr264, Tyr273, and Thr301 established hydrophobic contacts which were found to be common residues with the docking-based study. Moreover, H-bond interactions between SARS-CoV-2 and compound PC000361 were also found to be interesting in terms of revealing the almost similar residues (Arg166 and Asp302) involvement in intermolecular interaction formation even after a long run of the simulation. However, a new H-bond interaction was found for compound PC000361 with residue Ser245. In the dynamic state, intermolecular interaction for compound PC000558 revealed H-bond interaction with the residue Tyr268, which was slightly different from the association of other residues (Asp164 and Tyr273) found in the initial binding obtained in molecular docking. However, with a new residue (i.e. Asp164) and other common or similar residues (Tyr264 and Tyr268) involvements were found for hydrophobic contacts formation with the compound PC000558. At the simulation end i.e. at 100 ns, the molecular binding interactions for the compound PC000573 with SARS-CoV-2 PLpro protein was critically checked and interestingly it was found a few newer residue involvement. Compound PC000573 created hydrophobic contacts with Asp164 and Tyr268 and H-bond interactions with residues Glu167 and Tyr273 of SARS-CoV-2 PLpro protein. Moreover, salt bridge interaction with residue Asp164 was also found to be common for the compound PC000573. Certainly, it's worth noting that such new intermolecular binding interactions arise in MD simulation for compound PC000573, however, weren't all that much different from the initial binding orientation. During MD simulation, flexibility imposed on the compound PC000573 as well as to the PLpro protein might generate such a new conformational state of the PC000573-SARS-CoV-2 PLpro protein complex. Overall, the post-MD based intermolecular interaction analysis and conformational arrangement of all proposed compounds revealed quite strong interactions similarity with SARS-CoV-2 PLpro protein, as observed in molecular docking studies. The most fascinating finding was observed in terms of identification of many key residues (Asp164, Arg166, Pro248, Tyr264, Tyr268, Tyr273, and Asp302) of SARS-CoV-2 PLPro protein engagement in several types of intermolecular interaction formations, which might possibly trigger to modulate the activity of the studied protein. Moreover, for better understanding of the conformational binding organization of all identified compounds, superimposed snapshot of each compound's orientation during MD simulation at different time intervals like 20 ns, 40 ns, 60 ns, 80 ns, and 100 ns, over 0 ns has been deduced and displayed in Fig. 15 . It was truly interesting to observe that most of the proposed compounds have not largely deviated from their initial binding regions, except the compound PC000573. However, it is also interesting to see that during 40–60 ns, the compound PC000573 slightly deviated its original position, however, in long run, it again re-organized and re-gain its initial binding orientation, indicating its binding potentiality with SARS-CoV-2 PLpro protein.
Fig. 14.
Post-MD simulated interaction analyses for all proposed inhibitor/modulator compounds bound with the SARS-CoV-2 PLpro protein.
Fig. 15.
Molecular binding mode or orientation of all proposed compounds (displayed in sticks) in different time intervals (0, 20, 40, 60, 80, and 100 ns) obtained during MD simulations with SARS-CoV-2 PLpro protein (displayed in surface view presentation).
3.8. In silico ADMET profile analyses of proposed inhibitor/modulator compounds
Results of the predicted ADMET profile for all proposed inhibitor/modulator compounds have been given in Table 5 . No proposed compounds were found to violate the RoF, hence signifying the potentiality of the proposed compounds for being the potential drug-like candidate molecules. In general, it is reported that compounds having a TPSA value less than 140 Å might be potential for orally active in nature [98]. The predicted TPSA value of compounds PC000550, PC000361, PC000558, and PC000573 was found to be 129.62, 37.3, 37.3, and 16.13 Å2, respectively clearly suggested the orally active potentiality of all proposed molecules. Moreover, all identified molecules were found to be soluble in nature and also possessed to be highly absorbable in the gastrointestinal tract. The number of rotatable bonds for the compounds PC000550, PC000361, PC000558, and PC000573 was found to be 1, 0, 4, and 5, respectively. The above observation suggested that compound PC000558 and PC000573 is slightly flexible in comparison to the others two compounds. Moreover, the synthetic accessibility (SA) of the proposed compounds was also checked. SA parameter explains how easy or challenging to synthesize a molecule. Higher the SA value more difficult to synthesis. The SA of all four proposed compounds was found to be less than 4, and hence suggested the ease and feasibility of synthesis of the proposed compounds. AMES toxicity profile exhibited no indication of mutagenicity of any proposed compounds. Estimated maximum tolerated toxic dose of compounds was found to be 0.03, −0.49, 1.00, and −0.34 log(mg/kg/day) for compounds PC000550, PC000361, PC000558, and PC000573, respectively. As per recommended maximum tolerated toxic dose for any administered compound considered to be low at ≤ 0.477 log(mg/kg/day), otherwise high when value > 0.477 log(mg/kg/day) [99]. The majority of the proposed compounds were also revealed as non-inhibitor of hERG (human ether-a-go-go gene) – I and II, which is considered to be an important indicator for measuring the possible cardiotoxicity nature of any compound. Moreover, no indication of skin sensitization was also found for all proposed compounds, except compound PC000558, hence indicating that there will be no potential skin irritation or allergic effect that might occur on the administration of these compounds, except the compound PC000558. Another important parameter is the oral acute toxicity (LD50) was found to be 2.17, 1.19, 2.37, and 1.57 mol/kg for compound PC000550, PC000361, PC000558, and PC000573, respectively. Predicted values for oral acute toxicity profiles of all proposed compounds were found within the recommended range i.e. LD50 < 2.5 mol/kg. Overall predicted ADMET profiles signify that all the screened food constituents or metabolites compounds showed potential drug-candidate like characteristics and therefore, can be further assessed for better evaluation of SARS-CoV-2 PLpro protein modulation/inhibition activity. Hence, from the above discussion, it is clear that the final proposed molecules are potential enough for SARS-CoV-2 PLpro inhibition/modulation.
Table 5.
Predicted toxicity properties of selected SARS-CoV-2 PLpro inhibitors/modulators.
| Parameters | PC000550 | PC000361 | PC000558 | PC000573 |
|---|---|---|---|---|
| aMW(g/mol) | 253.26 | 270.37 | 206.28 | 274.79 |
| bNHA | 19 | 20 | 15 | 19 |
| cNAHA | 16 | 6 | 6 | 12 |
| dNRB | 1 | 0 | 4 | 5 |
| eMR | 73.78 | 80.06 | 62.18 | 80.82 |
| fTPSA(Åb) | 129.62 | 37.3 | 37.3 | 16.13 |
| gLogS | −2.58 | −3.71 | −3.36 | −3.82 |
| hSC | Soluble | Soluble | Soluble | Soluble |
| iGI | High | High | High | High |
| jvRoF | 0 | 0 | 0 | 0 |
| kBS | 0.55 | 0.55 | 0.85 | 0.55 |
| lSA | 2.51 | 3.27 | 1.92 | 2.7 |
| LogP | 1.45 | 2.4 | 2.17 | 3.17 |
| Toxicity Parameters | ||||
| AMES toxicity | No | No | No | No |
| Max. tolerated dose (human) | 0.03 | −0.49 | 1.00 | −0.34 |
| hERG I/hERG II inhibitor | No/Yes | No/Yes | No/No | No/No |
| Oral Rat Acute Toxicity (LD50) | 2.17 | 1.19 | 2.37 | 1.57 |
| Oral Rat Chronic Toxicity (LOAEL) | 1.64 | 2.00 | 2.04 | 1.04 |
| Skin Sensitization | No | No | Yes | No |
Molecular weight.
No of heavy atoms.
No of aromatic heavy atoms.
No of rotatable bonds.
Molar refractivity.
Topological polar surface area.
Solubility.
Solubility class.
Gastrointestinal absorption.
Violation of Lipinski's rule of five.
Bioavailability score.
Synthetic accessibility.
3.9. Rational of different pharmacophoric and chemical features of identified compounds targeting SARS-CoV-2 PLpro protein
The presence of some common chemical functional groups and/or different important pharmacophoric features in any small molecule is crucial for explicating the effective binding interactions with biologically important amino acid residues at the binding site of the respective protein. Therefore, the propensity of the presence of different chemical functional groups and pharmacophoric features in the proposed molecules and standard GLR0617 has been explored through the Python RDKit. Specifically, the presence or absence of a few important pharmacophoric features like HB acceptor (HBA), HB donor (HBD), hydrophobic (HY) and ring aromatic (RA) features has been investigated, and their possible active role has been correlated in terms of observed intermolecular connectivity with those identified features predicted through molecular docking analyses for implicating better binding affinity. The number of each pharmacophoric feature found in the proposed molecules is given in Table 6 . It can be seen that all proposed molecules were found to have a comparable number of pharmacophoric features to the standard molecule, GRL0617. In detail, compound PC000550 revealed a total of 4 and 3 HBA and HBD features, respectively that indicated the possibility and potentiality of formation a large number of HB interactions with SARS-CoV-2 PLpro amino acids. PC000361 was found to have more hydrophobic features that raised the possibility of hydrophobic binding interactions formation with the studied protein. Likewise compound PC000361, the higher number of hydrophobic features were also found for compound PC00558. Compound PC000573 was found to have 1, 1, 4 and 2 HBA, HBD, HY and RA feature, respectively. On the other hand, the standard molecule, GRL0617 was found to have 1, 2, 5 and 3 HBA, HBD, HY and RA feature, respectively. In the binding interaction analysis, it was also found the importance of a number of chemical groups to interact SARS-CoV-2 PLpro. Hence, explored pharmacophoric features and observed binding interaction analyses efficaciously corroborated the potentiality of the molecules. Moreover, it can be postulated that the above-identified pharmacophoric features and important chemical groups might be helpful to design new potential and effective molecules for the successful inhibition of SARS-CoV-2 PLpro.
Table 6.
Different important pharmacophoric features present in the identified proposed compounds targeting the SARS-CoV-2 PLpro protein.
| Compounds | aHBA | bHBD | cHY | dRA |
|---|---|---|---|---|
| PC000550 | 4 | 3 | 1 | 3 |
| PC000558 | 1 | 0 | 5 | 1 |
| PC000361 | 2 | 1 | 6 | 1 |
| PC000573 | 1 | 1 | 4 | 2 |
| GRL0617 | 1 | 2 | 5 | 3 |
HB acceptor.
HB donor.
Hydrophobic.
Ring aromatics.
4. Conclusion
In the present study, NIB model based screening was performed against a total of 1039 food compounds or metabolites from the FoodComEx database for impeding the role of the SARS-CoV-2 PLpro protein. A number of advanced pharmacoinformatics methods were implemented to find out promising PLpro inhibitors/modulators for successful application in recently pandemic COVID-19 management. Exhaustive analyses based on NIB screening, molecular docking and MD simulation, and binding free energy estimations of four potentially active dietary compounds PC000550, PC000361, PC000558, PC000573 as SARS-CoV-2 PLpro inhibitors or modulators have been identified. Initially, based on ShaEP similarity_score and binding affinity score, the chemical space was reduced. Interestingly, all three generated NIB models highlighted that all proposed dietary compounds were capable of engaging negative images suitably at the ligand's binding cavity site. Further, extensive binding interactions analysis was carried and compared with the standard compound GRL0617, which suggested better binding affinity of proposed dietary compounds. In molecular docking, significant numbers of intermolecular binding interactions were found between the various functional chemical groups of the identified dietary compounds and the close proximity catalytic site amino acid residues of SARS-CoV-2 PLpro protein. The majority of the bonding interactions were also highlighted through important residues Asp164, Pro247, Pro248, Tyr264, Tyr268 and Gln269 which were also reported in several studies confirmed by computational simulations studies in a dynamic state. Precisely, the dynamic behavior of each molecule inside the PLpro active site was assessed through MD simulation study. A number of statistical parameters such as RMSD and RMSF, RoG, SASA, H-bond interaction profile etc., were obtained from MD simulation to explain the greater conformational stability for all SARS-CoV-2 PLpro-dietary compound complexes. Moreover, the lower binding free energy score of each proposed food compound was determined by MM-GBSA based application, which also suggests that greater potentiality of most of the dietary compounds' binding affinity for the SARS-CoV-2 PLpro than the standard compound GRL0617. Although, among all the proposed inhibitors/modulators, the compound PC000558 showed however comparable, but relatively lower MM-GBSA binding energy, entropy contribution and per residue decomposition energies than the standard compound GRL0617, which suggest that the compound PC000558 might need further structural modification or optimization for exhibiting better binding affinity for the studied target protein. However, structural stability of the compound PC000558 found in MD simulation analysis reflected a solid degree of robustness for the said protein-ligand complex indicates that an entropic penalty that counters complex formation. Collectively, the strong negative ΔG values were obtained for all proposed compounds bound with SARS-CoV-2 PLpro protein ranging from −25.948 to −31.637 kcal/mol. The computationally exhaustive entropy terms was calculated which also revealed a strong energetic contribution for all proposed compounds necessary for maintaining stable interaction affinity with the SARS-CoV-2 PLpro protein. ADMET profile analyses also suggested potential drug-like characteristics of all compounds. Overall, the present exhaustive multiple computational study outcomes explained that all proposed compounds could be potential inhibitors/modulators of SARS-CoV-2 PLpro protein, for suppressing or blocking the viral replication process upon binding with the studied protein, however, more extensive in-vitro and in-vivo research is needed for better evaluation of the efficacy of the proposed compounds.
Availability of data and material
Not availability.
CRediT authorship contribution statement
Shovonlal Bhowmick: Conceptualization, Funding acquisition, Data curation, Formal analysis, Writing – original draft, Conception and design of the study, acquisition of data, analysis and interpretation of data and drafting the article. Nora Abdullah AlFaris: Formal analysis, Data curation, Writing – original draft, Analysis and interpretation of data and drafting the article. Jozaa Zaidan ALTamimi: Formal analysis, Data curation, Writing – original draft, Analysis and interpretation of data and drafting the article. Zeid A. ALOthman: Writing – original draft, conception and design of the study and drafting the article. Pritee Chunarkar Patil: Funding acquisition, Data curation, Formal analysis, conception and design of the study, acquisition of data, analysis and interpretation of data. Tahany Saleh Aldayel: Funding acquisition, Data curation, Formal analysis, conception and design of the study, acquisition of data, analysis and interpretation of data. Saikh Mohammad Wabaidur: Writing – original draft, conception and design of the study and drafting the article. Achintya Saha: Writing – original draft, Conception and design of the study, drafting the article, final approval and submission.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgment
This work was funded by the Deanship of Scientific Research at the Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia, through the Research Groups Program Grant No. (RGP-1440–0021).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105474.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P.G.T., Fu H., Dighe A., Griffin J.T., Baguelin M., Bhatia S., Boonyasiri A., Cori A., Cucunubá Z., FitzJohn R., Gaythorpe K., Green W., Hamlet A., Hinsley W., Laydon D., Nedjati-Gilani G., Riley S., van Elsland S., Volz E., Wang H., Wang Y., Xi X., Donnelly C.A., Ghani A.C., Ferguson N.M. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhowmick S., Saha A., Osman S.M., Alasmary F.A., Almutairi T.M., Islam M.A. Structure-based identification of SARS-CoV-2 main protease inhibitors from anti-viral specific chemical libraries: an exhaustive computational screening approach. Mol. Divers. 2021;25:1979–1997. doi: 10.1007/s11030-021-10214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steuten K., Kim H., Widen J.C., Babin B.M., Onguka O., Lovell S., Bolgi O., Cerikan B., Neufeldt C.J., Cortese M., Muir R.K., Bennett J.M., Geiss-Friedlander R., Peters C., Bartenschlager R., Bogyo M. ACS infectious diseases; 2021. Challenges for Targeting SARS-CoV-2 Proteases as a Therapeutic Strategy for COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu G., Lee J.H., Parker Z.M., Acharya D., Chiang J.J., van Gent M., Riedl W., Davis-Gardner M.E., Wies E., Chiang C., Gack M.U. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 2021;6:467–478. doi: 10.1038/s41564-021-00884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Z., Huang B., Tang J., Liu S., Liu M., Ye Y., Liu Z., Xiong Y., Zhu W., Cao D., Li J., Niu X., Zhou H., Zhao Y.J., Zhang G., Huang H. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021;12:488. doi: 10.1038/s41467-020-20718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden van Noort G.J., Ovaa H., Muller S., Knobeloch K.P., Rajalingam K., Schulman B.A., Cinatl J., Hummer G., Ciesek S., Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiti B.K. Can papain-like protease inhibitors Halt SARS-CoV-2 replication? ACS Pharmacol. Transl. Sci. 2020;3:1017–1019. doi: 10.1021/acsptsci.0c00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y., Chen K., Pan J., Lei Y., Zhang D., Fang L., Tang J., Chen X., Ma Y., Zheng Y., Zhang B., Zhou Y., Zhan J., Xu W. Repurposing clinically approved drugs for COVID-19 treatment targeting SARS-CoV-2 papain-like protease. Int. J. Biol. Macromol. 2021;188:137–146. doi: 10.1016/j.ijbiomac.2021.07.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach, Biochimica et biophysica acta. Mol. Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti-COVID-19 drug design. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden van Noort G.J., Ovaa H., Müller S., Knobeloch K.-P., Rajalingam K., Schulman B.A., Cinatl J., Hummer G., Ciesek S., Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho C.-C., Li S.G., Lalonde T.J., Yang K.S., Yu G., Qiao Y., Xu S., Ray Liu W. Drug repurposing for the SARS-CoV-2 papain-like protease. ChemMedChem. 2022;17 doi: 10.1002/cmdc.202100455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osipiuk J., Azizi S.-A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A., Kang S., Kathayat R.S., Kim Y., Lisnyak V.G., Maki S.L., Nicolaescu V., Taylor C.A., Tesar C., Zhang Y.-A., Zhou Z., Randall G., Michalska K., Snyder S.A., Dickinson B.C., Joachimiak A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021;12:743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osipiuk J., Azizi S.A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A., Kang S., Kathayat R.S., Kim Y., Lisnyak V.G., Maki S.L., Nicolaescu V., Taylor C.A., Tesar C., Zhang Y.A., Zhou Z., Randall G., Michalska K., Snyder S.A., Dickinson B.C., Joachimiak A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021;12:743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson J.A., Verma N., Harris R.C., Liu R., Shen J. Assessment of proton-coupled conformational dynamics of SARS and MERS coronavirus papain-like proteases: implication for designing broad-spectrum antiviral inhibitors. J. Chem. Phys. 2020;153:115101. doi: 10.1063/5.0020458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alamri M.A., Tahir ul Qamar M., Mirza M.U., Alqahtani S.M., Froeyen M., Chen L.-L. Discovery of human coronaviruses pan-papain-like protease inhibitors using computational approaches. J. Pharmaceut. Anal. 2020;10:546–559. doi: 10.1016/j.jpha.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong S., Sun J., Mao Z., Wang L., Lu Y.L., Li J. A guideline for homology modeling of the proteins from newly discovered betacoronavirus, 2019 novel coronavirus (2019-nCoV) J. Med. Virol. 2020;92:1542–1548. doi: 10.1002/jmv.25768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vardhan S., Sahoo S.K. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput. Biol. Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W., Grohmann C., Shibata Y., Gan Z.Y., Cooney J.P., Doerflinger M., Au A.E., Blackmore T.R., van der Heden van Noort G.J., Geurink P.P., Ovaa H., Newman J., Riboldi-Tunnicliffe A., Czabotar P.E., Mitchell J.P., Feltham R., Lechtenberg B.C., Lowes K.N., Dewson G., Pellegrini M., Lessene G., Komander D. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39 doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosken Y.K., Cholko T., Lou Y.C., Wu K.P., Chang C.A. Insights into dynamics of inhibitor and ubiquitin-like protein binding in SARS-CoV-2 papain-like protease. Front. Mol. Biosci. 2020;7:174. doi: 10.3389/fmolb.2020.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welker A., Kersten C., Muller C., Madhugiri R., Zimmer C., Muller P., Zimmermann R., Hammerschmidt S., Maus H., Ziebuhr J., Sotriffer C., Schirmeister T. Structure-activity relationships of benzamides and isoindolines designed as SARS-CoV protease inhibitors effective against SARS-CoV-2. ChemMedChem. 2021;16:340–354. doi: 10.1002/cmdc.202000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thurakkal L., Singh S., Roy R., Kar P., Sadhukhan S., Porel M. An in-silico study on selected organosulfur compounds as potential drugs for SARS-CoV-2 infection via binding multiple drug targets. Chem. Phys. Lett. 2021;763 doi: 10.1016/j.cplett.2020.138193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouffouk C., Mouffouk S., Hambaba L., Haba H. Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CL(pro) and PL(pro)), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2) Eur. J. Pharmacol. 2021;891 doi: 10.1016/j.ejphar.2020.173759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delre P., Caporuscio F., Saviano M., Mangiatordi G.F. Repurposing known drugs as covalent and non-covalent inhibitors of the SARS-CoV-2 papain-like protease. Front. Chem. 2020;8:594009. doi: 10.3389/fchem.2020.594009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishra D., Suri G.S., Kaur G., Tiwari M. Comparative insight into the genomic landscape of SARS-CoV-2 and identification of mutations associated with the origin of infection and diversity. J. Med. Virol. 2021;93:2406–2419. doi: 10.1002/jmv.26744. [DOI] [PubMed] [Google Scholar]
- 29.Jamalan M., Barzegari E., Gholami-Borujeni F. Structure-based screening to discover new inhibitors for papain-like proteinase of SARS-CoV-2: an in silico study. J. Proteome Res. 2021;20:1015–1026. doi: 10.1021/acs.jproteome.0c00836. [DOI] [PubMed] [Google Scholar]
- 30.Malakar S. Bioactive food chemicals and gastrointestinal symptoms: a focus of salicylates. J. Gastroenterol. Hepatol. 2017;32(Suppl 1):73–77. doi: 10.1111/jgh.13702. [DOI] [PubMed] [Google Scholar]
- 31.Koziolek M., Alcaro S., Augustijns P., Basit A.W., Grimm M., Hens B., Hoad C.L., Jedamzik P., Madla C.M., Maliepaard M., Marciani L., Maruca A., Parrott N., Pávek P., Porter C.J.H., Reppas C., van Riet-Nales D., Rubbens J., Statelova M., Trevaskis N.L., Valentová K., Vertzoni M., Čepo D.V., Corsetti M. The mechanisms of pharmacokinetic food-drug interactions – a perspective from the UNGAP group. Eur. J. Pharmaceut. Sci. 2019;134:31–59. doi: 10.1016/j.ejps.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Vernocchi P., Del Chierico F., Putignani L. Gut microbiota metabolism and interaction with food components. Int. J. Mol. Sci. 2020;21:3688. doi: 10.3390/ijms21103688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burton-Freeman B.M., Sandhu A.K., Edirisinghe I. Red raspberries and their bioactive polyphenols: Cardiometabolic and neuronal health links. Adv. Nutr. 2016;7:44–65. doi: 10.3945/an.115.009639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mozaffarian D. Dietary and policy priorities for cardiovascular disease, diabetes, and obesity: a comprehensive review. Circulation. 2016;133:187–225. doi: 10.1161/CIRCULATIONAHA.115.018585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cory H., Passarelli S., Szeto J., Tamez M., Mattei J. The role of polyphenols in human health and food systems: a mini-review. Front. Nutr. 2018;5 doi: 10.3389/fnut.2018.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konstantinidi M., Koutelidakis A.E. Functional foods and bioactive compounds: a review of its possible role on weight management and obesity's metabolic consequences. Medicines (Basel) 2019:6. doi: 10.3390/medicines6030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nediani C., Ruzzolini J., Romani A., Calorini L. Oleuropein, a bioactive compound from olea europaea L., as a potential preventive and therapeutic agent in non-communicable diseases. Antioxidants. 2019;8:578. doi: 10.3390/antiox8120578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung J., Ho C.-T., Wang Y. Preventive mechanism of bioactive dietary foods on obesity-related inflammation and diseases. Food Funct. 2018;9:6081–6095. doi: 10.1039/c8fo01561a. [DOI] [PubMed] [Google Scholar]
- 39.Sidhu J.S., Zafar T.A. Bioactive compounds in banana fruits and their health benefits. Food Qual. Saf. 2018;2:183–188. [Google Scholar]
- 40.Kris-Etherton P.M., Hecker K.D., Bonanome A., Coval S.M., Binkoski A.E., Hilpert K.F., Griel A.E., Etherton T.D. Bioactive compounds in foods: their role in the prevention of cardiovascular disease and cancer. Am. J. Med. 2002;113:71–88. doi: 10.1016/s0002-9343(01)00995-0. [DOI] [PubMed] [Google Scholar]
- 41.Kris-Etherton P.M., Hecker K.D., Bonanome A., Coval S.M., Binkoski A.E., Hilpert K.F., Griel A.E., Etherton T.D. Bioactive compounds in foods: their role in the prevention of cardiovascular disease and cancer. Am. J. Med. 2002;113(Suppl 9B):71S–88S. doi: 10.1016/s0002-9343(01)00995-0. [DOI] [PubMed] [Google Scholar]
- 42.Natarajan T.D., Ramasamy J.R., Palanisamy K. Nutraceutical potentials of synergic foods: a systematic review. J. Ethnic Foods. 2019;6:27. [Google Scholar]
- 43.Wallace T.C., Bailey R.L., Blumberg J.B., Burton-Freeman B., Chen C.O., Crowe-White K.M., Drewnowski A., Hooshmand S., Johnson E., Lewis R., Murray R., Shapses S.A., Wang D.D. Fruits, vegetables, and health: a comprehensive narrative, umbrella review of the science and recommendations for enhanced public policy to improve intake. Crit. Rev. Food Sci. Nutr. 2019:1–38. doi: 10.1080/10408398.2019.1632258. [DOI] [PubMed] [Google Scholar]
- 44.Silva L., Pinheiro-Castro N., Novaes G.M., Pascoal G.F.L., Ong T.P. Bioactive food compounds, epigenetics and chronic disease prevention: focus on early-life interventions with polyphenols. Food Res. Int. 2019;125 doi: 10.1016/j.foodres.2019.108646. [DOI] [PubMed] [Google Scholar]
- 45.Ulaszewska M.M., Weinert C.H., Trimigno A., Portmann R., Andres Lacueva C., Badertscher R., Brennan L., Brunius C., Bub A., Capozzi F., Cialiè Rosso M., Cordero C.E., Daniel H., Durand S., Egert B., Ferrario P.G., Feskens E.J.M., Franceschi P., Garcia-Aloy M., Giacomoni F., Giesbertz P., González-Domínguez R., Hanhineva K., Hemeryck L.Y., Kopka J., Kulling S.E., Llorach R., Manach C., Mattivi F., Migné C., Münger L.H., Ott B., Picone G., Pimentel G., Pujos-Guillot E., Riccadonna S., Rist M.J., Rombouts C., Rubert J., Skurk T., Sri Harsha P.S.C., Van Meulebroek L., Vanhaecke L., Vázquez-Fresno R., Wishart D., Vergères G. Nutrimetabolomics: an integrative action for metabolomic analyses in human nutritional studies. Mol. Nutr. Food Res. 2019;63 doi: 10.1002/mnfr.201800384. [DOI] [PubMed] [Google Scholar]
- 46.Milner J.A. Molecular targets for bioactive food components. J. Nutr. 2004;134:2492S–2498S. doi: 10.1093/jn/134.9.2492S. [DOI] [PubMed] [Google Scholar]
- 47.Choi S.W., Friso S. Epigenetics: a new bridge between nutrition and health. Adv. Nutr. 2010;1:8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hardy T.M., Tollefsbol T.O. Epigenetic diet: impact on the epigenome and cancer. Epigenomics. 2011;3:503–518. doi: 10.2217/epi.11.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tiffon C. The impact of nutrition and environmental epigenetics on human health and disease. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahinko M., Kurkinen S.T., Niinivehmas S.P., Pentikäinen O.T., Postila P.A. A practical perspective: the effect of ligand conformers on the negative image-based screening. Int. J. Mol. Sci. 2019;20:2779. doi: 10.3390/ijms20112779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jokinen E.M., Postila P.A., Ahinko M., Niinivehmas S., Pentikäinen O.T. Fragment- and negative image-based screening of phosphodiesterase 10A inhibitors. Chem. Biol. Drug Des. 2019;94:1799–1812. doi: 10.1111/cbdd.13584. [DOI] [PubMed] [Google Scholar]
- 52.Kurkinen S.T., Lätti S., Pentikäinen O.T., Postila P.A. Getting docking into shape using negative image-based rescoring. J. Chem. Inf. Model. 2019;59:3584–3599. doi: 10.1021/acs.jcim.9b00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurkinen S.T., Niinivehmas S., Ahinko M., Lätti S., Pentikäinen O.T., Postila P.A. Improving docking performance using negative image-based rescoring. Front. Pharmacol. 2018;9 doi: 10.3389/fphar.2018.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu Z., Huang B., Tang J., Liu S., Liu M., Ye Y., Liu Z., Xiong Y., Zhu W., Cao D., Li J., Niu X., Zhou H., Zhao Y.J., Zhang G., Huang H. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021;12 doi: 10.1038/s41467-020-20718-8. 488-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niinivehmas S.P., Salokas K., Lätti S., Raunio H., Pentikäinen O.T. Ultrafast protein structure-based virtual screening with Panther. J. Comput. Aided Mol. Des. 2015;29:989–1006. doi: 10.1007/s10822-015-9870-3. [DOI] [PubMed] [Google Scholar]
- 60.Vainio M.J., Puranen J.S., Johnson M.S. ShaEP: molecular overlay based on shape and electrostatic potential. J. Chem. Inf. Model. 2009;49:492–502. doi: 10.1021/ci800315d. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen N.T., Nguyen T.H., Pham T.N.H., Huy N.T., Bay M.V., Pham M.Q., Nam P.C., Vu V.V., Ngo S.T. Autodock vina adopts more accurate binding poses but Autodock4 forms better binding affinity. J. Chem. Inf. Model. 2020;60:204–211. doi: 10.1021/acs.jcim.9b00778. [DOI] [PubMed] [Google Scholar]
- 62.Shinde P.B., Bhowmick S., Alfantoukh E., Patil P.C., Wabaidur S.M., Chikhale R.V., Islam M.A. De novo design based identification of potential HIV-1 integrase inhibitors: a pharmacoinformatics study. Comput. Biol. Chem. 2020;88 doi: 10.1016/j.compbiolchem.2020.107319. [DOI] [PubMed] [Google Scholar]
- 63.Bhowmick S., Alissa S.A., Wabaidur S.M., Chikhale R.V., Islam M.A. Structure-guided screening of chemical database to identify NS3-NS2B inhibitors for effective therapeutic application in dengue infection. J. Mol. Recogn. 2020;33 doi: 10.1002/jmr.2838. [DOI] [PubMed] [Google Scholar]
- 64.Gupta S., Parihar D., Shah M., Yadav S., Managori H., Bhowmick S., Patil P.C., Alissa S.A., Wabaidur S.M., Islam M.A. Computational screening of promising beta-secretase 1 inhibitors through multi-step molecular docking and molecular dynamics simulations - pharmacoinformatics approach. J. Mol. Struct. 2020;1205 [Google Scholar]
- 65.Kumari R., Dalal V. Identification of potential inhibitors for LLM of Staphylococcus aureus: structure-based pharmacophore modeling, molecular dynamics, and binding free energy studies. J. Biomol. Struct. Dyn. 2021:1–15. doi: 10.1080/07391102.2021.1936179. [DOI] [PubMed] [Google Scholar]
- 66.Kumari N., Dalal V., Kumar P., Rath S.N. Antagonistic interaction between TTA-A2 and paclitaxel for anti-cancer effects by complex formation with T-type calcium channel. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1839558. [DOI] [PubMed] [Google Scholar]
- 67.van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A.M.J.J. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 68.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 69.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 70.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Träg J., Zahn D. Improved GAFF2 parameters for fluorinated alkanes and mixed hydro- and fluorocarbons. J. Mol. Model. 2019;25:39. doi: 10.1007/s00894-018-3911-5. [DOI] [PubMed] [Google Scholar]
- 72.Case D.A., Cheatham T.E., III, Darden T., Gohlke H., Luo R., Merz K.M., Jr., Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expet Opin. Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xue W., Yang F., Wang P., Zheng G., Chen Y., Yao X., Zhu F. What contributes to serotonin–norepinephrine reuptake inhibitors' dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem. Neurosci. 2018;9:1128–1140. doi: 10.1021/acschemneuro.7b00490. [DOI] [PubMed] [Google Scholar]
- 75.Pang J., Gao S., Sun Z., Yang G. Discovery of small molecule PLpro inhibitor against COVID-19 using structure-based virtual screening, molecular dynamics simulation, and molecular mechanics/Generalized Born surface area (MM/GBSA) calculation. Struct. Chem. 2021;32:879–886. doi: 10.1007/s11224-020-01665-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Onufriev A., Bashford D., Case D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins: Struct. Funct. Bioinf. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 77.Weiser J., Shenkin P.S., Still W.C. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO) J. Comput. Chem. 1999;20:217–230. [Google Scholar]
- 78.Gohlke H., Kiel C., Case D.A. Insights into protein–protein binding by binding free energy calculation and free energy decomposition for the ras–raf and ras–RalGDS complexes. J. Mol. Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 79.Dalal V., Dhankhar P., Singh V., Rakhaminov G., Golemi-Kotra D., Kumar P. Structure-based identification of potential drugs against FmtA of Staphylococcus aureus: virtual screening, molecular dynamics, MM-GBSA, and QM/MM. Protein J. 2021;40:148–165. doi: 10.1007/s10930-020-09953-6. [DOI] [PubMed] [Google Scholar]
- 80.Genheden S., Kuhn O., Mikulskis P., Hoffmann D., Ryde U. The normal-mode entropy in the MM/GBSA method: effect of system truncation, buffer region, and dielectric constant. J. Chem. Inf. Model. 2012;52:2079–2088. doi: 10.1021/ci3001919. [DOI] [PubMed] [Google Scholar]
- 81.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pires D.E.V., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58:4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings1PII of original article: S0169-409X(96)00423-1. The article was originally published in Advanced Drug Delivery Reviews 23 (1997) 3–25.1. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 84.Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43:W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hosseini M., Chen W., Xiao D., Wang C. Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs. Precis. Clin. Med. 2021;4:1–16. doi: 10.1093/pcmedi/pbab001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rao P., Patel R., Shukla A., Parmar P., Rawal R.M., Saraf M., Goswami D. Molecular Diversity; 2021. Identifying Structural–Functional Analogue of GRL0617, the Only Well-Established Inhibitor for Papain-like Protease (PLpro) of SARS-CoV2 from the Pool of Fungal Metabolites Using Docking and Molecular Dynamics Simulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patel D., Athar M., Jha P.C. Computational investigation of binding of chloroquinone and hydroxychloroquinone against PLPro of SARS-CoV-2. J. Biomol. Struct. Dynam. 2020:1–11. doi: 10.1080/07391102.2020.1844804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alamri M.A., Tahir Ul Qamar M., Mirza M.U., Alqahtani S.M., Froeyen M., Chen L.L. Discovery of human coronaviruses pan-papain-like protease inhibitors using computational approaches. J. Pharmaceut. Anal. 2020;10:546–559. doi: 10.1016/j.jpha.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sohraby F., Aryapour H. Unraveling the unbinding pathways of SARS-CoV-2 Papain-like proteinase known inhibitors by Supervised Molecular Dynamics simulation. PLoS One. 2021;16 doi: 10.1371/journal.pone.0251910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rajpoot S., Alagumuthu M., Baig M.S. Dual targeting of 3CLpro and PLpro of SARS-CoV-2: a novel structure-based design approach to treat COVID-19. Curr. Res. Struct. Biol. 2021;3:9–18. doi: 10.1016/j.crstbi.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baildya N., Khan A.A., Ghosh N.N., Dutta T., Chattopadhyay A.P. Screening of potential drug from Azadirachta Indica (Neem) extracts for SARS-CoV-2: an insight from molecular docking and MD-simulation studies. J. Mol. Struct. 2021;1227:129390. doi: 10.1016/j.molstruc.2020.129390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ma C., Sacco M.D., Xia Z., Lambrinidis G., Townsend J.A., Hu Y., Meng X., Szeto T., Ba M., Zhang X., Gongora M., Zhang F., Marty M.T., Xiang Y., Kolocouris A., Chen Y., Wang J. Discovery of SARS-CoV-2 papain-like protease inhibitors through a combination of high-throughput screening and a FlipGFP-based reporter assay. ACS Cent. Sci. 2021;7:1245–1260. doi: 10.1021/acscentsci.1c00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rao P., Patel R., Shukla A., Parmar P., Rawal R.M., Saraf M., Goswami D. Identifying structural-functional analogue of GRL0617, the only well-established inhibitor for papain-like protease (PLpro) of SARS-CoV2 from the pool of fungal metabolites using docking and molecular dynamics simulation. Mol. Divers. 2021:1–21. doi: 10.1007/s11030-021-10220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patel R., Prajapati J., Rao P., Rawal R.M., Saraf M., Goswami D. Repurposing the antibacterial drugs for inhibition of SARS-CoV2-PLpro using molecular docking, MD simulation and binding energy calculation. Mol. Divers. 2021:1–21. doi: 10.1007/s11030-021-10325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parmar P., Rao P., Sharma A., Shukla A., Rawal R.M., Saraf M., Patel B.V., Goswami D. Meticulous assessment of natural compounds from NPASS database for identifying analogue of GRL0617, the only known inhibitor for SARS-CoV2 papain-like protease (PLpro) using rigorous computational workflow. Mol. Divers. 2021:1–19. doi: 10.1007/s11030-021-10233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ogunyemi O.M., Gyebi G.A., Ibrahim I.M., Olaiya C.O., Ocheje J.O., Fabusiwa M.M., Adebayo J.O. Dietary stigmastane-type saponins as promising dual-target directed inhibitors of SARS-CoV-2 proteases: a structure-based screening. RSC Adv. 2021;11:33380–33398. doi: 10.1039/d1ra05976a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bhowmick S., Saha A., AlFaris N.A., Altamimi J.Z., Alothman Z.A., Aldayel T.S., Wabaidur S.M., Islam M.A. Identification of potent food constituents as SARS-CoV-2 papain-like protease modulators through advanced pharmacoinformatics approaches. J. Mol. Graph. Model. 2022;111 doi: 10.1016/j.jmgm.2021.108113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kalbhor M.S., Bhowmick S., Alanazi A.M., Patil P.C., Islam M.A. Multi-step molecular docking and dynamics simulation-based screening of large antiviral specific chemical libraries for identification of Nipah virus glycoprotein inhibitors. Biophys. Chem. 2021;270:106537. doi: 10.1016/j.bpc.2020.106537. [DOI] [PubMed] [Google Scholar]
- 99.Savale R.U., Bhowmick S., Osman S.M., Alasmary F.A., Almutairi T.M., Abdullah D.S., Patil P.C., Islam M.A. Pharmacoinformatics approach based identification of potential Nsp15 endoribonuclease modulators for SARS-CoV-2 inhibition. Arch. Biochem. Biophys. 2021;700 doi: 10.1016/j.abb.2021.108771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not availability.