Abstract
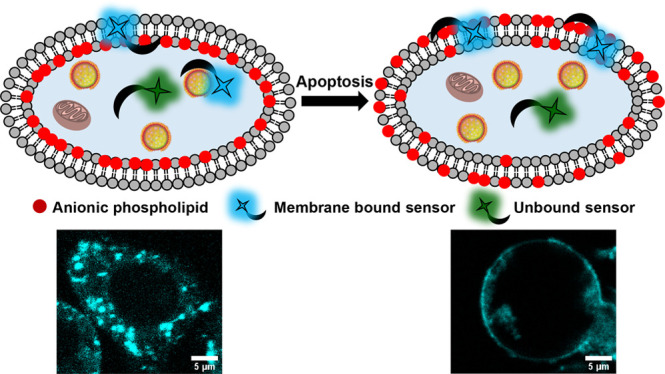
Anionic phospholipids are key cell signal mediators. The distribution of these lipids on the cell membrane and intracellular organelle membranes guides the recruitment of signaling proteins leading to the regulation of cellular processes. Hence, fluorescent sensors that can detect anionic phospholipids within living cells can provide a handle into revealing molecular mechanisms underlying lipid-mediated signal regulation. A major challenge in the detection of anionic phospholipids is related to the presence of these phospholipids mostly in the inner leaflet of the plasma membrane and in the membranes of intracellular organelles. Hence, cell-permeable sensors would provide an advantage by enabling the rapid detection and tracking of intracellular pools of anionic phospholipids. We have developed a peptide-based, cell-permeable, water-soluble, and ratiometric fluorescent sensor that entered cells within 15 min of incubation via the endosomal machinery and showed punctate labeling in the cytoplasm. The probe could also be introduced into living cells via lipofection, which allows bypassing of endosomal uptake, to image anionic phospholipids in the cell membrane. We validated the ability of the sensor toward detection of intracellular anionic phospholipids by colocalization studies with a fluorescently tagged lipid and a protein-based anionic phospholipid sensor. Further, the sensor could image the externalization of anionic phospholipids during programmed cell death, indicating the ability of the probe toward detection of both intra- and extracellular anionic phospholipids based on the biological context.
Introduction
Anionic phospholipids are essential structural and functional constituents of both cellular and intracellular membranes.1−3 A major anionic phospholipid on the eukaryotic cell membrane is phosphatidylserine (PS), which constitutes 10–15% of the total plasma membrane phospholipid population.1,4,5 PS is also present in lower percentages (∼1–5%) in intracellular organelles like the endoplasmic reticulum, Golgi complex, and mitochondria. Further, PS constitutes around 8.5% of total phospholipids in early endosomes and 2.5–3.9% in late endosomes.6,7 Another major anionic phospholipid, phosphatidylinositol (PI),8 which is a precursor of phosphoinositides (PIPs), is typically present in the endoplasmic reticulum and the Golgi complex while PIPs, which are also anionic lipids, constitute less than 2% of the plasma membrane phospholipids.9 Phosphatidic acid (PA) and phosphatidylglycerol (PG) are minor anionic phospholipids that are each present in ∼1–2% or less of the total cellular phospholipids.10,11 This repertoire of anionic phospholipids mediates key cell signaling, membrane trafficking, and nuclear events.3 Electrostatic interactions of negatively charged polar headgroups of anionic phospholipids with positively charged interfaces of cytoplasmic proteins form the basis of lipid–protein interactions at the membrane cytosol interface.12,13 These initial interactions lead to downstream signaling events that regulate cellular processes.9
Under physiological conditions, anionic phospholipids are almost exclusively present in the inner leaflet of the plasma membrane and in intracellular organelle membranes.14 The plasma membrane asymmetry arising due to the localization of anionic phospholipids in the inner leaflet is teleologically linked to their mediatory roles in the transmission of cellular signals from the membrane to intracellular compartments.4,8,9,12,13,15−17 To achieve molecular insights into the functional roles of anionic phospholipids in cell signaling and membrane trafficking, it is therefore necessary to develop cell-permeable, reversible, and fluorescent sensors that can track intracellular anionic phospholipids in an optical imaging platform.
To date, most fluorescent sensors for anionic phospholipids including both protein-based and small molecule-based sensors, except a few, are cell-impermeable.2,15,18−25 In an attempt to develop a cell-permeable fluorescent sensor for anionic phospholipids, we scanned short peptides that could bind PS, as PS constitutes the highest percentage among all anionic phospholipids in eukaryotic cells. We reckoned that short peptides derived from binding sites of proteins that interact with anionic phospholipids would be positively charged and hence might be inherently cell-permeable.26 We zeroed in on PSBP-6 (FNFRLKAGAKIRFG) a peptide derived from the protein PS decarboxylase.27,28 Previous studies had shown that this peptide could bind to PS on the extracellular leaflet of the plasma membrane during programmed cell death or apoptosis.27,29 This is because PS flips to the outer leaflet of the plasma membrane during apoptosis, and this process acts as an “eat me” signal for phagosomes to clear out the dying cell.30,31 However, we could not find any report on whether this peptide was cell-permeable and on whether the peptide was PS selective or could bind to other anionic phospholipids as well. We hypothesized that, since the charge on PSBP-6 is +4, it could serve as a scaffold for the development of a cell-permeable anionic phospholipid sensor.
We attached a ratiometric polarity sensitive dye, DAN, to the N-terminus of the peptide via a reaction of the dye precursor acrylodan with a cysteine residue. The resultant sensor afforded a distinct shift in emission from the green region of the visible spectrum to the blue region upon binding anionic phospholipids. The sensor had the highest binding affinity toward PS followed by PA, phosphatidylinositol-(4,5)-bisphosphate (PIP2), and PG. Since the levels of PA, PIP2, and PG are significantly lower than that of PS in eukaryotic cells, we expected that the probe would mostly detect PS over the other major anionic phospholipid PI. Importantly, our minimalistic dye–peptide conjugate-based probe was cell-permeable. The probe afforded punctate staining in the cytosol, and the PS sensing ability of the probe was validated by imaging PS externalization during apoptosis. Since the probe was taken up into the cells via an active pathway and possibly remained within the endolysosomal compartments thereby imaging PS within these vesicles, we could not directly image PS on the inner leaflet of the plasma membrane when the probe was simply incubated with cells. However, when the probe was incorporated into living cells via lipofection to bypass the active uptake pathway, it lighted up the plasma membrane along with the intracellular compartments, indicating its potential toward detecting different pools of intracellular anionic phospholipids depending on the mode of cellular uptake.
Results and Discussions
A cell-permeable, ratiometric, and fluorescent anionic phospholipid sensor was designed by conjugating a PS binding peptide, PSBP-6, to a polarity sensitive dye DAN (Figure 1A).
Figure 1.
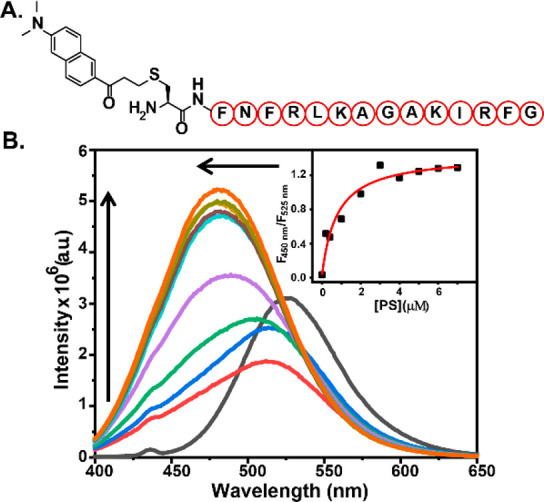
(A) Chemical representation of DAN-APS. (B) In vitro fluorescence response of DAN-APS with PS. Fluorescence emission spectra (λex: 380 nm) of individual solutions containing DAN-APS (1.5 μM) in 20 mM Na-HEPES, 100 mM NaCl (pH 7.4), added with either no lipid (black) or SUVs containing 1 mol % (red), 2 mol % (blue), 5 mol % (green), 10 mol % (purple), 15 mol % (yellow), 20 mol % (cyan), 25 mol % (brown), 30 mol % (pear), and 35 mol % (saffron) PS mixed with PC such that the total lipid concentrations were 20 μM. (Inset) Plot of the ratiometric response of the sensor versus PS concentration. To obtain the dissociation constant (Kd), the response curve was fitted to eq 6 in the Supporting Information.
The DAN dye undergoes a blue-shift in its emission maxima in nonpolar medium with respect to its emission in polar medium.20,32−34 Hence, we hypothesized that, once the peptide would bind to anionic phospholipids, the dye would come closer to the membrane35 and its emission maxima would shift, affording a ratiometric fluorescent sensor for anionic phospholipids. The sensor will be henceforth referred to as the DAN-anionic phospholipid sensor (DAN-APS). DAN-APS was synthesized in two steps (Scheme S1). The peptide sequence CFNFRLKAGAKIRFG, Cys-PSBP-6, was synthesized via solid-phase peptide synthesis. Acrylodan was then conjugated to the N-terminal cysteine residue of the peptide in solution phase to afford DAN-APS. The sensor was purified by HPLC and characterized by LC-ESI-MS and MALDI (Figures S1, S2, and S3). The probe was highly water-soluble with a log P value of −0.93 ± 0.03 based on the partition of DAN-APS between water and octanol layers. Since DAN-APS was water-soluble, all experiments were performed by making stock solutions of the sensor in water followed by dilution in aqueous buffer.
The DAN dye has a broad absorption maxima peak at 365 nm.36 Importantly, the dye has a two-photon absorption cross-section of 75 GM allowing excitation at 780 nm using a multiphoton setup.37,38 Multiphoton excitation at longer wavelengths affords greater detection sensitivity in living cells due to the low absorption of near-infrared radiation by biological molecules. The broad absorption of the dye peaked at 371 nm after conjugation to the peptide (Figure S4), indicating that the conjugation step did not significantly affect the spectral features of the dye.
We next examined the fluorescence response of DAN-APS in the presence of physiologically relevant phospholipids (Figures 1B and S12). Small unilamellar vesicles (SUVs) were prepared as membrane mimics (details of SUV preparation are provided in Table S1, along with SUV characterization in Figures S5–S11 and Tables S2 and S3), and the SUVs were used to determine the in vitro selectivity of the sensor. Phosphatidylcholine (PC) is the most abundant phospholipid in mammalian cells.3 Hence, SUVs were prepared by mixing phospholipids with PC in varying molar ratios. The fluorescence emission response of the sensor showed a clear blue shift in the presence of SUVs containing anionic phospholipids upon excitation at 380 nm. The emission maxima shifted from 525 to 480 nm with a concomitant increase in emission at 480 nm for PS upon titrating with SUVs containing increasing percentages of PS. The quantum yields for DAN-APS and PS bound DAN-APS were determined to be 0.07 and 0.20, respectively (Figure S13).
Similar blue shifts were also observed when SUVs containing increasing percentages of other anionic phospholipids including PA, PIP2, PG, and PI were titrated into DAN-APS solutions. However, the sensor did not afford any blue shift in emission in the presence of major zwitterionic phospholipids like PC and phosphatidylethanolamine (PE). A decrease in the emission of the sensor in the green region (525 nm) was observed for these lipids with no associated increase in the emission in the blue region (480 nm), indicating weak binding of the sensor to zwitterionic lipids. Since DAN-APS has an overall positive charge, in order to check whether the ratiometric response of DAN-APS toward anionic phospholipids might be altered in the presence of soluble anionic phosphates, the probe response to PS was measured in the presence of inositol phosphates. The ratiometric response of DAN-APS to PS remained unaltered even in the presence of eight times higher molar equivalents of these negatively charged sugars with respect to molar equivalents of PS (Figure S14a) further validating the specificity of our probe. Finally, upon addition of the unlabeled PS-binding peptide, Cys-PSBP-6, to DAN-APS bound to PS, we observed that the emission maxima shifted back to 518 nm from 476 nm (Figure S14b). Although the spectra did not shift back completely to that of the unbound probe (emission maxima at 526 nm), indicating the higher affinity of DAN-APS toward PS in comparison to that of Cys-PSBP-6 toward PS, the data clearly showed that the DAN-APS probe response was reversible. The in vitro titration experiments distinctly indicated that DAN-APS could selectively detect anionic phospholipids over neutral phospholipids.
Binding isotherms were generated by taking the ratio of emission intensities at 450 and 525 nm (Figures 1B, inset, and S12; equations used and the derivation are in Section 11 in the Supporting Information). On the basis of a single-site binding model39 where one molecule of the peptide-based probe would bind to one molecule of the lipid, the plots were fitted to eq 6 in the Supporting Information to obtain association constant (Ka) values for both anionic and neutral phospholipids (Figures 1B and S12 and Table 1). DAN-APS afforded the highest Ka value with PS, as expected. The Ka value of DAN-APS toward PS was 1.9 times higher than the Ka of DAN-APS toward PA. Other anionic phospholipids like PIP2 and PG had ∼2.5 times lower Ka values toward DAN-APS in comparison to PS. Importantly, the probe had a 24 times higher affinity toward PS when compared to the other major anionic phospholipid PI.
Table 1. Association Constant Values (Ka × 106 (M–1)) of DAN-APS for Different Phospholipids.
| PS | PA | PIP2 | PG | PI | PE | PC |
|---|---|---|---|---|---|---|
| 1.20 ± 0.28 | 0.63 ± 0.15 | 0.47 ± 0.14 | 0.43 ± 0.18 | 0.05 ± 0.02 | NBa | 0.04 ± 0.02 |
NB: No binding.
Considering the facts that DAN-APS showed the highest affinity toward PS and that PS is the major anionic phospholipid in eukaryotic cells, we imaged giant unilamellar vesicles (GUVs) with increasing levels of PS using multiphoton excitation (780 nm) in a confocal microscopy setup (Figure 2A). This experiment provided an assessment on the visual response of the sensor and would form the basis of experiments in living cells. Emission in the blue channel (λem of 420–460 nm) represented the response of the lipid bound sensor. An increase in the blue channel intensity was observed with increasing PS levels in the vesicles, indicating that the probe could be applied for sensing PS in a confocal microscopy setup (Figure 2B).
Figure 2.
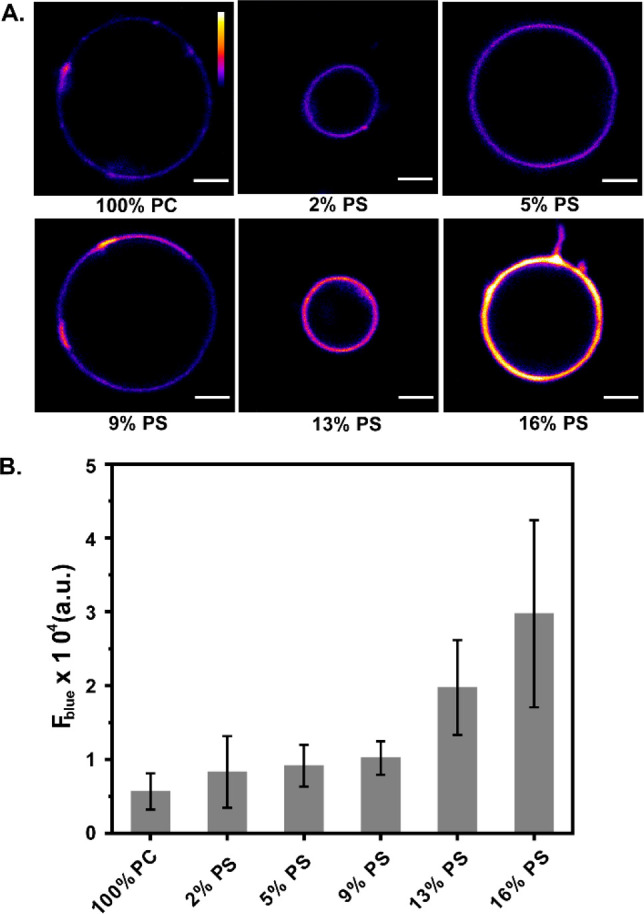
Confocal fluorescence images using multiphoton excitation (λex: 780 nm) showing the response of the probe DAN-APS in the presence of GUVs with increasing PS levels. (A) Blue channel images (λem: 420–460 nm) of GUVs composed of only PC, 2 mol % PS, 5 mol % PS, 9 mol % PS, 13 mol % PS, and 16 mol % PS in PC incubated with 1.5 μM DAN-APS in 20 mM Na-HEPES, 100 mM NaCl (pH 7.4). Scale bar, 5 μm; calibration bar, 0 to 65 535. (B) Bar plots representing the average blue channel fluorescence intensities of the GUVs. Mean fluorescence intensities have been plotted, and error bars indicate the standard deviation derived from the intensity analysis of ten GUVs of each composition.
We next evaluated the in-cell response of DAN-APS. In order to check if the response of DAN-APS might be affected in cellular media, the response of the probe was first recorded via an in vitro fluorescence titration experiment with SUVs in cell culture media (Figure S15). The data indicated that the probe response remained unaltered in cell culture media with a similar blue shift and fluorescence enhancement. Further, an MTT assay on DAN-APS indicated that ∼65% cells were viable at less than 10 μM incubation concentrations with up to a 1 h incubation with increased toxicity at higher concentrations (Figure S16). Hence, we decided to incubate cells with low μM levels of the probe for at most 15 min to check cellular uptake. Thus, living HeLa cells were incubated with the sensor for 15 min at 37 °C and imaged in a confocal microscopy setup using an excitation wavelength of 780 nm (Figure 3, top row). Emission was collected simultaneously in blue (λem: 420–460 nm) and green (λem: 560–590 nm) channels. The blue channel emission corresponded to the lipid-bound sensor while the green channel emission would account for lipid-bound and unbound sensor. We observed punctate staining of the cytoplasm and parts of the cell membrane. The z-stacks of the confocal images (Figure S19) distinctly indicated that DAN-APS was cell-permeable within the 15 min incubation time. While punctate staining within the cytoplasm was expected due to the presence of anionic phospholipids in intracellular organelles, the punctate staining of the plasma membrane was contrary to our expectation. This was because protein-based transfectable PS probes usually uniformly light up the plasma membrane, indicating uniform distribution of this lipid in the membrane.15,19 We suspected that the probe was being incorporated into cells via an active uptake pathway, which led to the probe being trapped within intracellular vesicles. When HeLa cells were incubated with DAN-APS at 4 °C40 (Figure 3, bottom row), the probe did not enter the cells, confirming our hypothesis regarding an active uptake pathway. In order to eliminate any possibility of probe aggregation leading to punctate staining at either 37 or 4 °C, we performed dynamic light scattering (DLS) experiments of DAN-APS solutions in cell culture media at both temperatures. The data (Figure S17) indicated no aggregation, further validating the endosomal uptake of the probe.
Figure 3.
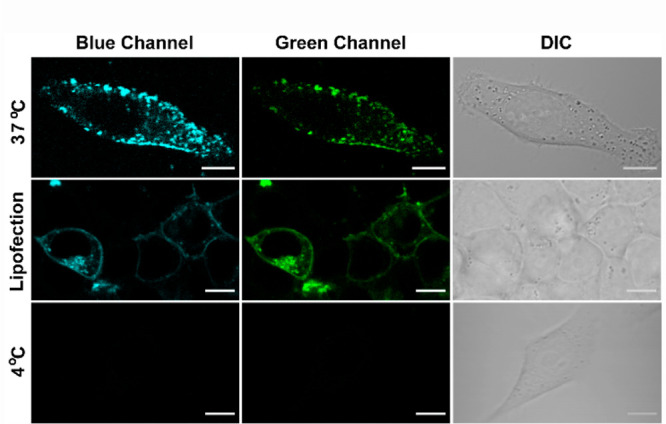
Representative confocal single z-plane images of living HeLa cells. Cells were incubated with 7 μM DAN-APS for 15 min at either 37 °C (first row) or 4 °C (third row) in serum-free DMEM media, pH 7.4. For the lipofection experiment, DAN-APS (10 μg) was incorporated into live HeLa cells via lipofection (second row, 37 °C) and washed. DAN-APS was excited with a two-photon laser (λex: 780 nm). Fluorescence emission: blue channel (λem: 420–460 nm); top row green channel (λem: 560–590 nm); middle and bottom row green channel (λem: 510–540 nm). Differential interference contrast (DIC) images in right column. Scale bar, 10 μm.
In order to test whether the probe could also light up anionic phospholipids in the inner leaflet of the plasma membrane, we then incorporated DAN-APS into HeLa cells via lipofection (Figure 3, middle row) as this method could bypass the active uptake mode.41 As an initial check, to ensure that the fluorescence response of the probe remained similar upon lipofection, we recorded the fluorescence emission of DAN-APS in the presence of lipofectamine (Figure S20). The spectral features of the probe in the presence of lipofectamine and the ratio of emission at 450 to 525 nm remained similar to that of free DAN-APS. Cells in which the probe was incorporated via lipofection showed uniform lighting up of the plasma membrane (λex: 780 nm; via multiphoton excitation), indicating that DAN-APS might be applicable for the detection of intracellular PS in the plasma membrane as well, when introduced into cells via lipofection.
We reasoned that the intracellular punctate staining observed when the probe was directly incubated in living cells could be intracellular pools of PS, which are reported to exist in endosomes and lysosomes.6,15,19 Hence, we performed colocalization studies with lysosomal and endosomal markers (Figure 4, first and second rows). We indeed observed colocalization with LysoTracker Red (tM1: 0.54 ± 0.14; tM2: 0.53 ± 0.11) and endosomal marker Dextran AlexaFluor 546 (tM1: 0.55 ± 0.18; tM2: 0.44 ± 0.12). The Mander’s coefficient values indicated partial colocalization with endolysosomal vesicles, signifying that the probe was taken up by an active pathway and was trapped in these vesicles. Minimal colocalization was observed with either mitochondrial tracker or a general plasma membrane marker (Figure S21).
Figure 4.
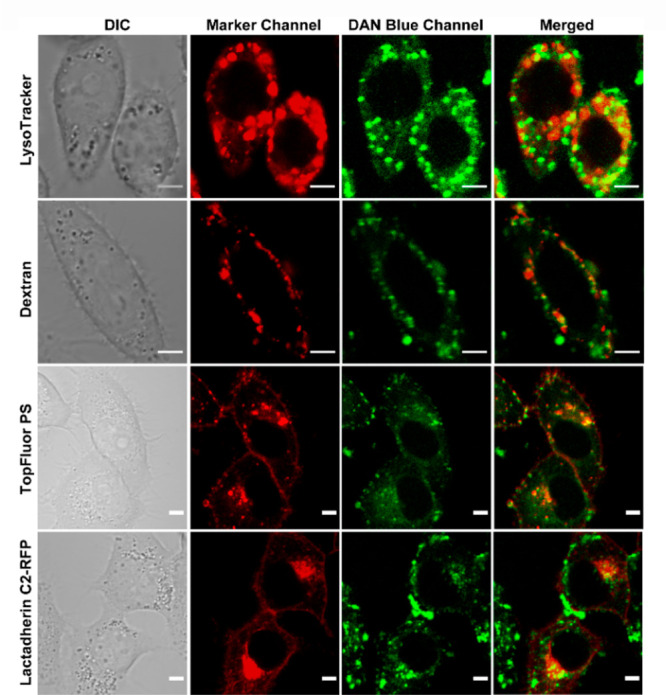
Representative confocal single z-plane images of living HeLa cells incubated with LysoTracker Red (200 nM, 30 min), Dextran AlexaFluor 546 (10 μM, 4 h), or TopFluor PS (2.7 μM, 15 min, 0 °C) or transfected with Lactadherin C2-RFP plasmid and incubated with DAN-APS (7 μM, 15 min) at 37 °C in DMEM, pH 7.4, with no phenol red and serum, and washed. The cells were irradiated with λex of either 561 nm (for Lysotracker, Dextran, and Lactadherin C2-RFP) or 488 nm (for TopFluor PS) and a two photon laser (λex: 780 nm) for DAN-APS. The fluorescence emissions at the marker channel (for Lysotracker and Dextran, λem: 570–650 nm; TopFluor PS, λem: 500–550 nm; Lactadherin C2-RFP, λem: 570–700 nm) and blue channel for DAN-APS (λem: 420–460 nm) were collected successively. Scale bar, 5 μm. The colocalization of organelle trackers and PS markers (red false-color) with DAN-APS (green false-color) is depicted by yellow spots in merged panel. DIC images in left column.
In order to directly confirm if the intracellular staining was due to the presence of PS on these vesicles, we performed colocalization studies of directly incubated DAN-APS with a fluorescently tagged PS analog, TopFluor PS,42 and a transfectable protein-based PS sensor, Lactadherin C2-RFP15 (Figure 4, third and fourth rows). Unlike these probes, directly incubated DAN-APS did not label the plasma membrane, but the intracellular staining of our probe closely resembled that of these orthogonal PS probes. Mander’s coefficient values (TopFluor PS, tM1: 0.39 ± 0.07, tM2: 0.45 ± 0.13; Lactadherin C2-RFP, tM1: 0.42 ± 0.26, tM2: 0.51 ± 0.20) indicated partial colocalization as expected since DAN-APS only lighted up intracellular vesicles and not the plasma membrane. Importantly, bleed-through control experiments indicated no overlap between DAN-APS emission and other tracker/orthogonal PS probe emissions (Figures S22 and S23). The colocalization data with fluorescently tagged PS and the protein-based PS sensor, taken together, showed that DAN-APS could image PS in intracellular vesicles upon direct incubation with cells.
Further, the live cell confocal images of lipofected DAN-APS were very similar to that of both TopFluor PS and Lactadherin C2-RFP (Figure S24) and showed lighting up of both the plasma membrane and intracellular vesicles. These studies distinctly showed that DAN-APS is a cell-permeable PS sensor that could image different pools of intracellular PS depending on the mode of uptake, i.e., incubation versus lipofection.
As a final validation of DAN-APS as an anionic phospholipid sensor and a potential PS sensor, we imaged apoptotic cells that show characteristic PS externalization.30,31,43 HeLa cells were treated with either cisplatin44,45 or H2O246 to induce apoptosis and incubated directly with DAN-APS (Figure 5A). Both DAN-APS and Annexin V-Alexa Fluor 647 (Anx V-AF 647), which is a protein-based apoptotic marker, labeled the membranes of apoptotic cells (Figure 5A), showing that DAN-APS could detect anionic phospholipids, especially PS in cellular systems. Importantly, flow cytometry experiments were performed to quantify the uptake of DAN-APS via direct incubation in both viable (Figure S18) and apoptotic cells (Figure 5B, a–c), and the data was compared to that of Anx V-AF 647 (Figure 5B, d–f). DAN-APS stained 97.7 ± 2.9% of cisplatin treated cells, which was similar to the results with Anx V-AF 647, which also stained 98.4 ± 2.0% of the cisplatin treated cells. Further coincubation of DAN-APS and Anx V-AF 647 in cisplatin treated cells distinctly showed that DAN-APS could mark the same pool of apoptotic cells marked by Anx V-AF 647 (Figure 5B, g–i). Hence, we conclude that DAN-APS can detect both intracellular PS (Figure 3) and PS externalization during cell death (Figure 5).
Figure 5.
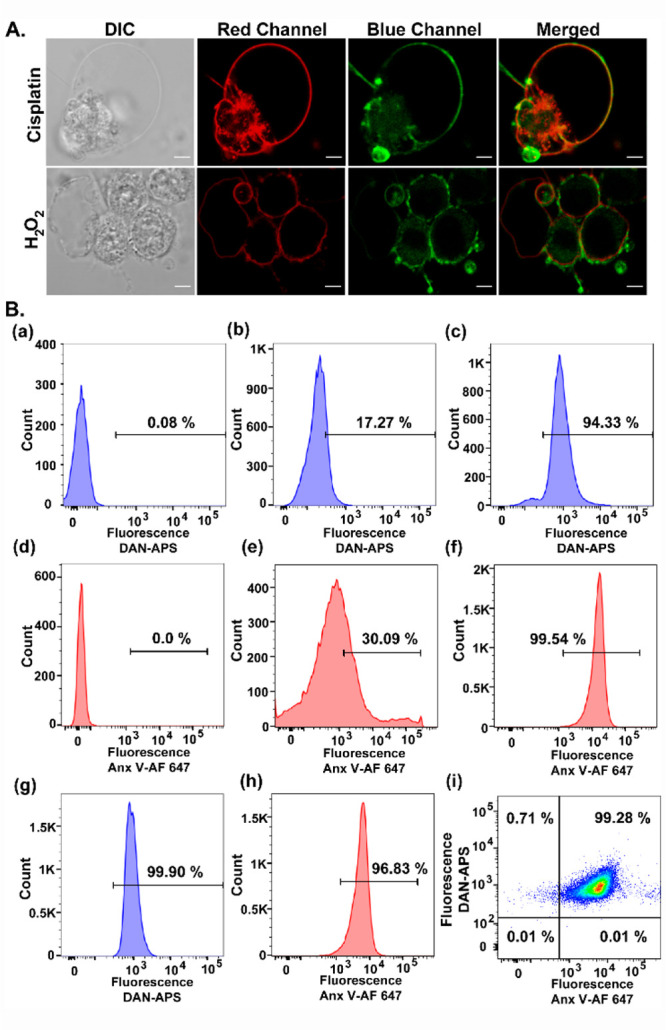
(A) Representative confocal single z-plane images of apoptotic HeLa cells incubated with Anx V-AF 647 (15 min) and DAN-APS (7 μM, 15 min) at 25 °C in buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2) and washed. For inducing apoptosis, HeLa cells were incubated with cisplatin (20 μM, 24 h) or H2O2 (0.2 mM, 4 h) in serum-free DMEM media. The cells were irradiated with λex of 633 nm and a two photon laser (λex: 780 nm), and fluorescence emissions at the red channel (λem: 650–700 nm) and blue channel (λem: 420–460 nm) were collected successively. DIC images in left column. Scale bar, 5 μm. (B) Representative flow cytometry results indicating the effectiveness of DAN-APS for the detection of both viable and apoptotic cells. Cells were treated with either DAN-APS (7 μM, 15 min) (b, c) or Anx V-AF 647 (e, f) or both DAN-APS and Anx V-AF 647 (g, h). Data for probe untreated control cells are shown (a) and (d). All measurements were performed at 25 °C in 10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2. For flow cytometry experiments, the following excitation sources/emission filters were used: Anx V-AF 647, 640 nm/670 nm; DAN-APS, 405 nm/450 nm. Cell count versus mean fluorescence intensities of either DAN-APS or Anx V-AF 647 are plotted as histograms (a–h). (i) Scatter plot of apoptotic cells treated with both DAN-APS and Anx V-AF 647.
Conclusions
We have developed a peptide-based, cell-permeable, water-soluble, ratiometric, and reversible anionic phospholipid sensor, DAN-APS. The sensor afforded the highest affinity toward PS and also responded to other anionic phospholipids like PA, PIP2, and PG. Intracellular staining of DAN-APS resembled PS localization in both living and apoptotic cells, indicating that the probe can be used to detect PS dynamics in living cells in the future. Our modular sensor design strategy based on a peptide-based PS detecting scaffold is amenable to easy synthetic modifications, which should allow the development of probes with lower toxicity, improved sensitivity, and selectivity along with tunable targetability.
Experimental Section
Synthesis of DAN-APS
Peptides were synthesized via solid-phase peptide synthesis on a fast-flow, solid-phase peptide synthesizer.47 Rink amide resin was used as the solid-phase for synthesis. After synthesis, peptides were cleaved from the resin beads, purified using HPLC, and characterized via LC-ESI-MS and MALDI-TOF-MS. For dye labeling, the peptide was mixed with acrylodan in anhydrous DMF and allowed to react for 6 h at room temperature. Following the labeling step, the reaction mixture was purified via HPLC. Purified fractions containing the DAN labeled peptide were characterized using LC-ESI-MS and MALDI-TOF-MS. Detailed procedures for synthesis and characterization data are included in Sections 1–5 in the Supporting Information.
Fluorescence Titrations with SUVs
Fluorescence measurements with DAN-APS were performed in 20 mM Na-HEPES buffer with 100 mM NaCl (pH 7.4). The concentrations of the DAN-APS stock solutions in deionized water were measured from absorption spectra on the basis of the extinction coefficient of Prodan at 365 nm.36 SUV solutions with different percentages of phospholipids (PS, PA, PIP2, PG, PI, PE) mixed with PC were prepared (details in Section 8 in the Supporting Information). Fluorescence spectra of individual solutions containing only the sensor or a mixture of the sensor and SUVs were recorded on a fluorescence spectrophotometer by exciting the sensor at 380 nm (details in Section 10 in the Supporting Information).
Confocal Fluorescence Imaging of GUVs
GUVs of specific phospholipid compositions were prepared and then incubated with the sensor solution (see Section 15 in the Supporting Information). For imaging, the GUV solutions were taken out and transferred into an 8 well glass bottomed imaging chamber. The GUVs were allowed to settle down to the bottom of the wells and imaged on a confocal fluorescence microscope (imaging details in Section 16 in the Supporting Information). GUVs were located by monitoring transmission images. The sensor was excited at 780 nm via two photon excitation, and the emission was collected at the blue channel (420–460 nm).
Live Cell Experiments with DAN-APS
Living HeLa cells were plated on glass bottomed imaging plates. The cells were serum starved for 4–5 h prior to imaging. Media was removed, and the cells were washed with DMEM (without phenol red and serum) and incubated with a DAN-APS solution in the same media for 15 min. After incubation, the cells were washed and taken for imaging (details of cell culture and imaging in Sections 17 and 19 in the Supporting Information).
For the lipofection experiments, the DAN-APS sensor was mixed with lipofectamine reagents for 15 min at room temperature. Cells were washed with Opti-MEM media and then incubated with the lipofection mixture at 37 °C in a CO2 incubator for 4.5 h. After incubation, the lipofection mixture was removed and the cells were washed with DMEM (without phenol red and serum) and taken forward for imaging in the same media (details of cell preparation and imaging in Section 19 in the Supporting Information).
Colocalization Studies with Organelle Trackers and Other PS Sensitive Probes
Serum starved HeLa cells, plated on glass bottomed imaging plates, were first incubated with solutions of organelle trackers. The organelle trackers used were LysoTracker Red, Dextran AlexaFluor 546, MitoTracker Red, and CellMask Green. The cells were washed with DMEM (without phenol red and serum) and incubated with the DAN-APS sensor for 15 min. The cells were then washed and taken for imaging (details in Section 24 in the Supporting Information).
For checking the colocalization of DAN-APS with TopFluor PS, the cells were incubated with TopFluor PS at 0 °C for 15 min. The cells were then washed with DMEM (without phenol red and serum) at 37 °C and incubated with the DAN-APS sensor for 15 min. The cells were then washed and taken for imaging (details in Section 25 in the Supporting Information).
For imaging the colocalization of DAN-APS with Lactadherin C2-RFP, the protein-based probe was expressed in HeLa cells via transfection. The transfected cells were incubated with the DAN-APS sensor for 15 min. The cells were then washed and taken for imaging (details in Section 26 in the Supporting Information).
Experiments in Apoptotic Cells with DAN-APS
HeLa cells were incubated with either cisplatin for 24 h or hydrogen peroxide for 4 h to induce apoptosis. The cells were then washed and incubated with an Anx V-AF647 solution for 15 min. The AnxV treated cells were washed and incubated with the DAN-APS sensor for 15 min. The cells were then washed again and taken for imaging (details in Section 30 in the Supporting Information).
Flow Cytometry Experiments
For flow cytometry experiments, apoptosis was induced in HeLa cells with cisplatin. Both cisplatin untreated (control) and apoptotic cells were incubated with either DAN-APS or Anx V-AF 647 for 15 min, leading to four sets of cells: (1) cisplatin untreated cells incubated with DAN-APS; (2) cisplatin treated cells incubated with DAN-APS; (3) cisplatin untreated cells incubated with Anx V-AF 647; (4) cisplatin treated cells incubated with Anx V-AF 647. Another set of cisplatin treated cells was first incubated with Anx V-AF 647, washed, and then incubated with DAN-APS. As controls for the flow cytometry experiments, cells not treated with cisplatin and without any dye incubation were used. The cells were washed and used for flow cytometry measurements (details in Section 21 in the Supporting Information).
Acknowledgments
A.D. acknowledges the support of the Department of Atomic Energy, Government of India, under Project Identification No. RTI4003. The authors acknowledge S. Panwar and A. Verma, TIFR, for help with cell culture; B. Parmar, TIFR, for help with confocal imaging; S. Chhatar, TIFR, for help in flow cytometry experiments; S. Grinstein, University of Toronto, Canada, and S. Mayor, NCBS, India, for sharing Lactadherin C2-RFP plasmid; R. Kundu and S. Kahali for help with data analysis. The authors thank the Cell Culture Facility and MALDI Facility at the Department of Chemical Sciences, TIFR.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c06981.
General procedures and materials; peptide synthesis and purification; HPLC, LC-ESI-ML, and UV-vis data; log P, binding constant, and quantum yield determination; size distributions; fluorescence responses; GUV preparation and imaging; cell culture and cell viability experiments; cell imaging; DLS measurements; flow cytometry experiments; confocal fluorescence images; colocalization studies and analysis; bleed-through control experiments; imaging apoptotic cell membranes (PDF)
Author Contributions
A.C.: investigation, data analysis, and writing; A.D.: conceptualization, supervision, and writing.
The authors declare no competing financial interest.
Supplementary Material
References
- Vance J. E. Phospholipid Synthesis and Transport in Mammalian Cells. Traffic 2015, 16 (1), 1–18. 10.1111/tra.12230. [DOI] [PubMed] [Google Scholar]
- Kundu R.; Chandra A.; Datta A. Fluorescent Chemical Tools for Tracking Anionic Phospholipids. Isr. J. Chem. 2021, 61 (3–4), 199–216. 10.1002/ijch.202100003. [DOI] [Google Scholar]
- van Meer G.; Voelker D. R.; Feigenson G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9 (2), 112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay J. G.; Fairn G. D. Distribution, dynamics and functional roles of phosphatidylserine within the cell. Cell Commun. Signaling 2019, 17 (1), 126. 10.1186/s12964-019-0438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J. E.; Steenbergen R. Metabolism and functions of phosphatidylserine. Prog. Lipid Res. 2005, 44 (4), 207–234. 10.1016/j.plipres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Leventis P. A.; Grinstein S. The Distribution and Function of Phosphatidylserine in Cellular Membranes. Annu. Rev. Biophys. 2010, 39 (1), 407–427. 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- van Meer G.; de Kroon A. I. P. M. Lipid map of the mammalian cell. J. Cell Sci. 2011, 124 (1), 5–8. 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- Lemmon M. A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008, 9 (2), 99–111. 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Di Paolo G.; De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443 (7112), 651–657. 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Morita S.-y.; Terada T. Enzymatic measurement of phosphatidylglycerol and cardiolipin in cultured cells and mitochondria. Sci. Rep. 2015, 5 (1), 11737. 10.1038/srep11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S.-y.; Ueda K.; Kitagawa S. Enzymatic measurement of phosphatidic acid in cultured cells. J. Lipid Res. 2009, 50 (9), 1945–1952. 10.1194/jlr.D900014-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay J. G.; Grinstein S.. Phosphatidylserine-Mediated Cellular Signaling. In Lipid-mediated Protein Signaling; Capelluto D. G. S., Ed.; Springer Netherlands: Dordrecht, 2013; pp 177–193. [DOI] [PubMed] [Google Scholar]
- Mondal S.; Chandra A.; Venkatramani R.; Datta A. Optically sensing phospholipid induced coil–helix transitions in the phosphoinositide-binding motif of gelsolin. Faraday Discuss. 2018, 207 (0), 437–458. 10.1039/C7FD00197E. [DOI] [PubMed] [Google Scholar]
- Lorent J. H.; Levental K. R.; Ganesan L.; Rivera-Longsworth G.; Sezgin E.; Doktorova M.; Lyman E.; Levental I. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 2020, 16 (6), 644–652. 10.1038/s41589-020-0529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T.; Gilbert G. E.; Shi J.; Silvius J.; Kapus A.; Grinstein S. Membrane Phosphatidylserine Regulates Surface Charge and Protein Localization. Science 2008, 319 (5860), 210. 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- Hirama T.; Lu S. M.; Kay J. G.; Maekawa M.; Kozlov M. M.; Grinstein S.; Fairn G. D. Membrane curvature induced by proximity of anionic phospholipids can initiate endocytosis. Nat. Commun. 2017, 8 (1), 1393. 10.1038/s41467-017-01554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Lee M.; Fairn G. D. Phospholipid subcellular localization and dynamics. J. Biol. Chem. 2018, 293 (17), 6230–6240. 10.1074/jbc.R117.000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiVittorio K. M.; Johnson J. R.; Johansson E.; Reynolds A. J.; Jolliffe K. A.; Smith B. D. Synthetic peptides with selective affinity for apoptotic cells. Org. Biomol. Chem. 2006, 4 (10), 1966–1976. 10.1039/b514748d. [DOI] [PubMed] [Google Scholar]
- Calderon F.; Kim H.-Y. Detection of intracellular phosphatidylserine in living cells. J. Neurochem. 2008, 104 (5), 1271–1279. 10.1111/j.1471-4159.2007.05079.x. [DOI] [PubMed] [Google Scholar]
- Yoon Y.; Lee P. J.; Kurilova S.; Cho W. In situ quantitative imaging of cellular lipids using molecular sensors. Nat. Chem. 2011, 3 (11), 868–874. 10.1038/nchem.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker V. E.; Oliveira B. L.; Yeo J. H.; Fraser S. T.; Bernardes G. J. L.; New E. J.; Jolliffe K. A. A Fluorogenic Probe for Cell Surface Phosphatidylserine Using an Intramolecular Indicator Displacement Sensing Mechanism.. Angew. Chem., Int. Ed. 2019, 58 (10), 3087–3091. 10.1002/anie.201812489. [DOI] [PubMed] [Google Scholar]
- Barth N. D.; Subiros-Funosas R.; Mendive-Tapia L.; Duffin R.; Shields M. A.; Cartwright J. A.; Henriques S. T.; Sot J.; Goñi F. M.; Lavilla R.; Marwick J. A.; Vermeren S.; Rossi A. G.; Egeblad M.; Dransfield I.; Vendrell M. A fluorogenic cyclic peptide for imaging and quantification of drug-induced apoptosis. Nat. Commun. 2020, 11 (1), 4027. 10.1038/s41467-020-17772-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanshaw R. G.; Smith B. D. New reagents for phosphatidylserine recognition and detection of apoptosis. Bioorg. Med. Chem. 2005, 13 (17), 5035–5042. 10.1016/j.bmc.2005.04.071. [DOI] [PubMed] [Google Scholar]
- Rice D. R.; Clear K. J.; Smith B. D. Imaging and therapeutic applications of zinc(ii)-dipicolylamine molecular probes for anionic biomembranes. Chem. Commun. 2016, 52 (57), 8787–8801. 10.1039/C6CC03669D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.-L.; Sheng R.; O’Connor M. J.; Cui Y.; Yoon Y.; Kurilova S.; Lee D.; Cho W. Simultaneous In Situ Quantification of Two Cellular Lipid Pools Using Orthogonal Fluorescent Sensors.. Angew. Chem., Int. Ed. 2014, 53 (52), 14387–14391. 10.1002/anie.201408153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A. E.; Shrestha R.; Webb L. J.; Elber R. Membrane Permeation of a Peptide: It Is Better to be Positive. J. Phys. Chem. B 2015, 119 (21), 6412–6420. 10.1021/acs.jpcb.5b02122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C.; Brewer K.; Song S.; Zhang R.; Lu W.; Wen X.; Li C. Peptide-Based Imaging Agents Targeting Phosphatidylserine for the Detection of Apoptosis. J. Med. Chem. 2011, 54 (6), 1825–1835. 10.1021/jm101477d. [DOI] [PubMed] [Google Scholar]
- Igarashi K.; Kaneda M.; Yamaji A.; Saido T. C.; Kikkawa U.; Ono Y.; Inoue K.; Umeda M. A Novel Phosphatidylserine-binding Peptide Motif Defined by an Anti-idiotypic Monoclonal Antibody: Localization of Phosphatidylserine-Specific Binding Sites on Protein Kinase C and Phosphatidylserine Decarboxylase. J. Biol. Chem. 1995, 270 (49), 29075–29078. 10.1074/jbc.270.49.29075. [DOI] [PubMed] [Google Scholar]
- Perreault A.; Richter S.; Bergman C.; Wuest M.; Wuest F. Targeting Phosphatidylserine with a 64Cu-Labeled Peptide for Molecular Imaging of Apoptosis. Mol. Pharmaceutics 2016, 13 (10), 3564–3577. 10.1021/acs.molpharmaceut.6b00666. [DOI] [PubMed] [Google Scholar]
- Birge R. B.; Boeltz S.; Kumar S.; Carlson J.; Wanderley J.; Calianese D.; Barcinski M.; Brekken R. A.; Huang X.; Hutchins J. T.; Freimark B.; Empig C.; Mercer J.; Schroit A. J.; Schett G.; Herrmann M. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 2016, 23 (6), 962–978. 10.1038/cdd.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-Y.; Kim I.-S. Engulfment signals and the phagocytic machinery for apoptotic cell clearance. Exp. Mol. Med. 2017, 49 (5), e331–e331. 10.1038/emm.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast F. G.; Meyer M.; Carlson G. L.; Iida S.; Potter J. D. Synthesis, spectral properties, and use of 6-acryloyl-2-dimethylaminonaphthalene (Acrylodan). A thiol-selective, polarity-sensitive fluorescent probe. J. Biol. Chem. 1983, 258 (12), 7541–7544. 10.1016/S0021-9258(18)32211-7. [DOI] [PubMed] [Google Scholar]
- Dupuis A.; Duszynski J.; Vignais P. V. A fluorescent derivative of the oligomycin-sensitivity conferring protein (acrylodan-OSCP). Evidence for polarity changes in the environment of CYS118 of OSCP upon binding to mitochondrial F1. Mol. Cell Biol. Res. Commun. 1987, 142 (1), 31–37. 10.1016/0006-291X(87)90447-5. [DOI] [PubMed] [Google Scholar]
- Mondal S.; Rakshit A.; Pal S.; Datta A. Cell Permeable Ratiometric Fluorescent Sensors for Imaging Phosphoinositides. ACS Chem. Biol. 2016, 11 (7), 1834–1843. 10.1021/acschembio.6b00067. [DOI] [PubMed] [Google Scholar]
- Ghosh A. K.; Rukmini R.; Chattopadhyay A. Modulation of Tryptophan Environment in Membrane-Bound Melittin by Negatively Charged Phospholipids: Implications in Membrane Organization and Function. Biochemistry 1997, 36 (47), 14291–14305. 10.1021/bi971933j. [DOI] [PubMed] [Google Scholar]
- Weber G.; Farris F. J. Synthesis and spectral properties of a hydrophobic fluorescent probe: 6-propionyl-2-(dimethylamino)naphthalene. Biochemistry 1979, 18 (14), 3075–3078. 10.1021/bi00581a025. [DOI] [PubMed] [Google Scholar]
- Klymchenko A. S. Solvatochromic and Fluorogenic Dyes as Environment-Sensitive Probes: Design and Biological Applications. Acc. Chem. Res. 2017, 50 (2), 366–375. 10.1021/acs.accounts.6b00517. [DOI] [PubMed] [Google Scholar]
- Kucherak O. A.; Didier P.; Mély Y.; Klymchenko A. S. Fluorene Analogues of Prodan with Superior Fluorescence Brightness and Solvatochromism. J. Phys. Chem. Lett. 2010, 1 (3), 616–620. 10.1021/jz9003685. [DOI] [Google Scholar]
- Grynkiewicz G.; Poenie M.; Tsien R. Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 1985, 260 (6), 3440–3450. 10.1016/S0021-9258(19)83641-4. [DOI] [PubMed] [Google Scholar]
- Chanaday N. L.; Kavalali E. T. Time course and temperature dependence of synaptic vesicle endocytosis. FEBS Lett. 2018, 592 (21), 3606–3614. 10.1002/1873-3468.13268. [DOI] [PubMed] [Google Scholar]
- Carter M.; Shieh J.. Chapter 11: Gene Delivery Strategies. In Guide to Research Techniques in Neuroscience, 2nd ed.; Carter M., Shieh J., Eds.; Academic Press: San Diego, 2015; pp 239–252. [Google Scholar]
- Kay J. G.; Koivusalo M.; Ma X.; Wohland T.; Grinstein S. Phosphatidylserine dynamics in cellular membranes. Mol. Biol. Cell 2012, 23 (11), 2198–2212. 10.1091/mbc.e11-11-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K.; Nagata S. An Apoptotic ‘Eat Me’ Signal: Phosphatidylserine Exposure. Trends Cell Biol. 2015, 25 (11), 639–650. 10.1016/j.tcb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Ormerod M. G.; O’Neill C.; Robertson D.; Kelland L. R.; Harrap K. R. cis-Diamminedichloroplatinum(II)-induced cell death through apoptosis in sensitive and resistant human ovarian carcinoma cell lines. Cancer Chemother. Pharmacol. 1996, 37 (5), 463–471. 10.1007/s002800050413. [DOI] [PubMed] [Google Scholar]
- Zamble D. B.; Lippard S. J. Cisplatin and DNA repair in cancer chemotherapy. Trends Biochem. Sci. 1995, 20 (10), 435–439. 10.1016/S0968-0004(00)89095-7. [DOI] [PubMed] [Google Scholar]
- Troyano A.; Sancho P.; Fernández C.; de Blas E.; Bernardi P.; Aller P. The selection between apoptosis and necrosis is differentially regulated in hydrogen peroxide-treated and glutathione-depleted human promonocytic cells. Cell Death Differ. 2003, 10 (8), 889–898. 10.1038/sj.cdd.4401249. [DOI] [PubMed] [Google Scholar]
- Simon M. D.; Heider P. L.; Adamo A.; Vinogradov A. A.; Mong S. K.; Li X.; Berger T.; Policarpo R. L.; Zhang C.; Zou Y.; Liao X.; Spokoyny A. M.; Jensen K. F.; Pentelute B. L. Rapid Flow-Based Peptide Synthesis. ChemBioChem. 2014, 15 (5), 713–720. 10.1002/cbic.201300796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


