Abstract
Current diagnostics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection heavily rely on reverse transcription-polymerase chain reaction (RT-PCR) or on rapid antigen detection tests. The former suffers from long time-to-result and high cost while the latter from poor sensitivity. Therefore, it is crucial to develop rapid, sensitive, robust, and inexpensive methods for SARS-CoV-2 testing. Herein, we report a novel optofluidic technology, a flow-virometry reader (FVR), for fast and reliable SARS-CoV-2 detection in saliva samples. A small microfluidic chip together with a laser-pumped optical head detects the presence of viruses tagged with fluorescent antibodies directly from saliva samples. The technology has been validated using clinical samples with high sensitivity (91.2%) and specificity (90%). Thanks also to its short time-to-result (<30 min) and small size (25 × 30 × 13 cm), which can be further reduced in the future, it is a strong alternative to existing tests, especially for point-of-care (POC) and low resource settings.
1. Introduction
From the beginning of the Coronavirus disease 2019 (COVID-19) pandemic, one of the most effective measures to control the spread of SARS-CoV-2 infection has been both individual and population-wide testing [1,2]. This strategy enables the identification of infected individuals, especially those asymptomatic and can silently spread the infection, and the tracing of their close contacts to limit the spread of the infection. Moreover, to evaluate the prevalence of new infections, it is also important to adopt additional control measures, such as the use of face masks, social distancing, or ultimately, to establish a lockdown.
The diagnosis of SARS-CoV-2 infection is based on the detection of viral RNA or viral antigens from active cases and indirectly by serology, detecting the antibodies elicited naturally against the viral antigens after infection. Among the different approaches for diagnosis of suspected infected cases [3], the reverse transcription polymerase chain reaction (RT-PCR) has become the gold standard testing assay [4,5,6,7]. Despite high specificity and sensitivity, this assay is expensive, time-consuming due to the need for a previous RNA extraction step, requires a specialized lab and highly trained personnel. These drawbacks may hamper its use in case of high infection rate, especially in developing countries with limited resources. As alternative, the antigen test is a less expensive method that allows rapid detection of SARS-CoV-2 in nasopharyngeal, nasal, or saliva samples. However, the antigen test shows a reduced sensitivity when compared to RT-PCR [3,8,9]. Therefore, the development of new methods that combine the advantages of both RT-PCR (high sensitivity) and antigen test (low cost and short time-to-result) is urgently needed. Optical biosensors are a promising alternative solution for SARS-CoV-2 diagnosis due to their high sensitivity, ease-of-use, short time-to-result, and point-of-care testing capabilities. Recently, much effort has been made in developing label-free optical sensors based on surface plasmon resonance (SPR), localised surface resonance (LSPR) and surface-enhanced Raman spectroscopy (SERS) [10]. Other emerging optical techniques involve spatial light-interference microscopy (SLIM) in combination with neural network or nano-interferometric biosensor for detection of SARS-CoV-2 [11,12]. Although preliminary results published in the literature suggest that these technologies have the potential to be used as a highly sensitive SARS-CoV-2 detection platform, much work remains to be done to operate outside the laboratory and bring them into the commercial phase.
SARS-CoV-2 can be consistently detected in samples obtained from the respiratory tract, including nasal/oropharyngeal swabs and sputum, but also from samples collected from the digestive tract such as saliva or stool [13]. Among those sample types, nasal or oropharyngeal swabs, alone or in combination, are commonly used for SARS-CoV-2 testing because of the presence of high number of viral particles. An important disadvantage of these routes is that sample collection requires skilled personnel to ensure proper sample collection and to minimize the discomfort caused during sampling, especially when several samples are collected over a short period of time. Saliva is an alternative to nasal and oropharyngeal swabs [13]. The levels of SARS-CoV-2 are similar or even higher than those described in nasopharyngeal swabs [3] or sputum [14] and the risk of a false negative result due to inaccurate sample collection is lower [3]. Furthermore, the use of saliva samples shows several clinical advantages: (1) quick collection and multiple sampling with minimal discomfort for the subject; (2) self-collection with simple instructions, minimizing the risk of viral transmission to healthcare personnel; (3) easy check of presence of the virus in asymptomatic individuals [15] and (4) possibility of large-scale SARS-CoV-2 testing, especially in combination with fast and sensitive detection method [16].
In this work, we demonstrate a novel portable flow-virometry reader (FVR) for fast detection of SARS-CoV-2 directly from saliva samples, which combines the short time to results of an antigen test and a high sensitivity, close to that of RT-PCR (Supporting Information Table S1). Our proposed diagnostic device relies on flow cytometry, which is a powerful laser-based cell analysis technique for counting and sorting cells by light scattering and/or fluorescence. Although flow-virometry has been previously proposed [17] for SARS-CoV-2 detection, the main drawbacks of conventional flow-cytometers are that these are bulky (∼ 49 × 58 × 43 cm) and expensive devices (∼ 30K€ to 300 K€), making them unsuitable for POC applications. To address these limitations of a traditional flow cytometer, we present a compact FVR that integrates a microfluidic chip, a laser-pumped optical head, and a multi-pixel photon counting (MPPC) system, specifically designed for sensitive detection of fluorescence from labelled SARS-CoV-2 viral spike proteins. The sensitivity and specificity of the technology were validated by performing a blind test with 54 clinical samples.
2. Results and discussion
2.1. Overview of the flow-virometry reader (FVR) for SARS-CoV-2 antigen detection
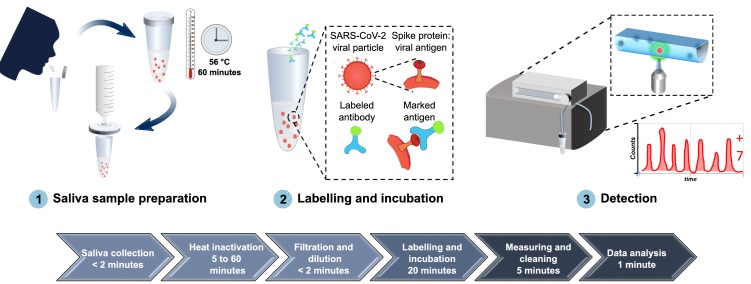
When addressing the need for faster, simple and accurate SARS-CoV-2 detection methods, we surmised that the combination of precise flow-virometry with a fast immunoassay test has the potential to improve the speed and precision of SARS-CoV-2 detection from saliva. Figure 1 shows the entire workflow of the developed diagnostic system, from sample collection to SARS-CoV-2 antigen detection.
Fig. 1.
A comprehensive overview of the developed diagnostic method for detection of SARS-CoV-2 antigen, from sample collection to SARS-CoV-2 antigen measurement. 1) Saliva is collected by spit, heat inactivated at 56°C for 1 hour, diluted in PBS and filtered with a 0.2 µm syringe filter. 2) The sample is then labelled with Alexa 488-anti-SARS-CoV spike antibodies and incubated for 20 minutes at room temperature. 3) The labelled sample is circulated inside the FVR and a fluorescence signal is detected every time antibody-virus complexes pass through the interrogation point.
2.2. Development and characterization of the flow-virometry reader (FVR)
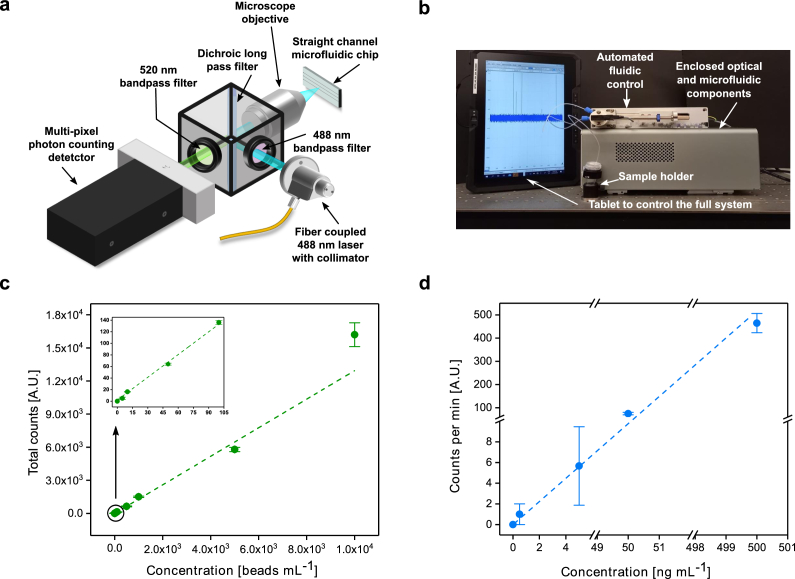
The FVR is a portable flow-virometer designed for the detection and quantification of fluorescence labelled micro and nanoparticles, including viruses, such as the SARS-CoV-2. It consists of an optical head for sample excitation and signal collection, coupled with a single straight channel microfluidic chip for sample circulation and interrogation. Figure 2(a) shows its schematic diagram with component details. A fibre coupled 488 nm laser is used as the light source. The laser beam is focused onto the microfluidic channel by a 100× microscope objective. As the sample flows, in the laser beam focus the fluorophores emit fluorescence, which is then directed to a multi-pixel photon counting (MPPC) detector after passing through a dichroic mirror and two optical filters. The optical filters ensure that the emitted fluorescence signal predominantly reaches the detector without any significant stray light from the excitation source, thus reducing the background noise and increasing system sensitivity. A lab-built prototype controlled by a tablet is shown in Fig. 2(b).
Fig. 2.
FVR characterization. (a) A schematic diagram of the FVR detection system. (b) A lab built FVR prototype with the tablet. (c) Standard curve of the polystyrene beads concentration vs. total counts, R2 = 0.99. (d) Standard curve of the fluorescent (Alexa 488) anti-SARS-CoV spike antibody concentration vs. counts per minute, R2 = 0.94. For visual representation purposes the y-axis is broken into two regions [0:10], [60:100] and the x-axis into three [0:6], [49:52] and [498:501]. The error bar represents the standard deviation.
The initial functional test and calibration of the FVR were performed using a known concentration of fluorescent polystyrene beads with a nominal diameter of 2 µm in phosphate-buffered saline (PBS). For the calibration of the FVR response, triplicate measurements of five 10-fold dilutions of the polystyrene beads solution were performed. Figure 2(c) shows the resulting calibration plot obtained for the polystyrene beads with concentrations ranging from 2.5 beads mL-1 to 2.5×104 beads mL-1. A linear response (R2 = 0.99) over the concentration range measured is evident, indicating that there is a good agreement between the total counted events in the 1 mL sample and the concentration of beads in the solution.
After the fluorescent bead's measurements, the FVR baseline was established by measuring a PBS solution of fluorescent antibodies. To determine the analytical response of anti-SARS-CoV spike antibody, four 10-fold serial dilutions with concentrations ranging from 0.5 ng mL-1 to 500 ng mL-1 were measured in triplicates. Figure 2(d) shows that there is a good correlation between the prepared anti-SARS-CoV spike antibody concentrations and the detected fluorescent events (R2= 0.94; p = 0.001).
2.3. Optimization of the FVR assay for SARS-CoV-2 antigen detection
Our detection method strictly relies on the effective binding between fluorescent anti-SARS-CoV spike antibody and SARS-CoV-2 spike proteins. The rate of this binding can be affected by several factors including sample composition, antibody/antigen concentration and temperature, among others. [18] Therefore, an optimization of these parameters was performed to facilitate the antigen-antibody complex formation. Saliva samples were collected from donors by spitting followed by a heat inactivation step at 56°C for 1 hour, mainly due to the constraints of performing the measurements in a Biosafety Level 2 laboratory. The inactivated samples were then filtered using 0.2 µm syringe filters to remove impurities such as large debris, cells and mucin to prevent the clogging of the FVR’s microfluidic channels and reduce the background of the samples (Supplementary Fig. S1, found in Supplement 1 (1.6MB, pdf) ). The filtered samples were diluted 100-fold in PBS. The filtration and dilution steps reduce the variability of salivary viscosity and enable a steady laminar flow in the microfluidic channel without the need to use high pressure (> 13 bar). The optimal antibody concentration was determined experimentally. For this, a SARS-CoV-2 positive sample with a viral load of 107 copies mL-1 was prepared in a 10-fold serial dilution in saliva from healthy uninfected donors (SARS-CoV-2 negative by RT-qPCR). The final dilution range varies from 107 copies mL-1 down to 100 copies mL-1. For each dilution, four different concentrations of the Alexa 488-anti-SARS-CoV spike antibody were assayed (25 µg mL-1, 5 µg mL-1, 500 ng mL-1 and 50 ng mL-1). We found 50 ng mL-1 to be the most effective antibody concentration (Supplementary Fig. 2a, Supplement 1 (1.6MB, pdf) ) and, therefore, it was used in our experiments for SARS-CoV-2 antigen detection by FVR.
Next, the incubation time for the detection of SARS-CoV-2 spike proteins was investigated and optimized with respect to signal increase and time-to-results. Two saliva samples with a SARS-CoV-2 viral load of 4×104 and 1.6×108 copies mL-1, respectively, and one SARS-CoV-2 negative saliva sample (confirmed by RT-qPCR) from a healthy donor, spiked with recombinant spike proteins at a concentration of 50 ng mL-1, were used (Supplementary Fig. S2b, Supplement 1 (1.6MB, pdf) ). Samples were incubated with a fixed concentration of Alexa 488-anti-SARS-CoV spike antibody (50 ng mL-1) and measured for different incubation time points (5 min, 10 min, 15 min, 20 min, 30min, 40 min and 50 min). The results showed that the signal increased after 15 min of incubation for all three analyzed samples (Supplementary Fig. S2b, Supplement 1 (1.6MB, pdf) ). After 20 min of incubation, the 4×104 copies mL-1 SARS-CoV-2 positive sample reached a plateau and showed a very slight signal increasing trend after a longer incubation time. This effect might be due to the steady antibody-antigen complex formation. On the contrary, in the sample with the highest viral load (1.6×108 copies mL-1) a decrease in the signal was observed over the same period of time, suggesting a Hook effect [19].
To confirm the increased signal due to the formation of aggregates between antibody and viral antigens, we used stimulated emission depletion (STED) microscopy to image labelled SARS-CoV-2 saliva samples and Alexa 488-anti-SARS-CoV spike antibody only. We observed a 5-fold increase in fluorescent signal of labelled viral particles compared to only 1-fold increase in that of antibodies with respect to the background (Supplementary Fig. S3, Supplement 1 (1.6MB, pdf) ).
It is worth highlighting here that our optimized labelling protocol combined with the simplified sample collection and purification via filtration has several advantages. Apart from FVR, it does not require special laboratory equipment and there is no need of sample purification after labelling, as the increased number of detected events with respect to the antibodies signal in a positive sample is significantly higher. Moreover, as opposed to the lateral flow based antigen tests, our FVR does not rely on the binding efficiency of the specific antibodies onto the surface, thus, eliminating any unspecific surface interactions, whilst the surface to volume ratio of the binding sites for antibody-antigen complex formation increases.
2.4. Limit of blank, limit of detection and standard curve for SARS-CoV-2 detection
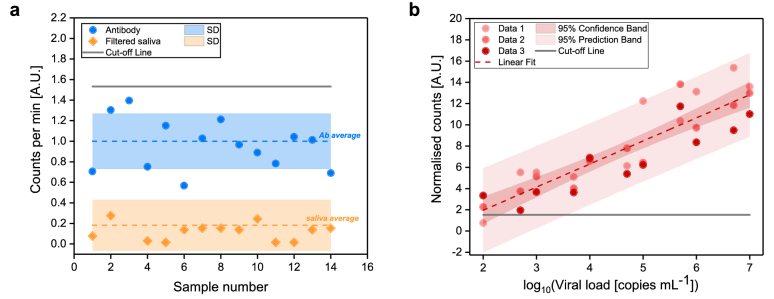
To investigate the potential of the developed FVR as a novel platform for fast detection of SARS-CoV-2 directly from saliva samples, we determined the Limit of Blank (LOB), the Limit of Detection (LOD), and built a standard curve to find the relationship between the FVR signal and viral load. First, we checked the signal obtained from the Alexa 488-anti-SARS-CoV spike antibody in PBS and from the filtered saliva samples (Fig. 3(a)). The results showed that the signal obtained from filtered saliva samples was lower than that of Alexa 488-anti-SARS-CoV spike antibody (p < 0.0001), indicating that the remaining saliva content after filtration did not affect the detection of SARS-CoV-2. The Alexa 488-anti-SARS-CoV spike antibody signal was used to determine the LOB of the FVR. The LOB represents the highest apparent SARS-CoV-2 concentration detected by the FVR when samples containing no SARS-CoV-2 antigens or viral particles are tested. Here, the LOB was calculated as the average of the blank plus 1.645 times its standard deviation [20]. We set the cut-off value for the detection of SARS-CoV-2 positive samples higher than the LOB and, specifically, as the average of the blank (antibody signal) plus two times its standard deviation (Fig. 3(a)). Importantly, LOB and cut-off values have to be established each time prior to the measurements of a new batch of the fluorescently labelled SARS-CoV-2 spike proteins. Then, a standard curve was built (Fig. 3(b)) by measuring labelled SARS-CoV-2 viral antigens from saliva samples with increasing viral load concentration. Three SARS-CoV-2 positive samples with an initial viral load of 109 copies mL-1 (determined by RT-qPCR) were first diluted 100-fold in PBS. From each of these samples further dilutions were prepared to give concentrations ranging from 102 to 107copies mL−1. Normalised counts here represent the counted events of each measured sample normalized with respect to the antibody average signal to take into account batch effects and user-to-user variability in sample handling. The standard curve shows good linearity (R2 = 0.94, p < 0.001). The sensitivity of the assay, reported from the slope of the calibration curve, equals 2.17 A.U. mL copies−1, highlighting the effectiveness of the developed FVR in screening of SARS-CoV-2 viral particles.
Fig. 3.
Determination of FVR cut-off value and standard curve for SARS-CoV-2 detection (a) Counted events per minute of the Alexa 488-anti-SARS-CoV spike antibody (blue) and of filtered saliva samples (orange). Mean values (dotted lines) and standard deviations (shaded area) are indicated. Cut-off, according to the antibody signal (blue), was calculated as mean plus two times its standard deviation and indicated as solid line. (b) Standard curve of the SARS-CoV-2 viral particles concentration vs. normalised counts; R2 = 0.94.
The reproducibility of the FVR to quantify the viral load was investigated over the entire linear range and data showed that the normalized root-mean-square error (NRMSE) was below 19% for three independent experiments. The LOD, calculated as the LOB plus 1.645 times the standard deviation of the sample with lowest concentration of the analyte tested [21], was found to be as low as 3834 copies mL-1, at least three orders of magnitude lower than that of commercially available rapid antigen tests [22].
Taking into consideration the obtained LOD, LOB and cut-off, our analytical range (monotonic behaviour) is from 4×103 to 107 viral load mL-1.
2.5. SARS-CoV-2 detection in saliva samples using FVR via blind test
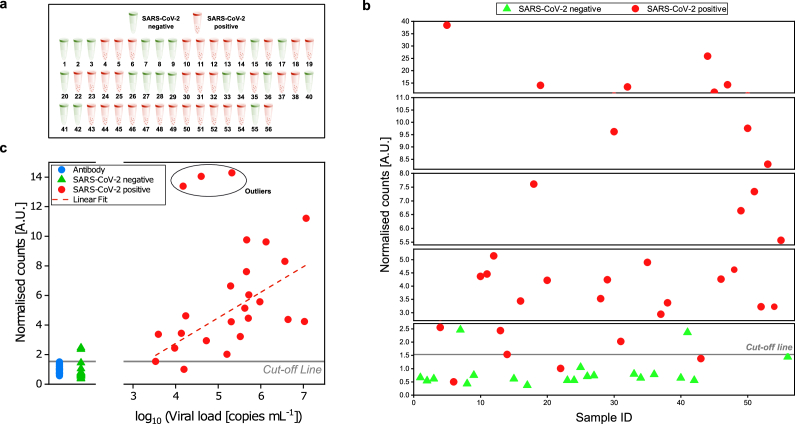
To evaluate the performance of the developed FVR for detection of SARS-CoV-2, we carried out a blind test using frozen saliva samples from both SARS-CoV-2 infected (n = 34) and uninfected individuals (n = 20). Viral load in the same saliva samples was also determined by RT-qPCR. For FVR measurements, each of the samples was diluted 1:100 in PBS to lower viscosity and to reach a sufficient volume for enabling filtration and minimal sample size for measurement. As described above, samples were filtered and labelled with the Alexa 488-anti-SARS-CoV spike antibody. The cut-off line was determined using the signal obtained with the fluorescent antibody solution, as previously mentioned. The results showed that the FVR was able to reliably detect 31 out of 34 SARS-CoV-2 positive patient samples and 18 out of 20 SARS-CoV-2 negative samples, previously analysed using RT-qPCR (Fig. 4(a) and (b)). This corresponds to a diagnostic sensitivity of 91.2% and a diagnostic specificity of 90% determined according to the medical device coordination group document MDCG 2020-21 [23].
Fig. 4.
Detection of SARS-CoV-2 in saliva samples from SARS-CoV-2 infected and uninfected individuals. SARS-CoV-2 was determined in saliva samples from both SARS-CoV-2 infected (n = 34) and uninfected (n = 20) individuals in a blind test. (a) Sample identification by color code (red: SARS-CoV-2 positive samples; green: SARS-CoV-2 uninfected samples) according to RT-qPCR. (b) Distribution of saliva samples showing the normalized count obtained with the FVR. (c) Correlation between normalized counts and RT-qPCR viral load [copies mL-1] of blind test samples. The dashed line indicates a linear fit (R2 = 0.43) to the data from positive samples that fall within our analytical range excluding the outliers marked with a circle. Here outliers are defined as data points whose residual values are greater than two times the standard deviation of all the data points.
When comparing the signal obtained from the blind samples with respect to the viral load determined by RT-qPCR, we confirmed a linear relationship (R2 = 0.43) between the normalized counts and copies mL-1 of the samples with concentration up to 107 copies mL−1 (Fig. 4(c)). Furthermore, from our obtained standard curve (Fig. 3(b)) we were able to predict the concentrations of the unknown samples (copies mL-1) and compare them with respect to the RT-qPCR results (Supplement 1 (1.6MB, pdf) , Fig. S4, S5). This enabled us to establish the viral load mL-1 of the measured samples with concentrations within the detection range of the FVR, allowing a normalized root-mean-square deviation (NRMSD) of 9.2%, after discarding from the quantification the samples that returned a normalized count over the upper limit of our calibration curve (3 out of 23). It is important to note here that we have chosen a polyclonal antibody for our experiments because of its commercial availability. This antibody is elicited against the spike glycoprotein of the SARS-CoV, which showed cross-reactivity with the spike of SARS-CoV-2. In future work, we will focus on selecting antibodies that form complexes only with the antigen of interest, for example, monoclonal rather than polyclonal, which may improve the specificity of the assay.
3. Conclusion
The main goal of this work was to develop a fast, reliable and inexpensive biosensor based on flow-virometry that combines the sensitivity of the RT-PCR with the quickness of the rapid antigen test. In addition, whereas RT-PCR and rapid antigen tests quite often use nasopharyngeal swab samples, the proposed technology and test use saliva samples that can be collected quickly with less discomfort to patients and can even be self-administered. Furthermore, the test can also be easily adapted to work with nasopharyngeal swabs samples if needed. Our highly sensitive sensor platform was easily adapted to SARS-CoV-2 detection in saliva samples from patients, with short time-to-result (<30 min), showing potential advantages over other currently available diagnostics. The measurements can also be performed at a low cost and in a fully portable and automated form factor, especially suitable to POC settings. The LOD obtained with the system is about 3800 viral copies mL−1, three orders of magnitude lower than that of commercial antigen tests, with high sensitivity (91%) and specificity (90%).
Given the results, we believe that our FVR in conjunction with saliva samples has great potential for becoming a fast, portable, user-friendly POC device, able to perform up to 2000 tests per day per single device. Its throughput can be significantly increased by multiplexing within the same unit and running several samples simultaneously. Furthermore, FVR is built with off-the-shelf components and currently has a bill of materials (BOM) of about 10 K€. The BOM and the overall cost of the system will be significantly lower when moving from current prototyping to large-scale production. The sample collection could be operated by providing a self-collection kit containing a tube pre-filled with reagents in which a filter is incorporated to clarify the saliva. The single test cost can have an operational price as low as 2.00 €. Interestingly, by the selection of proper antibodies, the technology can also be easily adapted to the detection of possible variants of the virus or other specific viruses, such as seasonal coronavirus or influenza virus. To this end, future work should also focus on cross-reactivity studies of antibodies with different viruses and antigens. Moreover, efforts will be devoted to developing an integrated FVR for simultaneous SARS-CoV-2 antigen and antibody analysis. This combined testing device will enable not only to detect an active infection in the body (antigen detection) but could also provide information on the immune response to the infection (antibody detection) or after vaccination. This will allow fast checking of vaccine efficiency and if additional vaccination boosters are needed.
4. Materials and methods
4.1. Design of the FVR
The basics of the design of the FVR is described in [24]. All the optomechanical parts were purchased from Thorlabs and the MPPC was purchased from Hamamatsu Photonics. The microfluidic chip (Dolomite Microfluidics) is connected to a syringe pump from CETONI GmbH. The signal from the MPPC is recorded using a USB oscilloscope (PicoScope). The data analysis for counting the number of events was performed by a custom MATLAB code using a threshold value (3.5mV) that was well above the background signal level of PBS, which was the aqueous solution for all samples measured in this work. In order to build a lab prototype, the optical setup together with the electronic modules were assembled into a 200 mm x 300 mm x 109 mm enclosure (EC2030B, Thorlabs).
4.2. FVR calibration
To calibrate the FVR system, 2 µm fluorescent polystyrene beads (Dragon Green, FSDG005, Bangs Laboratories, Inc.) at a concentration of 250 beads mL-1 were used. The green fluorescent beads have excitation centred at 480 nm and emission at 520 nm. In order to ensure proper alignment of the optical system with respect to the straight channel microfluidic chip, the beads solution was circulated inside the microfluidic channel at a flow rate of 1 mL min-1. The position of the chip was adjusted until a steady rate of at least 2 events per second was observed.
4.3. Saliva sample collection
Saliva samples were obtained from PCR-confirmed cases of COVID-19 individuals (n= 34) who attended the “Hospital Universitari Germans Trias i Pujol” (Badalona, Catalonia, Spain) between July 2020 and October 2020. As a control, saliva samples were collected from uninfected healthy donor also confirmed by RT-qPCR. All saliva samples were collected by spit in sterile tubes and stored at -80°C until use. The study was approved by the Institutional Review Board of the “Hospital Universitari Germans Trias i Pujol” (code PI-21-157, FlowsaCOVID, and code PI-20-217, KING cohort extension) and written informed consent for study participation was obtained from all individuals. Saliva samples were heat inactivated at 56 °C for 1 h, prior to performing the experiment in a Biosafety Level 2 laboratory [25].
4.4. FVR detection of SARS-CoV-2 in saliva samples
Saliva samples were thawed and treated at 56°C for 1 hour, for viral inactivation. The samples were then diluted 100-fold with PBS buffer (A9177, AppliChem GmbH) and filtered by passing through a 0.20 µm pore sterile Corning syringe filter (CLS431218-50EA, Sigma Aldrich) prior to labelling. Polyclonal Alexa 488-anti-SARS-CoV spike antibodies (ABIN914365, antibodies-online GmbH, Germany) were used for labelling the saliva samples. The Alexa Fluor 488 has an excitation peak at 499 nm and an emission peak at 520 nm. The antibodies were initially diluted to a concentration of 100 ng mL-1 in PBS and 500 µL were added to 500 µL of filtered saliva sample to obtain a final antibody concentration of 50 ng mL-1. When using a sample volume as low as 20 µL, the full sample preparation protocol calls for a 1:200 dilution factor. Next, the samples were incubated for 20 minutes at room temperature and immediately measured with the FVR. Labelled samples (1mL) were loaded into the FVR using an automatic syringe pump at 1 mL min-1 and the signal was recorded by an USB oscilloscope at 500 ms/div for 160 seconds. All samples were measured in triplicate and the system was cleaned between samples by performing cycles of circulation of 2 mL of PBS inside the tubing and the microfluidic chip at 8 mL min-1, until the fluorescent signal reached zero for 160 seconds. On average, six cleaning cycles were needed to ensure the complete cleaning of the system between samples.
SARS-CoV-2 recombinant protein (SinoBiological) was diluted in saliva samples collected from SARS-CoV-2 uninfected individuals (confirmed by RT-qPCR) at 50 ng mL-1 final concentration as a positive control for the labelling time optimization experiment.
4.5. RT-qPCR analysis of clinical saliva samples
RNA extraction from saliva samples was performed by using Viral RNA/Pathogen Nucleic Acid Isolation kit (Thermo Fisher Scientific), optimized for a KingFisher instrument (Thermo Fisher Scientific), following manufacturer's instructions. PCR amplification was based on the 2019-Novel Corona-virus Real-Time RT-qPCR Diagnostic Panel guidelines and protocol developed by the American Center for Disease Control and Prevention. Briefly, a 20 µl PCR reaction was set up containing 5 µL of RNA, 1.5 µL of N2 primers and probe (2019-nCov CDC EUA Kit, Integrated DNA Technologies) and 10 µl of GoTaq 1-Step RT-qPCR (Promega). Thermal cycling was performed at 50 °C for 15 min for reverse transcription, followed by 95 °C for 2 min and then 45 cycles of 95 °C for 10 sec, 56 °C for 15 sec and 72 °C for 30 sec in the Applied Biosystems 7500 or QuantStudio5 Real-Time PCR instruments (Thermo Fisher Scientific). For absolute quantification, a standard curve was built using 1:5 serial dilutions of a SARS-CoV2 plasmid (2019-nCoV_N_Positive Control, 200 copies µl-1, Integrated DNA Technologies) and run in parallel in all PCR determinations. Viral load of each sample was determined in triplicate and the mean viral load (in copies mL-1) was extrapolated from the standard curve and corrected by the corresponding dilution factor. RNaseP gene amplification was performed in duplicate for each sample as an amplification control.
4.6. Stimulated emission depletion (STED) imaging
Coverslips were firstly pre-coated with poly-L-Lysine at 0.01% in MiliQ water for 5 minutes at room temperature. Then, coverslips were left to dry out in the air. Next, 20 µL of labelled virus solution or anti-SARS-CoV spike antibody was spotted on the poly-L-coverslip, then left to adhere for around 10-15 min at RT. Afterwards the excess was washed off with PBS buffer (3 × 5 min). STED imaging was done using LEICA TCS SP8 X.
4.7. Statistical analysis
The statistical analysis was performed using Microsoft Excel 2016 (16.0.5173.1000). All results were showed as individual data or mean plus standard deviation. Groups were compared using the two-tailed Student’s t test and p value < 0.05 was considered as a significant statistical difference. Three technical repetitions were performed unless otherwise mentioned. OriginPro 8.0 (OriginLab Corp., Northhampton, MA) was used for all plots, including fitting and calculating 95% confidence and prediction band.
Acknowledgements
A.O., R.H., E.W. and V.P. acknowledge financial support from the Spanish Ministry of Economy and Competitiveness through the ‘Severo Ochoa’ Programme for Centres of Excellence in R&D CEX2019-000910-S [MCIN/ AEI/10.13039/501100011033], and project TUNA-SURF (PID2019-106892RB-I00), Fundació Cellex Mir-Puig, and from Generalitat de Catalunya through the CERCA program, from AGAUR 2017 SGR 1634. E.W. acknowledges the Beatriu de Pinos-3 Postdoctoral Pro-gramme (BP3) under grant agreement ID: 801370. This work has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 665884. We acknowledge the Super-resolution Light Microscopy and Nanoscopy (SLN) facility at ICFO, especially Dr. María Marsal, for her contribution in STED imaging. We also thank Prof. Lluís Torner for his help in initiating the project and continuous discussions.
Moreover, this work was supported by the crowdfunding project “YomeCorono”, the CERCA Program (Generalitat de Catalunya), the projects: SLD0015 and SLD0016 (Direcció General de Recerca i Innovació en Salut, Generalitat de Catalunya). J.B. is supported by the Health Department of the Catalan Government (Generalitat de Catalunya). We thank Foundation Dormeur for financial support for the acquisition of the QuantStudio-5 real-time PCR system. We would like to thank all participants and the Clinical Sample Management Team of IrsiCaixa for their outstanding work.
Funding
Severo Ochoa (CEX2019-000910-S, MCIN/ AEI/10.13039/501100011033, TUNA-SURF (PID2019-106892RB-I00)); Agència de Gestió d'Ajuts Universitaris i de Recerca10.13039/501100003030 (AGAUR 2017 SGR 1634); Fundación Cellex10.13039/100008050; Beatriu de Pinos-3 (801370); H2020 Marie Skłodowska-Curie Actions10.13039/100010665 (665884); Centres de Recerca de Catalunya10.13039/100015439 (SLD0015, SLD0016).
Disclosures
V.P. is one of the inventors of patent application WO2020212628A1, related to the FVR technology.
Data Availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
Supplemental document
See Supplement 1 (1.6MB, pdf) for supporting content.
References
- 1.Mercer T. R., Salit M., “Testing at scale during the COVID-19 pandemic,” Nat. Rev. Genet. 22(7), 415–426 (2021). 10.1038/s41576-021-00360-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen K., Leshem A., “Suppressing the impact of the COVID-19 pandemic using controlled testing and isolation,” Sci. Rep. 11(1), 6279 (2021). 10.1038/s41598-021-85458-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., Soni D., Das S., Hasan M., Patel M., Senan A.M., Gorantla S., McMillan J., Edagwa B., Eisenberg R., Gurumurthy C.B., Reid S.P.M., Punyadeera C., Chang L., Gendelman H.E., “Diagnostics for SARS-CoV-2 infections,” Nat. Mater. 20(5), 593–605 (2021). 10.1038/s41563-020-00906-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji T., Liu Z., Wang G., Guo X., Akbar S., “Detection of COVID-19: a review of the current literature and future perspectives,” Biosens. Bioelectron. 166, 112455 (2020). 10.1016/j.bios.2020.112455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park M., Won J., Choi B. Y., Lee C. J., “Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR,” Exp. Mol. Med. 52(6), 963–977 (2020). 10.1038/s12276-020-0452-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohn M.K., Lippi G., Horvath A., Sethi S., Koch D., Ferrari M., Wang C.B., Mancini N., Steele S., Adeli K., “Molecular, serological, and biochemical diagnosis and monitoring of COVID-19: IFCC taskforce evaluation of the latest evidence,” Clin. Chem. Lab. Med. 58(7), 1037–1052 (2020). 10.1515/cclm-2020-0722 [DOI] [PubMed] [Google Scholar]
- 7.Antibody , “Antigen and Molecular Tests for COVID-19,” Johns Hopkins Center for Health Security (2021).
- 8.Pilarowski G., Marquez C., Rubio L., Peng J., Martinez J., Black D., Chamie G., Jones D., Jacobo J., Tulier-Laiwa V., Rojas S., Cox C., Nakamura R., Petersen M., DeRisi J., Havlir D.V., “Field performance and public health response using the BinaxNOWTM Rapid SARS-CoV-2 antigen detection assay during community-based testing,” Clin Infect Dis 73(9), 1–27 (2020). 10.1093/cid/ciaa1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tobian F., Tanuri A., Lindner A.K., Gaeddert M., Köppel L., Tobian F., Brümmer L.E., Klein J.A.F., Lainati F., Schnitzler P., Nikolai O., Mockenhaupt F.P., Seybold J., Corman V.M., Jones T.C., Drosten C., Gottschalk C., Weber S.F., Weber S., Ferreira O.C., Mariani D., Dos Santos Nascimento E.R., Pereira Pinto Castineiras T.M., Galliez R.M., Faffe D.S., Leitão I.C., Dos Santos Rodrigues C., Frauches T.S., Nocchi K.J.C.V., Feitosa N.M., Ribeiro S.S., Polloc N.R., Knorr B., Welker A., de Vos M., Sacks J., Ongarello S., Denkinger C.M., Study Team , “Accuracy and ease-of-use of seven point-of-care SARS- CoV-2 antigen-detecting tests: a multi-centre clinical evaluation,” EBioMedicine 75, 103774 (2022). 10.1016/j.ebiom.2021.103774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asghari A., Wang C., Yoo K. M., Rostamian A., Xu X., Shin J. D., Dalir H., Chen R. T., “Fast, accurate, point-of-care COVID-19 pandemic diagnosis enabled through advanced lab-on-chip optical biosensors: opportunities and challenges,” Appl. Phys. Rev. 8(3), 031313 (2021). 10.1063/5.0022211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goswami N., He Y.R., Deng Y.H., Oh C., Sobh N., Valera E., Bashir R., Ismail N., Kong H., Nguyen T.H., Best-Popescu C., Popescu G., “Label-free SARS-CoV-2 detection and classification using phase imaging with computational specificity,” Light: Sci. Appl. 10(1), 176 (2021). 10.1038/s41377-021-00620-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz-vega G., Soler M., Lechuga L. M., “Nanophotonic biosensors for point-of-care COVID-19 diagnostics and coronavirus surveillance Nanophotonic biosensors for point-of-care COVID-19 diagnostics and coronavirus surveillance,” JPhys Photonics 3(1), 011002 (2021). 10.1088/2515-7647/abd4ee [DOI] [Google Scholar]
- 13.Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z., “Considerations for diagnostic COVID-19 tests,” Nat. Rev. Microbiol. 19(3), 171–183 (2021). 10.1038/s41579-020-00461-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung K. F., Sun Y. C., Chen B. H., Lo J. F., Cheng C. M., Chen C. Y., Wu C. H., Kao S. Y., “New COVID-19 saliva-based test: How good is it compared with the current nasopharyngeal or throat swab test?” J. Chin. Med. Assoc. 83(10), 891–894 (2020). 10.1097/JCMA.0000000000000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Ren B., Peng X., Hu T., Li J., Gong T., Tang B., Xu X., Zhou X., “Saliva is a non-negligible factor in the spread of COVID-19,” Mol. Oral Microbiol. 35(4), 141–145 (2020). 10.1111/omi.12289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sapkota D., Thapa S. B., Hasséus B., Jensen J. L., “Saliva testing for COVID-19?” Br. Dent. J. 228(9), 658–659 (2020). 10.1038/s41415-020-1594-7 [DOI] [PubMed] [Google Scholar]
- 17.Soni N., Pai P., Krishna Kumar G. R., Prasad V., Dasgupta S., Bhadra B., “A flow virometry process proposed for detection of SARS-CoV-2 and large-scale screening of COVID-19 cases,” Future Virol. 15(8), 525–532 (2020). 10.2217/fvl-2020-0141 [DOI] [Google Scholar]
- 18.Reverberi R., Reverberi L., “Factors affecting the antigen-antibody reaction,” Blood Transfus. 5(4), 227–240 (2007). 10.2450/2007.0047-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward G., Simpson A., Boscato L., Hickman P. E., “The investigation of interferences in immunoassay,” Clin. Biochem. 50(18), 1306–1311 (2017). 10.1016/j.clinbiochem.2017.08.015 [DOI] [PubMed] [Google Scholar]
- 20.Armbruster D. A., Pry T., “Limit of blank, limit of detection and limit of quantitation,” Clin. Biochem. Rev. 29(Suppl 1), S49–52 (2008). [PMC free article] [PubMed] [Google Scholar]
- 21.Shrivastava A., Gupta V., “Methods for the determination of limit of detection and limit of quantitation of the analytical methods,” Chronicles Young Sci. 2(1), 21 (2011). 10.4103/2229-5186.79345 [DOI] [Google Scholar]
- 22.Corman V. M., Haage V.C., Bleicker T., Schmidt M.L., Mühlemann B., Zuchowski M., Jo W.K., Tscheak P., Möncke-Buchner E., Müller M.A., Krumbholz A., Drexler J.F., Drosten C., “Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study,” Lancet Microbe 2(7), e311–e319 (2021). 10.1016/S2666-5247(21)00056-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.European Commission , MDCG 2021-21 Guidance on performance evaluation of SARS-CoV-2 in vitro diagnostic medical devices, 1–17 (2021). Available at: https://ec.europa.eu/health/md_sector/new_regulations/guidance_en.
- 24.Pruneri V., Jofre M. , Martinez Cordero P., “Fluidic apparatus for optical interrogation of individual microorganisms,” Patent No. WO2020212628A1 (22 October 2020).
- 25.Batéjat C., Grassin Q., Manuguerra J., “Heat inactivation of the severe acute respiratory syndrome coronavirus 2,” J. Biosaf. Biosecurity 3, 1 (2020). 10.1016/j.jobb.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.