ABSTRACT
Fertilization triggers physiological degradation of maternal-mRNAs, which are then replaced by embryonic transcripts. Ample evidence suggests that Argonaut 2 (AGO2) is a possible post-fertilization regulator of maternal-mRNAs degradation; but its role in degradation of maternal-mRNAs during oocyte maturation remains obscure. Fyn, a member of the Src family kinases (SFKs), and an essential factor in oocyte maturation, was reported to inhibit AGO2 activity in oligodendrocytes. Our aim was to examine the role of Fyn and AGO2 in degradation of maternal-mRNAs during oocyte maturation by either suppressing their activity with SU6656 – an SFKs inhibitor; or by microinjecting DN-Fyn RNA for suppression of Fyn and BCl-137 for suppression of AGO2. Batches of fifteen mouse oocytes or embryos were analyzed by qPCR to measure the expression level of nine maternal-mRNAs that were selected for their known role in oocyte growth, maturation and early embryogenesis. We found that Fyn/SFKs are involved in maintaining the stability of at least four pre-transcribed mRNAs in oocytes at the germinal vesicle (GV) stage, whereas AGO2 had no role at this stage. During in-vivo oocyte maturation, eight maternal-mRNAs were significantly degraded. Inhibition of AGO2 prevented the degreadation of at least five maternal-mRNAs, whereas inhibition of Fyn/SFK prevented degradation of at least five Fyn maternal-mRNAs and two SFKs maternal-mRNAs; pointing at their role in promoting the physiological degradation which occurs during in-vivo oocyte maturation. Our findings imply the involvement of Fyn/SFKs in stabilization of maternal-mRNA at the GV stage and the involvement of Fyn, SFKs and AGO2 in degradation of maternal mRNAs during oocyte maturation.
KEYWORDS: Fyn kinase, argonaut 2, oocyte maturation, src family kinases
Introduction
The follicle is the basic functional unit in the ovary; it is comprised of an oocyte surrounded by supporting somatic cells, which provide the appropriate environment required for oocyte growth and development [1]. The follicular oocyte, which is meiotically arrested at the prophase of the first meiotic division, is characterized by de-condensed chromosomes surrounded by an intact nuclear membrane termed the “germinal vesicle” (GV) [2]. During follicular growth, the size of the oocyte increases dramatically, and at the final stages, the oocyte undergoes a series of cytoplasmic changes, necessary for acquisition of meiotic competence. In response to the ovulatory stimulus, the fully grown oocyte, exits the meiotic arrest and progresses to the metaphase of the second meiotic division (MII), where it arrests again [3]. This series of events is defined as oocyte maturation and is proceeded by ovulation. An oocyte arrested at MII will resume meiosis upon fertilization. A fertilized oocyte develops into a zygote, which then continues through the first mitotic division to develop into a 2-cell embryo.
During oogenesis, mRNA is actively transcribed and accumulated in the growing oocyte. This transcription activity ceases in fully grown GV oocytes, coinciding with follicular antrum formation, and is accompanied by dynamic changes in chromatin organization. In the mouse, transcription of the embryonic genome is initiated only at the mid-to-late one-cell stage embryo [4], implying that oocyte maturation, fertilization, and early embryogenesis must rely on pre-synthesized maternal mRNAs [5]. The process, by which the maternal transcripts are degraded and replaced by zygotic transcripts, is termed the maternal-to-zygotic transition (MZT) [6]. The major reprogramming of gene expression occurs after fertilization upon the development of a 2-cell embryo; however, a massive degradation of maternal mRNAs begins as early as the resumption of the first meiotic division [7,8].
One of the primary mechanisms of gene repression in mammals is RNA silencing by small RNA molecules, such as microRNAs (miRNAs) and small interfering RNAs (siRNAs). A network of proteins process these small RNA molecules, thereby enabling their activity. Argonaut 2 (AGO2) is required for RNA-mediated gene silencing. Knockdown of AGO2 stabilizes maternal transcripts, which would normally be degraded by the late 2-cell stage, including those for fibroblast growth factor receptor 2 (Fgfr2) and leptin receptor 2 [9]. Phosphorylation of Y393 within AGO2 inactivates AGO2 by impairing the interaction between AGO2 and DICER, a central player in small RNA biogenesis [10,11], and preventing the association between AGO2 and small RNAs [12,13]. AGO2 has recently been identified as a downstream target for Fyn in oligodendrocytes; upon activation, Fyn co-localizes with AGO2 and phosphorylates its tyrosine residues [14].
Fyn, a member of the Src family of non-receptor tyrosine kinases (SFKs), is highly expressed in oocytes [15] and plays an essential role during the first and second meiotic divisions [16], as well as in early embryogenesis. We have recently reported post-transcription regulation of Fyn by miR-125a-3p in HEK 293 T cells [17], rodent granulosa cells [18] and mouse oocytes [19]. miR-125a-3p binds to Fyn 3ʹUTR in a specific manner, causing a decrease in the levels of Fyn mRNA and protein. In fact, miR-125a-3p and Fyn exhibit an inverse pattern of expression throughout oocyte maturation, where a decrease in the expression of miR-125a-3p, followed by an increase in the expression of Fyn protein, precede the resumption of the first meiotic division.
The ability of Fyn to phosphorylate and inhibit AGO2 activity combined with the pivotal role of AGO2 in MZT, led us hypothesize that Fyn and AGO2 participate in regulating the stability of maternal mRNAs prior to fertilization.
Methods
Animals
ICR female mice (Envigo RMS Limited, Jerusalem, Israel) were housed in temperature and humidity-controlled rooms under artificial illumination for 12 hours daily in the animal facilities of the Sackler Faculty of Medicine, Tel Aviv University. This study was approved by the Institutional Animal care and Use Committee (M-13-097) and performed in accordance with its guidelines.
Oocytes collection and treatment
Oocytes were recovered from large antral follicles of females’ ovaries (6–8 ovaries from 3 to 4 female mice) and denuded from the associated cumulus cells. Regular round oocytes, enclosed within the ZP, containing a central GV and clear cytoplasm were selected for microinjection. Microinjecting mice GV oocytes results in a high proportion of microinjected oocytes that are damaged. To be sure we will be left with at least 15 viable oocytes at each experimental group, even after an overnight maturation, we injected 60–80 oocytes for each treatment group (~300 at an experimental day). For analysis, we selected 15 oocytes from each treatment group, based on commonly employed morphological parameters: round oocytes with a clear homogenous cytoplasm presenting an intact GV (GV oocytes); or matured oocytes, with no GV and preferably with first polar body (MII oocyte). Oocytes with irregular shape, grainy cytoplasm and/or fragmentation, were excluded. We added this information to the revised manuscript
A. GV oocytes
A1. Cultured oocytes
GV oocytes were released from ovaries dissected from 4-week-old naive female mice (3–4 mice/experimental day) into pre-warmed (37°C) M2 medium (M-7167; Sigma-Aldrich, St. Louis, MO, USA) supplemented with penicillin (100 IU/ml), streptomycin (100 µg/ml; Biological Industries, Beit Ha’emek, Israel) with or without milrinone (1 µM; M-4659; Sigma-Aldrich). Milrinone prevents the spontaneous oocyte maturation, which takes place in vitro upon the release of the oocyte from the ovarian follicle, by inhibiting the oocyte-specific phosphodiesterase 3A, thus avoiding the drop in c’AMP (as performed by Grossman et al. [19]). The medium of the oocytes included in addition either 10 µM SU6656 or DMSO (diluent) and incubated for 2 hr. In the presence of milrinone the oocytes stayed meiotically arrested containing an intact GV. Fifteen of these oocytes were collected 2 hours after isolation. The remaining oocytes were kept in milrinone-free medium at 37°C in a humidified incubator, underwent in vitro maturation (IVM), reaching MII (IVM-MII), and collected in batches of fifteen oocytes on the following day. Equal numbers of GV and MII oocytes were subjected to qPCR analysis for detection of maternal mRNAs.
A2. Microinjected oocytes
GV oocytes, as in A1, were excised and transferred into L-15 tissue culture medium (L-1518; Sigma-Aldrich) supplemented with 5% fetal bovine serum (FBS. Life Technologies; Carlsbad, CA, USA), penicillin and streptomycin. Oocytes liberated from the ovaries were transferred into 10 µl droplets of the same culture medium with milrinone (1 μM; M-4659; Sigma-Aldrich), under paraffin oil. Oocytes with an intact GV were placed on the stage of an inverted DIC microscope (Axiovert 35; Zeiss, Oberkochen, Germany) and microinjected using an InjectMan NI2 micromanipulator and a FemtoJet autom (as performed by Grossman et al. [19],), with either inhibitor or GFP (diluent). These oocytes were further incubated for 6 hours in the presence of milrinone, to allow gene expression, while maintaining their meiotic arrest, thereafter were either collected or washed and incubated in milrinone-free medium and collected the next day. Batches of fifteen oocytes of GV or MII oocytes were subjected to qPCR analysis for detection of maternal mRNAs.
B. Ovulated MII oocytes
Oocytes were recovered from the oviductal ampullae 14 hours after administration of human chorionic gonadotrophin (hCG; 7 IU; Sigma-Aldrich) to pregnant mare serum gonadotropin (PMSG)-primed 4-weeks old female mice (5 IU; Syncro Part; Sanofi, Paris, France). Oocytes were collected into pre-warmed M2 medium supplemented with penicillin and streptomycin and then collected in batches of fifteen oocytes and subjected to qPCR analysis for detection of maternal mRNAs.
C. Embryos
Mice were primed with PMSG followed by hCG administration and allowed to mate over-night. Vaginal plugs were checked the following morning. One-cell zygotes were collected from the oviductal ampullae 12–16 hours after hCG administration. Two-, four-, and eight-cell stage embryos were flushed from the oviducts at 2.5 days post-coitus (dpc), 3.5 dpc, and 4.5 dpc, respectively. Three to four female mice were used for each developmental stage. Embryos of each developmental stage were collected in batches of fifteen oocytes and subjected to qPCR analysis for detection of maternal mRNAs. Zygotes, 2-cell, 4-cell and 8-cell embryos were collected on three separate experimental days; Other developmental stage were collected on five separate experimental days, each day from 3–4 mice.
Maternal mRNA characterization
We selected a set of maternal mRNAs involved in oocyte maturation, resumption of meiosis, and embryogenesis by choosing mRNAs that are completely destroyed by the 2-cell stage [20–22]. These mRNAs are also targeted by miRNAs, which are expressed when transcription activity re-occurs at the early zygote stage [23].
We cross referenced these candidates with genes that are modified in oocytes or embryos, retrieved from either DICER [23] or AGO2 [9] KO mice. Using several bioinformatics tools (TargetScan, miR.org and miRbase), we also screened for strong miRNA binding-sites within these maternal mRNAs and cross referenced them with embryonic miRNAs known to be transcribed by the embryo, already at the late zygote to 2-cell stage [23]. Because AGO2 does not exert its effect solely through miRNAs, but also through siRNAs, genes altered in DICER or AGO2 KO mice were not excluded, even if no binding-sites for embryonic miRNAs (e.g. Omt2b, C-mos; Table 1) were identified. Moreover, maternal mRNAs with no available data regarding their expression in DICER or AGO2 KO mice, but with a strong and conserved binding site to early transcribed embryonic miRNAs (e.g. H1Foo; Table 1), were also not excluded. Nobox and Bmp15 were selected in spite lack of data regarding their expression in DICER or AGO2 KO mice, and our inability to find binding sites for early transcribed embryonic miRNAs within their 3ʹUTR. These two genes are key players in oogenesis and embryogenesis and may potentially serve as controls for maternal mRNAs that are degraded by a mechanism other than the RNA interference machinery. As endogenous control genes, we selected CyclinA2, and Hprt1, which remain stable throughout maturation and MZT.
Table 1.
Selected maternal mRNAs involved in oocyte growth, resumption of meiosis, and embryogenesis
| Maternal mRNA |
Changes in AGO2 KO embryos |
Changes in DICER KO oocytes[23] |
Expression during MZT |
Early transcribed site directed miRNAs[23] |
| Fgfr2 | Stabilized[9] | upregulated | Downregulated [49,50] | miR-218-5b |
| Lepr | Stabilized[9] | n/a | downregulated[9] | miR-381 |
| Omt2b | Stabilized[9] | n/a | downregulated[9] | n/a |
| Plat | n/a | upregulated | downregulated[36,44] | miR-143 |
| C-mos | upregulated | downregulated[36] | n/a | |
| H1foo | upregulated | downregulated[36,50] | miR-328 | |
| H2afx | upregulated | downregulated[23] | miR-138-5p | |
| Nobox | n/a | downregulated[51] | n/a | |
| Bmp15 | n/a | downregulated[51] | n/a | |
| Gdf9 | upregulated | downregulated[36] | miR-322-3p | |
| Ldhb | stable[36] | n/a | ||
| CyclinA2 | upregulated | stable[35] | n/a | |
| Hprt1 | upregulated | stable[36] | n/a |
Inhibitors
Chemical
A selective SFKs inhibitor (SU6656; 10 µM, Calbiochem, San Diego, CA) was added to the mouse GV oocytes’ culture medium. SU6656 is known to inhibit Fyn, c-Src and c-Yes by diffusing into cells, attaching to the ATP binding pocket of SFKs and acting as a competitive inhibitor [16,24]. We have previously used SU6656 in a number of studies and showed that it inhibits mouse oocytes-activation [16,25]. A direct Argonaute-2 inhibitor (BCI-137; Catalog number 531,552, Calbiochem, Sigma-Aldrich; 10 µM) was microinjected into GV oocytes. BCI-137 is a small molecule that interferes with the interaction of Ago2-nucleic acid with the mRNA [26], by directly interacting with the miRNA binding domain of AGO2 [26].
Cloning
Fyn was inhibited by microinjecting DN-Fyn cRNA into the oocytes. The full-length sequences of human cDNAs encoding WT, dominant-negative (DN) or constitutively active (CA) forms of Fyn kinase (I.M.A.G.E. clone ID number 3,613,878, ResGen, Carlsbad, CA, USA), which were inserted into pCMV–Tag 4A (Stratagene, Cedar Creek, TX, USA) using the BamHI and EcoRI restriction enzymes (New England Biolabs, Ipswich, MA, USA), had already been constructed in our laboratory [27] and demonstrated specific inhibition [16,25]. GFP-encoding cDNA was inserted into pRN3-C3.
In vitro transcription
Plasmids were linearized using HpaI restriction enzyme (New England Biolabs; for pCMV–Tag 4A vector) or Sfi1 restriction enzyme (New England Biolabs; for pRN3-C3 vector), leaving a poly-A tail of about 250 nucleotides. Linear plasmids were loaded on a 1% agarose gel and purified using the PureLink® Quick Gel Extraction Kit) Invitrogen, Thermo-Fisher Scientific; Baltics, UAB). In vitro transcribed RNA transcripts were prepared using the mMESSAGE mMACHINE kit (AM-1340; Ambion, Austin, TX, USA) according to manufacturer’s instructions. All transcripts were extracted by phenol:chloroform, precipitated by isopropanol, eluted with nuclease-free water and diluted to a concentration of 250 ng/µl in PBS:di-ethylpyrocarbonate-treated water (Biological Industries) according to Grossman et al. [19].
RNA isolation, reverse transcription (RT) and quantitative PCR (qPCR)
To approximate the abundance of maternal mRNAs and housekeeping genes in oocytes, we determined the minimal number of oocytes required for a sufficient detection by qPCR analysis. Based on previous work conducted in our lab, where we used 30 GV oocytes to detect Fyn mRNA and miR-125a-3p [19,28]. We lysed batches of 10, 20, and 30 fully grown GV oocytes and subjected the sample to qPCR. The obtained Ct values represent the quantitative amount of maternal mRNA transcripts of each sample. We found that mRNA expression of 15 GV oocytes was in the linear part of the calibration curve (data not shown). Hence, we chose to work with 15 oocytes per experimental group. To adequately analyze our qPCR data, a constant threshold was applied. We selected the routinely used housekeeping genes CyclinA2, and Hprt1 as calibrators. Following the manufacturer's instructions, we imposed a ∆∆Ct analysis on the values of each gene and used the geometric mean of the two calibrators. In each experiment, the level of mRNA expression was normalized to that of the corresponding untreated GV oocytes.
Each batch of 15 GV or MII oocytes was analyzed as a single sample, subjected to qPCR analysis, using the “Cells-to-CT” kit to detect maternal mRNA genes.
A. Oocytes and embryos: Oocytes and embryos (15 in each experimental group) collected in 5 µl of nuclease-free water, were lysed using the lysis buffer supplied in the TaqMan MicroRNA Cells-to-CT Kit (4,391,848, Ambion) according to manufacturer’s instructions. Reverse transcription (RT) of miRNAs was performed using 5 µl of Cells-to-CT sample lysates and specific primers for miR-125a-3p (assay ID 2199; Applied Biosystems; Foster City, CA, USA) and U6 snRNA (Assay ID 001973; Applied Biosystems) according to Grossman et al. [19]. For qPCR, 2.7 µl of the RT product were used. Total RNA was reverse transcribed using 9 µl of the Cells-to-CT sample lysates and the High-Capacity cDNA Reverse Transcription Kit (4,368,814; Applied Biosystems). For the detection of mRNAs, 4 µl of the RT product was used together with specific primers (Gdf9 forward: 5′-CAGCAACCAGGTGACAGG-3′; reverse: 5′-ACAGTGGAGGAGGAAGAGG-3′, Nobox forward: 5′-TGGGTCCTGTTCAAACTCC-3′; reverse: 5′-GGTGAGGCGATACTAGTGC-3′, c-MOS forward: 5′-CTCGGTGTATAAAGCCACTTACC-3′; reverse: 5′-CAGTTCAGCCCAGAAACTCC-3′, Fgfr2 forward: 5′-AAGATGATGCCACAGAGAAGG-3′; reverse: 5′-TATGACGTAGAGAGGTCCATCC-3′, Plat forward: 5′-TGCCTGTCCGAAGTTGC-3′; reverse: 5′-ATCTATGTCACAGCGTTTCCC-3′, Ldhb forward: 5′-AGGAACTGAATCCAGAAATGGG-3′; reverse: 5′-GATGACTTCATAGGCACTGTCC-3′, H1foo forward: 5′-GCCAGCAAGGTCCAGAATAG-3′; reverse: 5′-CACTTTGGTCTCCTGTACTGTC-3′, Bmp15 forward: 5′-TACAAGGTCAGCTTCCACCA-3′; reverse: 5′-ATGGCATGGTTGGGTGAA-3′, H2afx forward: 5′-CATCCGCAACGACGAG-3′; reverse: 5′-CGCTGCTCTTCTTGG-3′, Omt2b forward: 5′-CGTTCTTATTACATCTGATGGTG-3′; reverse: 5′- CTCTGGCTCGATTTCGCT-3′, Fyn forward: 5′-ACCTCCATCCCGAACTAC-3′; reverse: 5′-CATAAAGCGCCACAAACA-3′, LepR: forward: 5′-TCTATGTGGTCCAGGTTCGCTG-3′; reverse: 5′- AGTGAGATTGGTCCGATTTCCC-3′, CyclinA2 forward: 5′-AGAAGACAAGCCAGTGAACG-3′; reverse: 5′-GGTTTACATTTAACCTCCATTTCCC-3′, Hprt1 forward: 5’-TCGTGATTAGCGATGATGAACC-3′; reverse: 5′-GCAAGTCTTTCAGTCCTGTCC-3′).
B. qPCR analysis: mRNAs level was examined in – (I) treated GV oocytes, compared to diluent-treated GV oocytes (DMSO or GFP), to examine the inhibitory effect at the GV stage; (II) diluent-treated in vitro matured MII oocytes, compared to in vivo matured MII oocytes, to examine the effect of the diluent on mRNA degradation during maturation; and (III) treated in vitro matured MII oocytes, compared to diluent-treated in vitro matured MII oocytes, to examine the inhibitory effect on mRNA degradation during maturation. Genes that presented different pattern of degradation following exposure to diluent or injection of GFP, which served as controls, were excluded.
Statistical analysis
Experiments were repeated at least three times, on separate days. In each experiment, the level of mRNA expression was normalized against that in the corresponding untreated GV oocytes (GV oocytes treated with diluent in Figures 2–4; Naïve GV oocytes in Figure 1).
Figure 2.
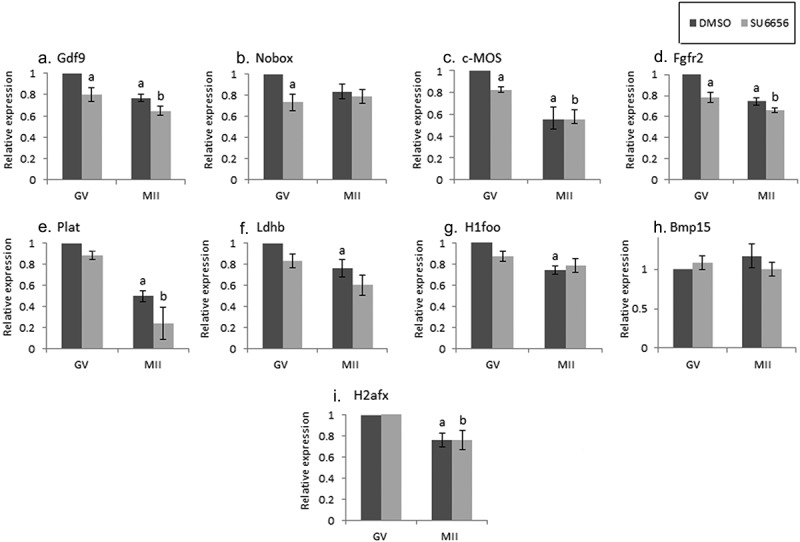
The effect of SU6656 on maternal mRNA degradation during oocyte maturation. Fully grown GV oocytes were isolated from ovaries of untreated mice and incubated in culture medium supplemented with 1 µM milrinone in the presence of either 10 µM SU6656, light gray, or DMSO, dark gray. GV oocytes (GV) were collected after 2 hours of incubation and the rest of the oocytes were allowed to mature in medium free of milrinone, SU6656, or DMSO. Mature MII oocytes (IVM-MII) were collected the following day. Batches of fifteen GV and IVM-MII oocytes from each experimental group were subjected to qPCR analysis for detection of maternal mRNAs, normalized to the geometric mean of Hprt1 and CyclinA2 endogenous controls. In each experimental group (SU6656, DMSO), results were normalized to the value of GV oocytes. Data were analyzed by one-way ANOVA followed by Tukey post hoc. Bars represent the mean ± SEM of five independent experiments. a,b,c p < 0.05; a significantly different from the value of DMSO-treated GV oocytes, b significantly different from the value of SU6656-treated GV oocytes, C significantly different from the value of MII oocytes derived from DMSO-treated GV oocytes.
Figure 3.
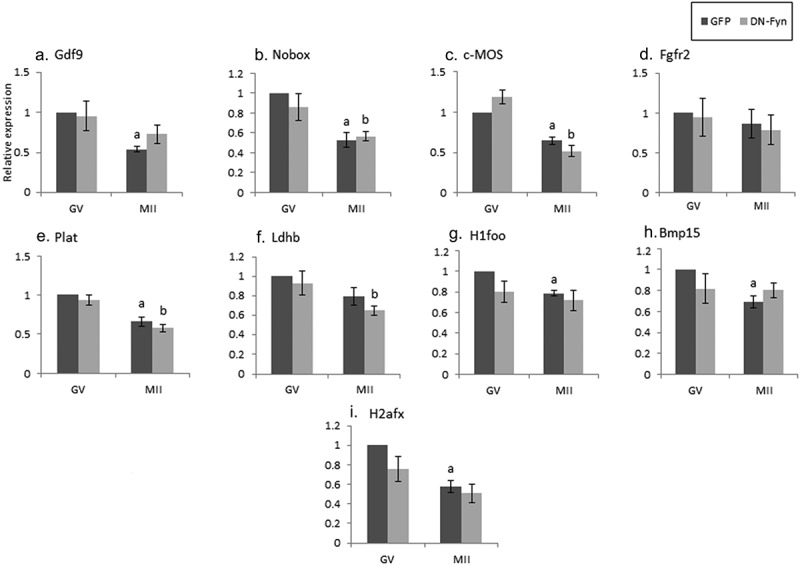
The effect of DN-Fyn on maternal mRNA degradation occurring during oocyte maturation. Fully grown GV oocytes were isolated from ovaries of untreated mice and microinjected with an in-vitro transcribed RNA construct encoding GFP (GFP), dark gray, or DN-Fyn, light gray, as described in “Methods”. Microinjected oocytes were incubated in medium supplemented with 1 µM milrinone. GV oocytes (GV) were collected after 6 hours of incubation and the rest of the oocytes were allowed to mature in medium devoid of milrinone. Mature MII oocytes (IVM-MII) were collected the following day. Batches of fifteen GV and IVM-MII oocytes from each experimental group were subjected to qPCR analysis for the detection of the maternal mRNAs and normalized to the geometric mean of the Hprt1 and CyclinA2 endogenous controls. In each experimental group (GFP, DN-Fyn), results were normalized to the value of GV oocytes. Data were analyzed by one-way ANOVA followed by Tukey post hoc. Bars represent the mean ± SEM of five independent experiments. a,b p < 0.05; a significantly different from GFP-microinjected GV oocytes, b significantly different from DN-Fyn-microinjected GV oocytes.
Figure 4.
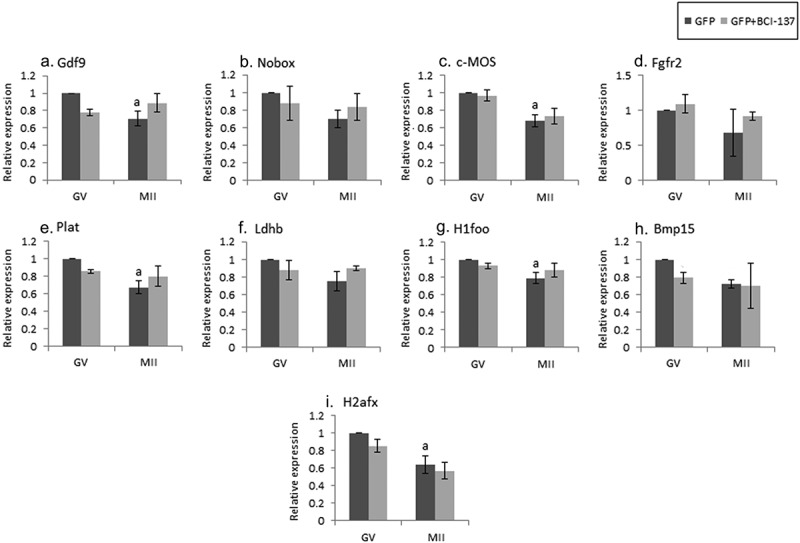
Inhibition of AGO2 results in maternal mRNA stabilization during oocyte maturation. Fully grown GV oocytes isolated from ovaries of untreated mice were co-microinjected with an in-vitro transcribed RNA encoding GFP and vehicle (GFP), dark gray, or with the same RNA transcript together with an AGO2 inhibitor (GFP + BCI-137), light gray, as described in “Methods”. Microinjected oocytes were incubated in culture medium supplemented with 1 µM milrinone. GV oocytes were collected after 6 hours of incubation and the rest of the oocytes were allowed to mature in medium devoid of milrinone. Mature MII oocytes (IVM-MII) were collected the following day. Batches of fifteen GV and IVM-MII oocytes from each experimental group were subjected to qPCR analysis for the detection of the maternal mRNAs, and the results were normalized to the geometric mean of Hprt1 and CyclinA2 as endogenous controls. Data in each experimental group (GFP, GFP+BCI-137) were normalized to the value of GV oocytes. Data were analyzed by one-way ANOVA followed by Tukey post hoc. Bars represent the mean ± SEM of three independent experiments. a p < 0.05; a significantly different from GV GFP value.
Figure 1.
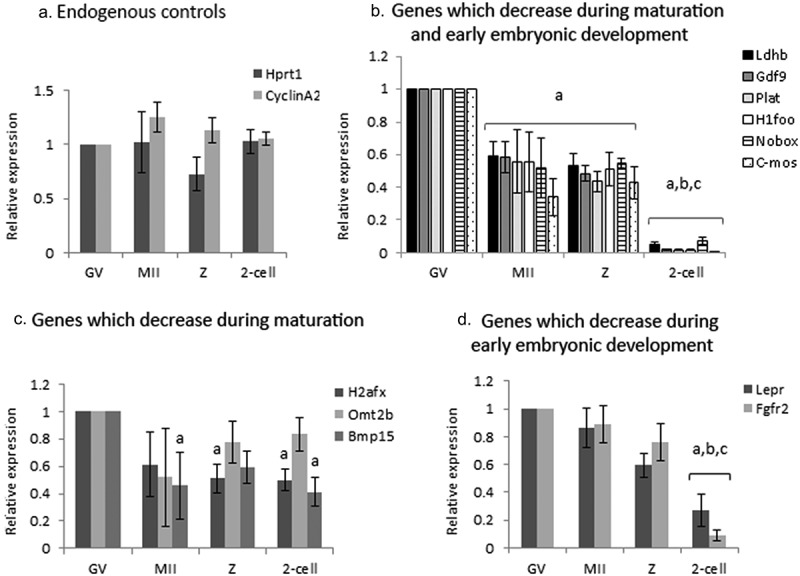
Expression of selected maternal mRNAs and endogenous controls during maturation and early embryonic development. GV oocytes (GV), in-vivo matured ovulated oocytes (MII) and embryos {zygotes (z), 2-cells (2-cell)} retrieved from mated, hormonally primed female mice, were isolated as described in “Methods”. Batches of fifteen oocytes and embryos from each experimental group) were lysed and subjected to qPCR analysis for the detection of endogenous controls (a) and maternal mRNAs (b-d), normalized to the geometric mean of endogenous controls. (A) Hprt1 (dark gray); CyclinA2 (light gray) (b) Ldhb (black); GDF1 (dark gray); Plat (light gray); H1foo (white); Nobox (stripes); C-mos (dots). (c) H2afx (dark gray); Omt2b (light gray); BMP15 (medium gray) (d) Lper (dark gray) Fgf2r (light gray). Data were normalized to the value of GV oocytes and analyzed by one-way ANOVA followed by Tukey post hoc. Bars represent the mean ± SEM of five independent experiments. a, b, c p < 0.05; a significantly different from the value of GV oocytes, bsignificantly different from the value of MII oocytes, c significantly different from the value of Z.
All statistical analyses were performed by SPSS (SPSS Inc.). Data normality was assessed using Shapiro–Wilk test. In all instances where raw data were not normally distributed, log transformations were applied to normalize data. Normally distributed data were then evaluated by one-way ANOVA followed by Tukey post hoc to evaluate significance. Data are presented as percent normalized to control. Data are expressed as the mean ± SEM. P < 0.05 was considered statistically significant.
Results
Maternal mRNA exhibits a distinct degradation pattern during oocyte maturation and early embryonic develo
pment
We screened the literature for mRNAs (“maternal mRNA”) that undergo degradation during oocyte maturation and early embryonic development, and could possibly reflect AGO2 activation (Table 1; see additional details in the Materials and Methods section). Some of the genes we examined: H1foo, Nobox, Bmp15, Gdf9, and C-mos, are transcripts specific to mammalian oocyte [29–32]. The degradation profile of the mRNAs was traced by qPCR analysis of batches of 15 isolated fully grown GV and MII oocytes, as well as late zygotes and 2-cell stage embryos. As the expression of Hprt1 and CyclinA2 remained stable during maturation and early embryonic development, these genes served as endogenous normalizing controls (Figure 1a). Our findings indicated that most of the studied genes showed two “waves” of maternal mRNAs degradation; the first occurring during oocyte maturation (~50%) and the second, a more dramatic decrease, during early embryonic development (Figure 1b). Other genes showed only one “wave” of degradation (Figure 1c,d). The mRNA level of Bmp15, H2afx, and Omt2b (Figure 1c), decreased by 40–60% during maturation (though only Bmp15 in a significant manner), but remained stable in the zygote and 2-cell stage embryos. Lepr and Fgfr2 mRNA levels (Figure 1d), were stable during maturation, but dropped dramatically in 2-cell embryos.
The role of SFKs in maternal-mRNAs degradation
In all the following experiments, we examined by qPCR analysis, maternal mRNAs level of batches of 15 oocytes, either cultured or injected with various inhibitory agents at the GV stage (see additional details in the Materials and Methods section). These oocytes were further incubated in the presence of milrinone, an inhibitor of oocyte maturation, to allow gene expression in the treated oocytes, while maintaining their meiotic arrest, and collected for analysis. The remaining oocytes were kept in milrinone-free medium overnight, underwent in vitro maturation (IVM), reaching MII (IVM-MII), and collected on the following day. We analyzed maternal mRNAs degradation in two developmental stages: degradation occurring already in GV oocytes, as well as degradation taking place in MII oocytes. Experiments for either SFKs inhibition or Fyn inhibition were repeated five times on five separate experimental days.
SFKs regulate degradation of maternal mRNA in oocytes
Some of the examined maternal mRNAs in GV oocytes exposed to the SFKs inhibitor SU6656 (2 h), underwent degradation (Figure 2, A-D; GDF9, Nobox, C-mos, Fgfr2; “GV,” light bars compared to dark bars), suggesting that SFKs take part in stabilizing the examined maternal mRNAs at the GV meiotic arrest. We allowed GV oocytes, incubated with or without the inhibitor, to mature in vitro and reached MII (IVM-MII), examined their mRNAs level and compared it to that occurred under physiological conditions (namely, isolated from the female tract; “in-vivo maturation; Figure 1). When we examined the mRNA level in MII oocytes that matured after exposure only to diluent (DMSO) prior to maturation, most mRNAs were degraded in a manner similar to in-vivo (Figure 2a, C-G, I dark bars). However, after SU6656 exposure (light bars), we detected two patterns of degradation: a) The expression of some genes (GDF9, C-mos and Plat; Figure 2a,C,E), was the same as the expression detected in control oocytes exposed to the diluent (resembling also their degradation in-vivo in Figure 1.) b) The expression of Ldhb, and H1foo was not significantly decreased, unlike the significant degradation in in-vivo matured oocytes (figure 2f,g); implying that SKFs play a role in mRNA degradation of these genes during oocyte maturation. As of the effect of the diluent on mRNA degradation, we could not conclude on SFKs involvement in Nobox, BMP15 or H2afx (light bars; Figure 2b,H,I).
Fyn regulates degradation of maternal mRNA in oocytes
Considering the fact that SU6656 is not Fyn-specific, thus inhibiting all members of SFKs, we microinjected GV oocytes with constructs encoding either DN-Fyn RNA or GFP (as control). Experiments were repeated five times on separate experimental days. As opposed to the degradation detected following SFKs inhibition, no significant degradation at the GV stage was detected after Fyn inhibition (Figure 3a-I, light bars compared to dark bars); thus, we cannot point at a role played by Fyn over that of Yes and Src in stabilizing maternal mRNAs at the GV stage. In MII oocytes that matured following Fyn inhibition (light bars), the degradation of GDF9, H1foo, BMP15 and H2afx was significantly different than that of the diluent (Figure 3a,G,H,I); implying that the degradation of these transcripts is regulated by Fyn. Nobox, C-mos and Plat (Figure 3b,C,E), were not affected by Fyn inhibition and degraded as in in-vivo maturation, and similar to the effect observed by inhibiting SFKs (Figure 2). As of the effect of the diluent on mRNA degradation, we could not conclude on Fyn involvement in Fgfr2 or Ldhb (light bars; Figure 3d,f).
AGO2 regulates maternal RNA degradation during maturation
Despite ample evidence indicating that AGO2 participates in maternal mRNAs degradation during the MZT [9], no reports were found on its possible role in the maternal mRNAs degradation in quiescent GV and maturing oocytes. We examined this possibility in GV oocytes microinjected with either an AGO2 inhibitor, which interferes with miRNA loading, or with its vehicle. Experiments were repeated three times on three separate experimental days. We showed that in most maternal mRNAs examined, no degradation was observed upon inhibition of AGO2 in oocytes at the GV stage (Figure 4; light bars compared to dark bars). However, in oocytes that were allowed to reach MII in vitro under the injection of AGO2 inhibitor, none of the maternal mRNAs examined presented the expected degradation compared to the genes of the in vivo matured MII oocytes (Figure 1).
We expected that the inhibition of AGO2 will prevent the degradation of certain mRNAs during oocyte maturation and observed this outcome in the following genes – GDF9, c-mos, Plat, H1foo, and H2afx, in which the expression levels during the transition from GV to MII oocytes did not decrease compared to the diluent. These findings indicate that AGO2 promotes their degradation during oocyte maturation, and when inhibited, their physiological degradation is prevented. However, as of the effect of the diluent on mRNA degradation, we could conclude that AGO2 is involved only in GDF9, c-mos, Plat, H1foo, and H2afx (light bars; Figure 4a,C,E,G,I).
Discussion
It is well established that fully grown GV mouse oocytes are transcriptionally quiescent. As transcription activity is reinitiated only at the zygote stage, oocytes must rely on pre-synthesized mRNA transcripts for the processes of maturation, fertilization, and early embryogenesis [33]. Thus, post-transcriptional regulation of existing transcripts is of great importance. Of the mechanisms of post-transcriptional regulation identified in mouse oocytes (reviewed by Kang and Han [34]), the one operated by maternal miRNAs appears to be necessary for both oocyte maturation and early embryogenesis, as embryos derived from Dicer null oocytes, in which miRNAs as well as siRNAs are depleted, show mis-regulated transcripts, lack of functional meiotic spindle and failure to develop beyond the first cell division [35]. Among the miRNA families, the miR-125 family has been linked to maternal mRNA clearance during MZT [36]. We have previously shown that miR-125a-3p binds directly to Fyn 3ʹUTR and regulates it post-transcriptionally in this HEK 293 T cell line [17], in follicles [37] as well as in oocytes [19]. We have also demonstrated the necessity of Fyn during GV breakdown (GVBD) and oocyte maturation [25,38] and that over-expression of miR-125a-3p in GV oocytes resulted in a decreased expression level of Fyn protein, and compromised GVBD [19].
Fyn is an upstream regulator of AGO2 that can mediate its action on gene silencing. We thus examined the expression of selected maternal mRNAs that are known to play a role in oocyte growth [29,39–41], maturation [40,41] and early embryogenesis [42–44]. Experiments conducted on HEK293T cells were used for preliminary examination of our hypothesis (data not shown), after which we followed and compared the levels of maternal mRNAs in treated and control GV and IVM-MII oocytes. Importantly, the degradation pattern in IVM-MII oocytes was similar to that seen in ovulated MII oocytes (in-vivo maturation), justifying the use of this in-vitro system.
We exposed GV oocytes to SU6656, an SFKs inhibitor, known to delay GVBD [25] and completion of the first meiotic division [16]. We excluded the genes that were affected by the diluent, or by the injection of GFP, which served as controls. The effect of inhibiting SFKs at the GV stage was prominent, as the examined maternal mRNAs were degraded already two hours after exposure to the inhibitor. To identify the specific role of Fyn in the process, we microinjected GV oocytes with a DN-Fyn RNA construct, which we have previously used and demonstrated its capability to inhibit endogenous Fyn activity [25]. All transcripts remained stable whereas only Fyn was inhibited, thus suggesting a role for SFKs in stabilizing maternal mRNA of these genes, but cannot state a specific role of Fyn itself probably as of Fyn compensation by other SFKs.
Gdf9, H1foo, BMP15, and H2afx, which were affected by Fyn inhibition, are expressed specifically in mammalian oocytes and thus may hold a special role in oocyte maturation; both BMP15 and GDF9 have an important role in the ovary, as well as in modulating oocyte quality [45]; H1foo has beneficial effects on generation of iPSC [46]. Taken together, these findings imply the participation of SFKs in stabilizing the maternal mRNAs during GV arrest, and point to a role in their degradation, during oocyte maturation. The inhibition of Fyn by the DN-Fyn RNA construct had a less profound effect than that of SU6656 at the GV stage; this was not surprising due to the ability of SFKs to compensate for one another [47]. Moreover, SU6656 acts by attaching to the ATP binding pocket of SFKs and acting as a competitive inhibitor, whereas the injected DN-Fyn RNA construct produces overexperssion of a nonactive Fyn, masking the activity of endogenous Fyn.
During IVM, the degradation of Gdf9, BMP15, and H2afx was prevented under Fyn inhibition, whereas under inhibition of SFKs they degraded, though Ldhb was stabilized; insinuating that Fyn may play a role in IVM and that at least one of the SFKs, other than Fyn, participates in the degradation of these genes during IVM. The different findings obtained by inhibiting either Fyn or SFKs could also result from the use of two different systems; SU6656, which inhibits the activity of the SFKs proteins, and DN-Fyn RNA that acts at the translational level.
It was reported that in oligodendrocytes, Fyn inactivates AGO2 by phosphorylation [14]. To elucidate the role of AGO2 during oocyte quiescence and maturation, we used an AGO2 inhibitor, interfering with its ability to load miRNAs and degrade them [26]. Inhibition of AGO2 did not compromise the stability of any of the maternal mRNAs during the GV stage, insinuating it has no effect on the transcripts during the quiescent period. However during oocytes maturation all the examined maternal mRNAs were not degraded as observed under physiological conditions. These findings support our hypothesis that AGO2 activity is involved in degradation of maternal mRNAs during oocyte maturation; this pattern of stabilization resembles the one seen in AGO2 RNAi and DICER knockout embryos [9,23]. It has been established that AGO2 is required for gene expression during early embryonic development from the two-cell embryos [9]. It has also been shown to be a key regulator of maternal mRNA degradation in mouse early embryos [48]. Here, we demonstrated its pivotal role in the degradation of maternal mRNAs during oocyte maturation as well.
Conclusions
In the current study, we found that SFKs and Fyn have an important role in maintaining the stability of the pre-transcribed mRNAs in quiescent oocytes during the GV stage, whereas AGO2 plays at this stage, only a very partial role, if at all. Conversely, during oocyte maturation, AGO2 plays a major role in mRNAs degradation, whereas SFKs/Fyn are less prominent. By monitoring the expression of maternal mRNAs in GV oocytes and in mature oocytes we can point at the importance of SFKs/Fyn in stabilizing maternal mRNAs in quiescent oocytes as well as in promoting maturation. We can also suggest that the role of AGO2 in degrading pre-transcribed maternal mRNAs is pivotal. However, the mechanism and regulation pattern of these processes needs to be further investigated.
Acknowledgments
The authors are grateful to Ruth Kaplan-Kraicer from Sackler Faculty of Medicine, Tel Aviv University, for her help in preparing the manuscript.
Funding Statement
This work was supported by the P.E.F. Israel Endowment Funds [No. 470/14].
Disclosure statement
No potential conflict of interest was reported by the authors.
Author contribution
N.G. performed this work in partial fulfillment of the requirements for a M.Sc. degree and wrote the manuscript. Along with H.G., she developed the concept, designed and carried out the experiments, organized the data and performed the statistical analyses. H.B-J participated in part of the experiments; I.M. and L.N. assisted in analyzing the data; N.N. performed the microinjections with the assistance of R.H. and D.G.; N.D. participated in discussing the results and assisted in manuscript preparation; R.S. conceived the study, participated in its design, coordination, manuscript preparation and supervised the study. All authors read the final manuscript and approved it.
References
- [1].Cortvrindt R, Smitz J.. In vitro follicle growth: achievements in mammalian species. Reprod Domest Anim. 2001;36(1):3–9. [DOI] [PubMed] [Google Scholar]
- [2].Dekel N. Protein phosphorylation/dephosphorylation in the meiotic cell cycle of mammalian oocytes. Rev Reprod. 1996;1(2):82–88. [DOI] [PubMed] [Google Scholar]
- [3].Gosden R, and Lee B. Portrait of an oocyte: our obscure origin. J Clin Invest. 2010;120(4):973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bouniol C, Nguyen E, and Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res. 1995;218(1):57–62. [DOI] [PubMed] [Google Scholar]
- [5].Schultz RM, Stein P, Svoboda P. The oocyte-to-embryo transition in mouse: past, present, and future. Biol Reprod. 2018;99(1):160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136(18):3033–3042. [DOI] [PubMed] [Google Scholar]
- [7].Schultz RM. Regulation of zygotic gene activation in the mouse. Bioessays. 1993;15(8):531–538. [DOI] [PubMed] [Google Scholar]
- [8].Conti M. Acquisition of oocyte competence to develop as an embryo : integrated nuclear and cytoplasmic events. Hum Reprod Update. 2018;24(3):245–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lykke-Andersen K, Gilchrist MJ, Grabarek JB, et al. Maternal argonaute 2 is essential for early mouse development at the maternal-zygotic transition. Mol Biol Cell. 2008;19(10):4383–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol. 2004;11(3):214–218. [DOI] [PubMed] [Google Scholar]
- [11].Bernstein E, Caudy AA, Hammond SM, et al. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. [DOI] [PubMed] [Google Scholar]
- [12].Yang M, Haase AD, Huang F-K, et al. Dephosphorylation of tyrosine 393 in argonaute 2 by protein tyrosine phosphatase 1B regulates gene silencing in oncogenic RAS-Induced senescence. Mol Cell. 2014;55(5):782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shen J, Xia W, Khotskaya YB, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nat. 2013;497(7449):383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Müller C, Schäfer I, Luhmann HJ, et al. Oligodendroglial argonaute protein AGO2 associates with molecules of the Mbp mRNA localization machinery and is a downstream target of Fyn kinase. Front Cell Neurosci. 2015;9:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lou J, McGinnis LK, Kinsey WH. FYn kinase activity is required for normal organization and functional polarity of the mouse oocyte cortex. Mol Reprod Dev. 2009;76(9):819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Levi M, Maro B, and Shalgi R. Fyn kinase is involved in cleavage furrow ingression during meiosis and mitosis. Reproduction. 2010;140(6):827–834. [DOI] [PubMed] [Google Scholar]
- [17].Ninio-Many L, Grossman H, and Shomron N, et al. microRNA-125a-3p reduces cell proliferation and migration by targeting Fyn. J Cell Sci. 2013. 126(Pt 13) 2867–2876. [DOI] [PubMed] [Google Scholar]
- [18].Grossman H, Chuderland D, and Ninio-Many L, et al. A novel regulatory pathway in granulosa cells, the LH/human chorionic gonadotropin-microRNA-125a-3p-Fyn pathway, is required for ovulation. FASEB J. 2015;29(8):3206–3216. [DOI] [PubMed] [Google Scholar]
- [19].Grossman H, Har-Paz E, and Gindi N, et al. Regulation of GVBD in mouse oocytes by miR-125a-3p and Fyn kinase through modulation of actin filaments. Sci Rep. 2017;7(1) 2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Alizadeh Z, Kageyama SI, Aoki F. Degradation of maternal mRNA in mouse embryos: selective degradation of specific mRNAs after fertilization. Mol Reprod Dev. 2005;72(3):281–290. [DOI] [PubMed] [Google Scholar]
- [21].Svoboda P, Franke V, Schultz RM. Chapter Nine - Sculpting the transcriptome during the oocyte-to-embryo transition in mouse. Curr Top Dev Biol. 2015;113:305–349. [DOI] [PubMed] [Google Scholar]
- [22].Su YQ, Sugiura K, Woo Y, et al. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302(1):104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tang F, Kaneda M, O’Carroll D, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21(6):644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Blake RA, Broome MA, Liu X, et al. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol Cell Biol. 2000;20(23):9018–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Levi M, Maro B, Shalgi R. The involvement of Fyn kinase in resumption of the first meiotic division in mouse oocytes. Cell Cycle. 2010;9(8):1577–1589. [DOI] [PubMed] [Google Scholar]
- [26].Masciarelli S, Quaranta R, Iosue I, et al. A small-molecule targeting the microRNA binding domain of argonaute 2 improves the retinoic acid differentiation response of the acute promyelocytic leukemia cell line NB4. ACS Chem Biol. 2014;9(8):1674–1679. [DOI] [PubMed] [Google Scholar]
- [27].Reut TM, Mattan L, Dafna T, et al. The role of Src family kinases in egg activation. Dev Biol. 2007;312(1):77–89. [DOI] [PubMed] [Google Scholar]
- [28].Grossman H, Har-paz E, and Levi M, et al. Regulation of GVBD in mouse oocytes by miR-125a-3p and Fyn kinase through modulation of actin filaments. Sci Rep. 2017;7(1):2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Suzumori N, Yan C, Matzuk MM, et al. Nobox is a homeobox-encoding gene preferentially expressed in primordial and growing oocytes. Mech Dev. 2002;111(1–2):137–141. [DOI] [PubMed] [Google Scholar]
- [30].West MF, Verrotti AC, and Salles FJ, et al. Isolation and characterization of two novel, cytoplasmically polyadenylated, oocyte-specific, mouse maternal RNAs. Dev Biol . 1996;175(1):132–141. [DOI] [PubMed] [Google Scholar]
- [31].Tanaka M, Hennebold JD, Macfarlane J, et al. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development. 2001;128:655–664. [DOI] [PubMed] [Google Scholar]
- [32].Dube JL, Wang P, Elvin J, et al. The bone morphogenetic protein 15 gene is X-linked and expressed in oocytes. Mol Endocrinol. 1998;12(12):1809–1817. [DOI] [PubMed] [Google Scholar]
- [33].Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol. 2005;283(1):40–57. [DOI] [PubMed] [Google Scholar]
- [34].Kang M-K, Han S-J. Post-transcriptional and post-translational regulation during mouse oocyte maturation. BMB Rep. 2011;44(3):147–157. [DOI] [PubMed] [Google Scholar]
- [35].Suh N, Baehner L, Moltzahn F, et al. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol. 2010;20(3):271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim K, Seo Y, Kim E, et al. The miR-125 family is an important regulator of the expression and maintenance of maternal effect genes during preimplantational embryo development. Open Biol. 2016;6(11):160181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Grossman H, Har-Paz E, Gindi N, et al. Pre-ovulatory intercellular regulation of miR-125a-3p within mouse ovarian follicles. Reproduction. 2020;159(2):215–225. [DOI] [PubMed] [Google Scholar]
- [38].Levi M, Kaplan-Kraicer R, Shalgi R. Regulation of division in mammalian oocytes: implications for polar body formation. Mol Hum Reprod. 2011;17(5):328–334. [DOI] [PubMed] [Google Scholar]
- [39].Tanaka M, Kihara M, and Hennebold JD, et al. H1FOO is coupled to the initiation of oocytic growth. Biol Reprod. 2005;72(1):135–142. [DOI] [PubMed] [Google Scholar]
- [40].Rajkovic A. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305(5687):1157–1159. [DOI] [PubMed] [Google Scholar]
- [41].Paulini F, Melo EO. The role of oocyte-secreted factors GDF9 and BMP15 in follicular development and oogenesis. Reprod Domest Anim. 2011;46(2):354–361. [DOI] [PubMed] [Google Scholar]
- [42].Mutter GL, Grills GS, Wolgemuth DJ. Evidence for the involvement of the proto-oncogene c-mos in mammalian meiotic maturation and possibly very early embryogenesis. EMBO J. 1988;7(3):683–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fuchimoto D-I. Posttranscriptional regulation of cyclin A1 and cyclin A2 during mouse oocyte meiotic maturation and preimplantation development. Biol Reprod. 2001;65(4):986–993. [DOI] [PubMed] [Google Scholar]
- [44].Haffner-Krausz R, Gorivodsky M, Chen Y, et al. Expression of Fgfr2 in the early mouse embryo indicates its involvement in preimplantation development. Mech Dev. 1999;85(1–2):167–172. [DOI] [PubMed] [Google Scholar]
- [45].Belli M, Shimasaki S. Molecular aspects and clinical relevance of GDF9 and BMP15 in ovarian function. Vitam Horm. 2018;107:317–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kunitomi A, Yuasa S, Sugiyama F, et al. H1foo has a pivotal role in qualifying induced pluripotent stem cells. Stem Cell Reports. 2016;6(6):825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stein PL, Soriano P. Combined deficiencies of Src, Fyn, and Yes tyrosine kinases in mutant mice. Genes Dev. 1994;8(17):1999–2007. [DOI] [PubMed] [Google Scholar]
- [48].Zhang JM, Hou WB, Du JW, et al. Argonaute 2 is a key regulator of maternal mRNA degradation in mouse early embryos. Cell Death Discov. 2020;6:133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bushati N, Stark A, and Brennecke J, et al. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18(7):501–506. [DOI] [PubMed] [Google Scholar]
- [50].Bachvarova R, De Leon V, Johnson A, et al. Changes in total RNA, polyadenylated RNA, and actin mRNA during meiotic maturation of mouse oocytes. Dev Biol. 1985;108(2):325–331. [DOI] [PubMed] [Google Scholar]
- [51].Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8(4):323–331. [DOI] [PubMed] [Google Scholar]


