ABSTRACT
Epigenetic regulation is one of the most precise and subtle ways of gene regulation, including DNA modification, histone modification, RNA modification, histone variants, chromatin remodeling, and long non-coding RNAs (lncRNAs). Chromatin modification is the most basic type of epigenetic regulation, which plays a key role in a myriad of developmental and physiological processes that have been thoroughly studied. These modifications are usually completed by a series of conserved chromatin modification complexes in eukaryotes. In recent years, a series of lncRNAs in organisms also have been described as having irreplaceable functions in biological environment adaptation, especially in biotic and abiotic stresses. Moreover, these molecules form a sophisticated regulatory network through mutual cross-regulation to achieve quantitative expression of key environmental response genes to external signals. For instance, the function of lncRNAs will directly or indirectly depend on the function of the chromatin modification complex. In this review, we mainly focus on chromatin modification, lncRNA, and their coordination mechanism to achieve the high adaptability of plants in low-temperature environments. We highlight recent findings and insights into lncRNA-mediated local chromatin environment changes during plant growth under low temperature via chromatin modification complexes, including target gene specificity for different lncRNA.
KEYWORDS: Lncrna, Histone modification, Low temperature, Local chromatin landscape
Introduction
It is harder for terrestrial plants to escape from unexpected damage than animals. Therefore, the growth of plants is susceptible to seasonal changes, among which temperature and light are the most important limiting factors.1 Generally speaking, the annual light and temperature change constantly, but in recent years, due to climate changes, there are frequent temperature fluctuations, which is a huge ordeal for the growth and development of plants. Principally in cold regions, plants are prone to cold stress, and their fitness is affected. Over the past two decades, the sophisticated mechanisms of plant resistance to cold temperature have been explored, including osmotic potential,2,3 proteins structural stability,4 ice crystal formation,5 cell membranes stability,6–9 and reactive oxygen species (ROS) scavenging.10 All those events are dependent on sophisticated gene regulation.
“Epigenetics” was proposed by Conrad Waddington in the 1940s (REF) as an energy-effective way to adjust gene expression, especially for terrestrial plants.11 Temperature is an important physical factor affecting the distribution of plants and their chromatin conformations by inducing chemical modification of biological macromolecules (DNA, proteins), thereby triggering biochemical and molecular reactions in each cell to respond to temperature changes. Post-translational histone modifications, such as histone acetylation, methylation, phosphorylation, ubiquitination, SUMOylation, and myristoylation are associated with gene expression levels in plants withstanding cold environments.12 Besides chemical modifications of histones, low temperature can induce lncRNAs in plants. LncRNAS are transcribed by RNA polymerase II and are not translated to proteins. Increasing evidence prove that lncRNAs also have a critical role in stress responses.13–19 Nowadays, researchers have found many lncRNAs function in conjunction with the histone modification complex.20–23
In this review, we highlight recent findings in plant chromatin modifications by lncRNAs at low temperatures, discuss the specificity of lncRNA mediated gene chromatin modification, and address questions that remain to be answered.
Histone modification provides the possibility of precise regulation to cold response in plants
Chromosome assembly is a multi-level highly ordered process. The most basic unit of the structure and function of eukaryotic chromosomes is the nucleosome. The nucleosome is a heterooctamer composed of H2A, H2B, H3, and H4, with 147 bp DNA wrapped around.24,25 The N-terminal and core regions of histones are susceptible to modifications, such as methylation, acetylation, phosphorylation, and ubiquitination.26–28 Histone modifications are involved in all plant development stage, such as, seed germination, hypocotyl elongation, and flowering time.29–32 Moreover, these histone modifications also play a crucial role in cold stress response.33–35 During Arabidopsis cold acclimation, after transferring to 4°C, histone modifications are associated with induction the transcription of CBF genes, which in turn activate the COR gene.36 In maize, histone deacetylases were significantly up-regulated during the cold acclimation, genome-wide H3 and H4 deacetylation, and local histone acetylation level of ZmDREB1 and ZmCOR413 were observed in cold-response.37 Also, a similar phenomenon was found in rice.38 Besides histone acetylation, histone methylation is also a tool for adjusting plant fitness through gene expression primarily when plants undergo cold stress. At low temperatures, H3K27me3 at COR15A and AtGolS3 were decreased.39
Since histone modifications are altered in cold stress, studies have identified the chromatin modification enzymes involved in the cold stress response. In Arabidopsis, long-term low temperature induces HDA6 transcription, changing histone acetylation levels.40 HOS1 (high expression of osmotically responsive gene 1), a RING finger E3 ligase, negatively regulates cold-responsive genes.41 Moreover, HOS1 helps HDA6 dissociate from the FLC locus to change chromatin status.42,43 HOS15, a WD40-repeat protein, can be induced by abiotic stresses such as freezing, deacetylating histone H4 and enhancing plants’ cold tolerance.44
Vernalization is a relatively precise adaptation mechanism produced by plants as a long-term evolutionary process responding to environmental seasonal temperature changes.45,46 This process is also a representative cold tolerance mechanism, that plants synchronizing flowering with spring and pollinators undergoing long-term cold temperatures, also helps plants complete their reproductive cycle as quickly as possible by shortening the growth cycle and sacrificing an amount of plant biomass to some extent. During vernalization, long-term cold conditions reduced the FLOWERING LOCUS C (FLC) gene expression, thereby promoting the early flowering of the plant. FLC encodes a MADS Box domain-containing protein, a key suppressor gene in the flowering regulatory network.47 FLC transcription level is mainly promoted by FRIGIDIA (FRI). FRI forms an FRI complex with FRL1, FES1, FLX and SUF4 to promote the FLC expression, thereby inhibiting flowering.48 Compared with Col-0 plants without FRI function, plants with functional FRI showed a clear late-flowering phenotype. In the initial stages of FLC transcription, the FRI complex forms a transcription activation super complex with SWR1, COMPASS, PAF1 (RNA polymerase II-associated complex) complexes, and EFS, and maintains the transcription activation markers H3K4me3 and H3K36me3 and the level and stability of the gene-loop at FLC site.49 After the vernalization, the recruitment of the PHD-PRC2 complex completes the histone H3 methylation at the 27th lysine (H3K27) at FLC locus, thereby inhibiting the FLC expression.50 When the cold signal ends, the stable inhibition of FLC in plants requires recognition of the cold memory element (CME) on the first intron of the FLC gene through VAL1 and VAL2 proteins and then recruits the PRC1 and PRC2 complex, thus the level of modification and expression of H3K27me3 at this site is stable.51
Dynamic histone modification as well as gene activation and genetic memory have become hot topics in plant responses to abiotic stress. However, the complete network of relationships between abiotic stress responses and epigenetic information, such as, stress-responsive histone-modifying enzymes, target stress-responsive genes, and specific histone-modification sites remains unclear.
LncRNA helps plants overcome the annual cold conditions
Besides chemical modifications on histones, lncRNAs are also a pivotal gene regulation level induce by low temperature in plants. Although lncRNAs related to mRNA and transcribed by RNA polymerase II, lncRNAs do not code for proteins.52,53 lncRNAs are more than 200 nt in length, and classified into natural antisense transcripts (NATs), overlapping lncRNAs (OT-lncRNAs), long intergenic non-coding RNAs (lincRNAs), and intronic non-coding RNAs (incRNAs).54,55 With the development of high-throughput sequencing technology, an increasing number of lncRNAs have been identified. Recently, lncRNAs have emerged as key epigenetic regulators of diverse cellular processes in mammals and plants. In the nucleus, lncRNAs can serve as transcription regulators or enhancers RNA, change chromosome construction, help spliceosome formation, and recruit the chromatin-modifying complex to target genes.56 While in the cytoplasm, lncRNA usually promotes mRNA stability, mRNA translation, and small peptides production.56 lncRNAs have a critical role in cold responses, and more importantly, function together with histone modifiers.57
In Medicago truncatula, 24,368 unique lncRNAs were identified, among which 983 and 1288 were responsive to cold treatment in the leaves and roots.58 The observations that transcript levels of the lncRNA MtCIR1 increased within 2 h of exposure to low temperature, followed by MtCBFs accumulation at 5 h may suggest a regulatory network between MtCBFs and MtCIR1.59 In wheat, the lncRNAs LncR9A, lncR117, and lncR616 indirectly regulate the CSD1 expression by competitively binding miR398, affecting the resistance of Dn1 (Dongnongdongmai 1) against cold.60
In Arabidopsis cold response, the SVALKA-asCBF1 cascade provides a CBF1 expression control mechanism that could be exploited to maximize freezing tolerance with mitigated fitness costs.61 Recently, the lncRNA AUXIN REGULATED PROMOTER LOOP (APOLO) was reported to directly identify multiple independent loci in the Arabidopsis genome and regulate its three-dimensional chromatin conformation, leading to transcriptional shift. After cold treatment, the APOLO gene transcriptional activity is higher in roots. The novel APOLO and WRKY42 ribonucleoprotein complex form a regulatory center. APOLO lncRNA directly regulates the RHD6 transcriptional activity by fine-tuning the epigenetic environment, such as local chromatin 3D conformation. The RHD6 activation further triggers the expression of RSL2 and RSL4, and it integrates signals that control the growth and development of root hairs.62 Analysis of the molecular mechanism of vernalization, three types of lncRNAs, including COOLAIR, COLDWRAP, and COLDAIR, were detected in FLC, a key gene of vernalization.63–66 In 2011, Jae Bok Heo and Sibum Sung reported the long intronic non-coding RNA COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA), required the vernalization-mediated epigenetic repression of FLC. COLDAIR is in the sense direction relative to FLC mRNA transcription, and it physically associates with subunit of PRC2 and targets PRC2 to FLC.67 Also, Dong-Hwan Kim identified another lncRNA can be bound by PRC2, COLDWRAP. COLDWRAP is derived from the repressed promoter of FLC and is necessary for establishing the stable repressed state of FLC by vernalization.65 Both COLDAIR and COLDWRAP are required to form a repressive intragenic chromatin loop at the FLC locus by vernalization.65 COOLAIR is another FLC antisense transcript with alternative polyadenylation and multiple splice variants linked to different FLC expression states. FCA directly binds to long and short COOLAIR transcripts, as well as it interacts with the PRC2 subunit CLF. Defects in COOLAIR and FCA result in reducing H3K27me3 and decreasing CLF enrichment at FLC.68 Fang et.al captured interactions between FLL2, FCA, the polymerase and nuclease modules of the RNA 3′-end processing machinery, this work provided an evidence that phase separation is involved in lncRNAs COOLAIR target chromatin modifications.69 Besides PRC2 recruitment, there are some examples of transcription activation complex recruitment. MAS, positively regulates the transcription of its cognate sense gene MAF4 through interacting with WDR5a, a core component of the COMPASS-like complexes, to MAF4.23 All above examples tell us lncRNAs play irrefutable role in helping plants overcome the cold conditions.
Perspectives and concluding remarks
LncRNAs have been proven to have a critical role in cold responses. This review examined the functions of histone modifications, lncRNAs, and lncRNA-assisted chromatin modification complex to target gene directly or indirectly. LncRNAs are key to local chromatin landscapes, especially under stress conditions. This has led to the hypothesis that the crop species may have more lncRNAs due to the selective pressures for stress tolerance. How are the enzymatic complexes guided to target these marks at a specific combination of sites under different cellular contexts? Many cis-elements recruit histone modification complexes to the modified target gene containing those elements. Short genomic fragments, such as GA repeats and telobox, known as Polycomb response elements, direct the Polycomb repressive complex 2 (PRC2) placement at developmental genes regulated by silencing in Arabidopsis thaliana.70,71 For example, PRC2 recruitment in Arabidopsis relies mainly on trans-acting binding factors to cis-localized DNA sequence motifs. Some works have identified transcription factor families that bind to these PREs, colocalize with PRC2 on chromatin, physically interact with and recruit PRC2, and are required for PRC2-mediated gene silencing in vivo.
Besides histone modifiers via specific cis-elements (as shown in Figure 1a), lncRNAs represent a key layer of epigenetic control, as non-coding RNA molecules may guide chromatin modification complexes to specific sites. LncRNAs can be primarily bound by RNA binding proteins containing one or more RNA recognition motifs, which may lead to the formation of “lncRNAs- RNA binding proteins-others chromatin modification factors” super-complex (as shown in Figure 1b). Studies in animals have shown similar results in the contribution of lncRNAs to changes in chromatin modification status in vivo. One is the function of XIST in X chromosome silencing. Xist adheres to a strip of X chromatin and spreads along with it, recruiting chromatin modification complexes to catalyze H3K27me3 modification to specifically silence a strip of X chromatin;72 Another example is HOTAIR. The 5ʹend of lncRNA directly binds to the catalytic subunit EZH2 of PRC2, thereby recruiting the PRC2 complex to the target gene site, silencing the target gene expression. However, when HOTAIR RNA was overexpressed in breast cancer cells, regardless of the presence of PRC2, few transcriptomic changes were detected, and the interaction of PRC2 with RNA in vitro and cultured cells lack specificity.73 Nowadays, phase separation is found to be another way for lncRNAs to maintain the local chromatin landscape (as shown in Figure 1c). In phase separation system, lots of lncRNAs and histone modifiers may be pulled together, thus this system promotes local epigenetic environment establish.
Figure 1.
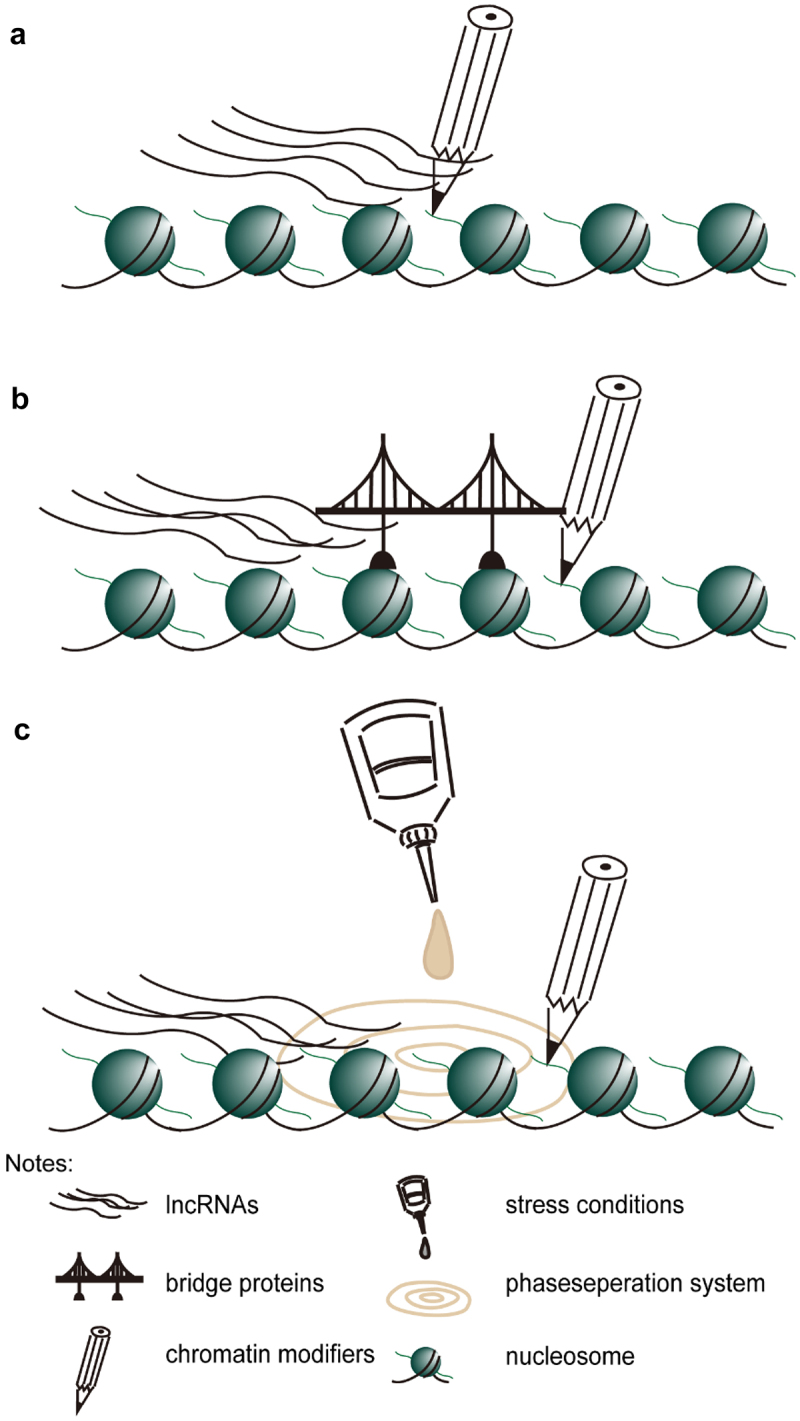
Possible work model for lncRNAs recruit chromatin modifiers. A. histone modifiers bind lncRNA directly. B. lncRNAs is found by a bridge protein (such as RNA-binding protein), which interacts with chromatin modifiers, then they form a super-complex maintain local chromatin landscape. C. Stress condition (as cold) prompt lncRNAs and chromatin modifiers form phase separation system, then they can easily maintain local chromatin landscape.
Although the current work in lncRNA has yielded some results, there are still many problems. For example, the biological functions of some lncRNA are not clarified, and it remains difficult to determine whether non-coding transcripts are functional. The mechanism of IncRNA is complex and diverse, and the results cannot be referred to each other. With the gradual maturity of research technology, the function of lncRNA in plants will be more thorough, which is of great significance to explore its role in plant life.
Acknowledgments
Also thank Core Facility of School of Life Science (Lanzhou University) for technical support.
Funding Statement
This work was supported by Key Laboratory of Superior Forage Germplasm in the Qinghai-Tibetan Plateau, Qinghai (2020-ZJ-Y03); The National Natural Science Foundation of China (No. 31872682, No. 32101244); Earmarked Fund for China Agriculture Research System (CARS-07-G14). Also thank Core Facility of School of Life Science (Lanzhou University) for technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Fournier-Level A, Perry, EO, Wang, JA, Braun, PT, Migneault, A, Cooper, MD, Metcalf, CJE, Schmitt, J.. Predicting the evolutionary dynamics of seasonal adaptation to novel climates in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 113, E2812–6, doi: 10.1073/pnas.1517456113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keunen E, Peshev D, Vangronsveld J, Van Den Ende W, Cuypers A. Plant sugars are crucial players in the oxidative challenge during abiotic stress: extending the traditional concept. Plant Cell Environ. 2013;36:1242–1255. doi: 10.1111/pce.12061. [DOI] [PubMed] [Google Scholar]
- 3.Zhuo C, Liang L, Zhao Y, Guo Z, Lu S. A cold responsive ethylene responsive factor from Medicago falcata confers cold tolerance by up-regulation of polyamine turnover, antioxidant protection, and proline accumulation. Plant Cell Environ. 2018;41:2021–2032. doi: 10.1111/pce.13114. [DOI] [PubMed] [Google Scholar]
- 4.Szabados L, Savoure A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Ruelland E, Vaultier MN, Zachowski A, Hurry V. Cold Signalling and Cold Acclimation in Plants. Adv Bot Res. 2009;49:35–150. doi: 10.1016/S0065-2296(08)00602-2. [DOI] [Google Scholar]
- 6.Alonso A, Queiroz CS, Magalhaes AC. Chilling stress leads to increased cell membrane rigidity in roots of coffee (Coffea arabica L) seedlings. Bba-Biomembranes. 1997;1323:75–84. doi: 10.1016/S0005-2736(96)00177-0. [DOI] [PubMed] [Google Scholar]
- 7.Murata N, Los DA. Membrane fluidity and temperature perception. Plant Physiol. 1997;115:875–879. doi: 10.1104/pp.115.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miquel M, James D, Dooner H, Browse J. Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc Natl Acad Sci U S A. 1993;90:6208–6212. doi: 10.1073/pnas.90.13.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okuley J, Jonathan L, Kenneth F, Yadav BN, Lark BE. Arabidopsis Fad2 gene encodes the enzyme that is essential for polyunsaturated lipid-synthesis. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nievola CC, Carvalho CP, Carvalho V, Rodrigues E. Rapid responses of plants to temperature changes. Temperature (Austin, Tex). 2017;4:371–405. doi: 10.1080/23328940.2017.1377812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jablonka E, Lamb MJ. The changing concept of epigenetics. Ann N Y Acad Sci. 2002;981:82–96. doi: 10.1111/j.1749-6632.2002.tb04913.x. [DOI] [PubMed] [Google Scholar]
- 12.Ding Y, Shi Y, Yang S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019;222:1690–1704. doi: 10.1111/nph.15696. [DOI] [PubMed] [Google Scholar]
- 13.Calixto CPG, Tzioutziou NA, James AB, Hornyik C, Guo W, Zhang R, Nimmo HG, Brown JWS. Cold-Dependent expression and alternative splicing of arabidopsis long non-coding RNAs. Front Plant Sci. 2019;10:235. doi: 10.3389/fpls.2019.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji H, Niu C, Zhan X, Xu J, Lian S, Xu B, Guo J, Zhen L, Yang H, Li S, Ma L. Identification, functional prediction, and key lncRNA verification of cold stress-related lncRNAs in rats liver. Sci Rep. 2020;10:521. doi: 10.1038/s41598-020-57451-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kindgren P, Ard R, Ivanov M, Marquardt S. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat Commun. 2018;9:4561. doi: 10.1038/s41467-018-07010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Q, Guo F, Xu Q, Cang J. LncRNA improves cold resistance of winter wheat by interacting with miR398. Funct Plant Biol. 2020;47:544–557. doi: 10.1071/fp19267. [DOI] [PubMed] [Google Scholar]
- 17.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 18.Wang P, Dai L, Ai J, Wang Y, Ren F. Identification and functional prediction of cold-related long non-coding RNA (lncRNA) in grapevine. Sci Rep. 2019;9:6638. doi: 10.1038/s41598-019-43269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao M, Wang T, Sun T, Yu X, Tian R, Zhang WH. Identification of tissue-specific and cold-responsive lncRNAs in Medicago truncatula by high-throughput RNA sequencing. BMC Plant Biol. 2020;20:99. doi: 10.1186/s12870-020-2301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 21.Kim DH, Sung S. Vernalization-Triggered intragenic chromatin loop formation by long noncoding RNAs. Dev Cell. 2017;40:302–312.e304. doi: 10.1016/j.devcel.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DH, Xi Y, Sung S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017;13:e1006939. doi: 10.1371/journal.pgen.1006939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Li J, Lian B, Gu H, Li Y, Qi Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat Commun. 2018;9:5056. doi: 10.1038/s41467-018-07500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fick RJ, Trievel RC. An overview of chromatin modifications. Biopolymers. 2013;99:95–97. doi: 10.1002/bip.22158. [DOI] [PubMed] [Google Scholar]
- 25.Gelato KA, Fischle W. Role of histone modifications in defining chromatin structure and function. Biol Chem. 2008;389:353–363. doi: 10.1515/bc.2008.048. [DOI] [PubMed] [Google Scholar]
- 26.Hirose S. Chromatin remodeling and transcription. J Biochem. 1998;124:1060–1064. doi: 10.1093/oxfordjournals.jbchem.a022220. [DOI] [PubMed] [Google Scholar]
- 27.Swygert SG, Peterson CL. Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochimica Et Biophysica Acta. 2014;1839:728–736. doi: 10.1016/j.bbagrm.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang R, Zeng X-L. ATP-dependent chromatin remodeling complex and its function in regulating chromatin structure. Yichuan. 2010;32:301–306. doi: 10.3724/sp.J.1005.2010.00301. [DOI] [PubMed] [Google Scholar]
- 29.Baroux C, Pien S, Grossniklaus U. Chromatin modification and remodeling during early seed development. Curr Opin Genet Dev. 2007;17:473–479. doi: 10.1016/j.gde.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Chen D-H, Huang Y, Jiang C, Si J-P. Chromatin-Based regulation of plant root development. Front Plant Sci. 2018:9. doi: 10.3389/fpls.2018.01509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farrona S, Mozgova I, Archacki R, Casas-Mollano JA. Editorial: chromatin stability and dynamics: targeting and recruitment of chromatin modifiers. Front Plant Sci. 2021:12. doi: 10.3389/fpls.2021.678702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao T, Zhan Z, Jiang D. Histone modifications and their regulatory roles in plant development and environmental memory. J Genetics Genomics. 2019;46:467–476. doi: 10.1016/j.jgg.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Kumar V, Thakur JK, Prasad M. Histone acetylation dynamics regulating plant development and stress responses. Cell Mol Life Sci. 2021;78:4467–4486. doi: 10.1007/s00018-021-03794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K. Chromatin modifications and remodeling in plant abiotic stress responses. Biochimica Et Biophysica Acta. 2012;1819:129–136. doi: 10.1016/j.bbagrm.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Yuan L, Liu X, Luo M, Yang S, Wu K. Involvement of histone modifications in plant abiotic stress responses. J Integr Plant Biol. 2013;55:892–901. doi: 10.1111/jipb.12060. [DOI] [PubMed] [Google Scholar]
- 36.Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 37.Hu Y, Zhang L, Zhao L, Li J, He S, Zhou K, Yang F, Huang M, Jiang L, Li L. Trichostatin a selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. Plos One. 2011;6. doi: 10.1371/journal.pone.0022132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy D, Paul A, Roy A, Ghosh R, Ganguly P, Chaudhuri S. Differential acetylation of histone H3 at the regulatory region of OsDREB1b Promoter facilitates chromatin remodelling and transcription activation during cold stress. Plos One. 2014;9. doi: 10.1371/journal.pone.0100343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon CS, Lee D, Choi G, Chung W-I. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 2009;60:112–121. doi: 10.1111/j.1365-313X.2009.03938.x. [DOI] [PubMed] [Google Scholar]
- 40.Stockinger EJ, Mao YP, Regier MK, Triezenberg SJ, Thomashow MF. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 2001;29:1524–1533. doi: 10.1093/nar/29.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong C-H, Agarwal M, Zhang Y, Xie Q, Zhu J-K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proceedings of the National Academy of Sciences of the United States of America 103, 8281–8286, doi: 10.1073/pnas.0602874103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung J-H, Park C-M. HOS1-mediated activation of FLC via chromatin remodeling under cold stress. Plant Signal Behav. 2013;8:e27342–Article No.: e27342. doi: 10.4161/psb.27342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung JH, Park JH, Lee S, To TK, Kim JM, Seki M, Park CM. The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE Gene1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell. 2013;25:4378–4390. doi: 10.1105/tpc.113.118364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J, et al. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proceedings of the National Academy of Sciences of the United States of America 105, 4945–4950, doi: 10.1073/pnas.0801029105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dennis ES, Helliwell CA, Peacock WJ. Vernalization: spring into flowering. Dev Cell. 2006;11:1–2. doi: 10.1016/j.devcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Dennis ES, Peacock WJ. Vernalization in cereals. J Biol. 2009;8:57–Article No.: 57. doi: 10.1186/jbiol156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu X, Kong X, Wang C, Ma L, Zhao J, Wei J, Zhang X, Loake GJ, Zhang T, Huang J, et al. Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during Vernalization. Plant Cell. 2014;26:4763–4781. doi: 10.1105/tpc.114.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Jiang D, He Y. FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat Plants. 2018;4:836–846. doi: 10.1038/s41477-018-0250-6. [DOI] [PubMed] [Google Scholar]
- 50.De Lucia F, Crevillen P, Jones AME, Greb T, Dean C. A PHD-Polycomb Repressive Complex 2 triggers the epigenetic silencing of FLC during vernalization. Proceedings of the National Academy of Sciences of the United States of America 105, 16831–16836, doi: 10.1073/pnas.0808687105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan W, Luo X, Li Z, Yang W, Wang Y, Liu R, Du J, He Y. A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat Genet. 2016;48:1527–1534. doi: 10.1038/ng.3712. [DOI] [PubMed] [Google Scholar]
- 52.Chen -L-L. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Advan. 2017:3. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Y, Cheng T, Liu C, Liu D, Zhang Q, Long R, Zhao P, Xia Q. Systematic Identification and Characterization of Long Non-Coding RNAs in the Silkworm, Bombyx mori. Plos One. 2016;11. doi: 10.1371/journal.pone.0147147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, Gutti RK. Long non-coding RNA: classification, biogenesis and functions in blood cells. Mol Immunol. 2019;112:82–92. doi: 10.1016/j.molimm.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 56.Yang Z, Wang K, Aziz U, Zhao C, Zhang M. Evaluation of duplicated reference genes for quantitative real-time PCR analysis in genome unknown hexaploid oat (Avena sativaL.). Plant Methods. 2020:16. doi: 10.1186/s13007-020-00679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, Shen F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int J Mol Sci. 2019;20. doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang T-Z, Liu M, Zhao M-G, Chen R, Zhang W-H. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015:15. doi: 10.1186/s12870-015-0530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao M, Wang T, Sun T, Yu X, Tian R, Zhang WH. Identification of tissue-specific and cold-responsive lncRNAs in Medicago truncatula by high-throughput RNA sequencing. BMC Plant Biol. 2020;20. doi: 10.1186/s12870-020-2301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu Q, Guo F, Xu Q, Cang J. LncRNA improves cold resistance of winter wheat by interacting with miR398. FunctPlant Biol. 2020;47:544–557. doi: 10.1071/fp19267. [DOI] [PubMed] [Google Scholar]
- 61.Kindgren P, Ard R, Ivanov M, Marquardt S. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat Commun. 2018:9. doi: 10.1038/s41467-018-07010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moison M, Martinez Pacheco C, Lucero L, Fonouni-Farde C, Rodriguez-Melo J, Mansilla N, Christ A, Bazin J, Benhamed M, Ibanez F, et al. The lncRNA APOLO interacts with the transcription factor WRKY42 to trigger root hair cell expansion in response to cold. Mol Plant. 2021;14:937–948. doi: 10.1016/j.molp.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 63.Csorba T, Questa JI, Sun Q, Dean C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proceedings of the National Academy of Sciences of the United States of America 111, 16160–16165, doi: 10.1073/pnas.1419030111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heo JB, Lee Y-S, Sung S. Epigenetic regulation by long noncoding RNAs in plants. Chromosome Res. 2013;21:685–693. doi: 10.1007/s10577-013-9392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim D-H, Sung S. Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev Cell. 2017;40:302–312. doi: 10.1016/j.devcel.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim D-H, Xi Y, Sung S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017:13. doi: 10.1371/journal.pgen.1006939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heo JB, Sung S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 68.Tian Y, Zheng H, Zhang F, Wang S, Ji X, Xu C, He Y, Ding Y. PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci Advan. 2019;5. doi: 10.1126/sciadv.aau7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fang X, Wang L, Ishikawa R, Li YX, Fiedler M, Liu FQ, Calder G, Rowan B, Weigel D, Li PL, et al. Arabidopsis FLL2 promotes liquid–liquid phase separation of polyadenylation complexes. Nature. 2019;569:265–269. doi: 10.1038/s41586-019-1165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huo Y, Yan Z, Zhang B, Wang X. Recruitment of Polycomb Repressive Complex 2 is Essential to Suppress the Target Chromatin in Arabidopsis. CRC Crit Rev Plant Sci. 2016;35:131–145. doi: 10.1080/07352689.2016.1245055. [DOI] [Google Scholar]
- 71.Xiao J, Jin R, Yu X, Shen M, Wagner JD, Pai A, Song C, Zhuang M, Klasfeld S, He C, et al. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat Genet. 2017;49:1546-+. doi: 10.1038/ng.3937. [DOI] [PubMed] [Google Scholar]
- 72.Maclary E, Hinten M, Harris C, Sethuraman S, Gayen S, Kalantry S . PRC2 represses transcribed genes on the imprinted inactive X chromosome in mice. Genome Biol. 2017;18. doi: 10.1186/s13059-017-1211-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L, Zeng X, Chen S, Ding L, Zhong J, Zhao JC, Wang L, Sarver A, Koller A, Zhi J, et al. BRCA1 is a negative modulator of the PRC2 complex. Embo J. 2013;32:1584–1597. doi: 10.1038/emboj.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]


