ABSTRACT
Aberrant upregulation and oncogenic roles of UBE2T are revealed in several cancers. However, the expression, clinical significance, and functions of UBE2T have not been explored in ovarian cancer (OC). In this study, the expression of UBE2T and its relation with clinicopathological features and prognosis of OC patients were explored by analyzing online data and experimental data. Besides, the functions of UBE2T in OC cells were investigated by in vitro experiments, including CCK-8, plate clone formation, and Transwell assays. Finally, the underlying mechanism of UBE2T associated functions in OC was analyzed. The results indicated that UBE2T was significantly upregulated in OC tissues. UBE2T expression was notably correlated with clinical features, such as primary T stage and FIGO stage in OC patients. UBE2T, acting as an independent prognostic indicator, was inversely associated with the prognosis of OC patients. The UBE2T knockdown remarkably suppressed the growth, proliferation, and invasion of OC cells, indicated by impaired cell viability, fewer cell clones, and invasive cells. Mechanistically, UBE2T depletion suppressed epithelial–mesenchymal transition (EMT), which was caused by autophagy activation due to inactivation of AKT/mTOR in OC cells with UBE2T knockdown. Collectively, our findings confirm that UBE2T upregulation predicts poor prognosis and promotes malignant progression in OC. UBE2T upregulation suppresses autophagy and subsequently boosts EMT via activating the AKT/mTOR axis, which accounts for the underlying mechanism of oncogenic roles of UBE2T in OC.
KEYWORDS: Ovarian cancer, ubiquitin-conjugating enzyme E2T (UBE2T), epithelial–mesenchymal transition (EMT), autophagy, AKT/mTOR
Introduction
Ovarian cancer (OC) is one of the aggressive gynecologic cancers with the poor prognosis, whose five-year overall survival rate is less than 50%. Consistent with the circumstances for most carcinomas, the favorable therapeutic and prognostic outcomes are just achievable for OC patients at an early stage. However, the actual rate of OC patients diagnosed at early stage is as low as 20%, indicating the most OC patients are diagnosed at advanced stages, which largely accounts for the unsatisfactory prognosis of OC patients [1,2]. Obviously, the unfavorable outcomes suggest the defects of the current biomarkers and treatments of OC, and it is vital to explore novel diagnostic and prognostic biomarkers and therapeutic targets to improve the efficiency of diagnosis and treatment in OC.
The aberrant expressions and critical functions of ubiquitin-conjugating enzymes, being considered as a family of constitutive enzymes before, in tumorigenesis, have been widely indicated [3]. For example, ubiquitin-conjugating enzyme E2T (UBE2T), firstly identified in Fanconi anemia syndrome (FA) and with vital roles in the development of FA via catalyzing the mono-ubiquitination of FANCD2 and FANCI [4], is upregulated, exerts oncogenic roles, and serves as an unfavorable prognostic indicator in both solid tumors, such as gastric cancer [5,6], hepatocellular carcinoma [7], breast cancer [8], lung cancer [8,9], nasopharyngeal carcinoma [10], osteosarcoma [11], prostate cancer [12] and renal cell carcinoma [13], and non-solid tumor-like multiple myeloma (MM) [14]. Generally, UBE2T depletion notably suppresses cell proliferation, migration, invasion, stemness, and therapy resistance in cancers via regulating cell cycle, epithelial–mesenchymal transition (EMT), and apoptosis by modulating the activity of oncogenic signaling regulators, such as PI3K/AKT and GSK3β/Wnt/β-catenin [10,13,15] or regulating the stabilities of tumor suppressors including p53 [7], FOXO1 [16], GRP78 [17], and BRCA1 [18]. Although a recent bioinformatic study has reported the UBE2T upregulation in OC [19], the function and underlying mechanism of UBE2T in OC remain to be validated.
As a vital means in maintaining cellular homeostasis, autophagy plays vital roles in tumorigenesis [20]. The pro-tumor or anti-tumor functions of autophagy in carcinogenesis are mainly determined by the extent of autophagic flux and the degraded substrates, which are tightly regulated by upstream signaling regulators like AKT/mTOR, AMPK, and MAPK pathways [20,21]. Theoretically, UBE2T, severing a well-known activator of AKT [10], would regulate autophagy, which may contribute to the oncogenic roles of UBE2T in cancers. Therefore, in this study, we first examined the mRNA and protein expression, prognostic value of UBE2T in OC via mining the online data and immunohistochemistry assay in a local OC cohort. Moreover, we also explored the primary functions of UBE2T in OC by detecting the effects of UBE2T knockdown on cell phenotypes, including growth, proliferation, and invasion, analyzed the effects of UBE2T on EMT and autophagy, and explored the roles of autophagy in UBE2T-related functions. The outcomes reveal that UBE2T is upregulated and serves as an indicator for poor prognosis of OC patients. Furthermore, UBE2T knockdown significantly inhibits cell growth, proliferation, and invasion by suppressing AKT/mTOR and EMT via activating autophagy in OC.
Methods and materials
Tissue samples, ethical statement, and immunohistochemistry (IHC)
An OC tissue chip, containing 70 OC tissues and 10 normal tissues (#OVC1504) was purchased from the Shanghai Superbiotek Pharmaceutical Technology Inc. (Shanghai, China). The study was approved by the Academic Ethics Committee of the Second Xiangya Hospital of Central South University (OV_XXM_20191128) and performed under the instructions of Declaration of Helsinki. IHC and scoring were applied according to our previous description [22] with primary antibody UBE2T (dilution: 1:50, BBI, Shanghai, China).
Online databases
Oncomine (http://www.oncomine.org) and GEPIA (http://gepia.cancer-pku.cn/) were used to explore the expression of UBE2T in OC patients [23,24]. GEPIA was also applied to analyze the relation of UBE2T expression with clinical features and prognosis of OC patients [24]. UALCAN (http://ualcan.path.uab.edu/) was adopted to investigate the protein levels of UBE2T in OC [25]. The prognostic value of UBE2T was explored by Kaplan–Meier (K-M) plotter (http://kmplot.com/analysis/) [26].
Cell culture
Normal ovarian epithelial cell line, IOSE80, and OC cell lines, OVCAR3, SKOV3, SKOV3-ip1, HO8910, HO8910-PM, and A2780, were purchased from the American Type Culture Collection (ATCC, VA, USA) and Cell bank of the Committee on Preservation of Typical Cultures of the Chinese Academy of Sciences (CCPTC, Shanghai, China). SKOV3, SKOV3-ip1, H08910 and HO8910-PM were maintained in RPMI-1640 medium plus 10% fetal bovine serum (FBS, Thermo, MA, USA). IOSE80, OVCAR3 and A2780 were cultured in DMEM medium plus 10% FBS (Thermo, MA, USA). The cells were cultured in a humidified incubator at 37°C and 5% CO2. The chloroquine (CQ, Sigma-Aldrich, MO, USA) and MK2206 (Selleck, TX, USA) were added into the culture medium for functional experiments as indicated in figure legends.
Small interfering RNAs (siRNAs) and plasmids transfection
The UBE2T and BECN1 siRNA (si-UBE2T and si-BECN1) and scrambled siRNA (si-NC) were purchased from RiboBio Inc. (Guangzhou, China). The UBE2T expression plasmid, pENTER-UBE2T, and the control plasmid, pENTER-vector, were obtained from Vigene Inc. (Jinan, China). The si-UBE2T-1 sequences are 5’-GTCCTGGTTCATCTTAGTTAA-3’, which targets the 3’ untranslated region of UBE2T mRNA. The si-UBE2T-2 sequences are 5’-TGAGGAAGAGATGCTTGATAA-3’, which targets the coding sequence region of the UBE2T mRNA. The si-BECN1 sequences are 5’-CAGTTTGGCACAATCAATA-3’. The siNC sequences were not offered by the manufacturer. HO8910 and OVCAR3 cells were transfected with si-UBE2Ts or co-transfected with si-UBE2Ts and pENTER-UBE2T or si-UBE2Ts and si-BECN1 using LipofectamineTM 2000 (Thermo, MA, USA) according to our previous description [27].
Cell growth assay
The cell growth assay was performed as previously described [27]. Briefly, cells were seeded into a 96-well plate at 2 × 103 cells per well. Then, CCK-8 reagent was applied into the cell medium and incubated for 1 hour in successive four days. The absorbance at 450 nm was detected via a spectrophotometer (BioTek, WI, USA). At the end, the growth curves were plotted based on the absorbance of every day. The experiments were independently performed in triplicate.
Plate clone formation assay
Plate clone formation assay was carried out as previously described [27][. Briefly, cells were seeded into six-well plates at 1000 cells per well and cultured for 8 days. Then cell clones were fixed with methanol and stained by crystal violet. Clones containing more than 50 cells were counted under an inverse microscope (Leica, Solms, Germany). The experiments were independently performed for three times.
Transwell migration and invasion assay
Transwell migration and invasion assay was performed as in the previous description [27][]. Briefly, 8-μm pore size transwell chambers (Costar, ME, USA) with or without pre-coated with Matrigel (Corning, NY, USA) were carried out to analyze invasion and migration abilities of OC cells, respectively. Four hundred microliters pure DMEM or RPIM-1640 medium containing 2.5 × 104 cells were pipetted into the chamber, and the lower well was filled with 750 μl DMEM or RPIM-1640 medium plus 5% FBS to form a chemoattractant. Eighteen hours (invasion assay) or 14 hours (migration assay) later, the chambers were filled with methanol and stained with crystal violet. The cells above on the upper side of the membrane were swabbed and the invasive cells were photographed and counted with an inverse microscopic scope (Leica, Solms, Germany). The experiments were independently performed for three times.
Western blot
The Western blot was performed as in the previous description [][27]. Simply put, the total proteins were obtained from RIPA cell lysis via centrifugation at 4°C. The proteins were denatured and separated by SDS-PAGE and subsequently transmitted into 0.22 μm PVDF membrane. The membrane was blocked with Protein Free Rapid Blocking Buffer (Epizyme, Shanghai, China) for 30 minutes at room temperature and then subjected for antibody incubation overnight at 4°C. Primary antibodies, including UBE2T (BBI, Shanghai, China), MMP2 (BBI, Shanghai, China), Slug (Abclonal, Wuhan, China), E-cadherin (Abclonal, Wuhan, China), N-cadherin (Abclonal, Wuhan, China), BECN1(Abclonal, Wuhan, China), p62 (Abclonal, Wuhan, China), LC3A/B (Abclonal, Wuhan, China), p-AKT(S473) (CST, MA, USA), AKT (CST, MA, USA), mTOR (CST, MA, USA), and p-mTOR (S2448) (CST, MA, USA), and mouse anti-β-Actin (Proteintech, Wuhan, China), were used. One day layer, after incubated with anti-rabbit or anti-mouse IgG HRP-conjugated secondary antibodies (BBI, Shanghai, China), the protein levels were visualized by chemiluminescent HRP substrate (EpiZyme, Shanghai, China).
Statistical analysis
Statistical analyses and statistical charts were analyzed and produced using SPSS20.0 software and GraphPad Prism version 8. For comparisons between two groups, a Student t-test or Fisher exact test was carried out. Survival curves were obtained via Kaplan–Meier method, and the statistical analysis was evaluated by Log-rank test. The univariate and multivariate Cox regression was performed to analyze the relationship of among UBE2T expression, clinicopathological parameters, and survival in OC patients. For all analyses, P (two sides) <0.05 was considered statistically significant.
Results
UBE2T is upregulated in OC
First, we analyzed the expression of UBE2T via several web portals. As shown in Figure 1a, compared with the normal counterparts, the mRNA expression of UBE2T was significantly upregulated in OC tissues reflected by two data sets in Oncomine [23]. Accordingly, through analyzing the data in GEPIA [24], which integrated the data from TCGA and GTEx databases, upregulation of UBE2T mRNA in OC was further validated (Figure 1b). Furthermore, mass-spectrometry-based proteomic data from UALCAN [25] indicated upregulation of UBE2T protein in OC (Figure 1e). Finally, we analyzed the protein level of UBE2T in a cohort of OC via IHC staining. The results showed that the staining intensity of UBE2T was significantly stronger in OC specimens than in normal samples (Figure 1f). Thus, these results revealed that UBE2T was upregulated in OC both at mRNA and protein level.
Figure 1.
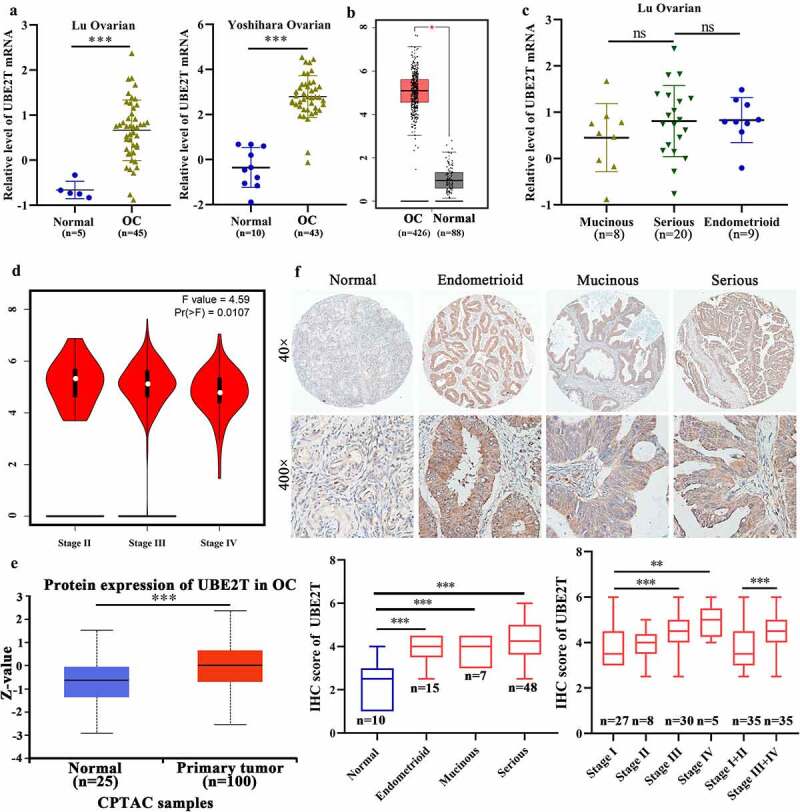
The expression of UBE2T in OC and its relation to clinical features of patients with OC. Oncomine data (a) and GEPIA data (b) indicated UBE2T mRNA upregulated in OC. C The Oncomine data showed no difference of UBE2T mRNA expression among mucinous, serious and endometrioid OC tissues. D GEPIA data showed UBE2T mRNA significantly correlated to stage of OC. E UALCAN data showed UBE2T protein was upregulated in OC tissues. F IHC results of UBE2T protein level in normal tissues and OC tissues with different pathological types and stages. *P < 0.05, ** P < 0.01, ***P < 0.001, ns, no significant statistical difference.
UBE2T correlates to the clinical features of patients with OC
Subsequently, we explored the relation between UBE2T expression and clinopathological variables. According to Oncomine data, the UBE2T mRNA level was comparative among different histological types including mucinous, serous, and endometrioid type of OC (Figure 1c), which was confirmed by the results of IHC (Figure 1f, Table 1). Interestingly, based on the data from GEPIA, UBE2T mRNA level was inversely associated with the stage of OC patients (Figure 1d). However, the IHC data revealed that although no significant difference of UBE2T protein among FIGO stage I, II, III, and IV, notable upregulation of UBE2T was observed in advance stage (III+IV) OC tissues than that in early stage (I+ II) (Figure 1f, Table 1). Moreover, the UBE2T level positively correlated with the primary T stages in OC (Table 1). However, possibly due to the limited sample volumes, UBE2T showed no significant relation with distant metastasis and lymph node metastasis (Table 1). These results revealed that UBE2T was associated with stages of OC patients indicating UBE2T may involve in the progression of OC.
Table 1.
Correlation between UBE2T expression and clinicopathological characteristics in ovarian cancer (n = 70, Chi-square or Fisher exact test)
| Variables | n | Expression level |
χ2 | P | |
|---|---|---|---|---|---|
| Low (1–3) | High (4–6) | ||||
| Age(years) | |||||
| ≥50 | 37 | 9 | 28 | 1.204 | 0.273 |
| <50 | 33 | 12 | 21 | ||
| Primary tumor (T) stage | |||||
| T1-2 | 36 | 18 | 18 | 14.118 | 0.000172*** |
| T3-4 | 34 | 3 | 31 | ||
| Lymph node(N) metastasis | |||||
| N0 | 67 | 21 | 46 | - | 0.549 |
| N1-3 | 3 | 0 | 3 | ||
| Distant metastasis (M) | |||||
| M0 | 65 | 21 | 44 | - | 0.313 |
| M1 | 5 | 0 | 5 | ||
| FIGO stage | 16.4682534 | 0.000091*** | |||
| I | 27 | 15 | 12 | ||
| II | 8 | 3 | 5 | ||
| III | 30 | 3 | 27 | ||
| IV | 5 | 0 | 4 | ||
| Pathological Classification | |||||
| Endometrioid | 15 | 6 | 9 | 1.836734 | 0.399170 |
| Mucinous | 7 | 3 | 4 | ||
| Serous | 48 | 12 | 36 | ||
| Pathological Stage | Fisher Test | ||||
| Endometrioid carcinoma | 2.468575 | 0.425574 | |||
| I | 3 | 2 | 1 | ||
| II | 6 | 3 | 3 | ||
| III | 6 | 1 | 5 | ||
| Serous cystadenocarcinoma | 0.027826 | 0.867518 | |||
| Low-grade | 12 | 6 | 6 | ||
| High-grade | 36 | 17 | 19 | ||
| Cytoreductive surgery | 6.024187 | 0.049188* | |||
| R0 | 45 | 13 | 32 | ||
| R1 | 14 | 4 | 10 | ||
| R2 | 11 | 4 | 7 | ||
* P < 0.05, ***P < 0.001, the difference was statistically significant.
UBE2T inversely associated with the prognostic outcomes and served as an independent indicator for OC patients.
Next, we investigated the relation between UBE2T and prognosis in OC via online and experimental data. The data from Kaplan-Meier Plotter portal [26] indicated UBE2T upregulation was negatively correlated with overall survival (OS), progression-free survival (PFS), and post-progression survival (PPS) of OC patients (Figure 2a). Besides, GEPIA data confirmed that higher UBE2T mRNA was significantly correlated poor OS of OC patients (Figure 2b). Consistently, upregulation of the UBE2T protein served as a poor indicator for OC patients as well (Figure 2c). The univariate Cox proportional hazards regression analysis revealed that primary T stage, FIGO stage, distant metastasis, and the level of UBE2T were significantly related to the OS of OC patients (Table 2). The multivariate Cox proportional hazards regression analysis indicated that UBE2T upregulation served as an independent predictor for poor OS for OC patients (Table 2). Thus, these outcomes confirmed that UBE2T was inversely related to the prognosis and served as an independent indicator for poor prognosis for OC patients.
Figure 2.
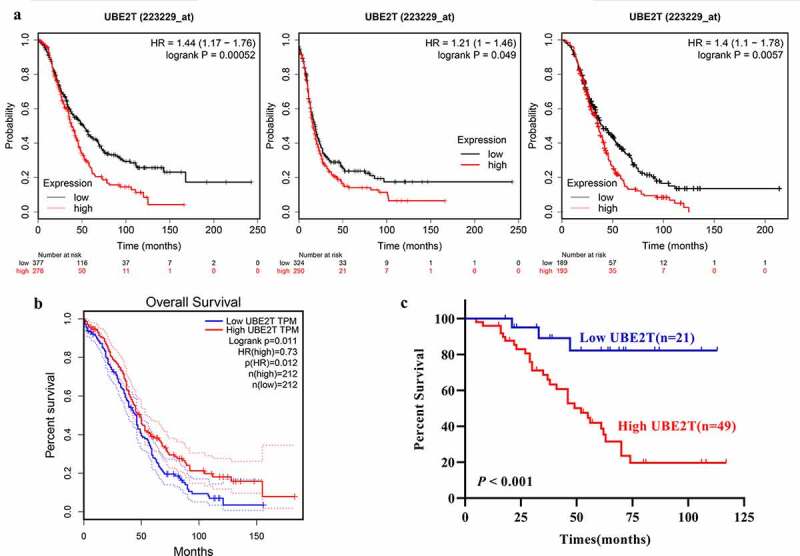
UBE2T upregulation predicts poor prognosis of patients with OC. A UBE2T mRNA level was inversely associated with OS (left panel), PFS (middle panel),and PPS (right panel) of OC patients indicated by Kaplan-Meier Plotter data. B UBE2T mRNA level was inversely associated with OS of OC patients indicated by GEPIA data. C UBE2T protein level was negatively related to OS of OC patients demonstrated by our data.
Table 2.
Univariate analysis and multivariate analysis of prognostic factors for overall survival using Cox proportional hazards regression model (N = 70)
| Variables | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR(95% CI) | P | HR(95% CI) | P | |
|
Age(years) <50 vs ≥50 |
1.292(0.642–2.602) | 0.473 | 1.069 (0.520–2.198) | 0.8558 |
|
Primary tumor (T) stage T1-2 vs T3-4 |
4.393(2.026–9.524) | 0.0002 | / | / |
|
Lymph node(N) metastasis |
4.547(0.530–39.010) | 0.1673 | 4.739(0.533–42.166) | 0.1630 |
| Distant metastasis (M) | 8.480(2.698–26.655) | 0.0003 | 6.494(1.986–21.239) | 0.0020 |
| TNM stage | 4.353(2.007–9.442) | 0.0003 | / | / |
| FIGO stage | 2.349(1.552–3.556) | 0.0005 | / | |
|
Pathological Classification |
1.423(0.898–2.255) | 0.1328 | 1.279(0.795–2.059) | 0.3103 |
|
Pathological Stage |
1.148(0.806–1.636) | 0.4433 | 0.980(0.679–1.414) | 0.9123 |
| Cytoreductive surgery |
2.927(1.847–4.640) | 0.0000 | / | / |
|
UBE2T High vs Low |
5.992(1.823–19.696) | 0.0032 | 4.994(1.477–16.885) | 0.0097a |
HR:hazard ratio; 95%CI:95% confidence interval;
UBE2T depletion inhibits growth, proliferation, and invasion of OC cells
Next, we further explored the functions of UBE2T in OC. Therefore, we checked the expression of UBE2T, depleted UBE2T expressions with si-UBE2Ts transfection, and subsequently detected the influences on OC cells. As shown in the Western blot results (Figure 3a), compared with IOSE80 cells, an immortalized normal ovarian epithelial cell line, UBE2T was remarkably higher in OC cells, especially in OVCAR3 and HO8910 cells. Thus, the expression of UBE2T was knocked down by transfected with
Figure 3.
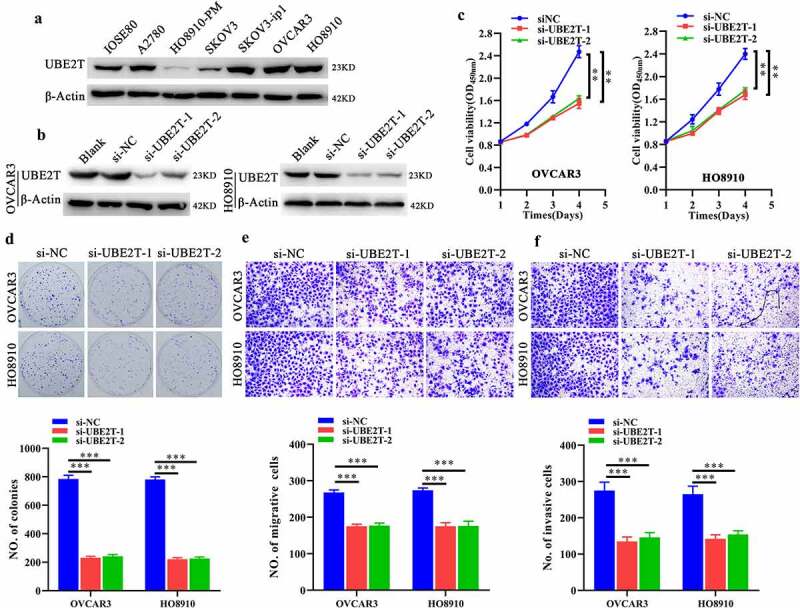
UBE2T depletion inhibits growth, proliferation and invasion of OC cells. A Western blot indicating the UBE2T expression among IOSE80, OVCAR3, HO8910, SKOV3, SKOV3-ip1 and HO8910-PM cells. B Western blot indicating the knockdown efficacy of siUBE2T-1 and −2 (50 nM) in OC cells. Analyzing the effects of UBE2T knockdown on growth, proliferation, migration, and invasion via CCK-8 (c), plate clone formation (d), transwell migration (e) and transwell invasion (f). *P < 0.05, ** P < 0.01, ***P < 0.001.
siUBE2Ts. As indicated by Figure 3b, UBE2T expression was successfully knocked down in OVCAR3 and SKOV3 cells by both si-UBE2T-1 and si-UBE2T-2, respectively. Then, CCK-8, plate clone formation and Transwell migration and invasion assays manifested UBE2T depletion significantly suppressed the growth, proliferation, and invasion of OC cells, demonstrated by impaired viability (Figure 3c), fewer cell clones (Figure 3d), migrative and invasive cells (Figure 3e,f). Herein, these results unveiled that UBE2T depletion could inhibit growth, proliferation and invasion of OC cells.
UBE2T knockdown suppresses EMT and malignant progression of OC cells via activating autophagy by inhibiting AKT/mTOR
The roles of UBE2T in the activation of EMT and AKT have been revealed in several studies [10,13,16]. We also detected the EMT and AKT activities in OC cells with UBE2T depletion. Accordingly, UBE2T silencing suppressed EMT and AKT/mTOR activities demonstrating by decreased Slug, N-cadherin (N-cad) and increased E-cadherin (Figure 4a), and decreased p-AKT (S473) and p-mTOR (S2448) (Figure 4b). Considering the regulatory effects of AKT/mTOR in autophagy [28], the influences of UBE2T alteration on autophagy were further analyzed. Indeed, the UBE2T knockdown activated the autophagy of OC cells indicated by upregulated BECN1, accumulated LC3A/B-II, and decreased p62 (Figure 4c). Moreover, the accumulative effects of LC3A/B-II were augmented under CQ (50 μM) treatment (Figure 4d), further confirming the enhanced autophagy flux induced by UBE2T depletion in OC cells. To explore the role of AKT/mTOR in UBE2T-related autophagy, ectopic UBE2T expression was rescued in OC cells with endogenous depletion of UBE2T (Figure 4e). UBE2T restoration rescued its inhibitory effects on autophagy, which was deprived by AKT inhibitor MK2206 (2.5 μM), demonstrating by the level of LC3A/B-II accumulation (Figure 4e) and suggesting UBE2T could inhibit autophagy via sustaining AKT/mTOR activity. Furthermore, we also explored the role of autophagy in EMT regulated by UBE2T. As Figure 4f shown, when autophagy was exogenously inhibited by depleting BECN1, the EMT was re-activated in OC cells with UBE2T knockdown, indicating by the level of Slug, N-cad, and E-cad. Besides, consistent phenotypes were observed indicating by CCK-8 (Figure 5a), plate clone formation (Figure 5b), Transwell migration (Figure 5c), and Transwell invasion assays (Figure 5d). Thus, we revealed that UBE2T knockdown suppresses EMT and malignant progression of OC cells via activating autophagy by inhibiting AKT/mTOR.
Figure 4.
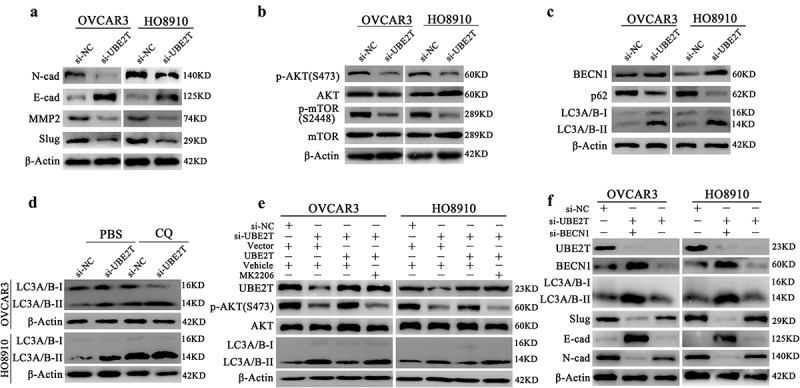
UBE2T depletion inhibited EMT process by inducing autophagy via suppressing AKT/mTOR in OC cells. A Western blot indicating the level of N-cad, E-cad, Slug, and MMP2 in OC cells with UBE2T silencing. B Western blot indicating the level of p-AKT (S473), AKT, p-mTOR (S2448) and mTOR in OC cells with UBE2T depletion. C Western blot showing the level of BECN1, p62, and LC3A/B-I/II in OC cells with UBE2T knockdown. D Western blot indicating the accumulated LC3A/B-II of OC cells with UBE2T knockdown under CQ (50 μM) treatment. E Western blot indicating the influences of MK2206 (2.5 μM) on the rescue effects of UBE2T on AKT and autophagy activity, reflected by the level of p-AKT(S473), AKT, and LC3A/B, in OC cells with UBE2T knockdown. F Western blot indicating the influences of BECN1 knockdown on BECN1 and LC3A/B-I/II, and the level of EMT related proteins, N-cad, E-cad, and Slug, with UBE2T knockdown.
Figure 5.
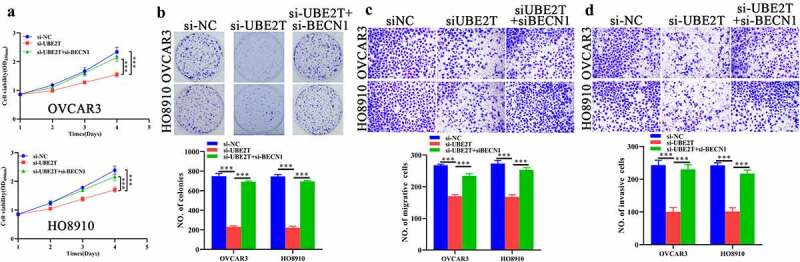
Autophagy suppression antagonizes the inhibitory effects of UBE2Tdepletion on malignant characteristics of OC cells. CCK-8 (a), plate clone formation (b), transwell migration (c), and transwell invasion (d) assays indicating the influences of BECN1 depletion on effects of UBE2T knockdown on growth, proliferation and invasion of OC cells. ***P < 0.001.
Discussion
Here, we demonstrated that the mRNA and protein level of UBE2T were notably upregulated in OC tissues and cells and validated the results of a recent bioinformatic study [19]. Moreover, our data confirmed that upregulation of UBE2T was inversely correlated with stages and prognosis of OC patients. UBE2T inhibition remarkably suppressed the growth, proliferation, and invasion of OC cells via suppressing EMT by activating autophagy by suppressing AKT/mTOR. Collectively, our findings suggest the promising values of UBE2T as a prognostic biomarker and therapeutic target in OC.
The roles of UBE2T have been primarily revealed in FA-related ubiquitin signaling [4]. UBE2T is critical for maintaining genome integrity in FA involving its regulatory roles in mono-ubiquitination of FANCD2 and FANCI. Notably, FA patients bearing related gene mutations are particularly prone to malignant outcomes, suggesting the vital roles of FA-related genes in tumorigenesis [4]. Indeed, aberrant upregulation of UBE2T has been observed in a host of cancers. For example, UBE2T is remarkably upregulated in MM cells, especially in the early stage. The UBE2T level is significantly associated with the IgG serotype of MM and the UBE2T upregulation predicts poor prognosis, including OS and event-free survival time, of patients with MM [29]. Furthermore, UBE2T is upregulated in gastric cancer tissues and cells, positively associated with poor differentiation, advanced T stage, and short OS time in patients with gastric cancer [5,6]. Accordingly, UBE2T upregulation, severing as a poor prognostic biomarker, has been confirmed in nasopharyngeal carcinoma [10][], osteosarcoma [11], lung cancer [8,9], breast cancer [8] [], prostate [12], and hepatocellular cancer [7,26]. Consistently, we demonstrated that UBE2T mRNA and protein were upregulated in OC tissues and cells and UBE2T upregulation was associated with stages and poor prognosis OC patients.
Although UBE2T is well known for catalyzing mono-ubiquitination of FANCD2 and FANCI in FA [4], the oncogenic roles of UBE2T seem to have little relation with FANCD2 and FANCI in cancers. The UBE2T knockdown exerts significantly inhibitory effects on cell characteristics via different mechanisms. For example, by inhibiting AKT and related pathways, UBE2T depletion remarkably suppresses cell proliferation, migration, and invasion of osteosarcoma [30], nasopharyngeal carcinoma [10], liver cancer [7], and renal cell carcinoma [13]. Besides, the suppressive roles of UBE2T knockdown on bladder cancer and hepatocellular carcinoma are mainly caused by apoptosis [31] and cell cycle arrest[32] []. Furthermore, the oncogenic roles of UBE2T closely depend on their enzymatic activity. UBE2T can inactivate p53, FOXO1, and BRAC1, promote proliferation and radiation resistance of hepatocellular carcinoma, lung cancer, and breast cancer cells via directly catalyzing their mono-ubiquitination [16,18][33]. Correspondingly, UBE2T knockdown inhibited AKT/mTOR and exerted notably inhibitory effects on growth, proliferation, and invasion of OC cells. Taken together, these outcomes suggest UBE2T servers as a common activator of AKT/mTOR axis; however, the detail regulatory mechanism remains to be revealed.
Autophagy plays a vital role in the regulation of physiopathologic processes, especially in cancers [20]. Carcinogenesis related autophagy is synergistically modulated by upstream signal including both oncogenic and tumor-suppressive regulators [20]. AKT/mTOR is the most common negative regulator of autophagy via control the activities of ULK1, BECN1, and IRS1/2 [34]. Besides, ubiquitin conjugating enzymes, such as UBE2L6, UBE2N, UBE2L3, and UBE2D2/3, can regulate autophagy process to exert their functions in physiopathologic processes, including cancers [4,35–37]. In line with, we also initially confirmed the negative regulatory effect of UBE2T on autophagy, which at least partially accounted for the oncogenic mechanism of UBE2T in OC. Additionally, autophagy play essential role in signal transduction by controlling the stabilities of specific substrates. For example, via selectively degrading Snail and Slug, autophagy could negatively regulate EMT, a key underlying reason for sustaining proliferation, stemness, metastasis, and therapy resistance in cancers, which is frequently activated by AKT/mTOR [38–40]. Consistent with available studies [16,17], UBE2T positively regulates EMT and subsequently promotes malignant progression of OC cell, mechanistically depending on its activation of AKT/mTOR that suppresses activation of autophagy.
In conclusion, we comprehensively explored the expression, clinical significance, fundamental functions, and underlying mechanisms of UBE2T in OC. The outcomes reveal that UBE2T is notably increased in OC, which serves as an indicator of poor prognosis in OC patients. The UBE2T knockdown suppresses AKT/mTOR activity, subsequently activates autophagy, then constrains EMT, and eventually inhibits growth, proliferation, and invasion of OC cells. These findings contribute a better understanding of the development and progression of OC and present UBE2T as a promising biomarker and therapeutic target for diagnosis and treatment of OC, respectively.
Funding Statement
The present study was granted by the National Natural Science Foundation of China [nos. 81702924, 81671437, 81801425, and 81771558]; the Natural Science Foundation of Hunan Province of China [nos. 2018JJ3811, 2020JJ4814; the Research Foundation of Education Bureau of Hunan Province of China [nos.19K101, 17K099]; Science and Technology Plan Project of Hunan Province [2020ZK4013, 2021ZK4209], and the Open Sharing Fund for the Largescale Instruments and Equipments of Central South University.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The online data used to support the findings of this study are from Oncomine (http://www.oncomine.org), GEPIA (http://gepia.cancer-pku.cn/), UALCAN (http://ualcan.path.uab.edu/), and Kaplan–Meier (K-M) plotter (http://kmplot.com/analysis/). The experimental data that support the findings of this study are available from the corresponding author, [XX], upon reasonable request.
References
- [1].Nash Z, Menon U.. Ovarian cancer screening: current status and future directions. Best Pract Res Clin Obstet Gynaecol. 2020;65:32–45. [DOI] [PubMed] [Google Scholar]
- [2].Chandra A, Pius C, Nabeel M, et al. Ovarian cancer: current status and strategies for improving therapeutic outcomes. Cancer Med. 2019;8:7018–7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hosseini SM, Okoye I, Chaleshtari MG, et al. E2 ubiquitin-conjugating enzymes in cancer: implications for immunotherapeutic interventions. Clin Chim Acta. 2019;498:126–134. [DOI] [PubMed] [Google Scholar]
- [4].Alpi AF, Chaugule V, Walden H. Mechanism and disease association of E2-conjugating enzymes: lessons from UBE2T and UBE2L3. Biochem J. 2016;473:3401–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Luo C, Yao Y, Yu Z, et al. UBE2T knockdown inhibits gastric cancer progression. Oncotarget. 2017;8:32639–32654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yu H, Xiang P, Pan Q, et al. Ubiquitin-conjugating enzyme E2T is an independent prognostic factor and promotes gastric cancer progression. Tumour Biol. 2016;37:11723–11732. [DOI] [PubMed] [Google Scholar]
- [7].Liu LP, Yang M, Peng QZ, et al. UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53. Biochem Biophys Res Commun. 2017;493:20–27. [DOI] [PubMed] [Google Scholar]
- [8].Perez-Pena J, Corrales-Sanchez V, Amir E, et al. Ubiquitin-conjugating enzyme E2T (UBE2T) and denticleless protein homolog (DTL) are linked to poor outcome in breast and lung cancers. Sci Rep. 2017;7:17530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu ZH, Zhang YJ, Sun HY. High ubiquitin conjugating enzyme E2 T mRNA expression and its prognostic significance in lung adenocarcinoma: a study based on the TCGA database. Medicine (Baltimore). 2020;99:e18543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hu W, Xiao L, Cao C, et al. UBE2T promotes nasopharyngeal carcinoma cell proliferation, invasion, and metastasis by activating the AKT/GSK3beta/beta-catenin pathway. Oncotarget. 2016;7:15161–15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shen L, Zhao K, Li H, et al. Downregulation of UBE2T can enhance the radiosensitivity of osteosarcoma in vitro and in vivo. Epigenomics. 2019;11:1283–1305. [DOI] [PubMed] [Google Scholar]
- [12].Wen M, Kwon Y, Wang Y, et al. Elevated expression of UBE2T exhibits oncogenic properties in human prostate cancer. Oncotarget. 2015;6:25226–25239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hao P, Kang B, Li Y, et al. UBE2T promotes proliferation and regulates PI3K/Akt signaling in renal cell carcinoma. Mol Med Rep. 2019;20:1212–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alagpulinsa DA, Kumar S, Talluri S, et al. Amplification and overexpression of E2 ubiquitin conjugase UBE2T promotes homologous recombination in multiple myeloma. Blood Adv. 2019;3:3968–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu J, Liu X. UBE2T silencing inhibited non-small cell lung cancer cell proliferation and invasion by suppressing the wnt/beta-catenin signaling pathway. Int J Clin Exp Pathol. 2017;10:9482–9488. [PMC free article] [PubMed] [Google Scholar]
- [16].Yin H, Wang X, Zhang X, et al. UBE2T promotes radiation resistance in non-small cell lung cancer via inducing epithelial-mesenchymal transition and the ubiquitination-mediated FOXO1 degradation. Cancer Lett. 2020;494:121–131. [DOI] [PubMed] [Google Scholar]
- [17].Huang P, Guo Y, Zhao Z, et al. UBE2T promotes glioblastoma invasion and migration via stabilizing GRP78 and regulating EMT. Aging (Albany NY). 2020;12:10275–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ueki T, Park JH, Nishidate T, et al. Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res. 2009;69:8752–8760. [DOI] [PubMed] [Google Scholar]
- [19].Zou R, Xu H, Li F, et al. Increased expression of UBE2T predicting poor survival of epithelial ovarian cancer: based on comprehensive analysis of UBE2s, clinical samples, and the GEO database. DNA Cell Biol. 2021;40:36–60. [DOI] [PubMed] [Google Scholar]
- [20].Huang T, Song X, Yang Y, et al. Autophagy and hallmarks of cancer. Crit Rev Oncog. 2018;23:247–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang W, Zeng C, Liu J, et al. Sodium butyrate induces autophagic apoptosis of nasopharyngeal carcinoma cells by inhibiting AKT/mTOR signaling. Biochem Biophys Res Commun. 2019;514:64–70. [DOI] [PubMed] [Google Scholar]
- [22].Liu J, Huang W, Ren C, et al. Flotillin-2 promotes metastasis of nasopharyngeal carcinoma by activating NF-kappaB and PI3K/Akt3 signaling pathways. Sci Rep. 2015;5:11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nagy A, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huang W, Liu J, Hu S, et al. miR-181a upregulation promotes radioresistance of nasopharyngeal carcinoma by targeting RKIP. Onco Targets Ther. 2019;12:10873–10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhan L, Zhang Y, Wang W, et al. Autophagy as an emerging therapy target for ovarian carcinoma. Oncotarget. 2016;7:83476–83487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang W, Zhang Y, Yang Z, et al. High expression of UBE2T predicts poor prognosis and survival in multiple myeloma. Cancer Gene Ther. 2019;26:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang Y, Leng H, Chen H, et al. Knockdown of UBE2T inhibits osteosarcoma cell proliferation, migration, and invasion by suppressing the PI3K/Akt signaling pathway. Oncol Res. 2016;24:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gong YQ, Peng D, Ning XH, et al. UBE2T silencing suppresses proliferation and induces cell cycle arrest and apoptosis in bladder cancer cells. Oncol Lett. 2016;12:4485–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Guo J, Wang M, Wang J, Wu C. (2019). Ubiquitin-conjugating enzyme E2T knockdown suppresses hepatocellular tumorigenesis via inducing cell cycle arrest and apoptosis. World J Gastroenterol, 25(43), 6386–6403. 10.3748/wjg.v25.i43.6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu L, Yang M, Peng Q, Li M, Zhang Y, Guo Y, Chen Y, Bao S. (2017). UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53. Biochem Biophys Res Commun, 493(1), 20–27. 10.1016/j.bbrc.2017.09.091 [DOI] [PubMed] [Google Scholar]
- [34].Ikeda F. Ubiquitin conjugating enzymes in the regulation of the autophagy-dependent degradation pathway. Matrix Biol. 2021;100–101:23–29. [DOI] [PubMed] [Google Scholar]
- [35].Falvey CM, O’Donovan TR, El-Mashed S, et al. UBE2L6/UBCH8 and ISG15 attenuate autophagy in esophageal cancer cells. Oncotarget. 2017;8:23479–23491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Geisler S, Vollmer S, Golombek S, et al. The ubiquitin-conjugating enzymes UBE2N, UBE2L3 and UBE2D2/3 are essential for Parkin-dependent mitophagy. J Cell Sci. 2014;127:3280–3293. [DOI] [PubMed] [Google Scholar]
- [37].Kma L, Baruah TJ. The interplay of ROS and the PI3K/Akt pathway in autophagy regulation. Biotechnol Appl Biochem. 2021. DOI: 10.1002/bab.2104 [DOI] [PubMed] [Google Scholar]
- [38].Grassi G, Di Caprio G, Santangelo L, et al. Autophagy regulates hepatocyte identity and epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions promoting Snail degradation. Cell Death Dis. 2015;6:e1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Catalano M, D’Alessandro G, Lepore F, et al. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Mol Oncol. 2015;9:1612–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zada S, Hwang JS, Ahmed M, et al. Control of the epithelial-to-mesenchymal transition and cancer metastasis by autophagy-dependent SNAI1 degradation. Cells. 2019;8:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The online data used to support the findings of this study are from Oncomine (http://www.oncomine.org), GEPIA (http://gepia.cancer-pku.cn/), UALCAN (http://ualcan.path.uab.edu/), and Kaplan–Meier (K-M) plotter (http://kmplot.com/analysis/). The experimental data that support the findings of this study are available from the corresponding author, [XX], upon reasonable request.


