Abstract
Techniques for atomic-resolution structural biology evolved during the last several decades. Breakthroughs in instrumentation, sample preparation, and data analysis that occurred in the past decade have enabled characterization of viruses with unprecedented level of detail. Here we review the recent advances in magic-angle spinning (MAS) NMR spectroscopy for structural analysis of viruses and viral assemblies. MAS NMR is a powerful method that yields information on three-dimensional structures and dynamics in a broad range of experimental conditions. After a brief introduction, we discuss recent structural and functional studies of several viruses investigated with atomic resolution at various levels of structural organization, from individual domains of a membrane protein reconstituted into lipid bilayers to virus-like particles and intact viruses. We present examples showing the unique information revealed by MAS NMR about drug binding, conduction mechanisms, interactions with cellular host factors, and DNA packaging in biologically relevant environments, inaccessible by other methods.
Keywords: MAS NMR, solid-state NMR, virus assemblies, structure, capsids, dynamics
1. INTRODUCTION
The contemporary structural biology toolbox developed over the past 3–4 decades enables for a wide variety of systems to be studied with varying degrees of spatial and temporal resolution. The majority of atomic-resolution structures of biological molecules so far were determined by X-ray crystallography, while cryo-EM is considered to be a method of choice for large biological assemblies. Solution NMR spectroscopy is commonly used for determination of 3D structures and dynamics of proteins and characterization of functional interactions with binding partners. While a powerful technique, solution NMR is limited by size and solubility thus precluding characterization of large and/or insoluble systems, such as biological assemblies and intact viruses. Those kinds of systems can be studied by magic angle spinning (MAS) solid-state NMR spectroscopy, as illustrated in Figure 1. In MAS NMR, a rotor containing the solid sample is spun in the magnetic field at frequencies on the order of thousands of revolutions per second (kHz). In contrast to solution state, where narrow spectral lines are attained because molecules naturally rotate, in the specimens where fast molecular rotations are absent, e.g., large assemblies or generally any immobilized systems, mechanical rotation of the specimen in the NMR rotor during the course of an experiment is required for high spectral resolution, as discussed below. MAS NMR stands apart from other structural biology methods as it exhibits all of the following features: i) there is no requirement that specimens be soluble or crystalline; ii) there are no inherent size or molecular weight limitations, so both small molecules and megadalton-large complexes are amenable to atomic-level characterization; iii) the experiments can be performed in a wide range of experimental conditions including pH, ionic strength, concentrations, and temperatures, which can range from physiological to cryogenic; iv) information on both structure and dynamics can be obtained, with atomic resolution, on the same specimen; v) details of local environment, such as interactions with small-molecule inhibitors, lipids, solvent, and protonation states of functional residues, are readily available from the experiments. The advantages and limitations of the various atomic-resolution structural biology methods are summarized in Table 1. Given the unique capabilities of MAS NMR enumerated above, the technique is rapidly gaining popularity for structural studies of a wide range of viruses and viral protein assemblies, as summarized in Table 2.
Figure 1.
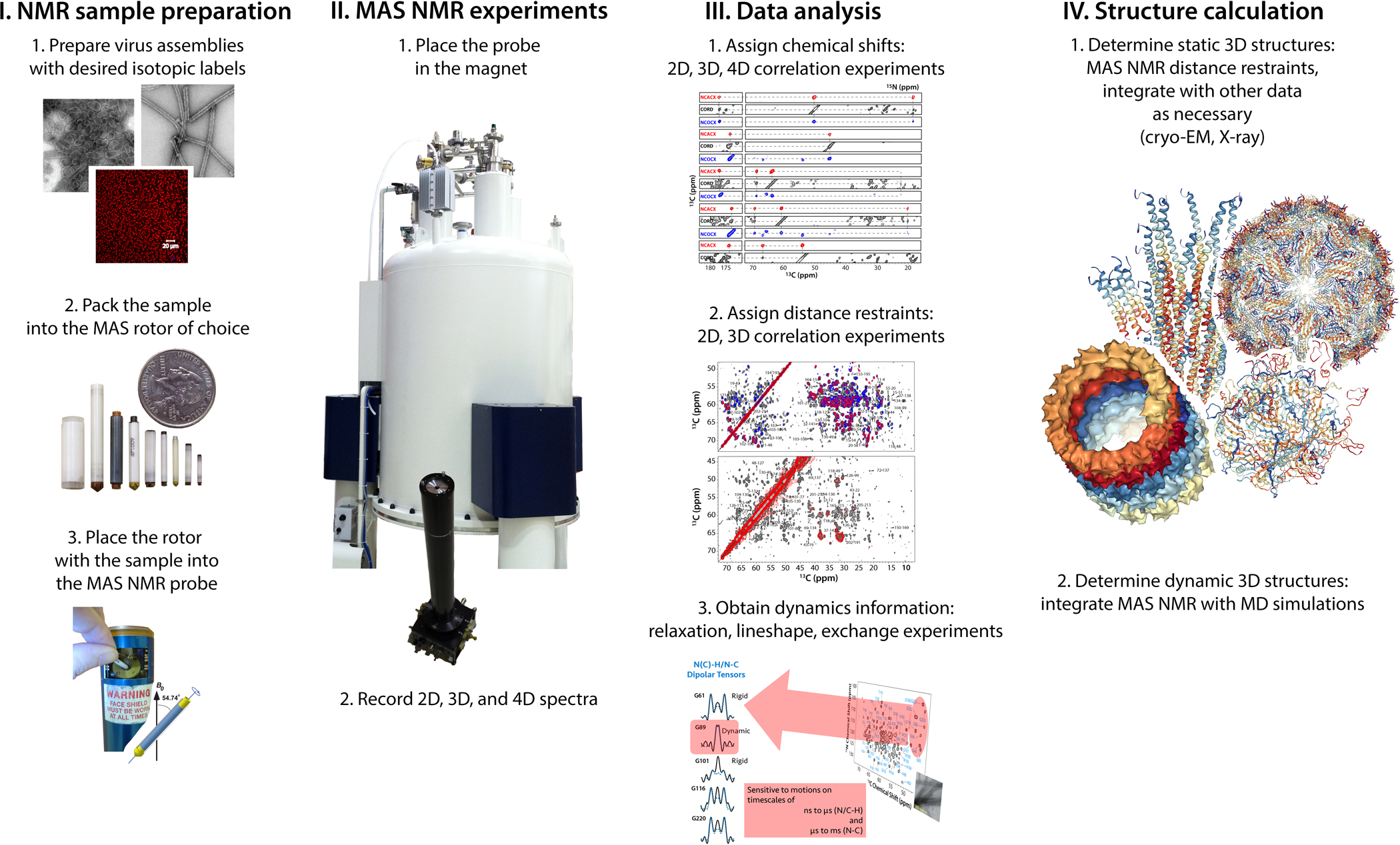
Schematic representation of the workflow for structure and dynamics investigations of viruses by MAS NMR.
Table 1.
Examples of Viruses Structurally Characterized by MAS NMR Spectroscopy, Grouped According to the Baltimore Classification (1)
| Class I | Class II | Class IV | Class V | Class VI | Class VII |
|---|---|---|---|---|---|
| T4a | Pf1a | Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)c | Measles virusb | HIV-1b,c | Hepatitis B virusb |
| T7a | fda | Hepatitis C virusb | Influenza Ac & Bc | Rous Sarcoma virus (RSV)b | |
| SPP1b | M13a | AP205b | Parainfluenza 5c |
Studied as an intact phage.
Studied as a viral protein assembly.
Studied as a viral protein embedded in lipid membranes.
Table 2.
Atomic-Resolution Techniques in Structural Biology
| X-ray crystallography | Cryo-EM | Solution NMR | MAS NMR | |
|---|---|---|---|---|
| Molecular weight requirement for atomic-resolution structural analysis | None | > 40 kDa | < 50 kDa | None |
| Specimen state | Single crystal | Vitrified | Solution, micelles | Crystalline, sedimented hydrated solutions, vitrified, powder |
| Accessibility of dynamics information | No/indirectly | No/indirectly | Yes | Yes |
| Temperature range | No limitations, determined by the sample stability | Cryogenic | Above freezing | No limitations, determined by the sample stability |
| Sample conditions (pH, salts, etc.) | Determined by crystallization conditions | No inherent limitations | No inherent limitations | No inherent limitations |
To introduce the reader to MAS NMR spectroscopy, we first present a brief overview of the following concepts and practical considerations: i) the requirement for fast mechanical rotation of the sample during the experiment; ii) the factors determining sensitivity of the measurements; iii) sample conditions; iv) information content of MAS NMR spectra. The essential steps in MAS NMR based structure characterization of viruses are illustrated in Figure 1 For the purposes of this review, we focus the most frequently studied biologically relevant nuclei, such as 1H, 13C, 15N, 31P whose nuclear spin is 1/2.
1.1. Mechanical rotation of the sample around a judiciously chosen axis, the “magic angle”, is required for high-resolution NMR spectroscopy of large biological assemblies.
The NMR frequencies measured in a typical experiment are determined by nuclear spin interactions, whose magnitudes are dependent on the orientation of the molecules comprising a sample, relative to the static magnetic field (Figure 1). In solution, rotate rapidly and isotropically (i.e., there is no preferred axis of rotation), and each atom in the specimen experiences an average field. Hence, a single NMR frequency is detected, corresponding to the average field experienced by the atom, and resulting in high spectral resolution (one narrow peak per atom). When molecules in a specimen do not rotate isotropically, such as in solid powders, crystals, large assemblies, frozen solutions, and molecules immobilized on solid supports, the NMR signals are broad (2).
If a sample is mechanically rotated around a single axis forming an angle of 54.74° with respect to the static magnetic field (dubbed “magic angle”), the orientational dependencies of the nuclear spin interactions are averaged out over a rotor period (3–5). If a sample is rotated rapidly enough, so that the spinning frequency exceeds the magnitude of the interaction, a complete averaging occurs and every atom in a specimen experiences an average field, resulting in a narrow signal, similar to solution NMR. In contrast to solution NMR, the widths of the individual peaks are independent of the molecular weight in the MAS NMR experiments. This is the basis for high-resolution MAS NMR spectroscopy of large assemblies and any specimens comprised of immobilized molecules.
MAS frequencies of at least 8–10 kHz are necessary for recording well-resolved spectra for 13C, 15N, and 31P nuclei. While this range of frequencies is sufficient as an entry point to biological MAS NMR, in practice the faster the specimen is spun, the higher the resolution and the sensitivity for the same rotor diameter. Attaining faster MAS frequencies requires NMR rotors of progressively smaller diameters and hence reduced sample amounts, as shown in Figure 1. The choice of the MAS frequency depends on the sample and experiment of interest, and the highest practically achievable MAS frequency in a commercial MAS NMR probe is currently 111 kHz.
1.2. Multiple factors determine the sensitivity of MAS NMR experiments in biological assemblies
Intrinsic sensitivity of an NMR experiment, S, can be expressed as:
where γ is the gyromagnetic ratio of the detected nucleus (a fundamental physical constant, a ratio of the magnetic moment of a nucleus to its angular momentum), B0 is the magnitude of the static magnetic field, and T is the temperature. Therefore, the higher magnetic field and the nucleus’ gyromagnetic ratio, and the lower the temperature, the higher the sensitivity. Of all stable isotopes, 1H possesses the highest gyromagnetic ratio, is nearly 100% naturally abundant and, hence, yields the highest sensitivity in NMR experiments. The sensitivity also depends on the hardware design, i.e., the spectrometer and the MAS NMR probe design (6).
In practice, MAS NMR experiments on large biological systems are usually performed at magnetic fields of 14.1 T to 23.5 T (1H Larmor frequency of 600 MHz and 1 GHz, respectively). Very recently, 28.2 T superconducting magnets (1H Larmor frequency of 1.2 GHz) have become commercially available (7). The choice of the MAS frequency and the experimental setup is determined by the type of sample under investigation, i.e., the amount and the isotopic labeling accessible for a given sample (see section 1.3), the molecular size and the number of nuclear spins that give rise to the NMR signal (in the case of isotopically diluted samples). Additionally, more considerations come into play when selecting a rotor, such as improved RF coil homogeneity of smaller rotors, available temperature range, and other sample dependent considerations.
1.3. Sample considerations for MAS NMR experiments
Since the NMR active isotopes of carbon and nitrogen, 13C and 15N, are only 1.11% and 0.37% naturally abundant, isotopic enrichment of one or both in the sample under investigation is required for in-depth structural and dynamic characterization of any biological samples. These isotopes can be introduced into viruses (8–12) or recombinantly generated viral proteins and/or nucleic acids (13–32) by adding them to the growth medium or synthetically (33, 34). Labels can be introduced either uniformly (i.e., every carbon and/or nitrogen atom will be enriched in the respective isotope) or into judiciously chosen sites (e.g., into selected amino acids types or following various sparse labeling schemes). Heterogeneous or mixed labels can also be incorporated, where each component in a complex may have a different labeling pattern or, alternatively, a single-component sample may contain differently labeled subpopulations. For example, by labeling the same protein with either 13C or 15N and mixing the two samples, one can obtain a sample in which the adjacent molecules contain different NMR active isotopes, which allows for investigation of intermolecular interactions and dynamics. As another approach, embedding a membrane protein containing one type of a label in lipid bilayers containing a different label can report on protein/membrane interactions. Deuterium (2H) labeling is also frequently used in MAS NMR studies of biological assemblies, to dilute the otherwise densely coupled proton networks, giving rise to large resolution enhancements. Alternatively, 2H itself is used to probe dynamics in proteins and oligonucleotides. For a review of the isotopic labeling schemes commonly used in MAS NMR of biological assemblies, we refer the interested reader to reference (35). Recently, 19F labeling has become popular for studies of virus assemblies (19, 36). Due to its favorable properties (very high gyromagnetic ratio, 100% natural abundance, and absence in biological systems) fluorine is an exquisitely sensitive NMR probe. Furthermore, interfluorine distances of up to 2 nm are detectable; these report on the quaternary structure of proteins in assemblies of multiple monomers (19, 36).
The sample quantities required for MAS NMR spectroscopy depend on the extent of isotopic labeling, i.e., whether the specimen is uniformly isotopically labeled or combines isotopically labeled and naturally abundant molecules or sites. Once the sample containing appropriate isotopic labels is prepared, it is centrifuged into an MAS NMR rotor, which is tightly sealed for the experiments. The specimens in the rotor are fully hydrated, their morphology is retained through the course of experiments, and they typically last through the entire duration of the measurements, which could be weeks or months (37).
1.4. Information content of MAS NMR experiments
The atomic-resolution 3D structure determination by MAS NMR relies mainly on two types of restraints: (i) backbone torsion angles calculated from chemical shifts (NMR frequencies) and more importantly, (ii) the availability of many hundreds of pairwise distance restraints between atoms in a biological assembly (38). These restraints are acquired in various types of correlation experiments, as shown in Figure 1 and reviewed in references (35, 39). While solution NMR experiments mostly correlate atoms connected by a covalent chemical bond, MAS NMR relies on through-space, dipolar interactions. These through-space correlations report uniquely on atom pairs which are close in space, but may not necessarily be linked through a covalent bond. Importantly, hydrogen atoms (1H or 2H) can be directly detected in MAS NMR experiments, providing exquisite chemical and structural details (such as hydrogen bonding, protonation, and tautomeric states), typically not accessible from X-ray or cryo-EM structures. The upper distance range practically detected in MAS NMR experiments involving 1H, 13C, and 15N nuclei is about 8 Å. Distances of up to 20 Å can be measured in 19F-19F correlation experiments.
Besides three-dimensional structures, MAS NMR can provide a wealth of structural information for viruses, over a broad range of length scales, from DNA packaging in viruses, to interactions with small molecules, all with atomic resolution. Furthermore, dynamic regions of the viruses can be uniquely investigated by MAS NMR integrated with molecular dynamics (MD) simulations. This information is available through various NMR parameters reporting on local dynamics spanning many decades of motional time scales. Furthermore, different conformers and conformational distributions can be detected directly in frozen samples (40, 41).
2. EXAMPLES OF VIRAL SYSTEMS STUDIED BY MAS NMR
In this section we discuss viral systems investigated by MAS NMR so far: membrane protein oligomers of Influenza and SARS-CoV-2 viruses, HIV-1 capsid protein assemblies, a Bacillus phage SPP1 flexible tail-tube assembly, Acinetobacter phage A205 virus like particles (VLPs), and intact filamentous bacteriophages Pf1, fd, and M13. We highlight the role of MAS NMR in yielding information into three-dimensional structure, functionally important dynamics and mechanisms, as well as interactions with cofactors and ssDNA packaging, inaccessible by other means.
2.1. Influenza A and B
The Orthomyxoviridae family viruses Influenza A and B cause seasonal flu. In the United States alone, the Center of Disease Control and Prevention (CDC) estimates a death toll rate between 12,000 and 79,000 individuals during every Influenza season since 2010 (42). Since a universal flu vaccine targeting all strains is not yet available, research focuses heavily on antiviral medication development. In the last quarter of the 20th century, two antiviral compounds were used to treat Influenza A infections, Amantadine (AMT) and Rimantadine (RMT). Unfortunately, most circulating Influenza A strains have evolved AMT and RMT resistance, rendering them ineffective.
AMT and RMT inhibit Influenza A replication by blocking the M2 transmembrane channel (AM2), but M2 proteins of low sequence homology and similar structure and function exist in Influenza B strains as well (BM2). M2 exclusively transfers protons into the virion under acidic conditions. The function of the channel is essential for both cell entry and maturation of the virus in the infected cell Golgi membrane. Blocking M2 severely halts virus replication. M2 oligomerizes into a four-helix bundle, with each subunit passing through the membrane once. Its transmembrane domain (TM) makes up the proton-selective channel, dependent on HxxxW motif (residues His-37 and Trp-41) that was found to be essential for proton selectivity (43, 44). M2 studies by MAS NMR allowed researchers to exploit membrane-mimetic environments to gain insights into the conduction mechanism and organization of the channel.
The conduction mechanism of AM2 was investigated by MAS NMR using His-37 as a probe of dynamics, protonation state, tautomerization (the process of low-barrier conversion from one structural isomer to another), and hydrogen bonding (23–25, 45–48). Two possible conduction models involve different histidine tautomers and charges and different hydrogen bonding partners (Figure 2c). The low-barrier hydrogen bond model suggests two histidines in the pore form a hydrogen bond. According to this model, at low pH hydronium ion protonates one of these residues, thus breaking the hydrogen bond. One of the protonated histidines then loses its proton to the less acidic viral interior, completing the proton transfer and reforming the inter-histidine hydrogen bond. Another model suggests a proton shuttling mechanism. Histidine’s sidechain is protonated in the acidic environment, flips, and protonates a water molecule in the viral interior. This model does not involve any interaction between adjacent histidine sidechains in the pore. The two models can be differentiated by MAS NMR through histidine chemical shifts, as well as by probing whether a hydrogen bond exists between two adjacent histidine sidechains.
Figure 2.
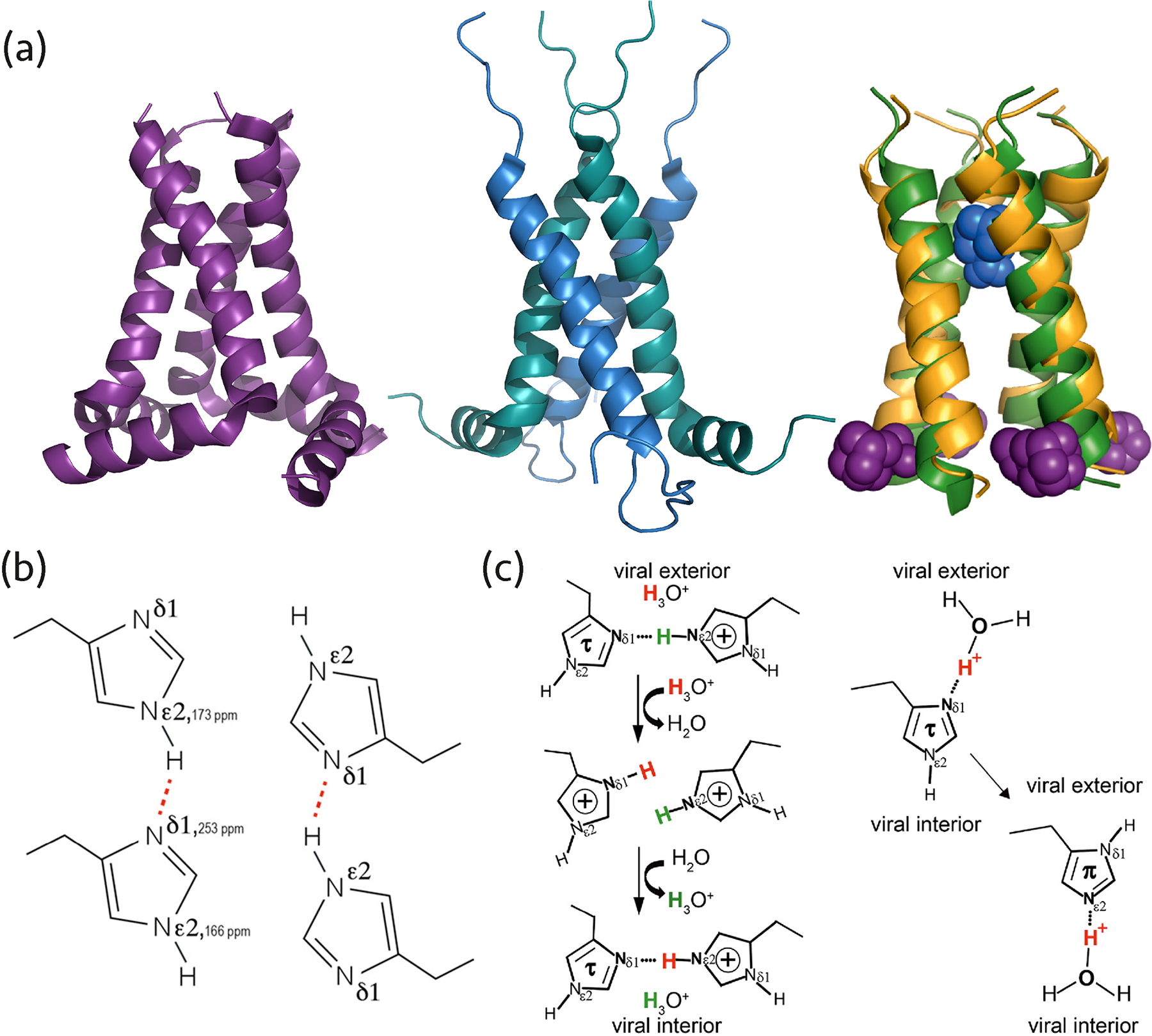
(a) AM2 structures solved by MAS NMR: left, homotetramer (PDB: 2L0J), middle, dimer of dimers (PDB: 2N70), right, superposition of AMT-bound AM2 (PDB: 2KQT) and RMT-bound AM2 (PDB: 2RLF) structures determined by MAS NMR (colored yellow/blue) and solution NMR (colored green/purple). (b) The dimeric arrangement of His-37 in the AM2 pore occurring due to hydrogen bonding, corresponding to the dimer-of-dimers organization of the M2 helices. Reprinted with permission from reference (24). Copyright 2020 American Chemical Society. (c) Two proton conduction models for AM2 His-37 involve different histidine tautomers, protonation states, and hydrogen bonds. One model entails hydrogen bond formation between two histidine residues (left), the other suggests proton shuttling by histidine tautomerization and hydrogen bonding with hydronium ions (right). Reprinted with permission from reference (25). Copyright 2020 American Chemical Society.
The imidazole-imidazolium and imidazole-imidazole hydrogen bonds of AM2 His-37 were probed at a low and high pH, respectively (24, 25). At low pH, the existence of a hydrogen bond was detected, suggesting a low-barrier hydrogen bond mechanism is operational (25). At high pH, increased dynamics of the histidine residues in the pore and breaking of the hydrogen bond were observed. Both also play a role in the RMT inhibitory mechanism, as discussed below (24).
Several groups also investigated the mechanism of AM2 inhibition by AMT and RMT using MAS NMR (21, 49–55). Two binding sites were identified, one inside the channel pore from the N-terminal side and the other at the cytoplasmic side of the pore (Figure 2a). Based on the data, an allosteric mechanism of inhibition was proposed (53, 54). Binding to the latter site was only observed at high inhibitor concentrations (21) or at cryogenic temperatures, by dynamic nuclear polarization (DNP) (55). Binding to the site within the channel pore was found to be more important for inhibition.
Interestingly, MAS NMR studies of AM2 and the drug-resistant mutant S31N AM2 in DPhPC (diphytanoyl-snglycero-3-phosphocholinestudies) bilayers revealed a dimer-of-dimers organization (20, 56), contrary to the homotetrameric arrangements found by solution NMR and X-ray diffraction (57, 58). Homotetrameric structure of AM2 was also observed in 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and other virus-mimetic lipid bilayers. The organization of the channel may be linked to the conduction mechanism, see above and Figure 2. Recent studies showed the full-length AM2 fold is affected by membrane composition, including cholesterol content, and drug binding (50, 59), possibly explaining that the channel can form different oligomeric structures depending on the lipid mixture used for the reconstitution (60).
MAS NMR studies of BM2 revealed a different proton conduction mechanism from the one found for AM2 (13). While AM2 was found to almost exclusively transfer protons inside the virion, BM2 allows bidirectional proton transfer (with yet stronger proton flow inside the virion than outside). Interhelical distance restraints from a mixture of fluorinated phenylalanine and 13C-labeled proteins aided in the determination of the BM2’s quaternary structure, in both open and closed states, at different pH. Interetingly, differences in pore volume were found between the two states. Furthermore, in contrast to the single histidine residue in AM2 TM helix, BM2 has an additional histidine (His-27) following the HxxxW motif (His-19 and Trp-23), forming HxxxWxxxH motif. MAS NMR revealed that the H27A mutant BM2 loses bidirectionality of proton transfer, resembling the AM2 conduction mechanism (61). Whether the functionally important histidine residues of BM2 form dimers through hydrogen-bonds as observed in AM2 His-37 remains an open question.
2.2. SARS-CoV-2
Viruses of the Coronaviridae family encode 4–5 structural proteins, making up about third of their ssRNA. The envelope (E) protein, the smallest Coronaviridae structural protein, is a homopentamer viroporin. Its functional roles and the ability of other non-structural proteins to compensate for its deletion from recombinant virions are under investigation (62). The deletion of E protein attenuated the growth of severe acute respiratory syndrome coronavirus (SARS-CoV) (63). Therefore, the pentameric E protein channel is a possible drug target.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for the COVID-19 pandemic, has been under intense investigation by various structural biology methods, including MAS NMR. Most recently, the E TM structure was solved in lipid bilayers (19).
The 3D structure revealed the backbone conformation, water accessibility, and drug binding. Exploiting the E protein pentameric conformation, differentially labeled samples were prepared, where 13C-labeled E protein monomers were mixed with fluorinated-phenylalanine labeled or 15N-labeled monomers. These preparations allowed for the collection of 87 13C-19F and 13C-15N interhelical distance restraints and permitted the calculation of the 3D structure of the pentamer.
Three phenylalanine residues in the middle of each chain appear to stack in the same helix and between helices, causing the formation of a non-ideal alpha-helical structure. This is in contrast to the structure reconstructed from solution-state NMR data for the SARS-CoV E protein in micelles (64). MAS NMR water accessibility experiments revealed a dehydrated pore, which agrees with the tight packing of the helices as indicated by the distance restraints based structure (19). The structure most likely resembles the channel’s closed-state due to observed minimal pore diameter of 2 Å.
The same study also characterized drug binding to the E protein. Chemical shift perturbation analysis revealed that hexamethylene amiloride and AMT, which inhibit the E protein of SARS-CoV (65, 66), bind to the N-terminus of the SARS-CoV-2 E protein’s TM helix, providing clues for future inhibitor design.
2.3. HIV-1
The Retroviridae HIV type 1 (HIV-1) is the causative agent of AIDS and the associated global pandemic that claimed more than 35 million lives so far. HIV-1 protein assemblies have been studied by several MAS NMR research groups focusing on the Gag polyprotein and mature capsid protein (CA) assemblies (15, 28, 36, 41, 67–81), viral protein “U” (Vpu) (18, 82–84), and glycoprotein 41 (gp41) (27, 85–87).
Mature HIV-1 conical capsid, which contains the viral genome, is assembled from 1000–1500 individual capsid CA proteins and plays multiple important roles in HIV-1 replication. Most of the MAS NMR studies to date focused on in-vitro CA assemblies of tubular morphologies, whose hexagonal lattice recapitulates the predominant symmetry of the mature cone-shaped capsid. MAS NMR investigations revealed information about structural details and dynamics of tubular CA assemblies (28, 41, 67, 69–72, 74, 76–78, 80, 81) inaccessible by other techniques. Integrated MAS NMR and MD investigations revealed that CA capsid is remarkably dynamic, with motions occurring over many timescales and being important for the HIV-1 maturation and capsid assembly (28, 71, 76). Recently, the atomic-resolution structure of CA tubular assemblies was determined by MAS NMR, in combination with low-resolution cryo-EM and data-guided MD simulations (Figure 3) (15). Notable differences from the crystallographic structure of flat hexamers were found and novel and unique atomic resolution information on flexible and functionally important regions of the capsid, that are inaccessible by other structural techniques, was obtained. Such regions comprise the cyclophilin A (CypA) binding loop, the interdomain linker and residues lining the hexamer pore (Figure 3c–e). These structural data can be exploited in complementary capsid-targeted therapeutic strategies.
Figure 3.
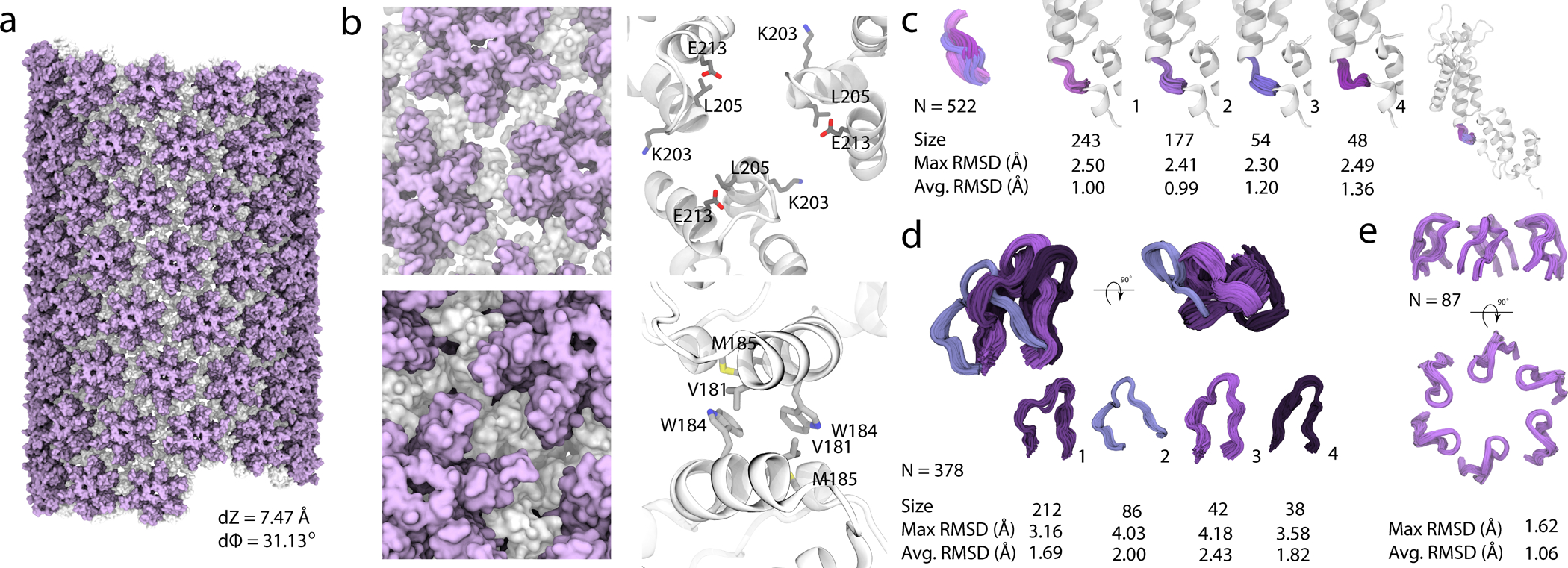
Structure of an HIV-1 CA tube generated by data-guided molecular dynamics. A, Molecular surface of a (−12,11) helical symmetry tube (helical parameters dZ = 7.47 Å and dϕ = 31.13°). The N- and C-terminal domains are colored purple and gray, respectively. B, Representative trimer and dimer interfaces, top and bottom, respectively, in surface (left) and ribbon (right) representation. K203, L205 and E213 side chains (trimer interface) and V181, W184 and M185 sidechains (dimer interface) are shown in ball-and-stick representations. C, Four representative conformations of the flexible linker region, colored and grouped by structural cluster. D, Four representative conformations of the CypA binding loop, colored and grouped by structural cluster. The size of each cluster and the RMSD within each cluster (Å), are listed. E, Arrangement of the β-hairpins in the hexamers. Reprinted with permission from reference (15). Copyright 2020 Springer Nature.
Several studies shed light on the interactions of CA assemblies with cellular host factors CypA (71, 76) and TRIM5α (67). CypA regulates viral infectivity by interacting directly with the capsid, and can either promote or inhibit viral infection, depending on host cell type and CA protein sequence. A MAS NMR investigation uncovered that capsid’s escape from CypA dependence is regulated allosterically through the modulation of the dynamics of the CypA-binding loop (76). Remarkably, the loop motions are greatly attenuated in the CypA escape mutants, A92E and G94D compared to the wildtype. Similarly, dynamic allosteric regulation was found to be critical for the interactions of capsid with TRIM5α (67). This restriction factor recognizes the capsid’s lattice and triggers an innate immune response causing capsid’s premature uncoating (88, 89). Binding of TRIM5α to capsid was found to induce global rigidification and perturb key intermolecular interfaces involved in higher-order capsid assembly. Remarkably, structural and dynamic changes occur throughout the entire CA in the assembly (67).
While CA assemblies recapitulating mature capsid have been the focus of multiple MAS NMR studies, Gag maturation intermediates and VLPs have also been investigated (28, 77, 80). Given the size of Gag polyprotein, its NMR characterization is challenging with uniform 13C,15N labeling. Segmental labeling was therefore explored to simplify the chemical shift assignments (68, 80). Therein, two segments of a single protein with different labels are individually expressed. The two segments are then purified and reattached using intein technology to form the full-length protein (90). Structural investigation of the CA-SP1 junction helices bound to the maturation inhibitor Bevirimat in HIV-1 VLPs showed a slight structural alteration that is apparently sufficient to inhibit the CA-SP1 cleavage (80).
2.4. Bacillus Phage SPP1 Flexible Tail Tube
Siphoviridae is a family of dsDNA viruses with an icosahedral nucleocapsid and a long flexible tail. The Siphoviridae SPP1 phage tail consists of protein gp17.1 and its translational frameshift, gp17.1*, in a ratio of 3:1. Nonetheless, gp17.1 can polymerize in solution and forms the tail tube even in absence of gp17.1*. This modification was shown not to affect phage assembly and infectivity, thus well representing the tail-tube assembly structure and function in vivo. While structures of phage tails were previously reconstructed by cryo-EM in a resolution of 3–4 Å, the flexibility of the Siphoviridae tail still poses a challenge, limiting the structural information available from cryo-EM.
To reveal the atomic-resolution structure of the Siphoviridae SPP1 phage flexible tail tube assembly, Lange and coworkers expressed, purified, and polymerized recombinant gp17.1 for MAS NMR investigations (14, 91, 92). The final calculated structure, consisting of 12 subunits, was refined using both NMR-derived distance restraints and a cryo-EM density map. The 12 subunits represent two rings of the tail tube assembly (Figure 4a–c). The structure revealed that the C-arm domain interacts with the suprajacent subunit to obscure hydrophobic chains of that subunit, and the loop region of one subunit participates in electrostatic interactions with five other subunits (Figure 4d–g).
Figure 4.
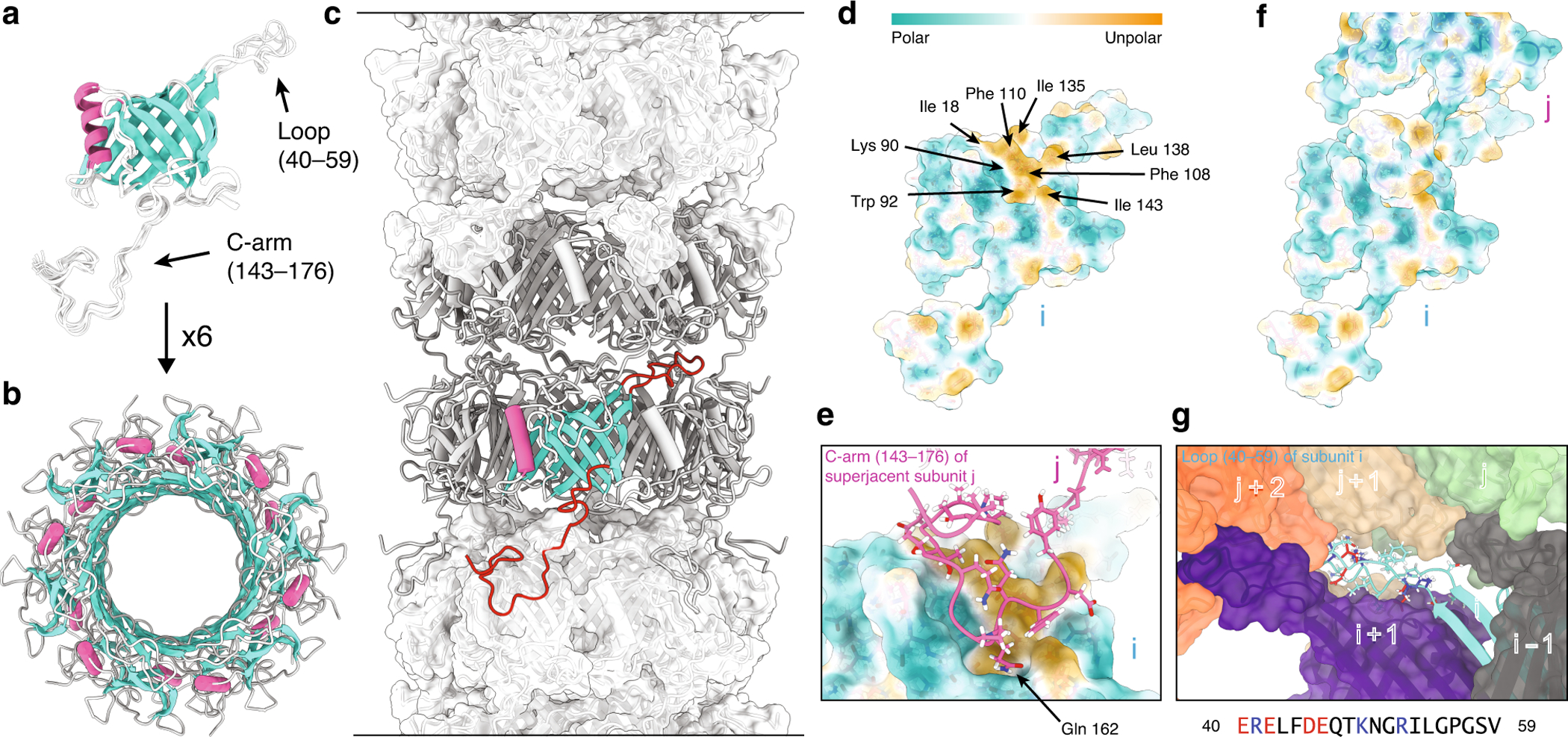
(a) Final ten lowest-energy structures of a gp17.1 subunit which consist of a central β-sandwich-type fold (turquoise) that is flanked by an α-helix (pink), a large loop and an extended C-terminal arm (C-arm). (b) Six gp17.1 monomers form a hexameric ring. The inner β-sheets of the β-sandwiches organize in a β-barrel motif that forms the lumen of the tube. (c) These hexameric rings stack onto each other in a helical fashion creating a hollow tube. Ring-to-ring contacts are mediated by the two loop regions (highlighted in red)—especially by the C-arm that folds onto the subjacent ring. (d) The molecular lipophilicity potential of gp17.1 reveals a hydrophobic patch on the surface of one subunit i. The color gradient represents the lipophilicity potential. (e, f) This unpolar area is obscured by the C-arm (pink) of the superjacent subunit j within the complex of the tail-tube—by anchoring the sidechain of Gln-162 into a pocket. (g) The loop of subunit I (turquoise) features mostly electrostatic interactions with five neighboring subunits (gray, purple, green, beige, and orange) within the complex. Charged amino acids are colored in red (negative) and blue (positive). The direction of the tail structure is baseplate upwards. Reprinted with permission from reference (14). Copyright 2020 Springer Nature.
2.5. Acinetobacter Phage AP205 Virus-like Particles
VLPs of ssRNA bacteriophages, such as AP205, are targets for antigen display vaccine development against cancer, viral infections, and more (93–98). Such VLPs are promising platforms for vaccine development, mostly due to their safety, relatively small size, and repetitive structure (99). AP205 has low sequence similarity with other ssRNA phages, and structural knowledge was lacking. To solve the structure of the entire AP205 VLP, MAS NMR and cryo-EM were combined (17). The available state-of-the-art MAS NMR hardware enabled the use of sub-milligram quantities of AP205 VLP crystals, instead of the routinely required yield of isotopically labeled sample on the order of tens of milligrams. Combining the 1.7-Å NMR-derived model of unassembled dimers with the cryo-EM low-resolution reconstruction yielded a high-resolution structure of the full AP205 capsid (Figure 5). Interestingly, comparison of the ssRNA phages MS2 and AP205 coat protein revealed a similar fold despite a change in the order of amino acids in the sequence.
Figure 5.
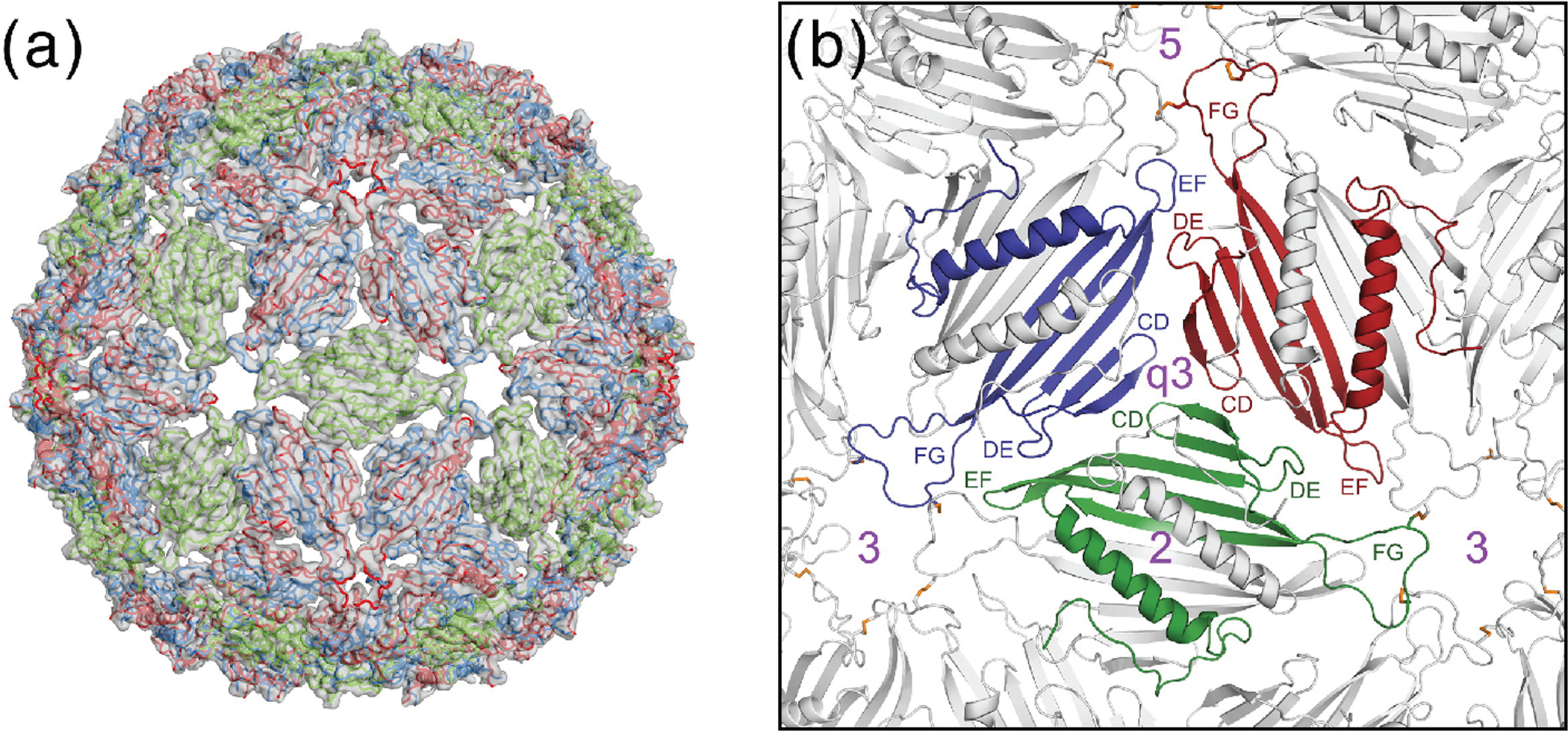
(a) The complete AP205 capsid model as derived from the low-resolution cryo-EM electron density map and MAS NMR derived coat protein dimer model. (b) A close-up look of interfaces between the coat protein dimers. Reprinted with permission from reference (17). Copyright 2016 Elsevier.
2.6. Intact Filamentous Bacteriophages
Filamentous bacteriophages are composed of a coat protein, which usually constitutes a major fraction of the virus. The number of copies of the coat proteins is three orders of magnitude larger than that of other structural and non-structural proteins in the virion. Such a large excess of the coat protein allows for MAS NMR studies of intact filamentous phages instead of the recombinantly expressed coat protein because other proteins are virtually invisible in the NMR spectra due to their low amounts. Several intact filamentous bacteriophages have been investigated by MAS NMR to date: Pf1 (8, 100–105), fd (10, 106–108), M13 (12, 106, 108), and fIKe (109).
The Pf1 coat protein was amongst the first proteins to be studied by MAS NMR and the first for which chemical shift assignments were reported in an intact virion (8) (Figure 6a). The results unequivocally revealed that all coat protein monomers adopt the same structure, in conflict with previous X-ray fiber diffraction model, where three distinct coat protein folds appeared to co-exist at high temperature structure of the virion. MAS NMR provided insights regarding the hydration of the different residues of the coat protein (101).
Figure 6.
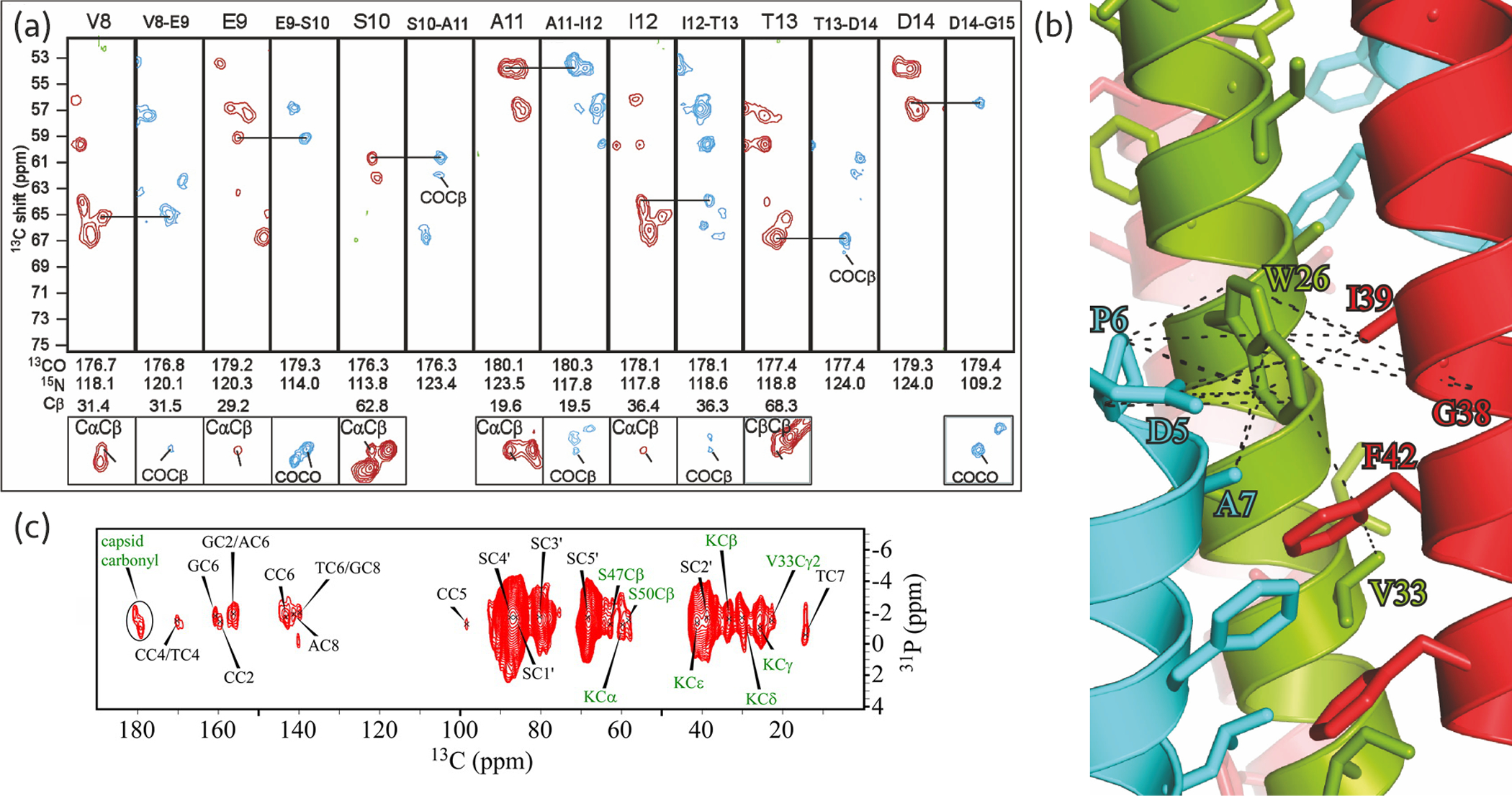
(a) Strip plot of the chemical shift assignment of the Pf1 coat protein in intact bacteriophages. NCOCX (blue) and NCACX (red) correlation spectra were used for sequential and intra-residue assignment, respectively. Adapted with permission from reference (8). Copyright 2007 American Chemical Society. (b) Tryptophan binding pocket of the M13 NMR-Rosetta model. Dashed lines represent distance restraints between tryptophan and other residues in the same chain (green) and adjacent chains (cyan and red) in the capsid. Reprinted with permission from reference (12). Copyright 2015 National Academy of Sciences. (c) 31P-13C through-space correlation spectrum of 13C- and 15N-labeled and aromatic-unlabeled intact fd phage. Green and black labels indicate coat protein and DNA bases correlations, respectively. Reprinted with permission from reference (107). Copyright 2014 American Chemical Society.
Additional DNP-based studies conducted at 100 K characterized ssDNA packaging in the Pf1 virion (102). The observed chemical shifts values indicate the ssDNA in the virion is devoid of hydrogen bonds and is relatively well-structured. Through-space correlation spectra showed that Tyr-40 is in close contact to the ssDNA inside the phage, possibly due to stacking of the bases with its aromatic sidechain. This study provided strong experimental evidence to corroborate the previous Pf1 ssDNA packaging model (110, 111), suggesting a stretched ssDNA strand with its bases facing outward.
The coat proteins of fd and M13 phages were also studied and chemical shifts were reported based on a combination of 2D and 3D experiments (10, 108). The coat proteins of the two phages only differ by a single mutation, and chemical shift perturbation and rigidity analysis revealed interesting atomic-level structural differences around the mutation sites. For example, the single D12N mutation of the fd coat protein caused perturbations to residue Lys-8 due to the removal of the negatively charged Asn-12 residue.
An M13 NMR-Rosetta model was built using interchain restraints obtained from the 13C-13C correlation spectra (12). This model revealed that each subunit hydrophobically interacts with four adjacent monomers in the capsid and these interactions mostly depend on the conserved tryptophan residue and other tyrosine and phenylalanine residues in the coat protein (Figure 6b).
Nucleotide-specific assignment of the fd phage, together with 31P correlation experiments, enabled characterization of its ssDNA packaging (107) (Figure 6c). An additional sample with unlabeled aromatic residues yielded the assignment of the ssDNA carbon nuclei that highly overlap with aromatic signals in the spectrum. Prior assignment of the coat protein assisted in identification of ssDNA base-, sugar-, and phosphate-capsid interactions. From the observed chemical shifts and correlations, as well as the previous models, the salient ssDNA packaging characteristics were inferred. Notable differences with respect to Pf1 model were observed, such as base pairing, and distinct ssDNA packaging. Of note, the virus remained infectious after being taken out of the MAS NMR rotor, which confirmed the biological relevance of the structural observations.
3. CONCLUSIONS AND OUTLOOK
We have reviewed the basic principles and applications of MAS NMR spectroscopy to study structure and dynamics of viruses. The remarkable progress in ultrahigh field superconducting magnet technology and the advent of high-frequency (60–150 kHz) MAS probes have opened doors for characterization of nanomole quantities of large viral assemblies and intact viruses. MAS NMR yields information inaccessible by other techniques, such as residue-specific drug-protein interactions, detailed DNA packaging characterization, specific hydrogen bonding, and functional dynamics, with atomic resolution. The recent notable advances also include integration of MAS NMR with other methods, such as MD simulations and/or cryo-EM. Further breakthroughs in experimental and data analysis protocols will enable application of MAS NMR to an even broader range of viruses.
ACKNOWLEDGMENTS
The authors acknowledge the support of the US-Israel Binational Science Foundation (Grant 2011077). TP acknowledges the support of the National Institutes of Health (NIH Grant P50AI1504817).
Terms and Definitions
- Gyromagnetic ratio
a property unique to particles possessing a magnetic spin; dictating how strong it is affected by a magnetic field
- Heterogenous labeling
mixing molecules that were produced by different isotopic labeling method to one sample (e.g., 50/50% 13C/15N-labeled proteins)
- Tautomerization
the process in which a molecule is interconverted to another structural isomer (e.g., proton transfer in histidine sidechain), where the interconversion barrier is low
- Chemical shift perturbation
a technique for assessing structural or electronic changes of a molecule caused by another molecule or change of experimental conditions
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Baltimore D 1971. Expression of animal virus genomes. Bacteriol. Rev 35: 235–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloembergen N, Rowland TJ. 1953. On the nuclear magnetic resonance in metals and alloys. Acta Metall. 1: 731–46 [Google Scholar]
- 3.Andrew ER, Bradbury A, Eades RG. 1958. Nuclear magnetic resonance spectra from a crystal rotated at high speed. Nature 182: 1659 [Google Scholar]
- 4.Lowe IJ. 1959. Free induction decays of rotating solids. Phys. Rev. Lett 2: 285–87 [Google Scholar]
- 5.Andrew ER, Bradbury A, Eades RG. 1959. Removal of dipolar broadening of nuclear magnetic resonance spectra of solids by specimen rotation. Nature 183: 1802–03 [Google Scholar]
- 6.Hoult DI, Richards RE. 1976. The signal-to-noise ratio of the nuclear magnetic resonance experiment. J. Magn. Reson 24: 71–85 [DOI] [PubMed] [Google Scholar]
- 7.Schwalbe H 2017. Editorial: new 1.2 GHz NMR spectrometers— new horizons? Angew. Chem. Int. Ed 56: 10252–53 [DOI] [PubMed] [Google Scholar]
- 8.Goldbourt A, Gross BJ, Day LA, McDermott AE. 2007. Filamentous phage studied by magic-angle spinning NMR: resonance assignment and secondary structure of the coat protein in Pf1. J. Am. Chem. Soc 129: 2338–44 [DOI] [PubMed] [Google Scholar]
- 9.Yu T-Y, Schaefer J 2008. REDOR NMR characterization of DNA packaging in bacteriophage T4. J. Mol. Biol 382: 1031–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abramov G, Morag O, Goldbourt A. 2011. Magic-angle spinning NMR of a class I filamentous bacteriophage virus. J. Phys. Chem. B 115: 9671–80 [DOI] [PubMed] [Google Scholar]
- 11.Abramov G, Goldbourt A. 2014. Nucleotide-type chemical shift assignment of the encapsulated 40 kbp dsDNA in intact bacteriophage T7 by MAS solid-state NMR. J. Biomol. NMR 59: 219–30 [DOI] [PubMed] [Google Scholar]
- 12.Morag O, Sgourakis NG, Baker D, Goldbourt A. 2015. The NMR–Rosetta capsid model of M13 bacteriophage reveals a quadrupled hydrophobic packing epitope. Proc. Natl. Acad. Sci. U.S.A 112: 971–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandala VS, Loftis AR, Shcherbakov AA, Pentelute BL, Hong M. 2020. Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism. Nat. Struct. Mol. Biol 27: 160–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zinke M, Sachowsky KAA, Öster C, Zinn-Justin S, Ravelli R, et al. 2020. Architecture of the flexible tail tube of bacteriophage SPP1. Nat. Commun 11: 5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu M, Russell RW, Bryer AJ, Quinn CM, Hou G, et al. 2020. Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR. Nat. Struct. Mol. Biol 27: 863–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeon J, Qiao X, Hung I, Mitra AK, Desfosses A, et al. 2017. Structural model of the tubular assembly of the Rous Sarcoma virus capsid protein. J. Am. Chem. Soc 139: 2006–13 [DOI] [PubMed] [Google Scholar]
- 17.Shishovs M, Rumnieks J, Diebolder C, Jaudzems K, Andreas LB, et al. 2016. Structure of AP205 coat protein reveals circular permutation in ssRNA bacteriophages. J. Mol. Biol 428: 4267–79 [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Lin EC, Das BB, Tian Y, Opella SJ. 2015. Structural determination of virus protein U from HIV-1 by NMR in membrane environments. Biochim. Biophys. Acta Biomembr 1848: 3007–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A, Hong M. 2020. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol 27: 1202–08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreas LB, Reese M, Eddy MT, Gelev V, Ni QZ, et al. 2015. Structure and mechanism of the influenza A M218–60 dimer of dimers. J. Am. Chem. Soc 137: 14877–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, DeGrado WF, Hong M. 2010. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 463: 689–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M, Yao H, Kwon B, Waring AJ, Ruchala P, et al. 2018. Conformation and trimer association of the transmembrane domain of the parainfluenza virus fusion protein in lipid bilayers from solid-state NMR: insights into the sequence determinants of trimer structure and fusion activity. J. Mol. Biol 430: 695–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu F, Luo W, Hong M. 2010. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science 330: 505–08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Movellan KT, Wegstroth M, Overkamp K, Leonov A, Becker S, Andreas LB. 2020. Imidazole–imidazole hydrogen bonding in the pH-sensing histidine side chains of influenza A M2. J. Am. Chem. Soc 142: 2704–08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu R, Miao Y, Qin H, Cross TA. 2020. Observation of the imidazole-imidazolium hydrogen bonds responsible for selective proton conductance in the influenza A M2 channel. J. Am. Chem. Soc 142: 2115–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovacs FA, Cross TA. 1997. Transmembrane four-helix bundle of influenza A M2 protein channel: structural implications from helix tilt and orientation. Biophys. J 73: 2511–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sackett K, Nethercott MJ, Zheng Z, Weliky DP. 2014. Solid-state NMR spectroscopy of the HIV gp41 membrane fusion protein supports intermolecular antiparallel β sheet fusion peptide structure in the final six-helix bundle state. J. Mol. Biol 426: 1077–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayro MJ, Ganser-Pornillos BK, Zadrozny KK, Yeager M, Tycko R. 2016. Helical conformation in the CA-SP1 junction of the immature HIV-1 lattice determined from solid-state NMR of virus-like particles. J. Am. Chem. Soc 138: 12029–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchanka A, Simon B, Althoff-Ospelt G, Carlomagno T. 2015. RNA structure determination by solid-state NMR spectroscopy. Nat. Commun 6: 7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchanka A, Stanek J, Pintacuda G, Carlomagno T. 2018. Rapid access to RNA resonances by proton-detected solid-state NMR at >100 kHz MAS. Chem. Commun 54: 8972–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asami S, Rakwalska-Bange M, Carlomagno T, Reif B. 2013. Protein–RNA interfaces probed by 1H-detected MAS solid-state NMR spectroscopy. Angew. Chem. Int. Ed 52: 2345–49 [DOI] [PubMed] [Google Scholar]
- 32.Lacabanne D, Boudet J, Malär AA, Wu P, Cadalbert R, et al. 2020. Protein side-chain–DNA contacts probed by fast magic-angle spinning NMR. J. Phys. Chem. B 124: 11089–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Fogeron ML, Schledorn M, Dujardin M, Penzel S, et al. 2019. Combining cell-free protein synthesis and NMR into a tool to study capsid assembly modulation. Front. Mol. Biosci 6: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fogeron M-L, Jirasko V, Penzel S, Paul D, Montserret R, et al. 2016. Cell-free expression, purification, and membrane reconstitution for NMR studies of the nonstructural protein 4B from hepatitis C virus. J. Biomol. NMR 65: 87–98 [DOI] [PubMed] [Google Scholar]
- 35.Kraus J, Sarkar S, Quinn CM, Polenova T. 2021. Solid-state NMR spectroscopy of microcrystalline proteins. Annu. Rep. NMR Spectrosc 102: 81–151 [Google Scholar]
- 36.Wang M, Lu M, Fritz MP, Quinn CM, Byeon I-JL, et al. 2018. Fast magic-angle spinning 19F NMR spectroscopy of HIV-1 capsid protein assemblies. Angew. Chem. Int. Ed 57: 16375–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiegand T, Lacabanne D, Torosyan A, Boudet J, Cadalbert R, et al. 2020. Sedimentation yields long-term stable protein samples as shown by solid-state NMR. Front. Mol. Biosci 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell RW, Fritz MP, Kraus J, Quinn CM, Polenova T, Gronenborn AM. 2019. Accuracy and precision of protein structures determined by magic angle spinning NMR spectroscopy: for some ‘with a little help from a friend’. J. Biomol. NMR 73: 333–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinn CM, Polenova T. 2017. Structural biology of supramolecular assemblies by magic-angle spinning NMR spectroscopy. Q. Rev. Biophys 50: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Havlin RH, Tycko R. 2005. Probing site-specific conformational distributions in protein folding with solid-state NMR. Proc. Natl. Acad. Sci. U.S.A 102: 3284–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta R, Zhang H, Lu M, Hou G, Caporini M, et al. 2019. Dynamic nuclear polarization magic-angle spinning nuclear magnetic resonance combined with molecular dynamics simulations permits detection of order and disorder in viral assemblies. J. Phys. Chem. B 123: 5048–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Centers for Disease Control and Prevention (CDC), National Center for Immunization and Respiratory Diseases (NCIRD). 2020. Past seasons estimated influenza disease burden. https://www.cdc.gov/flu/about/burden/past-seasons.html
- 43.Wang C, Lamb RA, Pinto LH. 1995. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys. J 69: 1363–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Y, Zaitseva F, Lamb RA, Pinto LH. 2002. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J. Biol. Chem 277: 39880–86 [DOI] [PubMed] [Google Scholar]
- 45.Sharma M, Yi M, Dong H, Qin H, Peterson E, et al. 2010. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science 330: 509–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong M, Fritzsching KJ, Williams JK. 2012. Hydrogen-bonding partner of the proton-conducting histidine in the influenza M2 proton channel revealed from 1H chemical shifts. J. Am. Chem. Soc 134: 14753–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu F, Schmidt-Rohr K, Hong M. 2012. NMR detection of pH-dependent histidine–water proton exchange reveals the conduction mechanism of a transmembrane proton channel. J. Am. Chem. Soc 134: 3703–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colvin MT, Andreas LB, Chou JJ, Griffin RG. 2014. Proton association constants of his 37 in the influenza-A M218–60 dimer-of-dimers. Biochemistry 53: 5987–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tzitzoglaki C, Wright A, Freudenberger K, Hoffmann A, Tietjen I, et al. 2017. Binding and proton blockage by amantadine variants of the influenza M2WT and M2S31N Explained. J. Med. Chem 60: 1716–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ekanayake EV, Fu R, Cross Timothy A. 2016. Structural influences: cholesterol, drug, and proton binding to full-length influenza A M2 protein. Biophys. J 110: 1391–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams JK, Tietze D, Wang J, Wu Y, DeGrado WF, Hong M. 2013. Drug-induced conformational and dynamical changes of the S31N mutant of the influenza M2 proton channel investigated by solid-state NMR. J. Am. Chem. Soc 135: 9885–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cady SD, Mishanina TV, Hong M. 2009. Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of ser31 in amantadine binding. J. Mol. Biol 385: 1127–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andreas LB, Eddy MT, Pielak RM, Chou J, Griffin RG. 2010. Magic angle spinning NMR investigation of influenza A M218−60: support for an allosteric mechanism of inhibition. J. Am. Chem. Soc 132: 10958–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cady SD, Hong M. 2008. Amantadine-induced conformational and dynamical changes of the influenza M2 transmembrane proton channel. Proc. Natl. Acad. Sci. U.S.A 105: 1483–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andreas LB, Barnes AB, Corzilius B, Chou JJ, Miller EA, et al. 2013. Dynamic nuclear polarization study of inhibitor binding to the M218–60 proton transporter from influenza A. Biochemistry 52: 2774–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andreas LB, Eddy MT, Chou JJ, Griffin RG. 2012. Magic-angle-spinning NMR of the drug resistant S31N M2 proton transporter from influenza A. J. Am. Chem. Soc 134: 7215–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnell JR, Chou JJ. 2008. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451: 591–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, et al. 2008. Structural basis for the function and inhibition of an influenza virus proton channel. Nature 451: 596–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao SY, Fritzsching KJ, Hong M. 2013. Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR. Protein Sci. 22: 1623–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mandala VS, Williams JK, Hong M. 2018. Structure and dynamics of membrane proteins from solid-state NMR. Annu. Rev. Biophys 47: 201–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwon B, Roos M, Mandala VS, Shcherbakov AA, Hong M. 2019. Elucidating relayed proton transfer through a his–trp–his triad of a transmembrane proton channel by solid-state NMR. J. Mol. Biol 431: 2554–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoeman D, Fielding BC. 2019. Coronavirus envelope protein: current knowledge. Virol. J 16: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeDiego ML, Álvarez E, Almazán F, Rejas MT, Lamirande E, et al. 2007. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol 81: 1701–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Surya W, Li Y, Torres J. 2018. Structural model of the SARS coronavirus E channel in LMPG micelles. Biochim. Biophys. Acta Biomembr 1860: 1309–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torres J, Maheswari U, Parthasarathy K, Ng L, Liu DX, Gong X. 2007. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 16: 2065–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Surya W, Claudine S, Torres J. 2014. Structure of a conserved golgi complex-targeting signal in coronavirus envelope proteins. J. Biol. Chem 289: 12535–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quinn CM, Wang M, Fritz MP, Runge B, Ahn J, et al. 2018. Dynamic regulation of HIV-1 capsid interaction with the restriction factor TRIM5α identified by magic-angle spinning NMR and molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A 115: 11519–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gupta S, Tycko R. 2018. Segmental isotopic labeling of HIV-1 capsid protein assemblies for solid state NMR. J. Biomol. NMR 70: 103–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang M, Quinn CM, Perilla JR, Zhang H, Shirra R Jr, et al. 2017. Quenching protein dynamics interferes with HIV capsid maturation. Nat. Commun 8: 1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bayro MJ, Tycko R. 2016. Structure of the dimerization interface in the mature HIV-1 capsid protein lattice from solid state NMR of tubular assemblies. J. Am. Chem. Soc 138: 8538–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu C, Perilla JR, Ning J, Lu M, Hou G, et al. 2016. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat. Commun 7: 10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu JX, Bayro MJ, Tycko R. 2016. Major variations in HIV-1 capsid assembly morphologies involve minor variations in molecular structures of structurally ordered protein segments. J. Biol. Chem 291: 13098–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gupta R, Lu M, Hou G, Caporini MA, Rosay M, et al. 2016. Dynamic nuclear polarization enhanced MAS NMR spectroscopy for structural analysis of HIV-1 protein assemblies. J.Phys. Chem. B 120: 329–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang H, Hou G, Lu M, Ahn J, Byeon I-JL, et al. 2016. HIV-1 capsid function is regulated by dynamics: quantitative atomic-resolution insights by integrating magic-angle-spinning NMR, QM/MM, and MD. J. Am. Chem. Soc 138: 14066–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suiter CL, Quinn CM, Lu M, Hou G, Zhang H, Polenova T. 2015. MAS NMR of HIV-1 protein assemblies. J. Magn. Reson 253: 10–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu M, Hou G, Zhang H, Suiter CL, Ahn J, et al. 2015. Dynamic allostery governs cyclophilin A–HIV capsid interplay. Proc. Natl. Acad. Sci. U.S.A 112: 14617–146122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han Y, Hou G, Suiter CL, Ahn J, Byeon I-JL, et al. 2013. Magic angle spinning NMR reveals sequence-dependent structural plasticity, dynamics, and the spacer peptide 1 conformation in HIV-1 capsid protein assemblies. J. Am. Chem. Soc 135: 17793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Byeon I-JL, Hou G, Han Y, Suiter CL, Ahn J, et al. 2012. Motions on the millisecond time scale and multiple conformations of HIV-1 capsid protein: implications for structural polymorphism of CA assemblies. J. Am. Chem. Soc 134: 6455–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Han Y, Ahn J, Concel J, Byeon I-JL, Gronenborn AM, et al. 2010. Solid-state NMR studies of HIV-1 capsid protein assemblies. J. Am. Chem. Soc 132: 1976–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gupta S, Louis JM, Tycko R. 2020. Effects of an HIV-1 maturation inhibitor on the structure and dynamics of CA-SP1 junction helices in virus-like particles. Proc. Natl. Acad. Sci. U.S.A 117: 10286–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bayro MJ, Chen B, Yau W-M, Tycko R. 2014. Site-specific structural variations accompanying tubular assembly of the HIV-1 capsid protein. J. Mol. Biol 426: 1109–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Do HQ, Wittlich M, Glück JM, Möckel L, Willbold D, et al. 2013. Full-length Vpu and human CD4(372–433) in phospholipid bilayers as seen by magic angle spinning NMR. Biol. Chem 394: 1453–63 [DOI] [PubMed] [Google Scholar]
- 83.Lu J-X, Sharpe S, Ghirlando R, Yau W-M, Tycko R. 2010. Oligomerization state and supramolecular structure of the HIV-1 Vpu protein transmembrane segment in phospholipid bilayers. Protein Sci. 19: 1877–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharpe S, Yau W-M, Tycko R. 2006. Structure and dynamics of the HIV-1 Vpu transmembrane domain revealed by solid-state NMR with magic-angle spinning. Biochemistry 45: 918–33 [DOI] [PubMed] [Google Scholar]
- 85.Kwon B, Mandal T, Elkins MR, Oh Y, Cui Q, Hong M. 2020. Cholesterol interaction with the trimeric HIV fusion protein gp41 in lipid bilayers investigated by solid-state NMR spectroscopy and molecular dynamics simulations. J. Mol. Biol 432: 4705–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee M, Morgan CA, Hong M. 2019. Fully hydrophobic HIV gp41 adopts a hemifusion-like conformation in phospholipid bilayers. J. Biol. Chem 294: 14732–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kwon B, Lee M, Waring AJ, Hong M. 2018. Oligomeric structure and three-dimensional fold of the HIV gp41 membrane-proximal external region and transmembrane domain in phospholipid bilayers. J. Am. Chem. Soc 140: 8246–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in old world monkeys. Nature 427: 848–53 [DOI] [PubMed] [Google Scholar]
- 89.Stremlau M, Perron M, Lee M, Li Y, Song B, et al. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. U.S.A 103: 5514–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Skrisovska L, Schubert M, Allain FHT. 2009. Recent advances in segmental isotope labeling of proteins: NMR applications to large proteins and glycoproteins. J. Biomol. NMR 46: 51–65 [DOI] [PubMed] [Google Scholar]
- 91.Zinke M, Fricke P, Lange S, Zinn-Justin S, Lange A. 2018. Protein−protein interfaces probed by methyl labeling and proton-detected solid-state NMR spectroscopy. ChemPhysChem 19: 2457–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zinke M, Fricke P, Samson C, Hwang S, Wall JS, et al. 2017. Bacteriophage tail-tube assembly studied by proton-detected 4D solid-state NMR. Angew. Chem. Int. Ed 56: 9497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tissot AC, Renhofa R, Schmitz N, Cielens I, Meijerink E, et al. 2010. Versatile virus-like particle carrier for epitope based vaccines. PLOS ONE 5: e9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu X, Deng Y, Chen X, Zhou Y, Zhang H, et al. 2017. Immune response of a novel ATR-AP205–001 conjugate anti-hypertensive vaccine. Sci. Rep 7: 12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Janitzek CM, Peabody J, Thrane S,HR Carlsen P,G Theander T, et al. 2019. A proof-of-concept study for the design of a VLP-based combinatorial HPV and placental malaria vaccine. Sci. Rep 9: 5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yenkoidiok-Douti L, Williams AE, Canepa GE, Molina-Cruz A, Barillas-Mury C. 2019. Engineering a virus-like particle as an antigenic platform for a Pfs47-targeted malaria transmission-blocking vaccine. Sci. Rep 9: 16833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Palladini A, Thrane S, Janitzek CM, Pihl J, Clemmensen SB, et al. 2018. Virus-like particle display of HER2 induces potent anti-cancer responses. OncoImmunology 7: e1408749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maurer P, Bachmann MF. 2007. Vaccination against nicotine: an emerging therapy for tobacco dependence. Expert Opin. Investig. Drugs 16: 1775–83 [DOI] [PubMed] [Google Scholar]
- 99.Mohsen MO, Augusto G, Bachmann MF. 2020. The 3Ds in virus-like particle based-vaccines: “design, delivery and dynamics”. Immunol. Rev 296: 155–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sergeyev IV, Itin B, Rogawski R, Day LA, McDermott AE. 2017. Efficient assignment and NMR analysis of an intact virus using sequential side-chain correlations and DNP sensitization. Proc. Natl. Acad. Sci. U.S.A 114: 5171–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sergeyev IV, Bahri S, Day LA, McDermott AE. 2014. Pf1 bacteriophage hydration by magic angle spinning solid-state NMR. J. Chem. Phys 141: 22D533. [DOI] [PubMed] [Google Scholar]
- 102.Sergeyev IV, Day LA, Goldbourt A, McDermott AE. 2011. Chemical shifts for the unusual DNA structure in Pf1 bacteriophage from dynamic-nuclear-polarization-enhanced solid-state NMR spectroscopy. J. Am. Chem. Soc 133: 20208–17 [DOI] [PubMed] [Google Scholar]
- 103.Goldbourt A, Day LA, McDermott AE. 2010. Intersubunit hydrophobic interactions in Pf1 filamentous phage. J. Biol. Chem 285: 37051–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lorieau JL, Day LA, McDermott AE. 2008. Conformational dynamics of an intact virus: order parameters for the coat protein of Pf1 bacteriophage. Proc. Natl. Acad. Sci. U.S.A 105: 10366–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goldbourt A, Day LA, McDermott AE. 2007. Assignment of congested NMR spectra: carbonyl backbone enrichment via the Entner–Doudoroff pathway. J. Magn. Reson 189: 157–65 [DOI] [PubMed] [Google Scholar]
- 106.Abramov G, Shaharabani R, Morag O, Avinery R, Haimovich A, et al. 2017. Structural effects of single mutations in a filamentous viral capsid across multiple length scales. Biomacromolecules 18: 2258–66 [DOI] [PubMed] [Google Scholar]
- 107.Morag O, Abramov G, Goldbourt A. 2014. Complete chemical shift assignment of the ssDNA in the filamentous bacteriophage fd reports on its conformation and on its interface with the capsid shell. J. Am. Chem. Soc 136: 2292–301 [DOI] [PubMed] [Google Scholar]
- 108.Morag O, Abramov G, Goldbourt A. 2011. Similarities and differences within members of the Ff family of filamentous bacteriophage viruses. J. Phys. Chem. B 115: 15370–79 [DOI] [PubMed] [Google Scholar]
- 109.Porat G, Lusky OS, Dayan N, Goldbourt A. 2021. Nonuniformly sampled exclusively-13C/15N 4D solid-state NMR experiments: assignment and characterization of IKe phage capsid. Magn. Reson. Chem 59: 237–46 [DOI] [PubMed] [Google Scholar]
- 110.Liu D, Day L. 1994. Pf1 virus structure: helical coat protein and DNA with paraxial phosphates. Science 265: 671–74 [DOI] [PubMed] [Google Scholar]
- 111.Tsuboi M, Tsunoda M, Overman SA, Benevides JM, Thomas GJ. 2010. A structural model for the single-stranded DNA genome of filamentous bacteriophage Pf1. Biochemistry 49: 1737–43 [DOI] [PMC free article] [PubMed] [Google Scholar]


