Abstract
Klebsiella pneumoniae ORI-1 was isolated in 1998 in France from a rectal swab of a 1-month-old girl who was previously hospitalized in Cayenne Hospital, Cayenne, French Guiana. This strain harbored a ca. 140-kb nontransferable plasmid, pTK1, that conferred an extended-spectrum cephalosporin resistance profile antagonized by the addition of clavulanic acid, tazobactam, or imipenem. The gene for GES-1 (Guiana extended-spectrum β-lactamase) was cloned, and its protein was expressed in Escherichia coli DH10B, where this pI-5.8 β-lactamase of a ca. 31-kDa molecular mass conferred resistance to oxyimino cephalosporins (mostly to ceftazidime). GES-1 is weakly related to the other plasmid-located Ambler class A extended-spectrum β-lactamases (ESBLs). The highest percentage of amino acid identity was obtained with the carbenicillinase GN79 from Proteus mirabilis; with YENT, a chromosome-borne penicillinase from Yersinia enterocolitica; and with L-2, a chromosome-borne class A cephalosporinase from Stenotrophomonas maltophilia (36% amino acid identity each). However, a dendrogram analysis showed that GES-1 clustered within a class A ESBL subgroup together with ESBLs VEB-1 and PER-1. Sequencing of a 7,098-bp DNA fragment from plasmid pTK1 revealed that the GES-1 gene was located on a novel class 1 integron named In52 that was characterized by (i) a 5′ conserved segment containing an intI1 gene possessing two putative promoters, P1 and P2, for coordinated expression of the downstream antibiotic resistance genes and an attI1 recombination site; (ii) five antibiotic gene cassettes, blaGES-1, aac(6′)Ib′ (gentamicin resistance and amikacin susceptibility), dfrXVb (trimethoprim resistance), a novel chloramphenicol resistance gene (cmlA4), and aadA2 (streptomycin-spectinomycin resistance); and (iii) a 3′ conserved segment consisting of qacEΔ1 and sulI. The blaGES-1 and aadA2 gene cassettes were peculiar, since they lacked a typical 59-base element. This work identified the second class A ESBL gene of a non-TEM, non-SHV series which was located in the plasmid and integron, thus providing it additional means for its spread and its expression.
Klebsiella pneumoniae is an important hospital- or community-acquired pathogen that is naturally susceptible to extended-spectrum cephalosporins. However, strains producing extended-spectrum β-lactamases (ESBLs) were described in the early 1980s and have now spread worldwide (34). Most of these ESBLs are derivatives of restricted-spectrum TEM- and SHV-type β-lactamases, with one or more amino acid substitutions surrounding their active site, thus explaining the extension of their hydrolysis profile (34). Plasmid-located ESBL genes are mostly found in K. pneumoniae but also in other Enterobacteriaceae species and have been recently described in Pseudomonas aeruginosa (34, 39).
In addition to these so-called classical ESBLs, non-SHV–non-TEM derivatives have been detected in clinical isolates, mostly in a variety of Enterobacteriaceae species with, in some cases, a specific geographical distribution: CTX-M-1 (also known as MEN-1) to CTX-M-6 in Europe, South America, and Mediterranean countries (4, 17; M. Galas, A. Petroni, R. Melano, A. Corso, A. Rodriguez, M. L. Cacace, A. M. Bru, and A. Rossi, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 174, 1998); TOHO-1, TOHO-2, and SFO-1 in Japan (23, 29, 33); PER-1, mostly in Turkey (39, 41, 62); PER-2 in South America (5; Galas et al., 38th ICAAC); and recently, VEB-1 in Vietnam and Thailand (36, 46). Similarly to SHV and TEM derivatives, these clavulanic acid-inhibited enzymes belong to the Bush 2be functional group (10) and to the Ambler molecular class A (2). Their genes are mostly located in plasmids (34).
Integrons are genetic elements that can integrate gene cassettes within their variable regions (11, 20, 54, 56). Integrons are divided into four classes based on the nature of the integrase gene and the overall structure. The greatest number of integrons described so far are those of class 1 (30, 49). Class 1 integrons possess a 5′ conserved segment (5′-CS) that contains an intI1 gene coding for an integrase which catalyzes a site-specific recombination, a recombination site (attI1), and most commonly, a 3′ conserved segment (3′-CS) that carries qacEΔ1, a functional deletion derivative of the qacE gene (disinfectant resistance), the sulI gene (sulfonamide resistance), and an open reading frame (ORF) of unknown function (ORF5) (42, 58). Within these two segments, the so-called variable region of the integron is made up of gene cassettes, usually antibiotic resistance genes. Each gene cassette is associated with a 59-base element (59-be) located downstream of the gene of the integrated gene cassette (19, 57). 59-be's vary in length from 57 to 141 bp, but they are all bounded by a core site (GTTRRRY) at the recombinant crossover point and an inverse core site (RYYYAAC) at the 3′ end of the inserted gene. Among the cassette-located β-lactamase genes, most of them encoded β-lactamases of Ambler class D (oxacillin-hydrolyzing β-lactamases), rarely those of class A (carbenicillin-hydrolyzing β-lactamases) or class B (such as IMP-1, a carbapenem-hydrolyzing β-lactamase) (3, 9, 27, 49). blaVEB-1 was the first gene cassette identified encoding a class A enzyme that possesses ESBL properties and that has been recently located in the integron in Escherichia coli, K. pneumoniae, and P. aeruginosa isolates (36, 46).
This report identifies biochemically and genetically a novel class A ESBL, GES-1, whose protein sequence significantly differs from the other class A β-lactamases. The K. pneumoniae isolate that produced GES-1 was from a child who was directly transferred to France from Cayenne Hospital, Cayenne, French Guiana. Detailed genetic analysis also characterized the integronic nature of this novel plasmid-located ESBL gene.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this work are listed in Table 1. K. pneumoniae ORI-1 isolate was identified with the API-20E system (bioMérieux, Marcy l'Etoile, France).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH10B | F′ mcrA Δ (mrr hsdRMS mrcBC)F80dlacZDM15 Δ lacX74 deoR recA1 endA1 ara Δ 139 Δ (ara leu)7697 galU galK1 rpsL nupG | Gibco BRL-Life Technologies |
| In vitro-obtained rifampin-resistant E. coli DH10B | Rifampin resistant | This study |
| E. coli NCTC 50192 | 154-, 66-, 48-, and 7-kb reference plasmids | 14 |
| K. pneumoniae ORI-1 | Extended spectrum cephalosporin resistant | This study |
| Plasmids | ||
| pBK-CMV phagemid | Neor Kanr | Stratagene |
| pTK1 | Natural plasmid from K. pneumoniae ORI-1 containing blaGES-1 conferring the studied expanded-resistance phenotype | This study |
| pC1 | Recombinant plasmid with a 4.5-kb HindIII insert from pTK1 containing blaGES-1 into pBK-CMV | This study |
Susceptibility testing.
Antibiotic disks were used for routine antibiograms (Sanofi-Diagnostics Pasteur, Marnes-la-Coquette, France). The antimicrobial agents were obtained from standard laboratory powders and were used immediately after their solubilization. The agents and their sources were as follows: amoxicillin, clavulanic acid, and ticarcillin (SmithKline Beecham, Nanterre, France); amikacin, aztreonam and cefepime (Bristol-Myers Squibb, Paris-La Défense, France); ceftazidime (Glaxo, Paris, France); cephalothin and moxalactam (Eli Lilly, Saint-Cloud, France); piperacillin and tazobactam (Lederle, Oullins, France); sulbactam (Pfizer, Orsay, France); cefotaxime, cefuroxime, and cefpirome (Hoechst-Roussel, Paris, France); cefoxitin and imipenem (Merck Sharp & Dohme-Chibret, Paris, France); kanamycin, rifampin, and chloramphenicol (Sigma, Saint-Quentin Falavier, France).
MICs were determined by an agar dilution technique on Mueller-Hinton agar (Diagnostics Pasteur) with an inoculum of 104 CFU (38). All plates were incubated at 37°C for 18 h. MICs of β-lactams were determined alone or in combination with a fixed concentration of clavulanic acid (2 μg/ml), tazobactam (2 μg/ml), sulbactam (4 μg/ml), or imipenem (0.01 μg/ml).
PCR and hybridization experiments.
Under standard PCR conditions (51), sets of primers were designed for the detection of class A β-lactamase genes and their extended-spectrum derivatives: TEM-A (5′-GAGTATTCAACATTTCCGTGTC-3′) and TEM-B (5′-TAATCAGTGAGGCACCTATCTC-3′) for blaTEM-1 (59), PER-A (5′-ATGAATGTCATTATAAAAGC-3′) and PER-B (5′-AATTTGGGCTTAGGGCAGAA-3′) for blaPER-1 (40), VEB1-A (5′-CGACTTCCATTTCCCGATGC-3′) and VEB1-B (5′-GGACTCTGCAACAAATACGC-3′) for blaVEB-1 (46), TOHO-A (5′-ATGTGCAGTACCAGTAA-3′) and TOHO-B (5′-TAGGTCACCAGAACCAG-3′) for blaTOHO-1 (23), SFO1-A (5′-GGTTATGCCGCTGCTGTTC-3′) and SFO1-B (5′-TCGTCGGTTGTGCAGAAACG-3′) for blaSFO-1 (33), and CTX-M2-A (5′-CGGAATTCATGATGACTCAGAGCATTCG-3′) and CTX-M2-B (5′-GCTCTAGATTATTGCATCAGAAACCGTG-3′) for blaCTX-M2 (4). Southern hybridizations were performed as described by the manufacturer using the ECL nonradioactive labeling and detection kit (Amersham Pharmacia Biotech, Orsay, France). The natural plasmid pTK1 (Table 1) was also hybridized with a probe consisting of the internal 860-bp PCR fragment for blaGES-1 (GES-1A, 5′-ATGCGCTTCATTCACGCAC-3′; GES-1B, 5′-CTATTTGTCCGTGCTCAGG-3′) from recombinant plasmid pC1 (see below and Fig. 1).
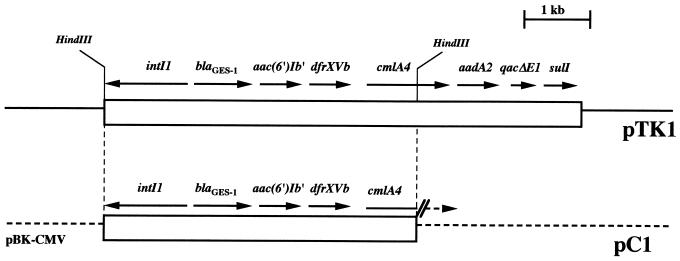
FIG. 1.
Schematic restriction endonuclease map of the natural plasmid pTK1 and the recombinant plasmid pC1 that contain blaGES-1. pC1 possesses a 4.5-kb HindIII fragment from pTK1 inserted into the HindIII site of pBK-CMV. The open boxes represents the sequenced part of plasmid pTK1 and the insert cloned in pC1, the dotted lines indicate the vector pBK-CMV, and the thin line represents the unsequenced part of pTK1. The GES-1 β-lactamase gene and the other ORFs are indicated.
Plasmid content, conjugation, and transformation experiments.
Plasmid DNAs of K. pneumoniae ORI-1 and of E. coli recombinant clones were extracted with the Qiagen plasmid DNA maxi kit (Courtaboeuf, France). DNAs were analyzed after their restriction digestion by electrophoresis on a 0.8% agarose gel (Gibco BRL-Life Technologies, Cergy-Pontoise, France) containing 0.15 μg of ethidium bromide per ml. Plasmid DNAs extracted from E. coli NCTC 50192 were used as size standards (14).
Conjugation experiments were performed between K. pneumoniae ORI-1 and in vitro-obtained rifampin-resistant E. coli DH10B in solid and liquid media at 37°C. Transconjugants were selected on Trypticase soy (TS) agar plates containing 150 μg of rifampin per ml and 100 μg of amoxicillin per ml. The extracted plasmid DNAs from K. pneumoniae ORI-1 were also subjected to electroporation into E. coli DH10B according to the manufacturer's instructions (Bio-Rad, Ivry-sur-Seine, France). Recombinant bacteria were plated onto TS agar plates containing 100 μg of amoxicillin per ml.
Cloning experiments and recombinant plasmid analysis.
Plasmid DNAs extracted from E. coli DH10B transformants displaying an ESBL phenotype were HindIII restricted and ligated into HindIII-restricted pBK-CMV phagemid (Stratagene, La Jolla, Calif.). Recombinant plasmids were selected onto amoxicillin-containing TS agar plates (100 μg/ml). A double-restriction digestion analysis allowed precise mapping of recombinant plasmids by comparison to the molecular weight marker 1-kb DNA ladder (Amersham Pharmacia Biotech).
β-Lactamase purification.
Cultures of E. coli DH10B harboring recombinant plasmid pC1 (see below) were grown overnight at 37°C in 1 liter of TS broth with amoxicillin (100 μg/ml). The bacterial suspension was pelleted, resuspended in 10 ml of 100 mM phosphate buffer (pH 7), disrupted by sonification (three times for 30 s at 40 kHz with a Vibra Cell 300 Phospholyser (BioBlock, Illkirch, France) and centrifuged for 1 h at 48,000 × g and 4°C. Nucleic acids were precipitated by the addition of 0.2 M spermin (7% [vol/vol]) (Sigma) overnight at 4°C. This suspension was ultracentrifuged at 100,000 × g for 1 h at 4°C and dialyzed overnight against 20 mM Bis-Tris {[bis(2-hydroxyethyl-imino]tris(hydroxymethyl)methane}, pH 6.5, at 4°C. The enzyme extract was loaded onto a preequilibrated Q-Sepharose column (1.6 by 5 cm; Amersham Pharmacia Biotech) with the same buffer. The resulting enzyme extract was recovered in the flowthrough and dialyzed against 20 mM Tris-HCl (pH 7.5) overnight at 4°C. This extract was then loaded onto a preequilibrated (20 mM Tris-HCl [pH 7.5]) Q-Sepharose column, and the proteins were eluted with a linear NaCl gradient (0 to 0.5 M). The β-lactamase activity was eluted with NaCl at a concentration of 200 mM in the same Tris-HCl buffer. The fractions presenting the highest β-lactamase activity were pooled and dialyzed against 100 mM phosphate buffer (pH 7.0), prior to a 10-fold concentration with a Centrisart-C30 microcentrifuge filter (Sartorius, Göttingen, Germany). The purified β-lactamase extract was immediately used for enzymatic determinations.
Isoelectric focusing.
Purified enzyme from E. coli DH10B harboring pC1 was subjected to analytical isoelectric focusing (IEF) on an ampholin polyacrylamide gel (pH 3.5 to 9.5, Ampholin PAG plates; Amersham Pharmacia Biotech) for 90 min at 30 W of constant power on a flatbed apparatus (Multiphor II; Amersham Pharmacia Biotech). Similarly, nonpurified β-lactamase extracts from K. pneumoniae ORI-1 and E. coli DH10B harboring natural plasmid pTK1 were submitted to IEF analysis. The focused β-lactamases were detected by overlaying the gel with 1 mM nitrocefin (Oxoid, Dardilly, France) in 100 mM phosphate buffer (pH 7.0). The pI values were determined and compared to those of known β-lactamases as previously described (44).
Determination of the β-lactamase relative molecular mass.
The relative molecular mass of the β-lactamase purified from E. coli DH10B(pC1) was estimated by sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis analysis (25). Enzyme extracts and marker proteins were boiled for 10 min in a 1% SDS–3% β-mercaptoethanol solution and then subjected to electrophoresis (25 mA for 4 h) at room temperature (51). Renaturation of the β-lactamase activity after denaturing electrophoresis and revelation with a benzylpenicillin-containing agar gel were performed as previously described (31).
Kinetic measurements.
Purified β-lactamase was used for kinetic measurements performed at 30°C in 100 mM sodium phosphate (pH 7.0). The initial rates of hydrolysis were determined with a Pharmacia ULTROSPEC 2000. The following wavelengths and absorption coefficients were used: for benzylpenicillin and amoxicillin, 232 and 240 nm, respectively, and Δɛ = −1,100 M−1 cm−1; for ticarcillin, 235 nm and Δɛ = −1,050 M−1 cm−1; for piperacillin, 235 nm and Δɛ = −1,070 M−1 cm−1; for cefepime, 264 nm and Δɛ = −8,240 M−1; for cephalothin, 262 nm and Δɛ = −7,960 M−1 cm−1; for cephaloridine, 255 nm and Δɛ = −9,360 M−1 cm−1; for cefotaxime, 265 nm and Δɛ = −6,260 M−1; for cefoxitin, 265 nm and Δɛ = −7,380 M−1 cm−1; for ceftazidime, 260 nm and Δɛ = −8,660 M−1 cm−1; for cefuroxime, (262 nm) and Δɛ = −7,800 M−1 cm−1; for imipenem, 297 nm and Δɛ = −9,210 M−1 cm−1; and for aztreonam (318 nm), Δɛ = −640 M−1 cm−1.
Kinetic parameters were determined by recording the initial rates at different substrate concentrations and by analyzing the results with the regression analysis program LEONARA, written by Cornish-Bowden (13). The kcat and Km values were estimated by using a nonlinear least-squares regression method with dynamic weights.
Fifty-percent inhibitory concentrations (IC50) were determined for clavulanic acid, tazobactam, and imipenem. Various concentrations of these inhibitors were preincubated with the purified enzyme for 3 min at 30°C to determine the concentrations that reduced the hydrolysis rate of 100 μM benzylpenicillin by 50%. Results were expressed in micromolar units.
The specific activity of the purified β-lactamase from E. coli DH10B harboring pC1 (GES-1) was obtained as described previously (44). One unit of enzyme activity was defined as the activity which hydrolyzed 1 μmol of benzylpenicillin per min per mg of protein. The total protein content was measured with the Bio-Rad DC protein assay kit.
DNA sequencing and protein analysis.
The 4,543-bp cloned DNA fragment from recombinant plasmid pC1 was sequenced on both strands with an Applied Biosystems sequencer (model ABI 373). Further sequencing analysis was performed on PCR products using laboratory designed sequencing primers and parts of the natural plasmid pTK1 as a template. The nucleotide sequences and the deduced protein sequences were analyzed by using the software available over the Internet at the National Center of Biotechnology Information website (http://www.ncbi.nlm.nih.gov) and at Pedro's BioMolecular Research Tools website (http://www.fmi.ch/biology/research_tools.html). Multiple protein sequence alignments were carried out with the ClustalW program available over the Internet at the University of Cambridge. Among the Ambler class A β-lactamases, 14 were compared to the identified β-lactamase: PER-1 from P. aeruginosa RNL-1 (40), VEB-1 from E. coli MG-1 (46), TEM-3 from K. pneumoniae CF104 (53), SHV-2 from Klebsiella ozaenae (22), CTX-M-1 (MEN-1) from Salmonella enterica serovar Typhimurium (4), L-2 from Stenotrophomonas maltophilia 1275 IID (63), TOHO-1 from E. coli TUH12191 (23), SME-1 from Serratia marcescens S6 (37), NMC-A from Enterobacter cloacae NOR-1 (35), OXY-2 from Klebsiella oxytoca (15), YENT from Yersinia enterocolitica (52), CARB from Proteus mirabilis GN79 (50), CITDI from Citrobacter diversus ULA27 (43), and SFO-1 from E. cloacae 8009 (33). A dendrogram was derived from the multiple sequence alignment by a parsimony method with the phylogeny package PAUP (Phylogenetic Analysis Using Parsimony), version 3.0 (16, 60).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the GenBank nucleotide database under accession no. AF156486.
RESULTS
Properties of the K. pneumoniae ORI-1 isolate.
ORI-1 was isolated in 1998 at the Hôpital de Bicêtre, Le Kremlin-Bicêtre (a southern suburb of Paris), France, as a result of systematic multiresistant-bacteria rectal screening of patients admitted into the pediatric intensive care unit. A 1-month-old girl born in Cayenne, French Guiana, South America, had developed a neonatal infection due to Morganella morganii, for which she had received 3 weeks of treatment with cefotaxime and netilmicin. She was then transferred to the intensive care unit of the Hôpital de Bicêtre where K. pneumoniae ORI-1 was isolated on the day of admission. Antibiotic susceptibility testing by disk diffusion suggested that the extended-spectrum cephalosporin resistance profile was due to the presence of an ESBL (data not shown). Synergies were observed between clavulanic acid-amoxicillin and ceftazidime, cefotaxime, aztreonam, and cefepime. K. pneumoniae ORI-1 was also resistant to gentamicin, kanamycin, tobramycin, streptomycin, spectinomycin, tetracycline, chloramphenicol, trimethoprim, and the sulfonamides; it was susceptible to amikacin, nalidixic acid, and the fluoroquinolones (data not shown). No other enterobacterial isolate with a similar ESBL resistance profile was identified in the same hospitalization unit concomitantly or during the following 4-month period (data not shown).
Preliminary PCR experiments, plasmid analysis, and cloning of the ESBL gene.
Preliminary PCR amplification experiments using primers designed to amplify several internal fragments of ESBL genes (TEM derivatives, CTX-M-2, PER-1, SFO-1, VEB-1, and TOHO-1) failed to give positive results. Conjugation performed in both solid and liquid media after mating K. pneumoniae ORI-1 and E. coli DH10B failed to give transconjugants. Extracted plasmid DNAs from K. pneumoniae ORI-1 were transformed by electroporation into E. coli DH10B. Sixty colonies were obtained after selection onto amoxicillin-containing plates. These strains conferred either a clavulanic acid-inhibited β-lactam-restricted resistant profile (resistance to amino-, carboxy-, and ureidopenicillins) or a resistance profile extended to oxyimino cephalosporins. This last resistance profile was retained for further analysis. One of these E. coli electroporants harbored a ca. 140-kb natural plasmid, named pTK1 (data not shown).
Plasmid pTK1 was HindIII restricted, and the resulting fragments were cloned into the HindIII site of pBK-CMV. A detailed restriction map was generated for one of the recombinant plasmids, pC1, that contained a 4.5-kb insert (Fig. 1).
Antibiotic susceptibility.
β-Lactam MICs for K. pneumoniae ORI-1 and E. coli DH10B harboring either the natural plasmid pTK1 or the recombinant plasmid pC1 were somewhat similar and might indicate the presence of an ESBL. In all cases, the ceftazidime MICs were higher than those of cefotaxime and aztreonam. β-Lactam MICs were always lowered by the addition of clavulanic acid or tazobactam, less so by sulbactam, and uncommonly by imipenem (Table 2). Oxyimino cephalosporin MICs were increased for E. coli DH10B(pC1) as compared to those for E. coli (pTK1). As found for K. pneumoniae ORI-1, E. coli DH10B(pTK1) was resistant to gentamicin, tobramycin, netilmicin, streptomycin, spectinomycin, tetracycline, trimethoprim, the sulfamides, and chloramphenicol and susceptible to amikacin (data not shown). E. coli DH10B(pC1) was resistant to gentamicin, tobramycin, and netilmicin but remained susceptible to amikacin, tetracycline, trimethoprim, the sulfonamides, and chloramphenicol.
TABLE 2.
MICs of β-lactams for K. pneumoniae ORI-1, E. coli DH10B harboring either natural plasmid pTK1 or recombinant plasmid pC1, and reference strain E. coli DH10B
| β-Lactam(s)a | MIC (μg/ml) for:
|
|||
|---|---|---|---|---|
| K. pneumoniae ORI-1b | E. coli DH10B (pTK1)c | E. coli DH10B (pC1)c | E. coli DH10B | |
| Amoxicillin | >512 | >512 | >512 | 4 |
| Amoxicillin + CLA | 64 | 64 | >128 | 4 |
| Ticarcillin | >512 | 256 | >512 | 4 |
| Ticarcillin + CLA | 64 | 8 | 64 | 4 |
| Ticarcillin + TZB | >512 | 8 | 256 | 2 |
| Ticarcillin + IPM | >512 | 32 | 256 | 4 |
| Ticarcillin + SUL | >512 | 8 | 512 | 4 |
| Piperacillin | 512 | 16 | 64 | 1 |
| Piperacillin + CLA | 128 | 2 | 8 | 1 |
| Piperacillin + TZB | 64 | 2 | 8 | 1 |
| Cephalothin | 64 | 32 | 256 | 2 |
| Cefuroxime | 32 | 32 | 256 | 2 |
| Cefoxitin | 4 | 8 | 8 | 1 |
| Ceftazidime | 4 | 8 | 128 | 0.5 |
| Ceftazidime + CLA | 0.5 | 1 | 8 | 0.5 |
| Ceftazidime + TZB | 0.5 | 0.5 | 8 | 0.5 |
| Ceftazidime + IPM | 1 | 0.5 | 16 | 0.5 |
| Ceftazidime + SUL | 1 | 0.5 | 16 | 0.5 |
| Cefotaxime | 0.5 | 0.5 | 4 | 0.06 |
| Cefotaxime + CLA | 0.06 | 0.06 | 0.5 | 0.06 |
| Cefotaxime + TZB | 0.06 | 0.06 | 2 | 0.06 |
| Cefotaxime + IPM | 0.06 | 0.12 | 1 | 0.06 |
| Cefotaxime + SUL | 0.12 | 0.06 | 2 | 0.12 |
| Cefepime | 0.25 | 0.25 | 0.5 | 0.03 |
| Cefepime + CLA | 0.06 | 0.06 | 0.06 | 0.03 |
| Cefepime + TZB | 0.12 | 0.06 | 0.12 | 0.03 |
| Aztreonam | 0.12 | 0.25 | 1 | 0.12 |
| Aztreonam + CLA | <0.03 | 0.12 | 0.25 | 0.12 |
| Aztreonam + TZB | 0.06 | 0.5 | 0.5 | 0.12 |
| Moxalactam | 0.06 | 0.06 | 0.12 | 0.06 |
| Imipenem | 0.12 | 0.25 | 0.25 | 0.12 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 2 μg/ml; IPM, imipenem at a fixed concentration of 0.01 μg/ml; SUL, sulbactam at a fixed concentration of 4 μg/ml.
K. pneumoniae ORI-1 produced a SHV-1-like penicillinase, an unidentified restricted-spectrum β-lactamase, and GES-1 β-lactamase.
E. coli DH10B harboring either natural plasmid pTK1 or recombinant multicopy plasmid pC1 produced GES-1 β-lactamase.
Biochemical properties of GES-1.
IEF analysis showed that K. pneumoniae ORI-1 had three β-lactamase activities with pI values of 5.8, 6.5, and 7.0. E. coli(pTK1) or E. coli(pC1) had only one β-lactamase activity with a pI value of 5.8, corresponding to that of the ESBL. β-Lactamase activity with a pI value of 7.0 in K. pneumoniae ORI-1 likely corresponded to an SHV-1 type penicillinase (positive hybridization with an internal probe for blaSHV-3; data not shown), although this value was somewhat lower than the common pI value of 7.6 for SHV-1 (21). β-Lactamase activity with a pI value of 6.5 in K. pneumoniae ORI-1 might correspond to an as-yet-unstudied restricted-spectrum penicillinase, as suggested by the obtainment of an E. coli electroporant-harboring plasmid that conferred a clavulanic acid-inhibited restricted-spectrum hydrolysis profile. The relative molecular mass of GES-1 expressed from E. coli DH10B(pC1) was estimated to be ca. 31 kDa by SDS-polyacrylamide gel electrophoresis analysis. The specific activity of the purified β-lactamase was 3.25 μmol · min−1 · mg−1 of protein, determined with 100 μM benzylpenicillin as substrate. Its overall recovery was 50% with a 30-fold purification.
Kinetic parameters of the purified β-lactamase GES-1 showed strong activity against most β-lactams tested except against aztreonam and imipenem (Table 3). High Km values were obtained for all cephalosporins that could be expressed in millimolar units and not, as usual for ESBL activity, in micromolar units (10). Ceftazidime was the best substrate for GES-1 activity, based on both kcat and kcat/Km measurements (Table 3). Inhibition studies, as measured by IC50 values with benzylpenicillin as the substrate, showed that GES-1 was inhibited by clavulanic acid (5 μM) and tazobactam (2.5 μM) and strongly inhibited by imipenem (0.1 μM).
TABLE 3.
Steady-state kinetic parameters of the purified GES-1 β-lactamasea
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (mM−1 · s−1) |
|---|---|---|---|
| Benzylpenicillin | 2.8 | 40 | 70 |
| Amoxicillin | 13 | 200 | 65 |
| Ticarcillin | 0.3 | 400 | 0.7 |
| Piperacillin | 8 | 900 | 9 |
| Cephalothin | 179 | 3,400 | 52 |
| Cephaloridine | 53 | 2,000 | 26 |
| Cefoxitin | 0.9 | 30 | 33 |
| Ceftazidime | 380 | 2,000 | 188 |
| Cefepime | 2.8 | 1,800 | 1.6 |
| Cefotaxime | 68 | 4,600 | 15 |
| Imipenem | 0.003 | 45 | 0.07 |
| Aztreonam | —b | — | — |
Values are means of three independent measures (standard deviations of the values were within 15%).
—, not determinable (the initial rate of hydrolysis was lower than 0.001 μM−1 · s−1).
Sequence analysis of blaGES-1 and its deduced protein sequence.
Analysis of the determined nucleotide sequence from recombinant plasmid pC1 (Fig. 1) revealed an 864-bp-long open reading frame (ORF) encoding a 288-amino-acid protein (named GES-1). The initiation codon (ATG) was preceded by two putative promoter regions named P1 (formerly Pc or Pant) (−35 [TGGACA]; −10 [TAAGCT]) and P2 (−35 [TTGTTA]; −10 [TACAGT]) (Fig. 2). The overall G+C content of blaGES-1 was 51.6%, which is within the range of G+C content for Enterobacteriaceae genes. Analysis of the immediate downstream regions of blaGES-1 revealed no palindromic sequences typical of Rho-independent transcriptional terminators. Hybridizations using a PCR fragment internal to blaGES-1 as a probe confirmed the presence of this gene on the natural plasmid pTK1 (data not shown).
FIG. 2.
Nucleotide sequence of a 7,098-bp fragment of pTK1 containing the GES-1 coding region. The deduced amino acid sequence is designated in single-letter code below the nucleotide sequence. The start codons of the ORFs are indicated by horizontal arrows, and the deduced proteins are reported below the nucleotide sequence. Stop codons for each ORF are indicated by asterisks. Dashes in several nucleotide sequences indicate where the reported sequence was identical to already-published sequences. The −35 and −10 sequences of the promoters P1, P2, P3, Pa, and Pb for cmlA4 are indicated; RBS indicates the putative ribosomal binding site for cmlA4. The conserved core and inverse core sites located at each cassette boundary are boxed, and the composite 59-be's are italicized. The cassette boundaries are indicated by vertical arrows. The attI1 site is underlined with a dotted line. Within the GES-1 protein, the conserved residues of Ambler class A β-lactamases are underlined. The amino acids of CMLA-1 that differ from those of CMLA-4 are indicated below the amino acid sequence of CMLA-4. The nucleotide sequence for putative translational attenuation regulation of cmlA4 (nucleotide positions 3521 to 3551) is underlined.
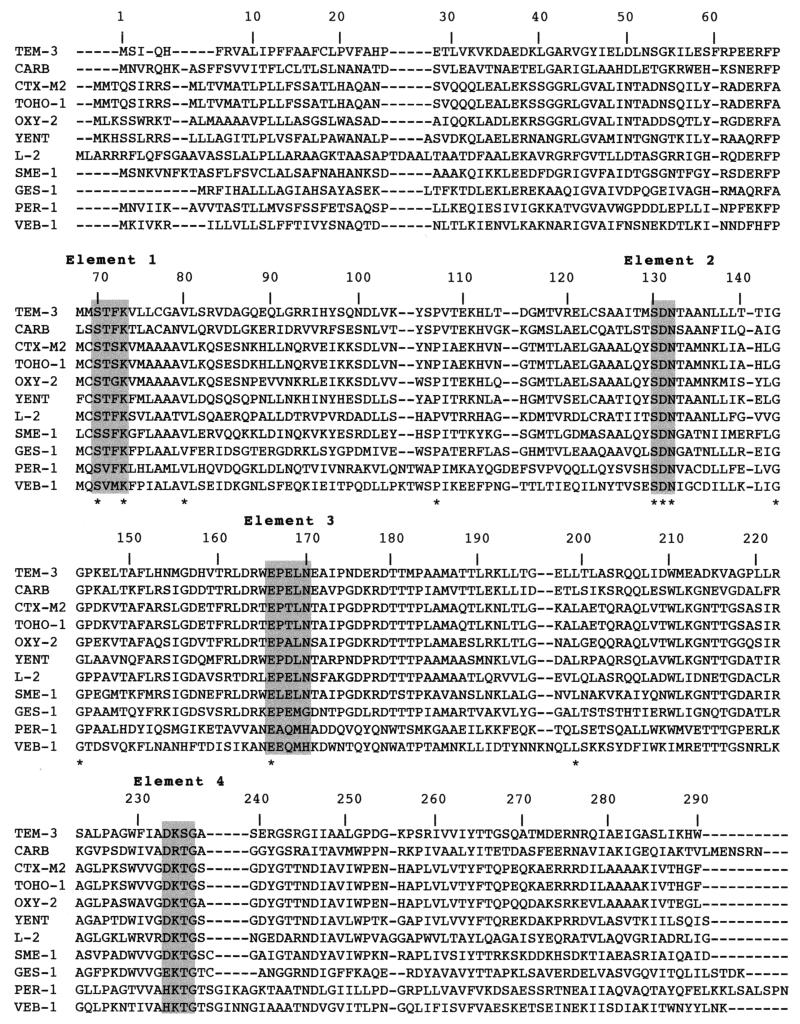
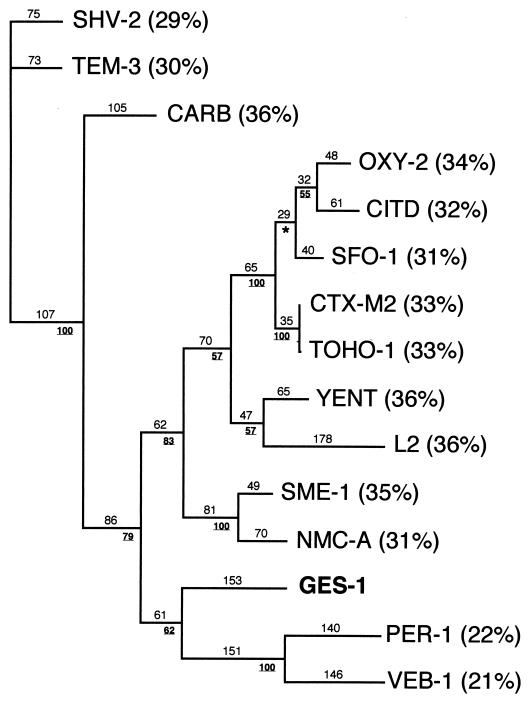
The nucleotide sequence of blaGES-1 had less than 24% DNA identity with other β-lactamase genes. Within the deduced protein, a serine-threonine-phenylalanine-lysine tetrad was found at Ambler positions 70 to 73 (Fig. 2), as well as several structural elements characteristic of Ambler class A β-lactamases (such as SDN) at positions 130 to 132 and a KTG motif at positions 234 to 236 (Fig. 2 and 3). GES-1 had less than 37% amino acid identity with all the class A β-lactamases, including the TEM and SHV derivatives (30% identity) (Fig. 4). It was only weakly related (less than 20% identity) to any other class A β-lactamases of gram-positive bacteria or anaerobes (data not shown). The highest percentage of identity was 36%, found with either the carbenicillinase GN79 from P. mirabilis (50) or YENT, a constitutive penicillinase from Y. enterocolitica (52), or L-2, a chromosomally-encoded class A cephalosporinase from S. maltophilia (63). A dendrogram analysis derived from a multiple sequence alignment of GES-1 with 14 β-lactamases showed that GES-1 is clearly a novel type of class A β-lactamase. Although weakly related to ESBLs of the non-TEM–non-SHV series, GES-1 clustered weakly within an ESBL subgroup of gram-negative bacteria that contains PER-1 and VEB-1 (Fig. 4).
FIG. 3.
Alignment of the amino acid sequence of GES-1 with those of the closest class A β-lactamases. Numbering is according to Ambler (2). Asterisks indicate the conserved amino acid residues among these class A sequences. Highlighted amino acids are those surrounding the active sites of class A sequences. The β-lactamases included in the alignment are TEM-3 from K. pneumoniae, CARB from P. mirabilis, CTX-M-2 from S. enterica serovar Typhimurium, TOHO-1 from E. coli, OXY-2 from K. oxytoca, YENT from Y. enterocolitica, L-2 from S. maltophilia, SME-1 from S. marcescens, GES-1 from K. pneumoniae, PER-1 from P. aeruginosa, and VEB-1 from E. coli. Dashes indicate gaps introduced to optimize the alignment.
FIG. 4.
Dendrogram obtained for 15 representative Ambler class A β-lactamases by the parsimony method (60). Branch lengths are drawn to scale and are proportional to the number of amino acid changes. The percentages at branching points (underlined) refer to the number of times a particular node was found in 100 bootstrap replications (the asterisk indicates uncertainty of nodes with bootstrap values of less than 50%). The distance along the vertical axis has no significance. Designations for β-lactamases are given in Materials and Methods. Percentages in parentheses are amino acid identities to GES-1.
Genetic environment of blaGES-1.
Sequence analysis of a 7,098-bp DNA fragment including blaGES-1 was performed first with recombinant plasmid pC1, then with PCR-amplified fragments from natural plasmid pTK1 with in-house-designed primers. It revealed the presence of the following class 1 integron features: (i) a 5′-CS containing an intI1 integrase gene with its own promoter region, (ii) an attI1 recombination site, (iii) antibiotic resistance genes with gene cassette features, and (iv) a 3′-CS containing qacEΔ1 and sulI (Fig. 2). The blaGES-1 gene cassette which was inserted at the attI1 recombination site has a core site (GTTAGAC) and an inverse core site (GTCTAAA) presenting a 1-bp mismatch. Surprisingly, downstream of this gene, the 59-be was only 19-bp long, which is unusual (Fig. 2). The second gene cassette encoded a 6′-N-aminoglycoside acetyl transferase [AAC (6′)-Ib′] of 185 amino acids (Fig. 2) that conferred resistance to gentamicin, tobramycin, and netilmicin and susceptibility to amikacin in K. pneumoniae ORI-1 and in E. coli DH10B harboring plasmid pTK1 or pC1. This gene structure shared 100% identity with the published sequence of this gene (26). The third gene cassette of this integron corresponded to a point mutant of the dfrXV gene (named dfrXVb) previously identified in E. coli (1), whose protein conferred resistance to trimethoprim in K. pneumoniae ORI-1 and E. coli DH10B(pTK1) (Fig. 2). The deduced DHFRXVb protein, as compared to DHFRXV, possessed a single serine-to-glycine change at position 133 (1). The fourth gene cassette, extending from base 3,442 to 4,890, contains an ORF of 1,260 nucleotides starting with a GTG initiation codon (position 3,649) (Fig. 2). This ORF is preceded by two putative promoters, Pa (−35 [TTGAAA]; −10 [TTCAAT]) and Pb (−35 [TTGTTA]; −10 [TGAGAT]) and a ribosome binding site (AGGAG) (Fig. 2). The deduced protein of 419 amino acids was similar to CMLA1, which is encoded by a gene within the class 1 integron In4 present in Tn1696 (6), differing by just 11 amino acid changes (98% identity) (Fig. 2). Like CMLA1, this novel protein (CMLA4) likely conferred resistance to chloramphenicol by a nonenzymatic mechanism. Among the 207 bp upstream of cmlA4, only four nucleotide changes were identified, compared to the sequence found upstream of cmlA1 (data not shown). Downstream from cmlA4, an inverse core site (GCCCAAC) was part of a composite 59-be of 70 bp. This 59-be was almost 100% identical to the downstream region of cmlA1, except for one nucleotide change (T to C in cmlA4) at the last position (position 4,997) (Fig. 2). The fifth gene cassette corresponded to aadA2, a cassette found in several integrons (9, 30), which codes for streptomycin and spectinomycin resistance. Interestingly, while the 59-be's of aadA gene cassettes are highly conserved in length and in nucleotides (7), the aadA2 cassette present on plasmid pTK1 lacked most of its 59-be. Together with the conserved 3′-CS sequence, this novel class 1 integron was named In52.
DISCUSSION
In this study, we report the identification of GES-1, an enzyme which expands the group of Ambler class A ESBLs. Indeed, GES-1 is only weakly related to the other Ambler class A β-lactamases, particularly to the plasmid-located ESBLs so far identified in Enterobacteriacae. As with most of the class A ESBLs, GES-1 was identified in K. pneumoniae, a species that remains the main reservoir of these worldwide-spread enzymes, for unknown reasons.
The profile of antimicrobial resistance conferred by GES-1 corresponded to those observed with structurally unrelated ESBLs. However, GES-1 may be classified rather as a ceftazidime-hydrolyzing enzyme as opposed to the recently described VEB-1 (46). The activity of GES-1 was inhibited by imipenem, as was VEB-1, and not by cephamycins (data not shown). Interestingly, Km values of cephalosporins for GES-1 were much higher than those for other ESBLs including the TEM or SHV series (10, 41, 46), indicating weak affinity of GES-1 for these β-lactams. Protein sequence alignment shows that GES-1 had the highest sequence identity with L-2, a chromosome-borne class A cephalosporinase from S. maltophilia, the narrow-spectrum carbenicillinase from P. mirabilis, and YENT from Y. enterocolitica (36% identity each). The majority of the ESBLs are plasmid located and are derivatives of either TEM-1, TEM-2, or SHV-1, differing from its ancestors by point mutations. GES-1 is distantly related to these enzymes sharing only 30% amino acid identity. Similarly, GES-1 is distantly related to the most recently described plasmid-mediated and clavulanic acid-inhibited ESBLs such as VEB-1, SFO-1, TOHO-1 and -2, CTX-M-1 to CTX-M-6, SME-1, NMC-A, IMI-1, and PER-1 and -2 (4, 5, 17, 23, 29, 33, 35, 41, 46, 48).
Taking into account that GES-1 is a novel class A ESBL belonging to the Bush 2be functional group without significant identity with any restricted-spectrum β-lactamase, it is difficult to draw definitive conclusions from analysis of the amino acid positions of GES-1. However, some residues may be important in catalytic activity, for instance, the threonine residue at position ABL 237 found in TEM-5, TEM-24, PER-1, and PER-2 (32). It may partially explain the expanded β-lactam resistance profile of GES-1 indicated recently by data resulting from experiments with in vitro-obtained TEM derivatives (8). At position ABL 165, GES-1 has a lysine that may also be involved in its expanded substrate profile, found in CITDI from C. diversus and in the naturally occurring β-lactamase from Proteus vulgaris (32, 43). It has been hypothesized that a potential hydrogen bond donor (Thr, Asn, or Lys) in some ESBLs of the non-TEM–non-SHV series (PER-1, OXY-2, CTX-M-2, and TOHO-1) may be involved in their expanded substrate profile (32).
Interestingly, GES-1 possesses cysteine residues at positions ABL 69 and ABL 238, as found in the carbapenem-hydrolyzing class A β-lactamases NMC-A, IMI-1, and SME-1 (35, 37, 48). Raquet et al. postulate that these cysteine residues may form a disulfide bridge in SME-1 that is involved in its carbapenem-resistance properties (48). The kinetic constants of GES-1 do not show any significant imipenem hydrolysis but rather a significant degree of inhibition by imipenem. A possible explanation is that GES-1 may be able to bind imipenem with strong affinity without being able to hydrolyze it.
Several interesting features emerged from the analysis of the surrounding sequences of blaGES-1. blaGES-1 is the second integron-located class A ESBL gene after blaVEB-1 that was first identified from E. coli (46). Upstream of blaGES-1, two putative promoter sequences named P1 and P2 were located in the structural integrase gene. Comparison of P1 with known promoters for which expression studies were performed (12, 28) identified P1 as a weak promoter. As described, the insertion of three guanosine molecules 119 bases downstream of the promoter P1 creates a secondary promoter, P2, for blaGES-1 expression. This three-nucleotide insertion brings the spacing between −35 and −10 regions of P2 to 17 bp. Therefore, P2 expression may be responsible for 90% of blaGES-1 transcription, as shown for other genes (12, 28). The combination of these two promoters is most commonly found in integrons such as those possessing aadA2 and dfrV genes (30). In contrast to blaVEB1, located at the second position downstream of the promoter region (46), blaGES-1 is located just downstream of P1 and P2 (Fig. 2). Thus, it is expected to be highly expressed in vivo. Gene cassette boundaries for blaGES-1 were determined by identification of the core site 5′ of the gene, the inverse core site 3′ of the gene, and another core site belonging to the next inserted gene cassette. Since the 59-be of blaGES-1 is only 19 bp long, it may not be functional in recombination and may therefore be unable to move by itself. Interestingly, this truncated 59-be is very similar to 59-be structures found at the end of the aadA1 cassette in Tn1331 (61) or at the end of blaOXA-11 or part of the sequence present in attI1 (18, 49). In fact, for blaGES-1, the same sequence (TAAAACAAAGTT) at nucleotide positions 1179 to 1290 and 2208 to 2219 (Fig. 2) is present at each side of blaGES-1 as if this gene was inserted in a manner similar to insertion sequences with a target site duplication.
Further analysis of In52 shows that cmlA4 has two putative promoter sequences (Pa and Pb) that are identical to two of three putative promoters (P2 and P3) found for cmlA1 in In4 (6). As described for cmlA1 and cmlA2 (6, 45), cmlA4 may be expressed from its own promoters as opposed to the other integron-located genes. As found for cmlA1 (55), regulation of expression of cmlA4 could be mediated by the same translational attenuation sequence (nucleotide positions 3521 to 3550) identified just downstream of the putative promoters of cmlA4 (Fig. 2). Interestingly, the nucleotide sequences upstream and downstream from cmlA4 are almost identical to those surrounding cmlA1 (6). Therefore, it is possible that evolutionary constraints are looser for the coding sequences than for the gene cassette. A similar observation was made for aadA gene cassettes where the 59-be's are highly conserved, whereas the coding sequences show some diversity (55). Amino acid sequence identities between CMLA-4 and CMLA-1, CMLA-2, and another CMLA-like protein from S. enterica serovar Typhimurium (9) that we named CMLA-3 are 96, 83, and 46%, respectively. This high percentage of amino acid identity, at least with CMLA-1 and CMLA-2, indicated a common function consisting of probable chloramphenicol efflux. The aminoglycoside resistance genes commonly found in other class 1 integrons and the dfrXVb gene identified in In52 are each associated with gene cassette features. Interestingly, aadA2 has a truncated 59-be, which again is fatal for recombination activity. This truncated 59-be is different in length and sequence from the truncated 59-be's of aadA1, blaGES-1, and blaOXA-11 (18, 61). Thus, In52 has two gene cassettes with truncated 59-be's. A possible explanation for these deletions is that nonspecific cassette excision may have occurred between a core site and a sequence present in the 59-be of the preceding cassette. It would be interesting to test the activity of these truncated 59-be's in cassette excision and integration.
This work sheds further light into the ongoing evolution of plasmid- and integron-located ESBL genes. It underlines that integron-located ESBL genes are not only part of class D (extended-spectrum oxacillin-hydrolyzing β-lactamases) or class B (IMP-1, a carbapenem-hydrolyzing β-lactamase) but may be also part of class A. This is of interest, since (i) most of the plasmid-mediated ESBLs that are spreading worldwide are of class A and (ii) their integron location may provide them additional potential for spreading. To our best knowledge, this is the first report of an ESBL-producing isolate from French Guiana, located in the northeastern part of South America. In South American countries, ESBLs of class A are known so far to be part of the TEM and SHV series, CTX-M-1 to CTX-M-6 or PER-2.
In addition, the identification of GES-1 is another illustration that structurally unrelated class A β-lactamases providing a similar β-lactam resistance phenotype may be recovered from geographically distinct areas. As with the distribution analysis of PER-1 in Turkey (62), an extended epidemiological study in South America would be of interest in determining the prevalence of enterobacterial and nonenterobacterial clinical isolates possessing GES-1.
ACKNOWLEDGMENTS
This work was financed by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES grant JE-2227), Université Paris XI, Paris, France.
We thank R. Labia for his contribution to the discussion.
REFERENCES
- 1.Adrian P V, Du Plessis M, Klugman K P, Amyes S G B. New trimethoprim-resistant dihydrofolate reductase cassette, dfrXV, inserted in a class 1 integron. Antimicrob Agents Chemother. 1998;42:2221–2224. doi: 10.1128/aac.42.9.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler R P. The structure of beta-lactamases. Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa Y, Murakami M, Suzuki K, Ito H, Wachacotyankun R, Ohsuka S, Kato N, Otha M. A novel integron-like element carrying the metallo β-lactamase gene blaIMP-1. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas J M. Sequences of beta-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Stemplinger I, Jungwirth R, Mangold P, Amann S, Akalin E, Ang O, Bal C, Casellas J M. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob Agents Chemother. 1996;40:616–620. doi: 10.1128/aac.40.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissonnette L, Champetier S, Buisson J P, Roy P H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol. 1991;173:4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bito A, Susani M. Revised analysis of aadA2 gene of plasmid pSa. Antimicrob Agents Chemother. 1994;38:1172–1175. doi: 10.1128/aac.38.5.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blazquez J, Negri M C, Morosini M I, Gomez-Gomez J M, Baquero F. A237T as a modulating mutation in naturally occurring extended-spectrum TEM type β-lactamases. Antimicrob Agents Chemother. 1998;42:1042–1044. doi: 10.1128/aac.42.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collis C M, Hall R M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collis C M, Hall R M. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornish-Bowden A. Fundamentals of enzyme kinetics. Seattle, Washington: Portland Press, Inc.; 1995. pp. 30–37. [Google Scholar]
- 14.Danel F, Hall L M C, Gur D, Livermore D. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1881–1884. doi: 10.1128/aac.39.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farzaneh S, Péduzzi J, Sofer L, Reynaud A, Barthélémy M, Labia R. Characterization and amino acid sequence of the OXY-2 group beta-lactamase of pI 5.7 isolated from aztreonam-resistant Klebsiella oxytoca strain HB60. J Antimicrob Chemother. 1997;40:789–795. doi: 10.1093/jac/40.6.789. [DOI] [PubMed] [Google Scholar]
- 16.Feng D F, Doolittle R F. Progressive alignment and phylogenetic tree construction of protein sequences. Methods Enzymol. 1990;183:375–387. doi: 10.1016/0076-6879(90)83025-5. [DOI] [PubMed] [Google Scholar]
- 17.Gazouli M, Tzelepi E, Sidorenko S V, Tzouvelekis L S. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob Agents Chemother. 1998;42:1259–1262. doi: 10.1128/aac.42.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall L M C, Livermore D M, Gur D, Akova M, Akalin H E. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:1637–1644. doi: 10.1128/aac.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-bp element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 20.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 21.Heritage J, M'Zali F H, Gascoyne-Binzi D, Hawkey P M. Evolution and spread of SHV extended spectrum β-lactamases in gram-negative bacteria. J Antimicrob Chemother. 1999;44:309–318. doi: 10.1093/jac/44.3.309. [DOI] [PubMed] [Google Scholar]
- 22.Huletsky A, Couture F, Levesque R C. Nucleotide sequence and phylogeny of SHV-2 beta-lactamase. Antimicrob Agents Chemother. 1990;34:1725–1732. doi: 10.1128/aac.34.9.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii Y, Ohno A, Taguchi H, Imajo S, Ishiguro M, Matsuzawa H. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A beta-lactamase isolated from Escherichia coli. Antimicrob Agents Chemother. 1995;39:2269–2275. doi: 10.1128/aac.39.10.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joris B, Ledent P, Dideberg O, Fonzé E, Lamotte-Brasseur J, Kelly J A, Ghuysen J M, Frère J M. Comparison of the sequences of class A beta-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991;35:2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Lambert T, Ploy M C, Courvalin P. A spontaneous point mutation in the aac(6′)-Ib′ gene results in altered substrate specificity of aminoglycoside 6′-N-acetyltransferase of a Pseudomonas fluorescens strain. FEMS Microbiol Lett. 1994;115:297–304. doi: 10.1111/j.1574-6968.1994.tb06654.x. [DOI] [PubMed] [Google Scholar]
- 27.Laraki N, Galleni M, Thamm I, Riccio M L, Amicosante G, Frère J-M, Rossolini G M. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother. 1999;43:890–901. doi: 10.1128/aac.43.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levesque C, Brassard S, Lapointe J, Roy P H. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 29.Ma L, Ishii Y, Ishiguro M, Matuzawa H, Yamaguchi K. Cloning and sequencing of the gene encoding TOHO-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob Agents Chemother. 1998;42:1181–1186. doi: 10.1128/aac.42.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Freijo P, Fluit A C, Schmitz F J, Verhoef J, Jones M E. Many class I integrons comprise distinct stable structures occurring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe. Antimicrob Agents Chemother. 1999;43:686–689. doi: 10.1128/aac.43.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massida O, Rossolini G M, Satta G. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-β-lactamases. J Bacteriol. 1991;173:4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matagne A, Lamotte-Brasseur J, Frère J M. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto Y, Inoue M. Characterization of SFO-1, a plasmid-mediated inducible class A β-lactamase from Enterobacter cloacae. Antimicrob Agents Chemother. 1999;43:307–313. doi: 10.1128/aac.43.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medeiros A A. Evolution and dissemination of β-lactamases accelerated by generation of β-lactam antibiotics. Clin Infect Dis. 1997;24(Suppl.):S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 35.Naas T, Nordmann P. Analysis of a carbapenem-hydrolyzing class A β-lactamase from Enterobacter cloacae and its LysR-type regulatory protein. Proc Natl Acad Sci USA. 1994;91:7693–7696. doi: 10.1073/pnas.91.16.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naas T, Poirel L, Karim A, Nordmann P. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1999;176:411–419. doi: 10.1111/j.1574-6968.1999.tb13691.x. [DOI] [PubMed] [Google Scholar]
- 37.Naas T, Vandel L, Sougakoff W, Livermore D, Nordmann P. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A β-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob Agents Chemother. 1994;38:1262–1270. doi: 10.1128/aac.38.6.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 39.Nordmann P, Guibert M. Extended-spectrum β-lactamases in Pseudomonas aeruginosa. J Antimicrob Chemother. 1998;42:128–131. doi: 10.1093/jac/42.2.128. [DOI] [PubMed] [Google Scholar]
- 40.Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum beta-lactamase from Pseudomonas aeruginosa and comparison with class A beta-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nordmann P, Ronco E, Naas T, Duport C, Michel-Briand Y, Labia R. Characterization of a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:962–969. doi: 10.1128/aac.37.5.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paulsen I T, Littlejohn T G, Radström P, Sundström L, Sköld O, Swedberg G, Skurray R A. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perrilli M, Franceschini N, Segatore B, Amicosante G, Ovatore A, Duez C, Joris B, Frère J M. Cloning and nucleotide sequencing of the gene encoding the β-lactamase from Citrobacter diversus. FEMS Microbiol Lett. 1991;83:79–84. doi: 10.1016/0378-1097(91)90448-j. [DOI] [PubMed] [Google Scholar]
- 44.Philippon L N, Naas T, Bouthors A T, Barakett V, Nordmann P. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ploy M C, Courvalin P, Lambert T. Characterization of In40 of Enterobacter aerogenes BM2688, a class 1 integron with two new gene cassettes, cmlA2 and qacF. Antimicrob Agents Chemother. 1998;42:2557–2563. doi: 10.1128/aac.42.10.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poirel L, Naas T, Guibert M, Chaibi E B, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raquet X, Lamotte-Brasseur J, Bouillenne F, Frère J M. A disulfide bridge near the active site of carbapenem hydrolyzing class A β-lactamases might explain their unusual substrate profile. Proteins. 1997;27:47–58. doi: 10.1002/(sici)1097-0134(199701)27:1<47::aid-prot6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 48.Rasmussen B A, Bush K, Keeney D, Yang Y, Hare R, O'Gara C, Medeiros A A. Characterization of IMI-1 β-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob Agents Chemother. 1996;40:2080–2086. doi: 10.1128/aac.40.9.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Recchia G D, Hall R M. Gene cassettes: a new class of mobile elements. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 50.Sakurai Y, Tsukamoto K, Sawai T. Nucleotide sequence and characterization of a carbenicillin-hydrolyzing penicillinase gene from Proteus mirabilis. J Bacteriol. 1991;173:7038–7041. doi: 10.1128/jb.173.21.7038-7041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Seoane A, Garcia-Lobo J M. Nucleotide sequence of a new class A β-lactamase gene from the chromosome of Yersinia enterocolitica: implications for the evolution of class A β-lactamases. Mol Gen Genet. 1991;228:215–220. doi: 10.1007/BF00282468. [DOI] [PubMed] [Google Scholar]
- 53.Sougakoff W, Goussard S, Courvalin P. The TEM-3 β-lactamase, which hydrolyses broad-spectrum cephalosporins, is derived from the TEM-2 penicillinase by two amino-acid substitutions. FEMS Microbiol Lett. 1988;56:343–348. [Google Scholar]
- 54.Stokes H W, Hall R M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 55.Stokes H W, Hall R M. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid. 1991;26:10–19. doi: 10.1016/0147-619x(91)90032-r. [DOI] [PubMed] [Google Scholar]
- 56.Stokes H W, Hall R M. The integron In1 in plasmid R46 includes two copies of the oxa2 gene cassette. Plasmid. 1992;28:225–234. doi: 10.1016/0147-619x(92)90054-e. [DOI] [PubMed] [Google Scholar]
- 57.Stokes H W, O'Gorman D B, Recchia G D, Parskhian M, Hall R M. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 58.Sundström L, Radström P, Swedberg G, Sköld O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 59.Sutcliffe J G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci USA. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swofford D L. PAUP (version 3.0): phylogenetic analysis using parsimony. Champaign, Ill: Illinois Natural History Survey; 1989. [Google Scholar]
- 61.Tolmasky M E, Crosa J H. Genetic organization of antibiotic resistance gene (aac(6′)Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid. 1993;29:31–40. doi: 10.1006/plas.1993.1004. [DOI] [PubMed] [Google Scholar]
- 62.Vahaboglu H, Öztürk R, Aygün G, Coşkunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik I̊, Ayolin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walsh T R, MacGowan A P, Bennett P M. Sequence analysis and enzyme kinetics of the L-2 serine beta-lactamase from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1997;41:1460–1464. doi: 10.1128/aac.41.7.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]