ABSTRACT
Acute respiratory distress syndrome (ARDS) can cause loss of alveolar-capillary membrane integrity and life-threatening immune responses. The underlying molecular mechanisms of ARDS remain unclear. N6-methyladenosine (m6A)-RNA modification plays an important part in many biological processes. However, it is not clear whether ARDS alters RNA methylation in lung tissue. We tried to investigate the changes of m6A-RNA methylation in lung tissues of lipopolysaccharide (LPS)-induced ARDS mice. Lung tissue samples were collected to detect the expression of m6A factors through hematoxylin and eosin (HE) staining, quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR), immunohistochemical analysis and western blot. The overall m6A levels in lung tissue of ARDS in mouse were detected by UPLC-UV-MS. HE staining showed that the degree of the inflammatory response was more severe in the LPS-3 h group. The mRNA expression of YTHDF1, YTHDC1 and IGFBP3 was remarkably up-regulated at, respectively, 6, 6 and 12 h after LPS treatment. The mRNA expression of METTL16, FTO, METTL3, KIAA1429, RBM15, ALKBH5, YTHDF2, YTHDF3, YTHDC2 and IGFBP2 was significantly down-regulated at 24 h after LPS treatment. The protein expression of METTL16 and FTO increased, YTHDC1, IGFBP3 YTHDF1 and YTHDF3 showed a down-regulation trend after LPS induction. Overall m6A-RNA methylation levels were significantly increased at 6 h after LPS induction. In ARDS mice, LPS-induced m6A methylation may be involved in the expression regulation of inflammatory factors and may play important roles in the occurrence and development of lung tissue. It is suggested that m6A modification may be a promising therapeutic target for ARDS.
KEYWORDS: Acute respiratory distress syndrome, lung, m6A, RNA methylation, inflammatory response
Graphical abstract
Introduction
Acute respiratory distress syndrome (ARDS), a serious life-threatening disease, occurs in both children and adults, which leads to respiratory dysfunction. The high rate of morbidity and the mortality rate is in the range of 35–60% [1,2]. The major hallmark of ARDS is diffuse alveolar damage characterized by lung edema, inflammation, hemorrhage and alveolar epithelial cell injury [3,4]. The clinical hallmarks of ARDS are hypoxemia and bilateral radiographic opacity, which is related to decreased lung compliance [5]. ARDS is related to a variety of pathological changes, including the destruction of pulmonary vascular endothelial cells, the release of inflammatory cytokines, the accumulation of lung fluid and the formation of excessive fibrosis [6]. In addition, ARDS could develop from direct lung injury (such as pneumonia, drowning, toxic inhalation or gastric content aspiration) or secondary to extra pulmonary conditions, including hemorrhage, sepsis, pancreatitis and blood transfusion [7]. It is worth mentioning that new pathogens may emerge that manifest in ARDS, such as the novel COVID-19 [8]. Clinically, mechanical ventilation and prone positioning are the only interventions proven to decrease mortality [9,10]. However, ARDS patients who receive mechanical ventilation are prone to develop lung fibrosis [11,12]. In addition, the treatment of ARDS is often challenging as it usually occurs in the clinical setting of multiple organ failure and can also lead to non-lung organ damage, such as acute kidney injury [13]. Therefore, it is needed to understand the pathological mechanism and find novel therapeutic targets of ARDS.
Recently, some new ways have been used to explore the pathological mechanism of ARDS, such as weighted gene co-expression network analysis and transcriptome profiling [14,15]. It is noted that DNA methylation is a critical regulator of gene transcription and affects the development of diseases [16]. Szilagyi KL et al. found that epigenetic variation of myosin light-chain kinase was associated with the pathogenesis of ARDS [17]. M6A (also known as N6-methyladenosine) refers to the methylation of the sixth nitrogen atom (N) of adenine, which is used to write, eraser or read methyl groups by methylase complexes. M6A methylation affects a variety of biological processes and plays an important role in regulating inflammatory responses [18]. It is shown that m6A methylation is associated with the pathogenesis of lung ischemia-reperfusion injury [19]. In addition, lung biopsy is one of the most common procedures to obtain a histologic diagnosis. Up to now, it is not clear whether ARDS alters RNA methylation in lung tissue, and whether m6A-RNA methylation is involved in the pathology of ARDS. In view of this, we tried to evaluate the changes of m6A-RNA methylation in lung tissues of lipopolysaccharide (LPS)-induced ARDS mice to uncover the potential role of m6A-RNA methylation in the development of ARDS.
Materials and methods
Construction of ARDS mouse model
The SPF C57BL/6 strain male mice (aged 3–8 weeks and weight 18–22 g) were purchased. The mice were fed freely and fed water adaptively for more than 3 days. All mice were divided into the following six groups: normal group (no treatment, NC), no load control group (normal saline modeling, VC), 3 h group after LPS treatment (LPS-3 h), 6 h group after LPS treatment (LPS-6 h), 12 h group after LPS treatment (LPS-12 h) and 24 h group after LPS treatment (LPS-24 h). LPS was purchased from Sigma Company. Physiological saline was used for ultrasonic solution at 60°C, and kept at 37°C for reserve. The NC group did not do any processing. Mice in the VC or LPS treatment group were administrated with invasive tracheotomy intubation (5 mg/kg per mouse). The mice were anesthetized with a small animal anesthesia machine. Then, the mice were supine on a wooden board with the tilt of the board at an angle of 50° to the horizontal plane. The submaxillary skin was incised, and the trachea was separated. One hundred microliters of air was pre-inhaled with a syringe. One hundred microliters of LPS solution or normal saline was extracted and slowly injected into the trachea. After the injection, the mice were upright and gently rotated vertically to make the drug evenly distributed in the lung. After the skin was sutured, the mice were placed on a thermal insulation plate to wake up. The mice were raised in a single cage for a certain time according to the group requirements. The study was approved by the Ethics Committee on Laboratory Animals of the First Affiliated Hospital of Anhui Medical University (LLS-C20210502).
Collection of lung tissue sample
After anesthesia, the mice were sacrificed. The chest was opened to collect the whole lung and irrigated twice with normal saline. The left lower lobe was fixed with paraformaldehyde solution. The lung tissue was fixed in 10% neutral formaldehyde. Left upper lobe/right upper lobe/right lower lobe was collected and added to TRNzol (TIANGEN), then pre-cooled with liquid nitrogen and cryopreserved at −80°C. Mice carcasses were treated innocuously.
Hematoxylin and eosin (HE) staining
The method of HE staining was referred from previous report [20]. Lung sections were dehydrated by automatic dehydrator as follows: 75% of alcohol for 4 h, 85% of alcohol for 2 h, 95% of alcohol for 1 h, 100% of alcohol for 0.5 h, 100% of alcohol for 0.5 h, 100% of alcohol for 0.5 h, 100% of alcohol for 0.5 h, 100% of alcohol for 0.5 h, 100% of alcohol for 0.5 h, xylene for 10 min for 2 times, paraffin for 1 h, 2 h and 3 h. Lung sections were dyed with HE for 10–20 min, rinsed with running water for 1–3 min, differentiated with alcohol hydrochloride for 5–10s and rinsed with running water for 1–3 min. Lung sections were put into 50°C warm water or weak alkaline aqueous solution to return blue. Lung sections were rinsed with running water for 1–3 min, added 85% of alcohol for 3–5 min, stained with Eosin for 3–5 min and rinsed for 3–5 s. Lung sections were dehydrated with gradient alcohol, transparent treatment with xylene and sealed with neutral gum. Pannoramic 250 digital section scanner was used for image collection of sections. All sections were firstly observed at 40 times to find gross lesions. In selected areas, images of 100 times and 400 times were collected to observe specific lesions.
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
Detailed operation method was based on previous research [21]. The lung tissue was collected into 2 ml of cryopreservation tube, added 1 ml of TRNzol solution and fully broken with a hand-held homogenizer. The homogenate samples were placed at room temperature for 5 min to separate the nucleic acid protein complex completely. The homogenate samples were centrifuged at 12000rpm at 4°C for 10 min to collect supernatant. The supernatant was added 0.2 ml of chloroform, swirled for about 15 s, mixed thoroughly, stand for 3 min at room temperature and centrifuged at 12000 rpm at 4°C for 15 min to collect the upper water phase. The upper water phase was added the same volume isopropyl alcohol, mixed upside down, stand at room temperature for 10 min and centrifuged at 12000 rpm at 4°C for 10 min to remove supernatant. The mixture was added 1 ml of 75% ethanol to wash and centrifuged at 10000 rpm at 4°C for 5 min. The liquid was poured out and blotted the remaining 75% ethanol with absorbent paper. Twenty microliters of RNAse-free H2O was added to dissolve the RNA precipitate. For the detection of m6A factors, the template RNA was thawed on ice. Genomic DNA removal system was thoroughly mixed, centrifuged briefly and incubated at 42°C for 3 min. The mixture of reverse transcription reaction system was prepared. The mixture was mixed with the genomic DNA removal system, incubated at 42°C for 15 min, incubated at 95°C for 3 min and then placed on ice. The cDNA obtained could be used for subsequent experiments or cryopreserved. 2× SuperReal PreMix Plus, 50× ROX Reference Dye, cDNA template, primer and RNAse-free ddH2O were balanced at room temperature and mixed thoroughly. The RT-PCR reaction was performed. The CT values were analyzed by 2−ΔΔCT method. For the analysis of overall m6A levels, 1.5 μl of buffer, 2 μl of alkaline phosphatase dilution, 4 μl of snake venom phosphatase I, deionized water were mixed with 15 μg of RNA and hydrolyzed at 37°C for 4 h. After enzymatic hydrolysis, 90 μl deionized water was added and diluted to 120 μl. One hundred microliters of chloroform was added and fully vortically mixed for 3 min, then centrifuged at 12000 rpm for 8 min. One hundred and twenty microliters of supernatant was collected and added another 100 μl of chloroform. The above process was repeated for 3 times. One hundred microliters of supernatant was collected and store at −20°C for UPLC-UV-MS analysis according to the previous method to detect the overall m6A levels in lung tissue of ARDS mouse [22]. The result was represented as m6A/A% (area m6A*Xm6A/Area A*100%). In the formula, A stands for A bases; Xm6A comes from a known concentration quality control sample (correction factor).
Immunohistochemical analysis
Paraffin sections of lung tissue were dewaxed to water as follows [21]: sections were placed in xylene I for 15 min, xylene II for 15 min, xylene III for 15 min and anhydrous ethanol I for 5 min, anhydrous ethanol II for 5 min, 85% of alcohol for 5 min, 75% of alcohol for 5 min and washed with distilled water. The sections were dipped into citric acid buffer, heat in microwave oven for 10 min on high heat, hold fire for 8 min and reheat in medium-high heat for 10 min. After cooling, the sections were washed for 3 times with PBS, 5 min each. The sections were placed in 3% of hydrogen peroxide at room temperature for 10 min and washed with PBS for 3 times, 5 min each. The sections were incubated with goat serum blocking solution at room temperature for 20 min. After incubation with primary antibody at 4°C overnight, the sections were washed with PBS for 3 times, incubated with secondary antibody at 37°C for 30 min and washed with PBS 3 times. Fresh DAB chromogenic solution was prepared and dropped onto the sections for chromogenic development at room temperature. The chromogenic time was controlled under the microscope, and the positive color was brownish yellow. The chromogenic effect was terminated by washing the sections in distilled water. After redyeing with hematoxylin for 3 min, the sections were washed with tap water until the emergence of blue color. The sections were soaked in 75%, 85%, 95% of anhydrous ethanol and xylene for 10 min, and sealed with neutral gum. The microphotography system was utilized to collect images of the sections. All lung tissues of each section were firstly observed at 100 time under the microscope. Those microscopic images at 400 time were collected. The percentage of positive staining in each image was calculated using Halo data analysis system. The nuclei stained with hematoxylin were blue. The positive expression of DAB was brownish yellow.
Western blot detection
A total of 32 μg of protein samples from lung tissue were slowly added to the well. Electrophoresis was performed with a stable voltage of 100 V for 15 min. Protein transmembrane process was performed at 200 mA for 1–2 h. The PVDF membrane was incubated with 5% skim milk and gently shaken on a shaker for 2 h. The PVDF membrane was incubated with the primary antibody, shaken gently on the shaker, incubated at 4°C overnight and washed with TBST for 3 times, 5 min each. The PVDF membrane was incubated with secondary antibody, shaken gently on a shaker, incubated at room temperature for 2–3 h and washed for 3 times with TBST, 10 min each. The PVDF membrane was laid on the exposure plate and mixed with the A and B reagents of ECL luminescence solution. After reaction for 1 min, the exposure plate with the film was put into the darkroom of chemiluminescence gel imager. The exposure time was adjusted according to the strength of the signal. The bands were scanned by exposure using TIeneng GIS box control software V2.0. The results were expressed as the relative expression of the target protein. Comparative expression of the target protein was the ratio of integrated optical density of target protein (IOD)/integrated optical density of internal reference (IOD). Above method was referred to the previous report [23].
Results
The underlying molecular mechanisms of m6A-RNA modification in ARDS remain unclear. In this study, we tried to investigate the changes of m6A-RNA methylation in lung tissues of LPS-induced ARDS mice. Lung tissue samples were collected to detect the expression of m6A factors through HE staining, qRT-PCR, immunohistochemical analysis and western blot.
HE staining in lung tissue of ARDS mouse
In the NC group, the bronchial structure of all levels of lung tissue was normal, and the bronchial ciliary epithelium was neatly arranged (Figure 1). The morphology of alveolar epithelial cells was basically normal, with no obvious degeneration and necrosis, no obvious thickening of alveolar diaphragm. A small number of macrophages were found in the alveolar cavity. No observable pathological changes were found. In the VC group, the bronchial structure of all levels of the lung was normal, and the bronchial ciliary epithelium was arranged neatly without obvious cell shedding. Alveolar epithelial cells were normal. The hemorrhage was observed in the alveolar cavity, and more red blood cells spilled. There was a minimal infiltration of inflammatory cells in the lung interstitium, mainly neutrophils. No other obvious pathological changes were observed. In the LPS-3 h group, the bronchial structure at all levels of lung tissue was normal, and the bronchial ciliary epithelium was neatly arranged. The morphology of alveolar epithelial cells was normal, without obvious degeneration necrosis. The alveolar diaphragm was thickened in some areas. More neutrophils infiltrated in alveolar stroma. The vascular lumen and perivascular interstitium were infiltrated by lymphocytes with round nuclei. The alveolar cavity showed a foam-like cellular infiltration of cytoplasm. No other obvious pathological changes were observed. In the LPS-6 h group, lung layer pleura structure was normal with a thin wall and without connective tissue hyperplasia and thickening. The bronchial structure of all levels was complete and clear. The morphology of epithelial cells was normal without obvious degeneration, necrosis or shedding. The morphology of type I and type II alveolar epithelial cells in the respiratory part of the lung was normal without obvious degeneration and necrosis. A small amount of inflammatory cells infiltrated the interstitium, mainly rod-shaped neutrophils. No other obvious pathological changes were observed. In the LPS-12 h group, the bronchial structure at all levels of lung tissue was normal, and the bronchial ciliary epithelium was neatly arranged. The morphology of alveolar epithelial cells was basically normal, with no obvious degeneration and necrosis, no obvious thickening of the alveolar diaphragm. More lymphocytes and neutrophils were observed in the alveolar stroma. There were more lymphocytes infiltrating in the interstitium around blood vessels. Macrophages were seen in the alveolar lumen. No other obvious pathological changes were found. In conclusion, compared with the NC group, different degrees of pathological changes (such as the inflammatory response) were observed in both the LPS-3 h and LPS-12 h groups. Significantly, the degree of pathological changes was more severe in the LPS-3 h group.
Figure 1.
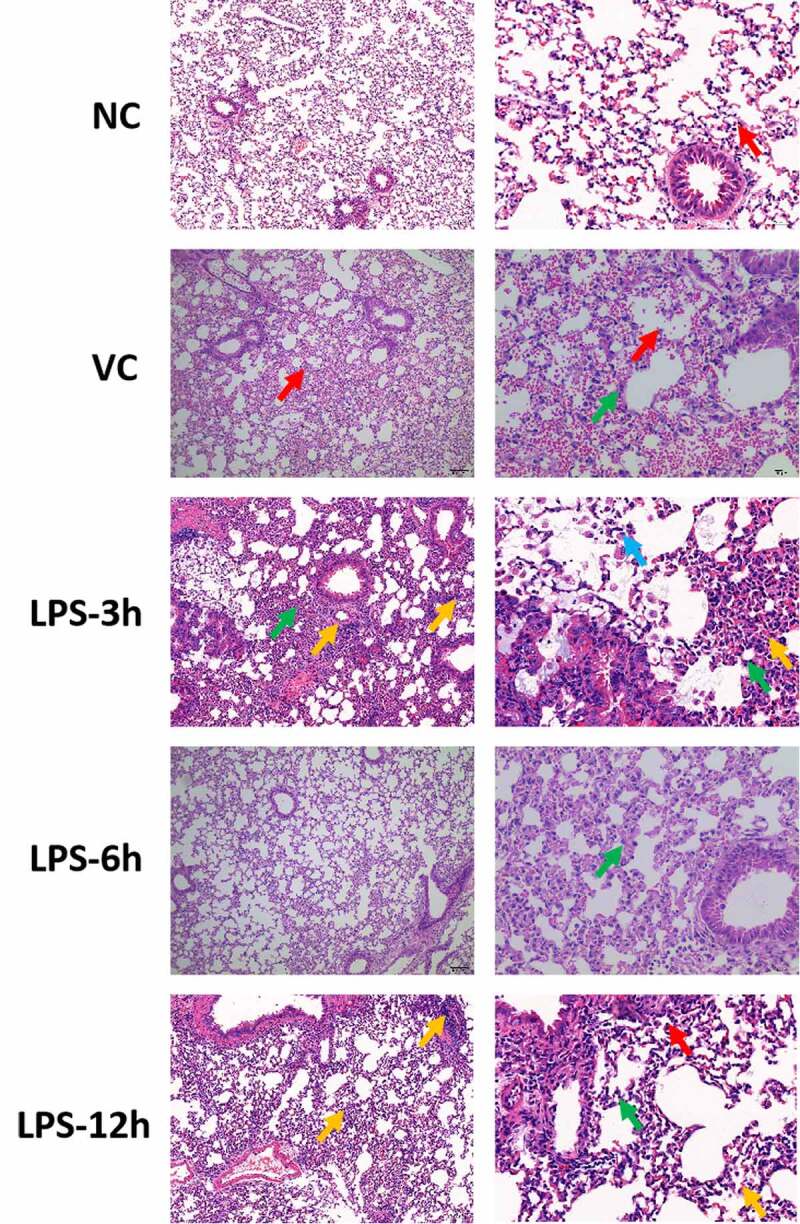
HE staining in lung tissue of ARDS mouse. Left and right side represent the picture with magnification × 100 and magnification × 400, respectively. In NC group, the red arrow represents macrophages; in VC group, red and green arrow represents red blood cells and neutrophils, respectively; in the LPS-3 h group, green, yellow and blue arrow represents lymphocyte, neutrophils and foam cells, respectively; in the LPS-6 h group, green arrow represents neutrophils; in the LPS-12 h group, green, yellow and red represents lymphocyte, neutrophils and macrophages, respectively.
The mRNA expression of m6A factors in lung tissue of ARDS mouse
In the lung tissue samples of ARDS mouse in NC, VC, LPS-3 h, LPS-6 h, LPS-12 h and LPS-24 h groups, the mRNA expression of 19 m6A-related factors were detected, including methylated transferases (METTL3, METTL14, METTL16, KIAA1429, RBM15, RBM15B and WTAP), demethylases (FTO and ALKBH5) (Figure 2) and recognition factors (YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, eIF3a, hnRNP A2/B1, IGFBP1, IGFBP2 and IGFBP3) (Figure 3). The primer sequences of above m6A factors are listed in Table 1. The expression of METTL16 and FTO showed a significant down-regulation at 3, 6, 12 and 24 h after LPS treatment. METTL3, KIAA1429, RBM15 and ALKBH5 were significantly down-regulated at 24 h after LPS treatment. The expression of YTHDF1, YTHDC1 and IGFBP3 was remarkably up-regulated at, respectively, 6, 6 and 12 h after LPS treatment. YTHDF2, YTHDF3, YTHDC2 and IGFBP2 were significantly down-regulated at 24 h after LPS treatment.
Figure 2.
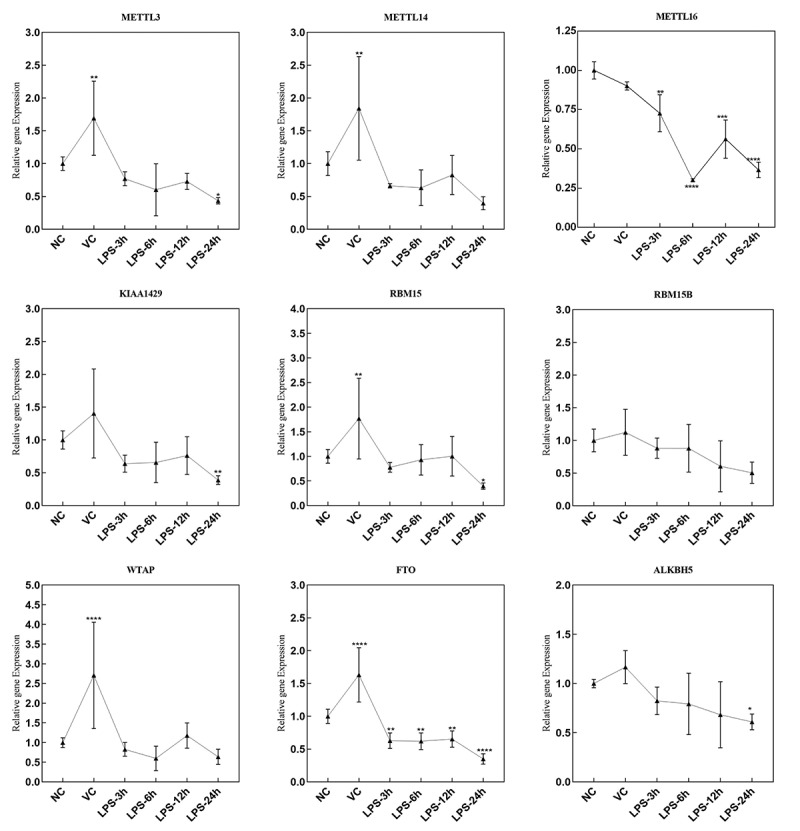
The mRNA expression of methylated transferases and demethylases in the lung tissue samples of ARDS mouse in NC, VC, LPS-3 h, LPS-6 h, LPS-12 h and LPS-24 h groups. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Figure 3.
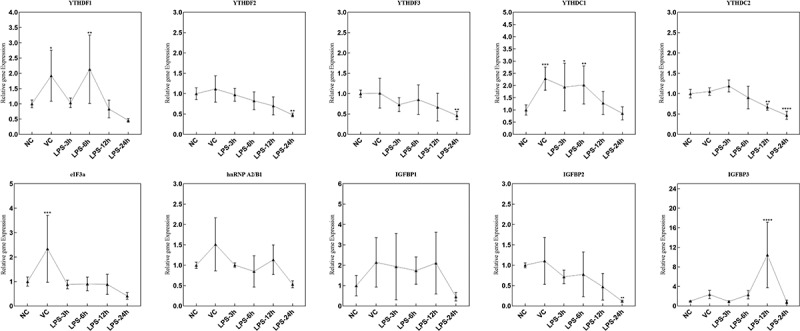
The mRNA expression of recognition factors in lung tissue samples of ARDS mouse in NC, VC, LPS-3 h, LPS-6 h, LPS-12 h and LPS-24 h groups. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Table 1.
The primer sequences of 19 m6A factors in the qRT-PCR
| Gene | Primer sequences (5’-3’) |
|---|---|
| METTL3 | Forward: ATCCAGGCCCATAAGAAACAG |
| Reverse: CTATCACTACGGAAGGTTGGG | |
| METTL14 | Forward: CTGAGAGTGCGGATAGCATTG |
| Reverse: GAGCAGATGTATCATAGGAAGCC | |
| ALKBH5 | Forward: CAGTGGGTATGCTGCTGATG |
| Reverse: GGGTCTCTGGTGTTTCCTGA | |
| FTO | Forward: GACACTTGGCTTCCTTACCTG |
| Reverse: CTCACCACGTCCCGAAACAA | |
| WTAP | Forward: GCCCCAACGTTTAAGTGCAG |
| Reverse: AGCATTCGACACTTCGCCAT | |
| RBM15B | Forward: CGTGGGAGGACAACTCACTC |
| Reverse: GAGTTGCTGCCTGACTCCTT | |
| METTL16 | Forward: ACAAACCACCTGACTTCGCA |
| Reverse: CTGACTGCTTCGGGGTCTTT | |
| eIF3a | Forward: GGAGAGATTCAAGTCGCCGT |
| Reverse: CTGTCATCAGTGCGTCTCCA | |
| HnRNP A2/B1 | Forward: CCGATAGGCAGTCTGGAAAG |
| Reverse: TATAGCCATCCCCAAATCCA | |
| IGFBP1 | Forward: GATCGCCGACCTCAAGAAATG |
| Reverse: CCTCTAGTCTCCAGAGACCCAG | |
| IGFBP2 | Forward: ACCCCTTGCCAGCAGGAGTTGGA |
| Reverse: TCCCTGGATGGGCTTCCCGGT | |
| IGFBP3 | Forward: GACAGAATACGGTCCCTGCC |
| Reverse: GGAGCATCTACTGGCTCTGC | |
| KIAA1429 | Forward: CAGTCGTCGGGAAGGAACAA |
| Reverse: TAGGGCGGTAACCCGTAGAA | |
| RBM15 | Forward: GCAGCATCCTCTCCCAAACT |
| Reverse: AGGTCACCCTGCAACAGATG | |
| YTHDF1 | Forward: CTGCAGTTAAGACGGTGGGT |
| Reverse: TAGCAATGGCTGCCCATGAA | |
| YTHDF2 | Forward: AGGCGGGTTCTGGATCTACT |
| Reverse: GATAGGCGGCATCCAGTCTC | |
| YTHDF3 | Forward: CAGAGACCTAAAGGGCAAGGA |
| Reverse: CATGCTGCTTCCCCAAGAGA | |
| YTHDC1 | Forward: CGTAGGAAGCTGAGTGGAGC |
| Reverse: TCCCCATCTTTCTCCTCCCG | |
| YTHDC2 | Forward: GGTCCGATCAATCATCTGT |
| Reverse: GAAGTAACGAATAGGCATGT | |
| GAPDH | Forward: TTCCAGTATGACTCTACCCACGGCA |
| Reverse: GCACCAGCATCACCCCATTTG |
Overall m6A levels in lung tissue of ARDS mouse
UPLC-UV-MS was used to detect the changes of m6A levels in the lung tissues of ARDS mouse. The overall expression level of m6A in all groups changed significantly and increased significantly at 6 h and 12 h after LPS induction, followed by a downward trend (Figure 4). Combined with the overall detection results, the expression of m6A factor changed significantly at 6 h after LPS induction. The methylation level of m6A showed a significant increase. The results showed that m6A-RNA methylation changed 6 h after LPS induction in the lung tissues of ARDS mouse.
Figure 4.
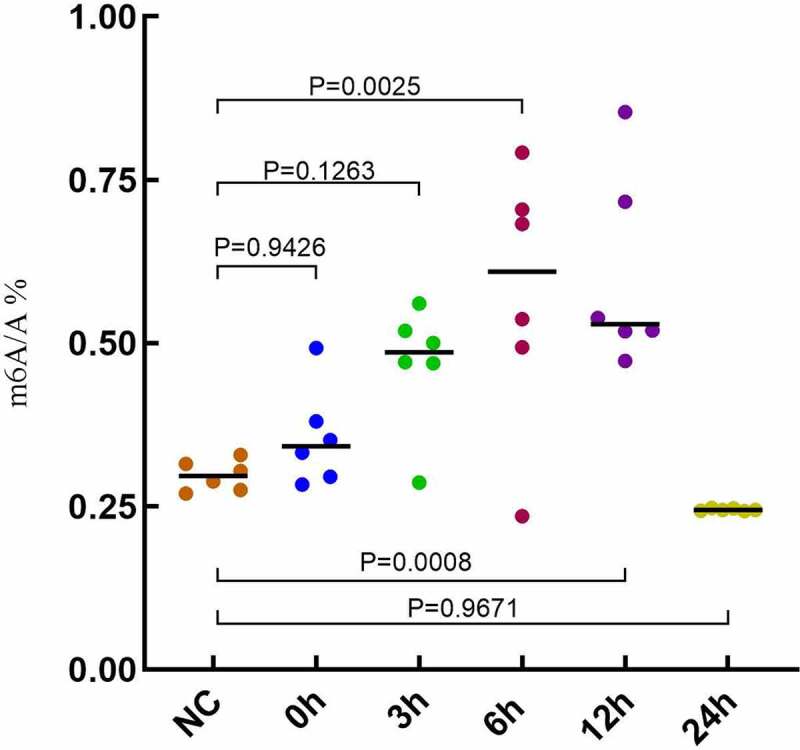
Overall m6A levels in lung tissue of ARDS mouse.
The protein expression of m6A factors in lung tissue of ARDS mouse
The protein expression of seven m6A factors was detected by Western blot, including (METTL16 and WTAP), demethylases (FTO) and recognition factors (YTHDF1, YTHDF3, YTHDC1 and IGFBP3) (Figure 5). In lung tissue of ARDS mouse, the expression of METTL16 and FTO increased, YTHDC1 and IGFBP3 showed a down-regulation trend. The protein expression of YTHDF1 and YTHDF3 decreased at 24 h after LPS induction, while WTAP did not change. The original images of western blots including weight markers are shown in supplementary Figure 1.
Figure 5.
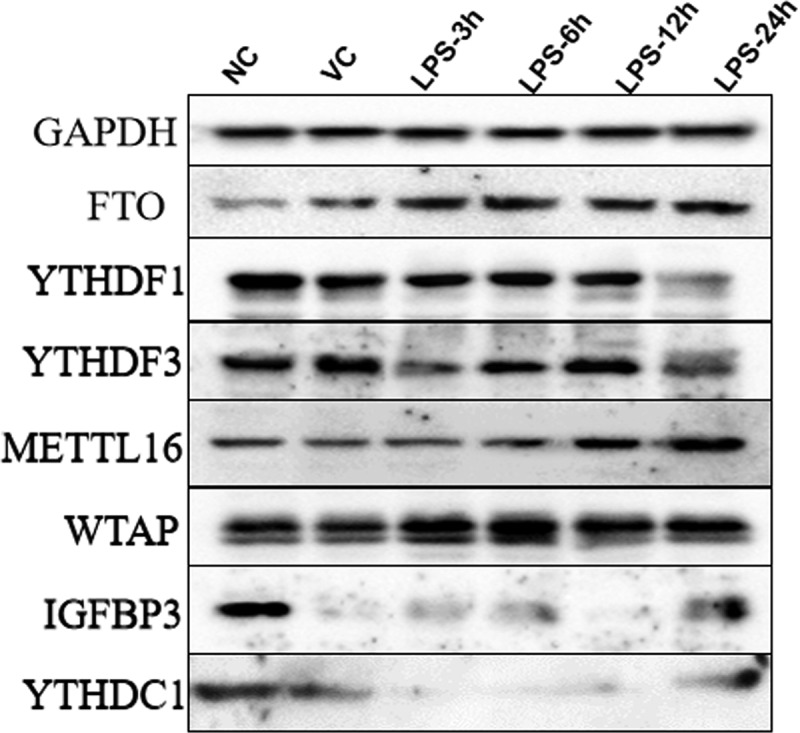
The protein expression of METTL16, WTAP, FTO, YTHDF1, YTHDF3, YTHDC1 and IGFBP3 in the lung tissue samples of ARDS mouse in NC, VC, LPS-3 h, LPS-6 h, LPS-12 h and LPS-24 h groups.
Immunohistochemical test of m6A factors in lung tissue of ARDS mouse
Immunohistochemical characteristics of five m6A proteins were detected in the right lung tissue of ARDS mouse, including FTO, METTL16, YTHDC1, YTHDF1 and YTHDF3 (Figure 6 and Figure 7). The result showed that the expression of FTO protein showed a significant upward trend after 3, 6, 12 and 24 h after LPS induction. YTHDC1 and YTHDF1 were significantly down-regulated after 3, 6, 12 and 24 h after LPS induction. METTL16 was significantly down-regulated at 3 h after LPS induction and up-regulated at 12 and 24 h after LPS induction. YTHDF3 was remarkably up-regulated at 12 h after LPS induction and down-regulated at 3, 6 and 24 h after LPS induction. The result was in line with those results of Western blot. Interestingly, the mRNA and protein expression of some m6A factors were inconsistent, which are worth further investigation.
Figure 6.
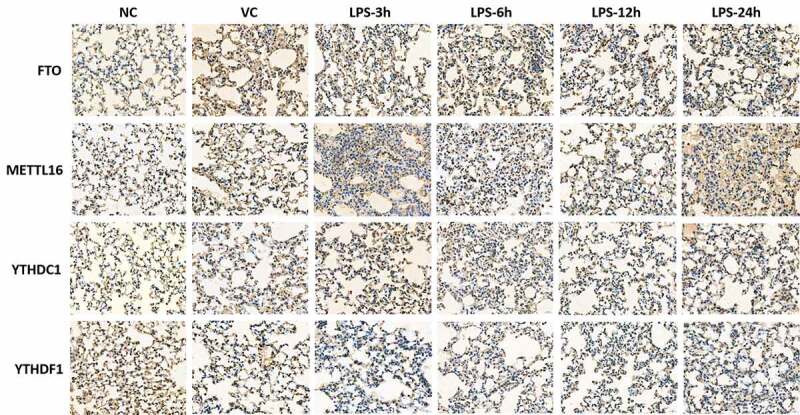
Immunohistochemical tests of FTO, METTL16, YTHDC1, YTHDF1 and YTHDF3 in right lung tissue of ARDS mouse. Magnification: 400 ×.
Figure 7.
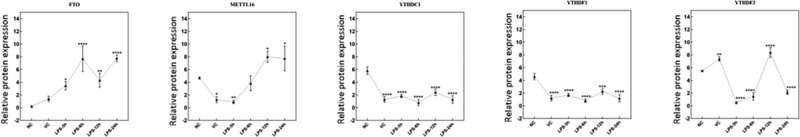
Immunohistochemical quantitative results of FTO, METTL16, YTHDC1, YTHDF1 and YTHDF3 in right lung tissue of ARDS mouse. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Discussion
A large amount of newly discovered evidence suggests that m6A modification is associated with sepsis (systemic inflammatory response syndrome) [24]. Changes in m6A modification are found in sepsis-induced myocardial dysfunction, which suggested that m6A modification may be a promising therapeutic target for sepsis [25]. Thus, it can be seen that m6A modification plays important roles in the inflammatory response. In this study, we found that compared with the NC group, different degrees of pathological changes (such as the inflammatory response) were observed in both the LPS-3 h and LPS-12 h groups in lung tissue of ARDS mouse. Significantly, the degree of pathological changes was more severe in the LPS-3 h group. Our result indicated that m6A methylation may be involved in the expression regulation of inflammatory factors in the lung tissues of ARDS in mice, which may be considered as a potential therapeutic target for ARDS.
YTHDF1 recruits translation initiation factors to facilitate translation [26]. In normal human bronchial epithelium cells, knockdown mRNA expression of YTHDF1 significantly abrogates hypoxia-induced cellular apoptosis [27]. YTHDC1 drives the binding of methylated mRNA targets to SRSF3 and NXF1 to form mRNA-protein complexes, which together promote nuclear export of RNA and regulate mRNA splicing [28,29]. IGFBP3 has been involved in fibrotic lung diseases and persistent ARDS because of profibrogenic and antiapoptotic activity [30]. Increased expression level of IGFBP3 is found in fibrotic lung and bronchoalveolar lavage fluid of patients with ARDS and patients with risk factors for ARDS [30,31]. In this study, we found that the mRNA expression of YTHDF1, YTHDC1 and IGFBP3 was remarkably up-regulated at, respectively, 6, 6 and 12 h after LPS treatment in ARDS mice. This indicated that these YTHDF1, YTHDC1 and IGFBP3 may be involved in the process of ARDS.
METTL3, is known as ‘writers,’ plays a crucial role in regulation of gene expression by influencing RNA stability and mRNA translation [26,32]. There is a significant correlation between the expression of METTL3 and the occurrence of chronic obstructive pulmonary disease [33]. METTL16 is differentially expressed in mild and severe pneumonia [34]. YTHDF2 recognizes dynamic m6A modification to influence the translation status and lifespan of mRNA [26,35]. YTHDF2 can promote influenza A virus republication and spread [36]. Additionally, YTHDF2 plays important roles in the intracellular regulation of inflammatory factors in cystic fibrosis lung disease [37,38]. YTHDF3, a direct m6A reader, promotes the translation of mRNAs and improve stability [39]. YTHDC2 promotes the ‘fast-track’ expression program for specific mRNAs by modulating the stability of mRNAs [40]. YTHDC2 is associated with some important genes that promote the development progression of chronic obstructive pulmonary disease [33]. FTO, a m6A eraser, impacts regulating transcription through removing methyl groups from nucleotides [41]. In non-small-cell lung cancer, FTO promotes disease progress by decreasing m6A level [42]. In lung adenocarcinoma, FTO can activate cell migration via mRNA demethylation and accelerate the disease progression [43]. KIAA1429 is a modification that plays important roles in the effectiveness of mRNA splicing and RNA processing [44]. In the lung adenocarcinoma, KIAA1429 is related to the formation of m6A methyltransferase complex, which promotes the formation of m6A [45–47]. RBM15 can mediate the formation of m6A in X-inactive specific transcripts and cellular mRNAs [48]. RBM15 is positively associated with the severity of COVID-19, implying that RBM15 has a key role in SARS-CoV-2 infection by influencing methylation levels of genes [49]. ALKBH5 can modulate mRNA splicing and stability. Dysfunctional demethylation of ALKBH5 reduces the expression of MALAT1, which induces apoptosis and reduces the production of inflammatory factors [50]. IGFBP2, a lineage marker for diverse human lung epithelial cell types, Alveolar Type I, is up-regulated during lung regeneration [51,52]. In this study, we found that the mRNA expression of METTL3, METTL16, YTHDF2, YTHDF3, YTHDC2, FTO, KIAA1429, RBM15, ALKBH5 and IGFBP2 were significantly down-regulated at 24 h after LPS treatment in ARDS mice. It is suggested that these m6A factors may be involved in the inflammation and lung pathology in the development of ARDS.
However, there are limitations to our study. Firstly, the protein expression of some m6A factors was inconsistent with the mRNA expression, which is needed for further investigation; Secondly, it is necessary to find target mRNAs (especially inflammatory cytokines) of these m6A factors to further study deeper pathological mechanism of ARDS.
Conclusion
We found that overall m6A-RNA methylation levels were significantly increased after LPS induction in ARDS. Furthermore, significant expression changes in some m6A factors were found, including YTHDF1, YTHDC1, IGFBP3, METTL16, FTO, METTL3, KIAA1429, RBM15, ALKBH5, YTHDF2, YTHDF3, YTHDC2 and IGFBP2. These m6A factors may be involved in regulating inflammatory cytokines in the process of ARDS, which could be regarded as promising therapeutic targets for ARDS.
Supplementary Material
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data are available in the article.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Bellani G, Laffey JG, Pham T, et al. Epidemiology, Patterns of Care, and Mortality for Patients With Acute Respiratory Distress Syndrome in Intensive Care Units in 50 Countries. Jama. 2016;315(8):788–800. [DOI] [PubMed] [Google Scholar]
- [2].Vincent JL, Sakr Y, Ranieri VM.. Epidemiology and outcome of acute respiratory failure in intensive care unit patients. Crit Care Med. 2003;31(Supplement):S296–9. [DOI] [PubMed] [Google Scholar]
- [3].Katzenstein AL, Bloor CM, Leibow AA. Diffuse alveolar damage–the role of oxygen, shock, and related factors. A review. Am J Pathol. 1976;85:209–228. [PMC free article] [PubMed] [Google Scholar]
- [4].Tomashefski JF Jr. Pulmonary pathology of acute respiratory distress syndrome. Clin Chest Med. 2000;21(3):435–466. [DOI] [PubMed] [Google Scholar]
- [5].Ranieri VM, Rubenfeld GD, Thompson BT, et al. Acute respiratory distress syndrome: the Berlin Definition. Jama. 2012;307(23):2526–2533. [DOI] [PubMed] [Google Scholar]
- [6].Metwaly S, Cote A, Donnelly SJ, et al. Evolution of ARDS biomarkers: will metabolomics be the answer? American Journal of Physiology. Lung Cellular and Molecular Physiology. 2018;315(4):L526–l34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dushianthan A, Cusack R, Goss V, et al. Clinical review: exogenous surfactant therapy for acute lung injury/acute respiratory distress syndrome–where do we go from here? Crit Care. 2012;16:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guan WJ, Ni ZY, Hu Y, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. J Integr Med. 2020;382(18):1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brower RG, Matthay MA, Morris A, et al. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N Engl J Med. 2000;342:1301–1308. [DOI] [PubMed] [Google Scholar]
- [10].Guérin C, Reignier J, Richard JC, et al. Prone positioning in severe acute respiratory distress syndrome. N Engl J Med. 2013;368(23):2159–2168. [DOI] [PubMed] [Google Scholar]
- [11].Cabrera-Benítez NE, Parotto M, Post M, et al. Mechanical stress induces lung fibrosis by epithelial-mesenchymal transition. Crit Care Med. 2012;40(2):510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang R, Pan Y, Fanelli V, et al. Mechanical Stress and the Induction of Lung Fibrosis via the Midkine Signaling Pathway. Am J Respir Crit Care Med. 2015;192(3):315–323. [DOI] [PubMed] [Google Scholar]
- [13].Williams GW, Berg NK, Reskallah A, et al. Acute Respiratory Distress Syndrome. Anesthesiology. 2021;134(2):270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fang Q, Wang Q, Zhou Z, et al. Consensus analysis via weighted gene co-expression network analysis (WGCNA) reveals genes participating in early phase of acute respiratory distress syndrome (ARDS) induced by sepsis. Bioengineered. 2021;12(1):1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ma J, Li Q, Ji D, et al. Predicting candidate therapeutic drugs for sepsis-induced acute respiratory distress syndrome based on transcriptome profiling. Bioengineered. 2021;12(1):1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].El Gazzar M, Yoza BK, Hu JY, et al. Epigenetic silencing of tumor necrosis factor alpha during endotoxin tolerance. J Biol Chem. 2007;282(37):26857–26864. [DOI] [PubMed] [Google Scholar]
- [17].Szilágyi KL, Liu C, Zhang X, et al. Epigenetic contribution of the myosin light chain kinase gene to the risk for acute respiratory distress syndrome. Transl Res. 2017;180:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Maity A, Das B. N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. The FEBS Journal. 2016;283(9):1607–1630. [DOI] [PubMed] [Google Scholar]
- [19].Xiao K, Liu P, Yan P, et al. N6-methyladenosine reader YTH N6-methyladenosine RNA binding protein 3 or insulin like growth factor 2 mRNA binding protein 2 knockdown protects human bronchial epithelial cells from hypoxia/reoxygenation injury by inactivating p38 MAPK, AKT, ERK1/2, and NF-κB pathways. 2021. [DOI] [PMC free article] [PubMed]
- [20].Bangaoil R, Santillan A, Angeles LM, et al. ATR-FTIR spectroscopy as adjunct method to the microscopic examination of hematoxylin and eosin-stained tissues in diagnosing lung cancer. PloS one. 2020;15(5):e0233626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nakajima N, Van Tin N, Sato Y, et al. Pathological study of archival lung tissues from five fatal cases of avian H5N1 influenza in Vietnam. Mod Pathol. 2013;26(3):357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chang JS, Lin ZX, Liu YJ, et al. Ultra performance liquid chromatography-tandem mass spectrometry assay for the quantification of RNA and DNA methylation. J Pharm Biomed Anal. 2021;197:113969. [DOI] [PubMed] [Google Scholar]
- [23].Punsawad C, Viriyavejakul P, Techarang T. Surfactant Protein D Is Altered in Experimental Malaria-Associated Acute Lung Injury/Acute Respiratory Distress Syndrome. J Trop Med. 2019;2019:9281605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang Y, Xu M, Yue P, et al. Novel Insights Into the Potential Mechanisms of N6-Methyladenosine RNA Modification on Sepsis-Induced Cardiovascular Dysfunction: an Update Summary on Direct and Indirect Evidences. Front Cell Dev Biol. 2021;9:772921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Han YC, Xie HZ, Lu B, et al. Lipopolysaccharide Alters the m6A Epitranscriptomic Tagging of RNAs in Cardiac Tissue. Front Mol Biosci. 2021;8:670160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161(6):1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shi Y, Fan S, Wu M, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nature Communications. 2019;10(1):4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elif. 2017;6(6) :e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Di Timoteo G, Dattilo D, Centrón-Broco A, et al. Modulation of circRNA Metabolism by m(6)A Modification. Cell Rep. 2020;31(6):107641. [DOI] [PubMed] [Google Scholar]
- [30].Ahasic AM, Zhai R, Su L, et al. IGF1 and IGFBP3 in acute respiratory distress syndrome. Eur J Endocrinol. 2012;166(1):121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schnapp LM, Donohoe S, Chen J, et al. Mining the acute respiratory distress syndrome proteome: identification of the insulin-like growth factor (IGF)/IGF-binding protein-3 pathway in acute lung injury. Am J Pathol. 2006;169(1):86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Huang X, Lv D, Yang X, et al. m6A RNA methylation regulators could contribute to the occurrence of chronic obstructive pulmonary disease. J Cell Mol Med. 2020;24(21):12706–12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Huang S, Feng C, Chen L, et al. Identification of Potential Key Long Non-Coding RNAs and Target Genes Associated with Pneumonia Using Long Non-Coding RNA Sequencing (lncRNA-Seq): a Preliminary Study. Med Sci Monit. 2016;22:3394–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shi H, Wei J, He C. Where, When, and How: context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol Cell. 2019;74(4):640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Feng Q, Zhao H, Xu L, et al. N6-Methyladenosine Modification and Its Regulation of Respiratory Viruses. Front Cell Dev Biol. 2021;9:699997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yu Y, Pan Y, Fan Z, et al. LuHui Derivative, A Novel Compound That Inhibits the Fat Mass and Obesity-Associated (FTO), Alleviates the Inflammatory Response and Injury in Hyperlipidemia-Induced Cardiomyopathy. Front Cell Dev Biol. 2021;9:731365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dang H, Polineni D, Pace RG, et al. Mining GWAS and eQTL data for CF lung disease modifiers by gene expression imputation. PloS one. 2020;15(11):e0239189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tan S, Li Z, Li K, et al. The Regulators Associated With N6-Methyladenosine in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma Reveal New Clinical and Prognostic Markers. Front Cell Dev Biol. 2021;9:741521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kretschmer J, Rao H, Hackert P, et al. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′–3′ exoribonuclease XRN1. RNA. 2018;24(10):1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang T, Kong S, Tao M, et al. The potential role of RNA N6-methyladenosine in Cancer progression. Mol Cancer. 2020;19(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shi H, Zhao J, Han L, et al. Retrospective study of gene signatures and prognostic value of m6A regulatory factor in non-small cell lung cancer using TCGA database and the verification of FTO. Aging (Albany NY). 2020;12(17):17022–17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ding Y, Qi N, Wang K, et al. FTO Facilitates Lung Adenocarcinoma Cell Progression by Activating Cell Migration Through mRNA Demethylation. OncoTargets and Therapy. 2020;13:1461–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8(1):284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bokar JA, Shambaugh ME, Polayes D, et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- [46].Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang Y, Li Y, Toth JI, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Moindrot B, Cerase A, Coker H, et al. A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell Rep. 2015;12(4):562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Meng Y, Zhang Q, Wang K, et al. RBM15-mediated N6-methyladenosine modification affects COVID-19 severity by regulating the expression of multitarget genes. Cell Death Dis. 2021;12(8):732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li C, Wang W, Xie SS, et al. The Programmed Cell Death of Macrophages, Endothelial Cells, and Tubular Epithelial Cells in Sepsis-AKI. Front Med (Lausanne). 2021;8:796724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Evans KV, Lee JH. Alveolar wars: the rise of in vitro models to understand human lung alveolar maintenance, regeneration, and disease. Stem Cells Translational Medicine. 2020;9(8):867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Paxson JA, Parkin CD, Iyer LK, et al. Global gene expression patterns in the post-pneumonectomy lung of adult mice. Respir Res. 2009;10(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the article.


