In this study, Barry et al. uncover the mechanism by which Rap1—a conserved telomere binding protein that also has been implicated in activation of the NF-κB signaling pathway, maintenance of metabolic function in vivo, and transcriptional regulation—modulates gene expression. Using a separation-of-function allele, they show that Rap1 transcriptional regulation is largely independent of TRF2-mediated binding to telomeres and does not involve direct binding to genomic loci.
Keywords: RAP1, telomere, TIP60, EPC1, 2C-like, ZSCAN4, MERVL
Abstract
In mammals, the conserved telomere binding protein Rap1 serves a diverse set of nontelomeric functions, including activation of the NF-kB signaling pathway, maintenance of metabolic function in vivo, and transcriptional regulation. Here, we uncover the mechanism by which Rap1 modulates gene expression. Using a separation-of-function allele, we show that Rap1 transcriptional regulation is largely independent of TRF2-mediated binding to telomeres and does not involve direct binding to genomic loci. Instead, Rap1 interacts with the TIP60/p400 complex and modulates its histone acetyltransferase activity. Notably, we show that deletion of Rap1 in mouse embryonic stem cells increases the fraction of two-cell-like cells. Specifically, Rap1 enhances the repressive activity of Tip60/p400 across a subset of two-cell-stage genes, including Zscan4 and the endogenous retrovirus MERVL. Preferential up-regulation of genes proximal to MERVL elements in Rap1-deficient settings implicates these endogenous retroviral elements in the derepression of proximal genes. Altogether, our study reveals an unprecedented link between Rap1 and the TIP60/p400 complex in the regulation of pluripotency.
Rap1 (repressor/activator protein 1) was first identified in Saccharomyces cerevisiae as a transcription factor that silences the alternate silent mating-type loci and maintains high transcriptional activity at genes encoding ribosomal proteins and glycolytic enzymes (Huet et al. 1985; Huet and Sentenac 1987; Shore and Nasmyth 1987; Shore et al. 1987; Vignais et al. 1987; Chambers et al. 1989). Subsequent studies found yeast Rap1 to be a primary telomere DNA binding protein that maintains proper telomere length, ensures subtelomere silencing, and inhibits telomere end–end fusions (Berman et al. 1986; Buchman et al. 1988a,b; Longtine et al. 1989; Lustig et al. 1990; Kyrion et al. 1993; Pardo and Marcand 2005). Upon senescence of budding yeast, Rap1 is evicted from telomeres and relocalizes to promoter targets, including histone-encoding genes (Platt et al. 2013; Song et al. 2020). In Schizosaccharomyces pombe, Rap1 performs a similar protective function at telomeres, albeit it relies on a protein–protein interaction with Taz1 to be recruited to chromosome ends (Kanoh and Ishikawa 2001).
Mammalian Rap1, encoded by TERF2IP, is a component of shelterin, a six-subunit protein complex that coats mammalian telomere DNA and prevents the activation of DNA damage signaling and repair at chromosome ends (de Lange 2005; Lazzerini-Denchi and Sfeir 2016). Whereas budding yeast Rap1 binds telomeric DNA directly, mammalian Rap1 relies on a stable interaction with the shelterin subunit TRF2 to be recruited to TTAGGG-bearing telomere DNA (Li et al. 2000; Hanaoka et al. 2001). Rap1 function at telomeres diverged from yeast, as revealed by genetic studies showing that deletion of mammalian Rap1 is largely dispensable for telomere end protection. Although loss of all other shelterin components is incompatible with life, Rap1 knockout (Rap1−/−) mice are alive and fertile and display no premature aging phenotypes (Karlseder et al. 2003; Chiang et al. 2004; Celli and de Lange 2005; Hockemeyer et al. 2006; Kibe et al. 2010; Sfeir et al. 2010). Loss-of-function analysis indicated that Rap1 is dispensable for chromosome end protection. However, in certain experimental settings, it was found to act with TRF2 to suppress NHEJ-mediated telomere fusions (Sarthy et al. 2009; Kabir et al. 2014; Lototska et al. 2020).
Analysis of Rap1−/− mice in vivo and ex vivo shed light into the function of mammalian Rap1 in regulating gene expression (Martínez et al. 2010, 2013; Teo et al. 2010; Yeung et al. 2013). At the organismal level, Rap1 deficiency leads to glucose intolerance, dyslipidemia, liver steatosis, and excess fat accumulation, ultimately manifesting as late-onset obesity. Rap1-null mice also display defective NF-κB activation (Teo et al. 2010). At the cellular level, Rap1 loss induces altered gene expression (Martínez et al. 2013; Yeung et al. 2013), pointing to an extratelomeric function of Rap1 in transcriptional control; however, the mechanistic basis of this regulation is unknown.
Regulation of gene expression is orchestrated by a complex interplay of transcription factors and chromatin regulators, including chromatin remodelers, histone chaperones, and epigenetic modifiers that write/erase histone post-translational modifications. Histone acetylation is one such modification that regulates the structure of chromatin and typically leads to activation of gene expression (Steunou et al. 2014). TIP60/p400 (also known as NuA4) is a histone acetyltransferase (HAT) complex composed of >17 subunits, including the catalytic subunit TIP60 (KAT5), two scaffold proteins (p400 [Ep400] and Trrap), Epc1/2, Dmap1, and Ing3 (Jacquet et al. 2016). TIP60/p400 plays a role in a diverse set of biological processes, including transcription, cell proliferation, and DNA repair by homologous recombination (Steunou et al. 2014; Jacquet et al. 2016; Sheikh and Akhtar 2019). TIP60/p400 regulates transcription through acetylation of histones H4 and H2A, catalyzed by the TIP60 subunit, and through incorporation of H2A.Z using the ATP-dependent chromatin remodeling activity of p400 (Allard et al. 1999; Pradhan et al. 2016). Recent studies revealed that TIP60/p400 is necessary for proper maintenance and renewal of embryonic stem cells (ESCs) (Fazzio et al. 2008; Acharya et al. 2017). As opposed to its well-characterized role in somatic cells to activate transcription, TIP60/p400 functions mainly as a transcriptional repressor in ESCs. This includes the silencing of endogenous retroviral MERVL elements that prevents the emergence of two-cell-like (2C-like) cells, a small population of cells in mouse ESC cultures that share characteristics with two-cell stage embryos (Rodriguez-Terrones et al. 2018). The underlying basis of this noncanonical and paradoxical TIP60/p400 function in suppressing transcription remains elusive.
Here, we uncovered a new mechanism by which endogenous Rap1 maintains native gene expression. We show that Rap1 transcriptional function in mammalian cells is largely independent of its telomere association and does not involve binding to DNA. Instead, our data reveal that nontelomeric Rap1 interacts with the TIP60/p400 complex and modulates TIP60 acetylation activity. Notably, loss of Rap1 in mESCs triggers a 2C-like state and augments the repressive activity of TIP60/p400 on a subset of 2C-stage genes, especially ones proximal to endogenous retroviral MERVL elements. In summary, our data uncover an unprecedented link between a conserved telomere binding protein and the TIP60/p400 complex in the regulation of pluripotency.
Results
Endogenous Rap1 maintains native transcription independent of telomeres
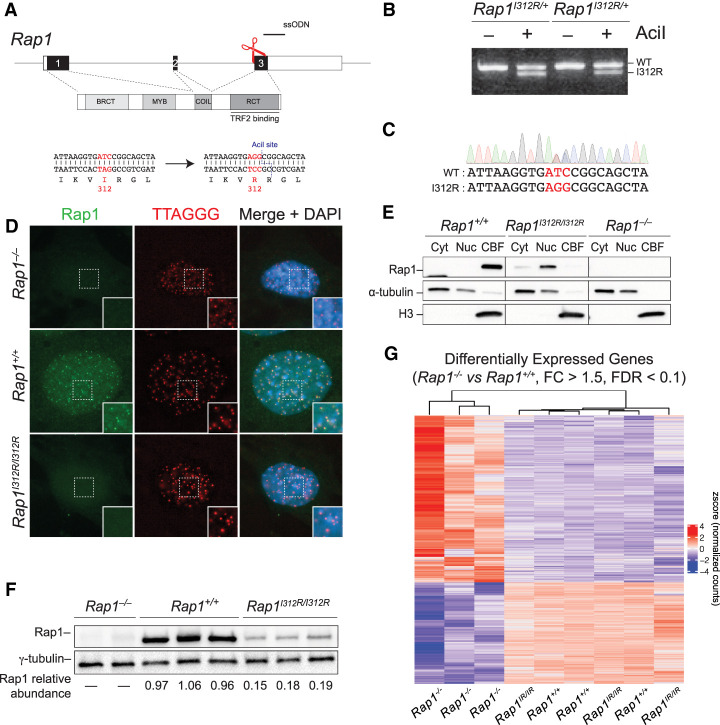
It has been proposed that the function of mammalian Rap1 in gene regulation is similar to budding yeast Rap1 and likely driven by its association with promoter elements. This is based on the direct binding of mammalian Rap1 to DNA in a sequence nonspecific manner (Arat and Griffith 2012), and ChIP-seq analysis that highlighted several genomic loci bound by Rap1 (Martínez et al. 2010; Yang et al. 2011). To gain insight into the mechanisms by which Rap1 regulates transcription, we used CRISPR/Cas9 to generate mice carrying Rap1I312R (Fig. 1A–C), an allele that is incapable of binding TRF2 (Chen et al. 2011).
Figure 1.
Rap1 maintains gene transcription independent of telomeres. (A) Schematic representation of CRISPR/Cas9 gene-editing strategy to substitute Rap1 isoleucine 312 with arginine. Successful targeting creates an AciI restriction site to be used during genotyping. Single-stranded oligo donor (ssODN, black line) and cut site (red scissors) are indicated. (B) PCR genotyping of tail tip DNA from two Rap1I312R/+ heterozygous mice following gene targeting. (C) Example Sanger sequencing of a Rap1I312R/+ heterozygous mouse. (D) Representative image of IF-FISH in Rap1+/+, Rap1−/−, and Rap1I312R/I312R MEFs for Rap1 (green) and telomeres (red) using Rap1 antibody and a TTAGGG PNA probe, respectively. DAPI (blue) was used as a counterstain. (E) Rap1 Western blot from Rap1−/−, Rap1+/+, and Rap1I312R/I312R MEFs following subcellular fractionation into cytoplasmic (Cyt), nucleoplasmic (Nuc), and chromatin-bound (CBFs) fractions. α-Tubulin and histone H3 were loading controls for Cyt and CBFs, respectively. The blot is representative of n = 2 biological replicates. (F) Immunoblot for Rap1 from whole-cell lysates obtained from Rap1−/− (n = 2), Rap1+/+ (n = 3), and Rap1I312R/I312R (n = 3) MEFs. Rap1 relative abundance was determined by normalizing to γ-tubulin. (G) Hierarchically clustered heat map representing RNA-seq data for differentially expressed genes (DEGs) between Rap1+/+ and Rap1−/− MEFs (FC > 1.5; FDR < 0.1; n = 3 biological replicates per genotype). (Rap1IR/IR) Rap1I312R/I312R.
To confirm the release of Rap1 from telomeres in Rap1I312R/I312R mouse embryonic fibroblasts (MEFs), we performed indirect immunofluorescence coupled with fluorescence in situ hybridization (IF-FISH). Consistent with previous reports (Chen et al. 2011; Yeung et al. 2013), Rap1 exhibited a diffuse nucleoplasmic staining pattern in Rap1I312R/I312R MEFs and contrasted with the punctate staining that colocalized with telomeres in Rap1+/+ cells (Fig. 1D). We then fractionated lysates into cytosolic, nucleoplasmic, and chromatin-bound fractions and showed that whereas wild-type Rap1 was strongly bound to chromatin, Rap1I312R lost its chromatin association (Fig. 1E). Furthermore, we observed an approximately fivefold reduction in overall Rap1 levels in Rap1I312R/I312R cells (Fig. 1F), consistent with the stabilization of Rap1 through binding to TRF2 (Celli and de Lange 2005; Sfeir et al. 2010). Along these lines, treatment with the proteasome inhibitor MG132 at 5 µM for 11 h increased Rap1 abundance by sixfold in Rap1I312R/I312R MEFs (Supplemental Fig. S1A,B). In addition, coimmunoprecipitation (co-IP) experiments in HEK293T cells showed that mouse Rap1 immunoprecipitated with K48Ub, providing evidence for its K48-linked ubiquitinylation (Supplemental Fig. S1C). Altogether, our results show that at telomeres, mammalian Rap1 is stabilized by its interaction with TRF2, while the extratelomeric Rap1 pool is tightly regulated by proteasomal degradation.
We then performed RNA sequencing (RNA-seq) on Rap1−/−, Rap1+/+, and Rap1I312R/I312R primary MEFs. Principal component analysis (PCA) showed close clustering of the Rap1+/+ and Rap1I312R/I312R samples apart from Rap1−/− samples (Supplemental Fig. S1D). Differential gene expression analysis using DESeq2 identified 654 differentially regulated genes in Rap1−/− versus Rap1+/+ MEFs (FC > 1.5, FDR < 0.1) (Supplemental Table S1), involving various pathways related to cell–cell signaling and developmental processes (Supplemental Fig. S1E; Supplemental Table S2). Interestingly, gene expression was predominantly maintained in Rap1I312R/I312R MEFs relative to wild-type cells (653 of 654, 99.8%) (Fig. 1G). Additionally, we observed no overlap with genes differentially expressed in Trf2-deleted mouse fibroblasts (Supplemental Fig. S1F; Markiewicz-Potoczny et al. 2021). Taken together, our data suggest that despite the overall reduction in Rap1 levels in Rap1I312R/I312R cells, a small pool of extratelomeric Rap1 is sufficient to fully maintain gene expression, and the function of mammalian Rap1 in gene expression is largely independent of TRF2 binding and telomere localization.
While transcriptional dysregulation due to Rap1 deficiency was largely rescued in Rap1I312R/I312R cells, in vivo analysis of targeted mice revealed partial rescue of weight gain and glucose tolerance (Supplemental Fig. S2A,B). These results point to telomere-dependent and -independent mechanisms by which Rap1 regulates metabolic function in vivo. However, we cannot rule out that the lower levels of Rap1 observed in Rap1I312R/I312R mice are insufficient to fully complement transcription regulation in critical populations of cells (Supplemental Fig. S2C,D).
Rap1-I312R does not directly associate with chromatin
In budding yeast, extratelomeric Rap1 binds directly to interstitial genomic sites, including several gene promoters (Bram and Kornberg 1985; Huet et al. 1985; Shore and Nasmyth 1987; Vignais et al. 1987; Chambers et al. 1989; Platt et al. 2013; Song et al. 2020). Similarly, in human and mouse cells, wild-type Rap1 was detected at many sites throughout the genome (Martínez et al. 2010, 2016; Yang et al. 2011) with partial overlap with TRF2 binding sites.
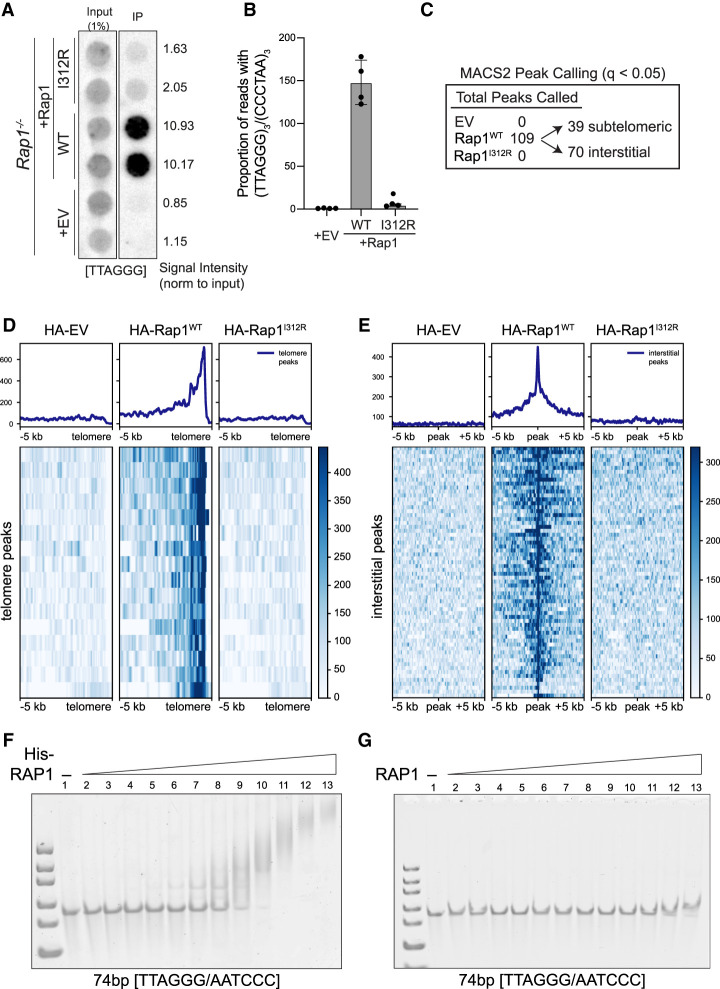
Given that Rap1I312R fully maintains transcriptional regulation in MEFs, we used this separation-of-function allele to identify TRF2-independent Rap1 binding sites that could be more relevant to its gene expression function. We transduced Rap1−/− MEFs with HA-Rap1 and HA-Rap1-I312R and noted that both alleles accumulated at similar levels when exogenously expressed and displayed the predicted subcellular localization (Supplemental Fig. S3A,B). As expected, chromatin immunoprecipitation (ChIP) using anti-HA antibody showed significant association of telomeric DNA with HA-Rap1 but not with HA-Rap1-I312R (Fig. 2A,B). We performed peak calling using MACS2 (q < 0.05) and identified 109 wild-type Rap1 binding sites: 39 subtelomeric and 70 other loci scattered throughout the genome (Fig. 2C–E; representative peaks in Supplemental Fig. S3C,D; Supplemental Table S3). This analysis is consistent with previous observations that Rap1 localizes to discrete sites throughout the genome (Martínez et al. 2010, 2016; Yang et al. 2011), albeit we failed to detect any enriched DNA motifs within the binding sites of Rap1 and saw no overlap between genes bound by Rap1 and those dysregulated in its absence (data not shown). Surprisingly, we did not identify any genomic binding sites for Rap1-I312R (Fig. 2C), either at subtelomeres (Fig. 2D) or at interstitial peaks bound by wild-type Rap1 (Fig. 2E).
Figure 2.
Rap1-I312R does not bind to genomic loci or DNA. (A) Dot blot for telomere repeats following ChIP using anti-HA antibody in cells expressing HA-tagged Rap1-WT, Rap1-I312R, or empty vector (EV) control (n = 2 biological replicates). Signal intensity was determined by normalizing to input. (B) High-throughput sequencing of ChIP samples as in A. Telomere binding was determined by calculating the proportion of reads with at least three telomere (TTAGGG)3/(CCCTAA)3 repeats. Data are the mean ± standard deviation of n = 4 biological replicates. (C) Summary of ChIP-seq peak calling analysis that identified 109 peaks in HA-Rap1-WT samples (MACS2; q < 0.05). Peaks were classified as subtelomeric if localized within 500 kb from the telomere. (D) Heat map representing ChIP-seq profiles of HA-Rap1-WT peaks at chromosome ends. (E) Heat map representing ChIP-seq profiles of HA-Rap1-WT interstitial peaks. (F) Electrophoretic mobility shift assay (EMSA) using His-tagged RAP (0–15 µM) and a 74-bp TTAGGG/AATCCC double-stranded DNA (100 nM). (G) EMSA using non-His-tagged RAP (0–15 µM) and a 74-bp TTAGGG/AATCCC double-stranded DNA (100 nM).
Having failed to detect Rap1-I312R binding to DNA by ChIP, we decided to revisit previous data suggesting in vitro binding of human RAP1 to DNA. We performed electrophoretic mobility shift assays (EMSAs) using purified protein and initially found that His-tagged human RAP1 binds to DNA with no sequence specificity and associates with purified nucleosomes (Fig. 2F; Supplemental Fig. S3E,F). Importantly, however, upon removal of the His tag used for purification purposes, the binding of RAP1 to free and nucleosome-bound DNA was abrogated (Fig. 2G; Supplemental Fig. S3G,H). The confounding impact of histidine tags on DNA-binding activity of purified proteins has been previously reported (Paul et al. 2020) and could well explain earlier studies that showed nonspecific binding of human RAP1 to DNA in vitro (Arat and Griffith 2012). Taken together, our data confirm that, unlike budding yeast Rap1, the mammalian counterpart is incapable of binding DNA directly, and therefore the mechanisms by which RAP1 regulates gene expression in higher eukaryotes diverged.
Proximity-based biotinylation reveals extratelomeric Rap1 interactors
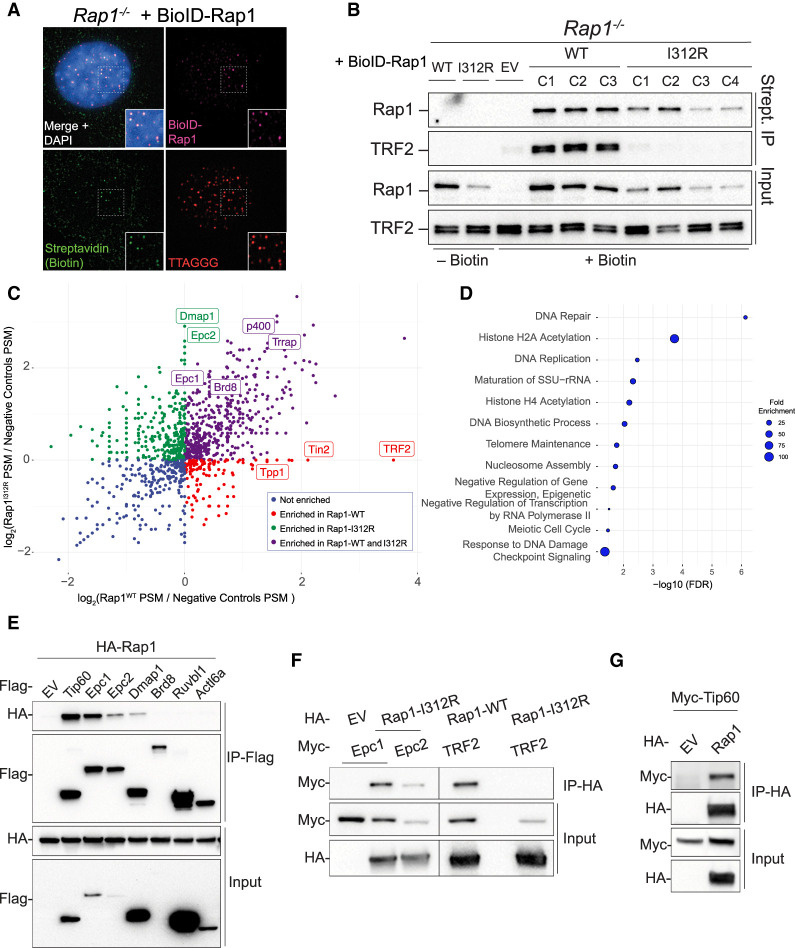
We next used proximity-based biotinylation to identify Rap1 protein interactions and provide insight into the potential mechanisms by which extratelomeric Rap1 regulates gene expression (Roux et al. 2012). To that end, we transduced Rap1−/− MEFs with BioID-tagged Rap1 and Rap1-I312R and established independent clonally derived cell lines with similar expression levels. As a control, we showed that BioID-Rap1 colocalized at telomeres and interacted with TRF2, whereas BioID-Rap1-I312R expressed diffusely throughout the nucleus and failed to associate with TRF2 (Fig. 3A,B; Supplemental Fig. S4A).
Figure 3.
Proximity-dependent biotinylation reveals extratelomeric Rap1 binding partners. (A) Representative image of IF-FISH in Rap1−/− MEFs expressing BioID-Rap1-WT stained for BioID-Rap1 (magenta), biotin (green), and telomeres (red) using anti-Flag antibody, streptavidin, and TTAGGG PNA probe, respectively. DAPI (blue) was used as a counterstain. (B) Immunoblot for Rap1 and TRF2 following streptavidin pull-down in Rap1−/− MEFs expressing BioID-Rap1-WT (n = 3 biological replicates; clones C1, C2, and C3), BioID-Rap1-I312R (n = 4 biological replicates; clones C1, C2, C3, and C4), and BioID alone (EV). Where indicated, cells were treated with 50 µM biotin for 20 h prior to harvest. (C) Scatter plot representing log2 fold change in peptide spectrum match (PSM) of proteins identified by BioID-Rap1-WT versus BioID-Rap1-I312R. Fold change values were calculated relative to no biotin and BioID-alone controls. Each dot represents a unique protein. Known Tip60/p400 (green and purple) and shelterin (red) complex members are indicated. (D) Graphical representation of the top 10 biological processes overrepresented in BioID-Rap1-I312R streptavidin pull-down (FC > 2) using the PANTHER classification system statistical overrepresentation test. (E) Co-IP of Flag-tagged Tip60/p400 subunits (Tip60, Epc1, Epc2, Dmap1, Brd8, Ruvbl1, and Actl6a) and HA-Rap1 following cotransfection in HEK293T cells. (F) Reciprocal co-IP of HA-Rap1-I312R and Myc-tagged Epc1 or Epc2. Co-IP of HA-Rap1 or HA-Rap1-I312R with TRF2 was used as a control. (G) Reciprocal co-IP of HA-Rap1 and Myc-Tip60.
We performed mass spectrometry analysis following streptavidin pull-down of nuclear extracts (Supplemental Fig. S4B; Supplemental Table S4) and detected unique Rap1 and Rap1-I312R interactors that were enriched more than twofold over the empty vector control (Supplemental Table S5), including several shelterin subunits that were exclusively retrieved in cells expressing wild-type Rap1 (Fig. 3C; Supplemental Fig. S4C). Notably, among factors associated with Rap1-I312R, we identified six members of the Tip60/p400 histone acetyltransferase complex: Epc1 and its paralog Epc2, Dmap1, p400, Trrap, and Brd8 (Fig. 3C; Supplemental Fig. S4C). Statistical overrepresentation test of biological processes using the PANTHER classification system revealed statistical significance for histone H2A acetylation, histone H4 acetylation, and DNA repair in Rap1-I312R interactors (Fig. 3D; Supplemental Table S6).
We validated the mass spectrometry data by performing co-IP experiments of Flag- and HA-tagged mouse proteins coexpressed in HEK293T and demonstrated stable interaction between Rap1 and Tip60, Epc1, Epc2, and Dmap1 (Fig. 3E–G). Importantly, the interaction between Rap1 and Epc1 was not diminished upon treatment with DNase (Supplemental Fig. S4D) and was evident when using purified recombinant human proteins (GST-EPC1 and His-RAP1) (Supplemental Fig. S4E), suggesting that the two proteins interact independent of DNA binding. In conclusion, our results reveal that extratelomeric Rap1 is a binding partner of the TIP60/p400 complex.
Rap1 associates with Epc1 and Tip60, enhancing histone acetyltransferase (HAT) activity
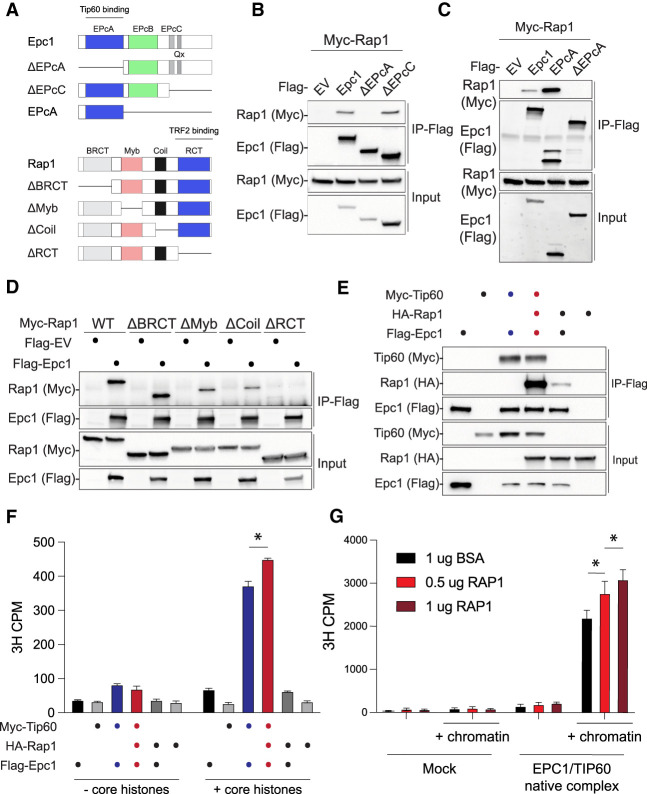
Tip60/p400 is composed of >17 subunits (Doyon and Côté 2004), with Trrap and p400 serving as scaffolds for complex assembly. Tip60 is the catalytic subunit that acetylates histones H2A, H4, and H2A.Z as well as other nonhistone proteins, including Tp53 (Steunou et al. 2014). Epc1 comprises four conserved domains (Fig. 4A), with the N-terminal EpcA portion promoting the assembly of the core catalytic module that includes Tip60 (also known as piccolo NuA4 or picNuA4). The central EpcB domain binds p400 and bridges the rest of the Tip60/p400 complex (Boudreault et al. 2003; Doyon et al. 2004; Selleck et al. 2005; Auger et al. 2008; Chittuluru et al. 2011; Xu et al. 2016; Setiaputra et al. 2018). Using mouse deletion mutants of the Epc1 subunit, we found that the EpcA domain is necessary and sufficient to interact with Rap1 (Fig. 4A–C), placing the telomere binding protein at the Epc1/Tip60 interaction interface. Through reciprocal mapping, we identified the C-terminal RCT domain of Rap1 to be necessary for binding to Epc1 (Fig. 4D). Notably, the Rap1 RCT domain also binds Trf2, suggesting that binding of Rap1 to Tip60/p400 and Trf2 is mutually exclusive. Consistent with this model, we found no evidence for Trf2 association with the Epc1/Rap1 complex. In contrast, the presence of Trf2 compromised Epc1–Rap1 interaction (Supplemental Fig. S5A). Taken together, these data link Rap1 to two separate complexes: being part of shelterin at telomeres and associating with Tip60/p400 during transcriptional regulation.
Figure 4.
Rap1 forms a complex with and modulates the histone acetyltransferase (HAT) activity of Tip60/Epc1. (A) Schematic illustration of mouse Epc1 and Rap1 protein domains and deletion mutant constructs used for co-IP assays. (B) Co-IP of Flag-Epc1 deletion mutants and Myc-Rap1 in cotransfected HEK293T cells. Western blot analysis for input and IP sample was performed with the indicated antibody. (C) Co-IP of the Flag-Epc1 EPcA domain and Myc-Rap1. (D) Co-IP of Flag-Rap1 deletion mutants and Myc-Epc1. (E) Co-IP of Flag-Epc1, Myc-Tip60, and HA-Rap1. (F) Histone acetyltransferase (HAT) activity assays performed using co-IPs from E on core histones. Data are the counts per minute (CPM) mean ± standard deviation of n = 2 technical replicates; two-way ANOVA, and Tukey's multiple comparison test. (*) P-value < 0.0001. (G) HAT activity assay using affinity-purified human EPC1/TIP60 and RAP1 (increasing amounts) on short oligonucleosome chromatin. Bovine serum albumin (BSA) was a negative control. Data are CPM mean ± standard deviation of n = 3 technical replicates; two-way ANOVA, and Tukey's multiple comparison test. (*) P-value < 0.01.
We then explored whether Rap1, Epc1, and Tip60 form a trimeric complex. Using Rap1 as bait, we found that Rap1–Epc1 and Rap1–Tip60 interactions were strongly enhanced in the presence of Tip60 and Epc1, respectively (Supplemental Fig. S5B). As expected, the effect was dependent on the EpcA domain of Epc1 (Supplemental Fig. S5C). In a parallel approach, we used Epc1 as bait and found that Tip60 greatly enhanced Rap1/Epc1 association (Fig. 4E). Notably, histone acetyltransferase (HAT) assays using mouse and human isoforms revealed that Rap1 enhances Epc1/Tip60 activity with a slight preference for H4 over H2A (Fig. 4F,G; Supplemental Fig. S5D,E). In summary, we conclude that extratelomeric Rap1 associates with the Tip60/p400 complex, where it cooperatively binds to and modulates the acetyltransferase activity of Tip60.
Rap1 suppresses the two-cell-like state in mouse embryonic stem cells
In addition to its well-established function as a transcriptional activator in somatic cells, the Tip60/p400 complex has a critical yet poorly understood role during stem cell self-renewal (Fazzio et al. 2008; Acharya et al. 2017). Specifically, members of the Tip60/p400 complex (e.g., Tip60, Dmap1, and Ep400) were shown to prevent the emergence of “two-cell-like” (2C-like) cells in mouse ESCs through unexpected and paradoxical repression of 2C genes, including MERVL, Zscan4, and others (Rodriguez-Terrones et al. 2018). Although the exact molecular mechanisms by which the Tip60/p400 complex represses gene expression in ESCs are completely unknown, they seem to be independent of Tip60-mediated acetylation of H4 and H2A (Acharya et al. 2017).
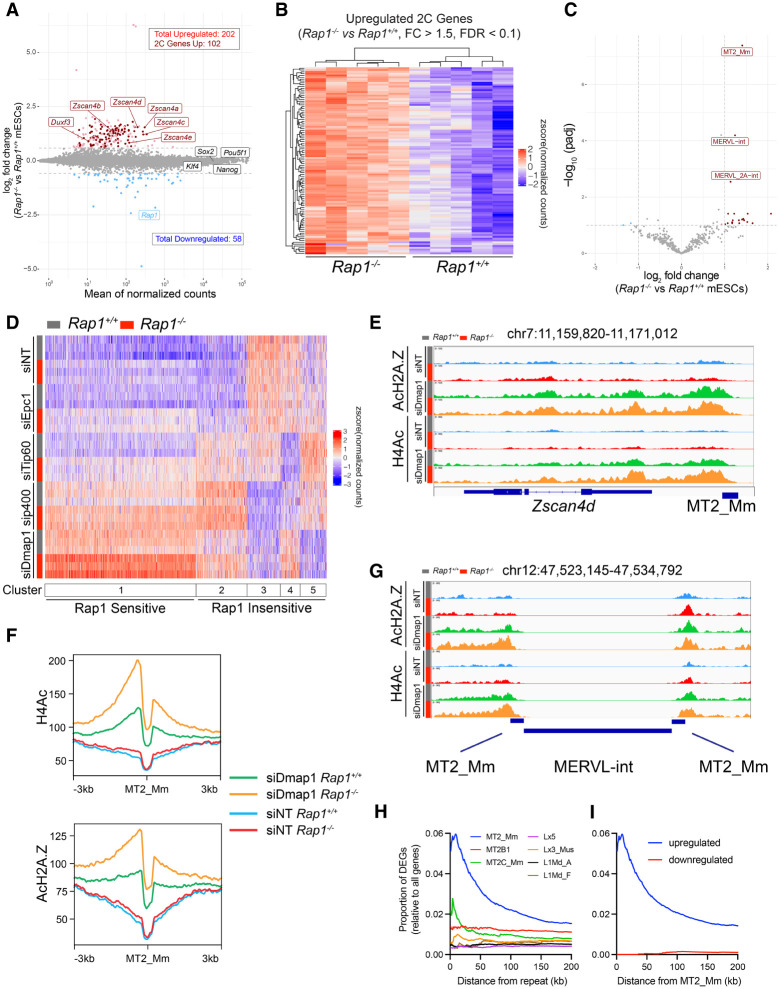
The direct binding of Rap1 to members of the Tip60 complex (Fig. 3E) and the recent link between telomere deprotection and induction of 2C gene expression (Markiewicz-Potoczny et al. 2021) led us to investigate whether Rap1 regulates 2C genes. To that end, we established independent mESC cultures from Rap1+/+ and Rap1−/− mouse preimplantation blastocysts (Supplemental Fig. S6A) and performed RNA-seq. Transcriptomic analysis identified 202 up-regulated genes (Fig. 5A; Supplemental Fig. S6B; Supplemental Table S7), half of which (102; 50.5%) were previously categorized as genes expressed in 2C-like cells (Fig. 5A,B; Fu et al. 2020), including the Zscan4 locus. We validated the up-regulation of Zscan4 using both RT-qPCR experiments and IF (Supplemental Fig. S6C,D). We also observed increased expression of the MERVL retrotransposon elements MT2_Mm, MERVL-int, and MERVL_2A-int in Rap1−/− mESCs (Fig. 5C; Supplemental Fig. S6E; Supplemental Table S8). In addition, to address whether Rap1 suppresses the 2C-like state independent of telomere binding, we derived mESC cultures from Rap1I312R/I312R mouse blastocysts and found similar expression of 2C-like genes and MERVL in Rap1I312R/I312R mESCs relative to wild type (Supplemental Fig. S6F–H). Last, given that deletion of the Rap1 telomere binding partner TRF2 also activated a 2C-like state in ESCs (Markiewicz-Potoczny et al. 2021), we compared RNA-seq data between Rap1- and Trf2-deficient ESCs and found minimal overlap in up-regulated 2C genes (Supplemental Fig. S6I,J). In summary, we uncovered an unexpected function for a highly conserved telomere binding protein in the regulation of pluripotency. Furthermore, our data are largely consistent with a TRF2-independent role for Rap1 in the regulation of 2C gene transcription.
Figure 5.
Rap1 suppresses the 2C-like state by enhancing Tip60/p400 repression of 2C genes. (A) MA plot of log2 fold changes in gene expression in Rap−/− versus Rap1+/+ mESCs. Up-regulated (light red) and down-regulated (light blue) genes are indicated (FC > 1.5; FDR < 0.1; n = 5 biological replicates). 2C-stage genes are highlighted in dark red. (B) Hierarchically clustered heat map representing RNA-seq data of 2C genes up-regulated in Rap−/− versus Rap1+/+ mESCs (n = 5 biological replicates). (C) Volcano plot of differential repeat expression analysis in Rap1−/− versus Rap1+/+ mESCs. Up-regulated (red) and down-regulated (blue) repeats are indicated (FC > 2; FDR < 0.1; n = 5 biological replicates). (D) Kmeans (k = 5) clustered heat map representing RNA-seq data of 2C genes in Rap1+/+ (gray bar) and Rap1−/− (red bar) mESCs treated with siRNAs targeting Tip60/p400 subunits (Epc1, Tip60, p400, and Dmap1) or nontargeting (siNT) control (n = 3 biological replicates were used per condition). (E) ChIP-seq profiles of H4Ac and AcH2A.Z at the Zscan4d gene locus in Rap1+/+ (gray bar) and Rap1−/− (red bar) mESCs transfected with siRNA targeting Dmap1 or nontargeting (siNT) control. Profiles plotted are representative of n = 3 biological replicates. (F) Density plot centered at all MT2_Mm sites (n = 2667) for H4Ac and AcH2A.Z ChIP-seq data. Density plots are representative of n = 3 biological replicates. (G) ChIP-seq profiles of H4Ac and AcH2A.Z at a representative MERVL site in Rap1+/+ (gray bar) and Rap1−/− (red bar) mESCs transfected with siRNA targeting Dmap1 and nontargeting control (siNT). (H) Plot of the proportion of differentially expressed genes (DEGs) in Rap1+/+ versus Rap1−/− mESCs (FC > 1.5; FDR < 0.1) at fixed distances from the indicated MERVL (MT2_Mm, n = 2667; MT2B1, n = 7248; and MT2C_Mm, n = 1982) and LINE1 (Lx5, n = 16339; Lx3_Mus, n = 12651; L1Md_A, n = 16844; and L1Md_F n = 4016) repeat sequences. Proportions were calculated by dividing the number of DEGs by the total number of genes. (I) Plot of the proportion of up-regulated (blue) and down-regulated (red) DEGs in Rap1+/+ versus Rap1−/− mESCs (FC > 1.5; FDR < 0.1) at fixed distances from MT2_Mm repeat sequences.
The interplay between Rap1 and the Tip60/p400 complex on mESC gene expression
Having established that Rap1 suppresses the 2C-like state in mESCs, and given its interaction with Epc1 and Tip60 (Fig. 4; Supplemental Fig. S7A), we sought to better understand the interplay between Rap1 and the Tip60/p400 complex during repression of 2C genes. We depleted Tip60 by siRNA in Rap1+/+ and Rap1−/− mESCs (Supplemental Fig. S7B) and performed RNA-seq. We first investigated the overlap between genes differentially expressed upon Rap1 loss and those altered in cells depleted of Tip60. Interestingly, while down-regulated genes displayed minimal overlap between the two genotypes, >50% of genes up-regulated in the absence of Rap1 were also up-regulated upon Tip60 depletion (Supplemental Fig. S7C,D), with common genes being enriched for 2C genes (65%, 73 of 112).
Given the minimal understanding of the repressive function of the Tip60/p400 complex on 2C-stage genes, we further interrogated the effect of depleting additional subunits of this complex, including Epc1, Dmap1, and p400, in Rap1+/+ and Rap1−/− mESCs (Supplemental Fig. S7B; Fu et al. 2020). Kmeans clustering highlighted five distinct 2C-state gene clusters (Fig. 5D). Depletion of Tip60/p400 subunits showed the expected reduction in transcription at three gene clusters (Fig. 5D, clusters #3, #4, and #5). As for clusters #1 and #2, depletion of Tip60, Ep400, and Dmap1 led to enhanced 2C gene expression (Fig. 5D, clusters #1 and #2), thus corroborating a critical repressive function for this complex in mESC transcription (Fazzio et al. 2008; Rodriguez-Terrones et al. 2018). Of note, the various 2C gene clusters were not equally sensitive to inhibition of the various Tip60 members. Overall, this analysis revealed a high degree of complexity in how the Tip60/p400 complex regulates 2C-stage genes in mESCs, whereby certain subunits act as negative repressors while others exert a positive effect on gene transcription.
We next overlaid the impact of Rap1 deletion on Tip60/p400 dysregulation of 2C gene expression and noted that half of all 2C genes (53.5%; 571 of 1068) were sensitive to Rap1 loss (Fig. 5D, cluster #1). Strikingly, Rap1-sensitive genes were further up-regulated upon Tip60/p400 inhibition, including Zscan4c and MERVL (Supplemental Fig. S7E,F). Instead, gene clusters that were down-regulated upon Tip60/p400 knockdown were largely insensitive to Rap1 (clusters #3, #4, and #5). Taken together, these results implicate Rap1 in the noncanonical function of Tip60/p400 during 2C genes suppression.
Repression of 2C genes in Rap1- and Tip60-depleted cells is potentially driven in cis by MT2 elements
We next aimed to gain insight into the paradoxical repression of 2C gene expression by Rap1 and the Tip60/p400 complex. Examination of the Zscan4 locus and the MERVL LTR MT2_Mm by ChIP-seq revealed a minimal increase in H4Ac and AcH2A.Z abundance in Rap1−/− cells that was synergistically enhanced upon depletion of Dmap1 (Fig. 5E–G; Supplemental Fig. S7G). Strikingly, we noticed that genes up-regulated in Rap1−/− mESCs are preferentially localized proximal to MERVL MT2_Mm elements. Instead, no such proximity was observed when comparing Rap1-regulated genes with unrelated retroviral elements such as LINE1 (Lx5, Lx3_Mus, L1Md_A, and L1Md_F) (Fig. 5H,I). While future experiments are necessary to unravel the mechanistic basis for the repressive function of Tip60/p400 on 2C genes, our observations potentially implicate MERVL regulatory dysfunction as a possible driver of 2C gene activation following Rap1 loss. These results are consistent with recent studies that showed that MERVL elements serve as enhancers and sites of cryptic transcription initiation to drive expression of nearby 2C genes (Macfarlan et al. 2012). In conclusion, derepression of 2C genes upon Tip60/p400 and Rap1 loss, while unrelated to the canonical H2A and H4 acetylation, could likely be driven by cis effects through activation of MERVL repeat elements.
Discussion
Our study sheds light on a mechanism by which Rap1 regulates gene expression in mammals by establishing that Rap1 maintains native transcription independent of its association with telomeres. We found that extratelomeric Rap1 responsible for its transcriptional function does not bind to discrete genomic DNA loci. Instead, this conserved telomere binding protein is part of the Tip60/p400 complex. Furthermore, we uncovered an unanticipated role for mammalian Rap1 in regulating the repressive activity of the Tip60/p400 complex on 2C-stage genes and endogenous retroviruses in mESCs.
Transcriptional regulation: an ancestral function for the most conserved telomere binding protein
Rap1 is the most highly conserved telomere binding protein and in unicellular eukaryotes serves an essential function in telomere maintenance by regulating telomere length, preventing telomere fusions, and suppressing telomere–telomere recombination events (Kanoh and Ishikawa 2001; Pardo and Marcand 2005; Nanavaty et al. 2017). When compared with its chimpanzee counterpart, the gene encoding human RAP1 shows little divergence relative to other shelterin subunits (Kabir et al. 2014). However, Rap1 is largely dispensable for telomere function and cell viability in mammals (Sfeir et al. 2010; Kabir et al. 2014), raising questions related to the selective pressure for its conservation in higher eukaryotes. We speculate that the function of Rap1 in gene regulation might underlie such conservation. In budding yeast, Rap1 maintains high expression of genes encoding ribosomal proteins and glycolytic enzymes (Bram and Kornberg 1985; Huet et al. 1985; Shore and Nasmyth 1987; Vignais et al. 1987; Chambers et al. 1989). Additionally, in both yeast and T. brucei, Rap1 maintains repression of telomere/subtelomere regions, preventing aberrant expression of genes proximal to chromosome ends, such as T. brucei variant surface glycoprotein (VSG) genes (Yang et al. 2009). Notably, Rap1 transcriptional activity in budding yeast is linked to telomeres. Specifically, as telomeres shorten in budding yeast, the abundance of extratelomeric Rap1 increases, facilitating its localization to additional target genes (Platt et al. 2013; Song et al. 2020). In contrast, transcriptional activity of mammalian Rap1 is largely uncoupled from telomere binding, with independent pools of Rap1 associating with either TRF2 or Tip60. We show that mutant Rap1 that cannot bind TRF2 or localize to telomeres complements gene expression (Fig. 1G; Supplemental Fig. S6G,H). Furthermore, whereas yeast Rap1 binds directly to DNA sequences at gene promoters to regulate transcription, we found that the extratelomeric pool of mammalian Rap1 lacks DNA binding activity (Fig. 2E,G; Supplemental Fig. S3G,H). Thus, although Rap1 function in transcription regulation is conserved in higher eukaryotes, the mechanism by which Rap1 regulates transcription has diverged.
RAP1 as a regulator of the TIP60/p400 complex
We established that extratelomeric Rap1 associates with TIP60/p400, a histone acetyltransferase complex with a fundamental role in the regulation of gene transcription, as demonstrated by proximity labeling as well as coimmunoprecipitation with the Tip60, Epc1/Epc2, and Dmap1 subunits of the complex (Fig. 3C,E–G; Supplemental Figs. S4E, S7A). Our data suggest that Rap1 binding to TIP60/p400 is mutually exclusive with TRF2, which is consistent with the observation that Rap1 maintains native transcription independent of TRF2 and provides evidence that Rap1 associates with multiple regulatory protein complexes through its RCT domain. This is similar to budding yeast, whereby Rap1 uses its RCT domain to recruit Rif1/Rif2 at chromosome ends to regulate telomere length (Hardy et al. 1992; Wotton and Shore 1997) and Sir3/Sir4 at HML/HMR to maintain transcriptional silencing (Moretti et al. 1994; Moretti and Shore 2001). Notably, despite the association of Rap1 with the TIP60/p400 complex, we failed to detect binding of Rap1-I312R to chromatin by ChIP-seq (Fig. 2C–E). One possible explanation is that the experimental conditions that we used for ChIP preferentially retrieved proteins closely associated with chromosomal DNA and failed to capture factors more indirectly associated through protein–protein interactions with larger-order protein complexes. Additionally, we cannot rule out that Rap1 regulates Tip60/p400 acetyltransferase activity on nonchromatin substrates, including previously identified targets—such as p53 or other unknown proteins—that would indirectly impact gene expression.
Importantly, we found that loss of Rap1 synergizes with Tip60/p400 depletion to increase H4 and H2A.Z acetylation at MT2_Mm elements near up-regulated 2C genes (Fig. 5E–G). The noncanonical activity of TIP60/p400 on mESC gene expression has been previously noted (Fazzio et al. 2008; Chen et al. 2013) and contrasts the case in somatic cells, where TIP60/p400 is primarily a transcriptional activator through histone acetylation. So far, the underlying molecular mechanism of TIP60-mediated repression of gene expression remains completely unknown, and our data implicate Rap1 in the noncanonical function of TIP60. Furthermore, our results suggest that the majority of Rap1 dysregulated genes are proximal to MERVL repeat elements (Fig. 5H,I). This proximity-dependent effect on gene expression has also been observed upon depletion of the FACT and CAF-1 histone chaperone complexes, contexts in which cryptic transcription generates MERVL fused chimeric transcripts (Ishiuchi et al. 2015; Chen et al. 2020). Taken together, we propose a model whereby the activation of endogenous retroviral element MERVL induces the transcription of proximal genes in the context of Rap1 deficiency. However, we cannot rule out that the effect of Rap1 on 2C genes is downstream from Zscan4 activation (Zhang et al. 2019). Additionally, Rap1 could modulate TIP60 activity upstream of MERVL activation by regulating acetylation of nonhistone targets or, possibly, completely independent of TIP60/p400 catalytic activity, supported by the observation that Tip60/p400 lysine acetyltransferase activity is dispensable for ESC self-renewal (Acharya et al. 2017).
Telomere binding proteins and totipotency
An unlikely link between telomeres, telomere binding proteins, and pluripotency emerged in recent literature. Loss of TRF1 in stem cells leads to increased TERRA levels that in turn modulated the recruitment of the PRC2 repressive complex to alter pluripotency gene expression (Marión et al. 2019). More recently, depletion of TRF2 in mESCs was shown to induce the expression of 2C-stage genes, including Zscan4, which contributed to telomere protection in the absence of TRF2 (Markiewicz-Potoczny et al. 2021). The latter study highlights an unexpected link between totipotency and telomere protection mechanisms. Our study unravels yet another connection between telomere biology and pluripotency, as we found that Rap1 suppresses a 2C-like state in mESCs by enhancing the repressive activity of TIP60/p400 on MERVL and other 2C genes (Fig. 5A–D). In effect, our data highlight how the telomere binding protein Rap1 interfaces with chromatin-modifying complexes in the control of pluripotency. Despite Rap1 transcriptional regulation seeming largely independent of TRF2-mediated binding to telomeres, it is possible that telomere stress resulting from loss of either Rap1 or TRF2 serves as a common mechanism by which 2C genes are activated. This may explain the apparent minor increase in 2C gene and MERVL expression in Rap1I312R/I312R mESCs relative to wild type (Supplemental Fig. S6G), even if the majority of genes tested, including Zscan4 and MERVL, did not reach statistical significance. It is tempting to speculate that the regulation of transcription at retroviral repeat elements constitutes an ancestral function for Rap1 and possibly other shelterin subunits prior to being co-opted as a telomere binding protein. Ultimately, further studies are necessary to explore the impact of this telomere binding protein on other classes of endogenous retroviruses and in different biological settings.
Materials and methods
Animal studies
Rap1 knockout mice were generated by crossing the Rap1 floxed allele to Rosa-Cre mice, as previously described (Sfeir et al. 2010). Rap1I312R knock-in mice were generated by CRISPR/Cas9 editing via injection of fertilized oocytes with Cas9 mRNA, sgRNA targeting the Rap1 locus (5′-TAGCTGCCGGATCACCTTAA-3′), and a single-stranded DNA donor (ssODN) carrying the mutated allele (Supplemental Table S9). Two mice heterozygous for the mutation were obtained using this method and were crossed to produce homozygous mice. Genotyping was performed using the following primers: Fwd: 5′-CTCTCACACACACACCATGCATTC-3′ and Rev: 5′-GACTCTAAGAAGGAGGACGTGG-3′. AciI restriction digest produced a 734-bp band for the Rap1-WT allele and two bands at 636 bp and 98 bp for Rap1-I312R.
For mouse embryonic fibroblast (MEF) isolation, pregnant mice were sacrificed at E13.5 and embryos were harvested and dissected to remove limb, liver, and neural tissue. The remaining tissue was diced, treated with trypsin, and then cultured in serum-containing media. Mouse embryonic stem cell (mESC) cultures were derived from the inner cell mass of preimplantation embryos.
For body weight analysis, animals were maintained on a regular chow diet (5053 Lab Diet) and weighed at the indicated time points. For glucose tolerance testing, 8-wk-old mice were fasted for 6 h before experiment and then given an intraperitoneal injection of a 10% glucose solution (1 U/g body weight). Blood samples were collected from the tail vein at 0, 15, 30, 60, 90, 120, and 150 min and glucose levels were measured using a glucometer.
Plasmids
Mouse Rap1 variants (wild type and Rap1-I312R) were cloned into the retroviral constructs pLPC-N-Flag-2xHA-MCS-Puro for ChIP-seq and pLPC-N-Flag-BirA*-13xGGGGS-Myc-MCS-Puro for proximity-based biotin labeling. For co-IP, Rap1 variants (wild type, Rap1-I312R, Rap1-ΔBRCT, Rap1-ΔMyb, Rap1-ΔCoil, and Rap1-ΔRCT) were subcloned into the retroviral constructs pLPC-N-Myc-MCS, pLPC-N-Flag-MCS, and pcDNA-N-3xHA-MCS. Mouse Tip60, Epc1, Epc2, Dmap1, Ruvbl1, and Actl6a cloned into pCMV6-MCS-Myc-Flag were purchased from Origene and used for co-IPs in Figure 3, E–G. Mouse Tip60 and Epc1 variants (wild type, Epc1-ΔEPcA, Epc1-ΔEPcC, and Epc1-EPcA) were subcloned by restriction enzyme digest of PCR products from pCMV6-MCS-Myc-Flag into pLPC-N-Flag-MCS and pLPC-N-Myc-MCS for co-IPs in Figure 4, B–E. Human RAP1 was expressed from pTriEx4 for protein affinity purification used in Figure 4G. For K48-linked ubiquitin co-IP, pRK5-HA-ubiquitin-K48 was a gift from Ted Dawson (Addgene 17605).
Cell culture
Primary MEFs were cultured in 15% fetal bovine serum (FBS), 1× MEM nonessential amino acids (Gibco 11140050), 100 U/mL Pen/Strep (Gibco 15140122), 2 mM L-glutamine (Gibco 25030149), and 50 µM 2-mercaptoethanol in DMEM. SV40LT immortalized MEFs were immortalized by retroviral infection with pBabeSV40LT (a gift from Greg Hannon) and cultured in 10% bovine calf serum (BCS), 1× MEM nonessential amino acids (Gibco 11140050), 100 U/mL Pen/Strep (Gibco 15140122), and 2 mM L-glutamine (Gibco 25030149) in DMEM. mESCs were grown on feeders or on gelatinized plates in 2i/LIF media composed of 15% FBS, 1× NEAA, 100 U/mL Pen/Strep, 2 mM L-glutamine, 50 µM 2-mercaptoethanol, 500 U/mL LIF protein (Millipore Sigma ESG1106), 3 µM CHIR99021/GSK-3 inhibitor (Millipore Sigma 361571), and 1 µM PD0325901/MEK1/2 inhibitor (Millipore Sigma 444968) in DMEM. Cells were tested for mycoplasma using the LookOut mycoplasma PCR detection kit (Sigma MP0035) following the manufacturer's instructions and using the JumpStart Taq DNA polymerase (Sigma D9307). For proteasome inhibitor studies, cells were treated with 5 µM MG132 (Invivogen TLRL-MG132) or with dimethyl sulfoxide (DMSO) as control.
Western blot analysis
Cells or tissues were either lysed directly in Laemmli buffer (Bio-Rad 1610747) or in RIPA buffer (25 mM Tris at pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS), sonicated to shear chromatin, and spun at high-speed to pellet cellular debris. If lysed in RIPA, protein concentration was determined by BCA assay (Thermo Scientific 23225). Equal amounts of protein were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibody. After incubation with secondary antibody (Millipore Sigma GENA931 for mice and GENA934 for rabbits), immunoblots were developed with enhanced chemiluminescence (Bio-Rad 1705060). Protein abundance was quantified by measuring the integrated intensity of bands using Fiji ImageJ and normalizing to loading controls. The following primary antibodies were used: mouse Rap1 (1252, rabbit polyclonal), TRF2 (Novus Biologicals NB110-57130, rabbit polyclonal), Tip60 (Cell Signaling 12058, rabbit polyclonal), γ-tubulin (Millipore Sigma T6557, mouse monoclonal), α-tubulin (Abcam ab7291, mouse monoclonal), histone H3 (Abcam ab1791, rabbit polyclonal), Flag (Cell Signaling 14793, rabbit monoclonal), c-Myc (Santa Cruz Biotechnology sc-789, rabbit polyclonal), HA (Abcam ab9110, rabbit polyclonal), and streptavidin-HRP (Invitrogen 1953050).
IF-FISH
MEFs and mESCs grown on coverslips were fixed with 4% paraformaldehyde for 10 min, washed with PBS, and permeabilized with 0.5% Triton X-100 buffer (0.5% Triton X-100, 20 mM HEPES at pH 7.9, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose). Cells were then incubated in blocking solution (1 mg/mL BSA, 3% goat serum, 0.1% Triton X-100, 1 mM EDTA in PBS) for 30 min at room temperature, followed by incubation with primary antibody diluted in blocking solution for 2 h at room temperature. After washing with PBST (0.1% Tween 20 in PBS) three times for 5 min each, cells were incubated with Alexa Fluor-labeled secondary antibody (Thermo Fisher Scientific) for 45 min at room temperature. After washing with PBST three times for 5 min each, if FISH was performed, cells were dehydrated with ethanol series (70%, 95%, and then 100%) and hybridized with TAMRA-OO-(TTAGGG)3 PNA probe (Applied Biosystems) in formamide hybridization solution (70% formamide, 0.5% blocking reagent [Roche], 10 mM Tris-HCl at pH 7.2) for 5 min at 80°C. Cells were allowed to cool for 2 h at room temperature and then washed four times for 10 min each with formamide washing solution (70% formamide, 10 mM Tris-HCl at pH 7.2). Cells were then washed with PBS three times for 5 min each and counterstained with DAPI, and coverslips were mounted on slides with antifade reagent (Prolong Gold, Invitrogen). Images were captured using a Nikon Eclipse Ti or DeltaVision microscope. The primary antibodies used were mouse Rap1 (1252, rabbit polyclonal), HA (Abcam ab9110, rabbit polyclonal), Flag M2 (Millipore Sigma F1804, mouse monoclonal), Alexa Fluor 488 streptavidin (Thermo Fisher Scientific S11223), and Zscan4 (Millipore Sigma AB4340, rabbit polyclonal).
2C::tdTomato reporter live-cell imaging
The 2C::tdTomato reporter was a gift from Samuel Pfaff (Addgene 40281). To establish stable cell lines, mESCs were transfected with the 2C::tdTomato reporter followed by selection with hygromycin for 7 d to select for cells with stable integrations. To compare 2C::tdTomato positivity across cell lines, each cell line was seeded at equal density (200,000 cells per well in a gelatin-coated six-well plate). After 24 h in culture, live-cell imaging and analysis were performed using Incucyte (Sartorius). Phase contrast and red fluorescence images were captured. Confluence and red object masks were then applied, and the proportion of 2C::tdTomato-positive cells was determined by normalizing the number of red objects to cell confluency.
RNA sequencing
Cell pellets were submitted to Genewiz for RNA extraction, polyA library preparation, and high-throughput sequencing using their pipelines. For bioinformatic analysis of RNA sequencing (RNA-seq) data, the Seq-N-Slide workflow (source code available at https://github.com/igordot/sns) was used. Under this workflow, FASTQ files were trimmed using Trimmomatic (Bolger et al. 2014) to remove adapters and low-quality bases and aligned to the mm10 reference genome using RNA-STAR (Dobin et al. 2013). Alignments to other species and common contaminants were removed by FastQ-Screen (source code available at https://github.com/StevenWingett/FastQ-Screen). Gene–sample count matrices were generated using featureCounts (Liao et al. 2019). Differential gene expression analysis was performed by DESeq2 (Love et al. 2014). Differentially expressed genes (DEGs) were determined based on a fold change (FC) >1.5 and false discovery rate (FDR) <0.1. Hierarchically clustered heat maps were generated using the ComplexHeatmaps package (Gu et al. 2016). Kmeans clustering was performed using Morpheus (https://software.broadinstitute.org/morpheus).
Differential repeat expression analysis was performed as previously described (Ishiuchi et al. 2015). Trimmed RNA-seq reads were aligned to the reference genome using RNA-STAR (Dobin et al. 2013), allowing multimapping of reads. Nonuniquely mapped reads were mapped to the annotation of repeat sequences in the mouse genome using RepeatMasker (http://www.repeatmasker.org). Differential gene expression analysis was performed by DESeq2 (Love et al. 2014). Differentially expressed repeats were determined based on a FC >2 and FDR <0.1. To quantify proportion of DEGs near MERVL MT_Mm sites, BEDtools (Quinlan and Hall 2010) window function was used to count the number of DEGs and total number of genes at increasing distances from MT2_Mm sites as annotated by RepeatMasker
Chromatin immunoprecipitation (ChIP)
For Rap1 ChIP, SV40LT immortalized Rap1−/− MEFs were transduced with pLPC-Flag-2xHA-EV, pLPC-Flag-2xHA-Rap1, and pLPC-Flag-2xHA-Rap1-I312R retrovirus produced from transfected Phoenix cells. Cells (108) were harvested by trypsinization, washed twice with PBS, and double-fixed with 2 mM ethylene glycol bis (succinimidylsuccinate) (EGS; Thermo Fisher Scientific 21565) in PBS for 45 min at room temperature and then with 1% formaldehyde for 20 min at room temperature. The reaction was quenched with 125 mM glycine and samples were spun at 240g for 5 min at 4°C. Pellets were washed twice with cold PBS, resuspended in 10 mL of cold lysis buffer 1 (100 mM HEPES-KOH, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, protease inhibitor cocktail), and incubated with rocking for 10 min at 4°C. Samples were spun at 950g for 2 min at 4°C, and pellets were resuspended in 10 mL of cold lysis buffer 2 (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA 10 mM Tris-HCl, protease inhibitor cocktail) and incubated with rocking for 10 min at 4°C. Samples were spun at 1500g for 2 min at 4°C, and pellets were resuspended in 3 mL of cold lysis buffer 3 (1 mM EDTA, 0.5 mM EGTA, 10 mM Tris-HCl at pH 8, 100 mM NaCl, 0.1% Na-deoxycholate, 0.5% N-lauroyl sarcosine, protease inhibitor cocktail) and run through a 27-gauge needle 10 times. Samples were sonicated for 10 cycles for 30 sec on/30 sec off in a Bioruptor Pico. After sonication, 1/10 vol of 10% Triton X-100 was added and samples were spun at 18,400g for 10 min at 4°C. Supernatants were precleared with 300 µL of ChIP-grade Protein G magnetic beads (Cell Signaling 9006) and then incubated while rotating overnight at 4°C with 10 µg of anti-HA ChIP-grade antibody (Abcam ab9110). The following morning, 30 µL of ChIP-grade Protein G magnetic beads was added and samples were incubated while rotating for 2 h at 4°C. Beads were washed a total of five times with cold wash buffer (50 mM HEPES at pH 7.6, 1 mM EDTA, 0.7% Na-deoxycholate, 1% NP-40, 0.5 M LiCl, protease inhibitor cocktail) and once with PBS. ChIP DNA was eluted in 250 µL of elution buffer (0.5% SDS, 100 mM NaHCO3) by incubating for 15 min at room temperature on a roller. A second elution was performed by adding 250 µL of elution buffer and incubating for 20 min at 65°C while mixing. Eluates were combined and reverse cross-linked by adding 20 µL of 5 M NaCl and then incubated for 4 h at 65°C. Samples were then purified by phenol-chloroform-isoamyl alcohol followed by ethanol precipitation and resuspension in water.
For mESC histone marks, 30 × 106 cells were cross-linked in fixation buffer (1% formaldehyde, 15 mM NaCl, 0.15 mM EDTA, 0.075 mM EGTA, 10 mM HEPES at pH 7.6 in DMEM) for 10 min at room temperature, and then quenched by addition of 0.125 M glycine. Cells were then washed with PBS, harvested by scraping, and centrifuged at 2500g for 5 min at 4°C. Cell pellets were resuspended in 5 mL of lysis buffer 1 (50 mM HEPES at pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton X-100, 10 mM sodium butyrate, 0.2 mM PMSF, protease/phosphatase inhibitor cocktail), incubated while rocking for 10 min at 4°C, and then centrifuged at 1350g for 5 min at 4°C. Cell pellets were then resuspended in 5 mL of lysis buffer 2 (10 mM Tris at pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM odium butyrate, 0.2 mM PMSF, protease/phosphatase inhibitor cocktail), incubated while rocking for 10 min at room temperature, and then centrifuged at 1350g for 5 min at 4°C. The resulting nuclear pellet was resuspended in 1.2 mL of lysis buffer 3 (10 mM Tris at pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 0.5% N-lauroylsarcosine, 10 mM sodium butyrate, 0.2 mM PMSF, protease/phosphatase inhibitor cocktail) and sonicated for 15 cycles of 30 sec on/30 sec off. Sonicated chromatin was spun at high speed for 30 min at 4°C to pellet insoluble debris, and supernatant was used for IP. Depending on the antibody used, 100 µg of chromatin was aliquoted to a tube, lysis buffer 3 was added to bring the volume to 200 µL, and then the volume was brought to 300 µL by addition of 100 µL of incubation buffer (3% Triton X-100, 0.3% sodium deoxycholate, 15 mM EDTA, 10 mM sodium butyrate, 0.2 mM PMSF, protease/phosphatase inhibitor cocktail). Eight micrograms of antibody (anti-acetyl-histone H4, Millipore Sigma 06-598; and anti-acetyl-histone H2A.Z, Millipore Sigma Abe1363) was then added, and tubes were rotated overnight at 4°C. The next day, Dynabeads Protein A magnetic beads (Thermo Fisher 1001D) were added and incubated for 2 h at 4°C while rotating. Beads were then washed five times with RIPA buffer (50 mM HEPES at pH 7.5, 0.7% sodium deoxycholate, 1 mM EDTA, 1% NP40, 500 mM lithium chloride, 10 mM sodium butyrate, 0.2 mM PMSF, protease/phosphatase cocktail) and once with 1× TE + 50 mM NaCl. Antibody/DNA complexes were then eluted by adding 125 µL of elution buffer (50 mM Tris at pH 8.0, 10 mM EDTA, 1% SDS) and incubating for 20 min at 65°C with shaking. The eluate was then moved to a new tube and cross-links were reversed by incubating with shaking overnight at 65°C. Then, 4 µL of 20 mg/mL Proteinase K was added and tubes were incubated for 2 h at 55°C with shaking. ChIP DNA was then purified by QIAquick PCR purification kit.
ChIP dot blot
ChIP DNA was denatured for 5 min at 95°C and dot-blotted onto Hybond nylon membranes in 2× SSC. Membranes were treated with 1.5 M NaCl/0.5 N NaOH for 10 min, and with 1 M NaCl/0.5 M Tris-HCl (pH 7.0) for 10 min. DNA was then cross-linked to the membrane using a Stratagene UV cross-linker. End-labeled telomere probe was prepared by incubating (CCCTAA)4 oligo with T4 PNK enzyme and 32P-γ-ATP for 45 min at 37°C. The probe was then filtered through a G-25 column and hybridized to the cross-linked membrane overnight at 55°C. Membranes were washed three times in 2× SSC for 5 min, exposed overnight to PhosphorImager screen, and imaged on a Typhoon gel scanner. The quantification of DNA precipitated was performed by measuring integrated intensity using Fiji ImageJ and normalizing to input samples.
ChIP sequencing library preparation, sequencing, and analysis
Sequencing libraries were prepared using the NEBNext Ultra II DNA library preparation kit for Illumina (NEB E7645S). For HA-Rap1 ChIP, sequencing was performed on an Illumina NextSeq 500 (SE 150 cycles). For histone marks in mESCs, sequencing was performed on an Illumina NovaSeq 6000 (PE 100 cycles).
For bioinformatic analysis of ChIP sequencing (ChIP-seq) data, the Seq-N-Slide workflow (source code available at https://github.com/igordot/sns) was used. Under this workflow, reads were aligned to the mm10 reference genome using Bowtie2 (Langmead and Salzberg 2012), duplicate reads were removed using Sambamba (source code available at https://github.com/biod/sambamba), and genome browser tracks representing ChIP-seq profiles were generated using IGV (Robinson et al. 2011). MACS2 (Zhang et al. 2008) was used for peak calling. Deeptools (Ramírez et al. 2016) was used to generate heat maps and density plots.
For telomere binding analysis using high-throughput sequencing, FASTQ files were processed using grep to count the number of reads containing three or more consecutive telomere repeats (TTAGGG/CCCGAA) and normalized to the total number of reads.
qRT-PCR
RNA was purified from cells using the NucleoSpin RNA mini kit (Macherey-Nagel 740955.50). One microgram of RNA was reverse-transcribed using the iScript gDNA clear cDNA synthesis kit (Bio-Rad 1725034), and cDNA libraries were diluted 1:5. RT-PCR was performed using ssoAdvanced SYBR Green supermix (1725270) on a Roche LightCycler 480 II. qPCR primers are in Supplemental Table S9.
Subcellular fractionation
Cells (1 × 107) were resuspended in 200 µL of buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail). After addition of 2 µL of 10% Triton X-100, the resuspension was incubated for 8 min on ice. Samples were centrifuged at 1300g for 5 min at 4°C. Supernatants (S1) were clarified at 20,000g for 5 min at 4°C to yield the cytoplasmic fraction (S2). The pellet was washed once with buffer A and then lysed in 100 µL of buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail) for 30 min at 4°C. Samples were centrifuged at 1700g for 5 min at 4°C, the supernatant was kept as the nucleoplasmid (S3) fraction, and pellets were the chromatin-bound fraction (P3). Chromatin was washed once with buffer B and then resuspended in Laemmli buffer, sonicated, and boiled for 10 min at 70°C.
Nucleosome reconstitution
The Widom 601 nucleosome positioning sequence was obtained from a plasmid containing eight copies of the fragment, each flanked by the EcoRV restriction site (Armache et al. 2011). Wild-type Xenopus histone was expressed individually in E. coli cells, extracted from inclusion bodies, and purified by size exclusion and anion chromatography (Dyer et al. 2004). Purified histones were lyophilized (Sentry, VirTis) and stored at −80°C for further use. Nucleosome reconstitutions were performed as described (Dyer et al. 2004; Armache et al. 2011). Briefly, recombinant histone octamers were assembled by mixing equimolar amounts of each of the four histones and dialyzing against refolding buffer (10 mM Tris at pH 7.5, 2.0 M NaCl, 1 mM EDTA, 5 mM BME). Assembled octamers were purified by size exclusion chromatography (Superdex 200, GE healthcare) in refolding buffer. Then, histone octamers and purified Widom 601 DNA were combined and dialyzed overnight with salt gradient dialysis (Rapid Pump, Gilson). Finally, assembled nucleosomes were purified through ion exchange chromatography (Resource Q, GE Healthcare). Purified nucleosomes were dialyzed into TCS buffer (20 mM Tris-HCl at pH 7.5, 1 mM EDTA, 1 mM DTT), concentrated, and stored at 4°C.
Proximity-based biotinylation (BioID) and streptavidin pull-down
SV40LT immortalized Rap1−/− MEFs were transduced with pLPC-Flag-BirA*-13xGGGGS-Myc-EV, pLPC-Flag-BirA*-13xGGGGS-Myc-Rap1, and pLPC-Flag-BirA*-13xGGGGS-Myc-Rap1-I312R retrovirus produced from transfected Phoenix cells. Single-cell clones were isolated, and independent clonal cell lines were screened by IF to identify those that expressed at high levels. Cells grown in 15-cm2 dishes to be 90% confluency at time of harvest were treated with 50 µM biotin for 20 h followed by biotin-free media for 1 h. For each condition, 10 × 106 cells were harvested by trypsinization, lysed for 1 h with rocking at 4°C in NP40 lysis buffer (10 mM Tris at pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1 mM PMSF, protease inhibitor cocktail), and then centrifuged at 3300g for 10 min at 4°C to pellet nuclei. Nuclei were resuspended in 500 µL of SDS lysis buffer (50 mM Tris at pH 8.0, 1% SDS, 10 mM EDTA, 1 mM PMSF, protease inhibitor cocktail), and incubated for 10 min on ice, followed by boiling for 5 min at 95°C. Samples were then sonicated for 10 cycles of 30 sec on/30 sec off and clarified by centrifuging at 15,000g for 10 min at 4°C. Fifty microliters of MyOne Streptavidin beads (Thermo Fisher 65001) was then added and incubated while rotating overnight at 4°C. Beads were then washed twice with RIPA buffer, twice with 2 M urea in 10 mM Tris (pH 8.0), twice more with RIPA, and then twice with buffer 4 (50 mM Tris at pH 7.4, 50 mM NaCl). Proteins were then eluted from magnetic beads into Laemmli buffer + 2 mM biotin by boiling for 5 min at 95°C. Eluates were used for Western blot, silver stain (Thermo Fisher Scientific 24612), and mass spectrometry analysis as previously described (Pinzaru et al. 2020).
Coimmunoprecipitation (co-IP)
HEK293T cells were cotransfected with the indicated plasmid constructs using polyethylenimine (PEI). Forty-eight hours after transfection, cells were harvested in cold PBS, washed once with PBS, and lysed for 15 min on ice in 500 µL of lysis buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.05% SDS, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail). Salt boost was performed by adding 25 µL of 5 M NaCl and incubating for an additional 5 min on ice. Then, 500 µL of cold H2O was added and lysates were spun at maximum speed for 10 min to pellet debris. The antibody for the epitope to be precipitated (anti-Flag, Millipore Sigma F1804; anti-HA, BioLegend 901533; and anti-c-Myc Millipore Sigma M4439) was added to supernatants and incubated for 3 h while rotating at 4°C. Protein G Sepharose (Millipore Sigma GE17-0618-01) preblocked in 5% BSA was added and incubated for one additional hour while rotating at 4°C. Beads were washed four times in wash buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS) and proteins were eluted in Laemmli buffer by boiling for 5 min at 95°C for Western blot or with 150 µg/mL 3xFlag peptide (Sigma) for 1 h at 4°C for HAT assay. For co-IP of Rap1 with K48-Ub, the protocol was the same as above except no salt boost was performed, TNE (10 mM Tris at pH 7.8, 1% NP-40, 0.15 M NaCl, 1 mM EDTA, protease inhibitor cocktail) was used as lysis and wash buffer, and 4 mM N-ethylmaleimide (Sigma E3876) and 4 mM 1,10-phenanthroline (Sigma 131377) were included in the buffers. For DNase sensitivity assays, lysis buffer did not include EDTA or DTT. Lysates were supplemented with 2.5 mM MgCl2 and incubated with 10 µg/mL DNase for 1 h at room temperature. DNase was then inactivated by addition of EDTA and DTT before addition of antibody for IP. For co-IP of Flag-Rap1 and endogenous Tip60, Rap1−/− mouse ESCs were transduced with pHAGE2-Flag-EV and pHAGE2-Flag-Rap1. The protocol was then the same as above starting at cell harvest (no transfection).
Affinity purification of Rap1 protein
Recombinant Rap1 with N-terminal His6 tags was expressed from the pTriEx4 vector in BL21(DE3) RIPL bacterial cells as described elsewhere (Vizlin-Hodzic et al. 2009; Nora et al. 2010). After a 3-h induction of protein expression with 0.5 mM isopropyl β-D-thiogalactoside, cells were harvested and resuspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 0.5% Tween 20 at pH 8.0) containing protease inhibitor cocktail (Roche). Sonicated cell extracts were cleared by centrifugation and subsequently filtered (0.45-µm SterivexTM filter, Millipore). The supernatant containing Rap1 was further purified by immobilized metal ion affinity chromatography using TALON metal affinity resin (Clontech) containing Co2 + cations as described (Yanez et al. 2005). Rap1-containing fractions were eluted in 200 mM imidazole. For His6 tag removal, samples were digested by thrombin protease and purified using gel filtration chromatography to remove cleaved tags and residual protease. The fractions containing pure protein were dialyzed into buffer composed of 50 mM sodium phosphate with 50 mM NaCl (pH 7.0) and, subsequently, proteins were concentrated by ultrafiltration (Amicon 30K, Millipore). The concentration of purified proteins was determined using the Bradford assay and purity was verified by electrophoresis in sodium dodecyl sulfate (SDS)-polyacrylamide gel that was subsequently stained using Bio-Safe Coomassie G 250 (Bio-Rad).
Electrophoretic mobility shift assay (EMSA)
Increasing amounts of 6xHis-tagged Rap1 or tag-free Rap1 proteins (0–15 µM) (lanes 1 = 0.0, 2 = 7.3, 3 = 14.6, 4 = 29.3, 5 = 58.6, 6 = 117.2, 7 = 234.4, 8 = 468.8, 9 = 937.5, 10 = 1875, 11 = 3750, 12 = 7500, 13 = 15,000 in Figs. 2F,G, 3E–H) were incubated with 100 nM DNA (74-bp-long telomeric or 601 sequence) or nucleosome substrate in EMSA buffer (10 mM Tris at pH 7.5, 100 mM NaCl, 2.5% glycerol, 1 mM DTT) for 30 min at room temperature. The binding reactions were resolved on native polyacrylamide gels (6% PAGE, 0.25× TBE). Runs were performed at 170 V for 1.5 h in a cold room (4°C), and gels were stained with ethidium bromide and visualized on a Typhoon Trio+ scanner (Molecular Dynamics). Purified nucleosome substrates were a generous gift from Dr. Karim-Jean Armache.
Histone acetyltransferase (HAT) assays
Core histones and short oligonucleosome substrates for HAT assays were purified as previously described (Cȏté et al. 1995). Indicated substrates were incubated in a 15-µL reaction containing 50 mM Tris-HCl (pH 8.0), 10% glycerol, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 10 mM sodium butyrate, and 0.125 µCi of 3H-labeled acetyl-CoA (Perkin Elmer). Samples were then spotted on phosphocellulose paper (St Vincent's Institute) and counts per minute (CPM) were quantified using a Beckman Coulter LS6500 multipurpose scintillation counter. Alternatively, for in-gel assays, SDS-PAGE was performed followed by Coomassie staining (0.25% Brilliant Blue G [Sigma-Aldrich], 45% methanol, 10% acetic acid), and then destaining overnight (30% methanol, 10% acetic acid). The gel was incubated in EN3HANCE solution (Perkin Elmer) for 1 h (for autoradiography signal amplification/conversion of 3H activity to fluorescence), and then dried on high vacuum (Bio-Rad HydroTech vacuum pump, Savant SGD2000 slab gel dryer) for 2.5 h at 60°C. Exposure was done for 72 h at −80°C.
Tandem affinity purification (TAP) of native TIP60/p400 complex for HAT assays
Native TIP60/p400 complex was affinity-purified as previously described (Dalvai et al. 2015). Briefly, nuclear extracts (Abmayr et al. 2006) were prepared from 1 × 109 to 3 × 109 K562 cells expressing EPC1-3xFLAG-2xStrep from the AAVS1 locus (empty vector for mock preparations). Tween-20 was added to 0.1%, and extracts were centrifuged at 17,000 rpm in a Beckman JA-20 rotor for 1 h at 4°C. After preclearing with 300 µL of Sepharose CL-6B (Sigma), extracts were incubated with 250 µL of anti-Flag M2 affinity resin (Sigma) for 2 h at 4°C. Beads were then washed in Poly-Prep columns (Bio-Rad) with 40 column volumes of buffer 1 (20 mM HEPES-KOH at pH 7.9, 10% glycerol, 300 mM KCl, 0.1% Tween 20, 1 mM DTT, Halt protease, phosphatase inhibitor cocktail without EDTA [Pierce]), followed by 40 column volumes of buffer 2 (20 mM HEPES-KOH at pH 7.9, 10% glycerol, 150 mM KCl, 0.1% Tween 20, 1 mM DTT, Halt protease, phosphatase inhibitor cocktail without EDTA [Pierce]). Protein complexes were eluted with 200 µg/mL 3xFlag peptide (Sigma) in five column volumes of buffer 2 for 1 h at 4°C. The eluted fraction was then mixed with 125 µL of Strep-Tactin Sepharose (IBA) affinity matrix for 1 h at 4°C. Beads were then washed again in in Poly-Prep columns (Bio-Rad) with 40 column volumes of buffer 2. Complexes were then eluted in two fractions with 2.5 mM D-biotin in four column volumes of buffer 2 and flash-frozen in liquid nitrogen.
siRNA transfection
Mouse ESCs were reverse-transfected into gelatin-coated plates using Lipofectamine RNAiMAX (Thermo Fisher Scientific 13778100) according to the manufacturer's instructions and a final concentration of 30 nM for each siRNA. Forty-eight hours after transfection, RNA was purified from cells using the NucleoSpin RNA mini kit (Macherey-Nagel 740955.50). A list of siRNAs used in this study is in Supplemental Table S9.
RAP1-EPC1 interaction in vitro
One microgram of soluble His-RAP1 was precleared on glutathione Sepharose 4B beads (GE Healthcare) for 1 h at 4°C and then incubated with either 3 µg (measured by Bradford protein assay) of GST-tagged EPC1 (amino acids 1–581) or GST alone immobilized on glutathione Sepharose beads for 2.5 h at 4°C in 200 µL of pull-down buffer (200 mM NaCl, 25 mM HEPES at pH 7.5, 10% glycerol, 100 µg/mL BSA, 1 mM PMSF, 0.5 mM DTT, 0.1% Tween-20, 2 µg/mL leupeptin, 2 µg/mL pepstatin, 5 µg/mL aprotinin). Beads were washed twice with pull-down buffer and run on SDS-PAGE followed by Western blotting (anti-His, Clontech 631212).
Supplementary Material
Acknowledgments
We thank Mike Al-Kareh for technical assistance, and members of the Sfeir laboratory for feedback and comments on the manuscript. pRK5-HA-ubiquitin-K48 was a gift from Ted Dawson (Addgene 17605). We thank Karim-Jean Armache for guidance on RAP1 gel shift assays. Catherine Lachance is acknowledged for purified NuA4/TIP60 and chromatin fractions. We acknowledge Beatrix M. Ueberheide and the Proteomics Laboratory as well as Sang Y. Kim and the Rodent Genetic Engineering Laboratory at New York University School of Medicine. We thank the Genome Technology Center at New York University School of Medicine and the Integrated Genomics Operation at Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center. This work was supported by a grant from the National Institutes of Health (NIH; R01 DK102562) to A.S., and from the Canadian Institutes of Health Research (FDN-143314) to J.C. R.M.B. is supported by a training grant from the NIH (1F30 DK118901). C.H.’s laboratory is funded by internal institutional support of the Institute of Biophysics of the Czech Academy of Sciences (68081707), the Czech Science Foundation (project 19-18226S), and the Ministry of Education, Youth, and Sports of the Czech Republic (project LTAUSA19024). A.M. held studentships from the National Science and Engineering Council of Canada and the Fonds de Recherche du Québec-Nature et Technologie.
Author contributions: A.S. and R.M.B. conceived the experimental design. R.M.B. performed experiments with the help of P.S., O.S., and W.K. M.S. in C.H.’s laboratory performed EMSA and RAP1 purification. A.M. in J.C.’s laboratory performed HAT assays. P.D.I. prepared nucleosomes for EMSA. A.S. and R.M.B. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349039.121.
Competing interest statement
Agnel Sfeir is a cofounder of, consultant for, and shareholder in Repare Therapeutics.
References
- Abmayr SM, Yao T, Parmely T, Workman JL. 2006. Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr Protoc Mol Biol. 75: 12.1.1–12.1.10. 10.1002/0471142727.mb1201s75 [DOI] [PubMed] [Google Scholar]
- Acharya D, Hainer SJ, Yoon Y, Wang F, Bach I, Rivera-Pérez JA, Fazzio TG. 2017. KAT-independent gene regulation by Tip60 promotes ESC self-renewal but Not pluripotency. Cell Rep 19: 671–679. 10.1016/j.celrep.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cȏté J. 1999. Nua4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18: 5108–5119. 10.1093/emboj/18.18.5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arat NO, Griffith JD. 2012. Human Rap1 interacts directly with telomeric DNA and regulates TRF2 localization at the telomere. J Biol Chem 287: 41583–41594. 10.1074/jbc.M112.415984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. 2011. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334: 977–982. 10.1126/science.1210915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger A, Galarneau L, Altaf M, Nourani A, Doyon Y, Utley RT, Cronier D, Allard S, Côté J. 2008. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol 28: 2257–2270. 10.1128/MCB.01755-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J, Tachibana CY, Tye BK. 1986. Identification of a telomere-binding activity from yeast. Proc Natl Acad Sci 83: 3713–3717. 10.1073/pnas.83.11.3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Côté J. 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev 17: 1415–1428. 10.1101/gad.1056603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram RJ, Kornberg RD. 1985. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci 82: 43–47. 10.1073/pnas.82.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman AR, Kimmerly WJ, Rine J, Kornberg RD. 1988a. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol 8: 210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman AR, Lue NF, Kornberg RD. 1988b. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol 8: 5086–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T. 2005. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 7: 712–718. 10.1038/ncb1275 [DOI] [PubMed] [Google Scholar]
- Chambers A, Tsang JS, Stanway C, Kingsman AJ, Kingsman SM. 1989. Transcriptional control of the saccharomyces cerevisiae PGK gene by RAP1. Mol Cell Biol 9: 5516–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, et al. 2011. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol 18: 213–221. 10.1038/nsmb.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PB, Hung JH, Hickman TL, Coles AH, Carey JF, Weng Z, Chu F, Fazzio TG. 2013. Hdac6 regulates Tip60-p400 function in stem cells. Elife 2: e01557. 10.7554/eLife.01557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang W, Xie D, Gao T, Dong Z, Lu X. 2020. Histone chaperone FACT represses retrotransposon MERVL and MERVL-derived cryptic promoters. Nucleic Acids Res 48: 10211–10225. 10.1093/nar/gkaa732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YJ, Kim SH, Tessarollo L, Campisi J, Hodes RJ. 2004. Telomere-associated protein TIN2 is essential for early embryonic development through a telomerase-independent pathway. Mol Cell Biol 24: 6631–6634. 10.1128/MCB.24.15.6631-6634.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittuluru JR, Chaban Y, Monnet-Saksouk J, Carrozza MJ, Sapountzi V, Selleck W, Huang J, Utley RT, Cramet M, Allard S, et al. 2011. Structure and nucleosome interaction of the yeast NuA4 and piccolo-NuA4 histone acetyltransferase complexes. Nat Struct Mol Biol 18: 1196–1203. 10.1038/nsmb.2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cȏté J, Utley R, Workman J. 1995. Basic analysis of transcription factor binding to nucleosomes. Meth Mol Genet 6: 108–128. 10.1016/S1067-2389(06)80009-9 [DOI] [PubMed] [Google Scholar]
- Dalvai M, Loehr J, Jacquet K, Huard CC, Roques C, Herst P, Côté J, Doyon Y. 2015. A scalable genome-editing-based approach for mapping multiprotein complexes in human cells. Cell Rep 13: 621–633. 10.1016/j.celrep.2015.09.009 [DOI] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110. 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Côté J. 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev 14: 147–154. 10.1016/j.gde.2004.02.009 [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Côté J. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol 24: 1884–1896. 10.1128/MCB.24.5.1884-1896.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. 2004. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol 375: 23–44. 10.1016/S0076-6879(03)75002-2 [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134: 162–174. 10.1016/j.cell.2008.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Djekidel MN, Zhang Y. 2020. A transcriptional roadmap for 2C-like-to-pluripotent state transition. Sci Adv 6: eaay5181. 10.1126/sciadv.aay5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32: 2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Hanaoka S, Nagadoi A, Yoshimura S, Aimoto S, Li B, de Lange T, Nishimura Y. 2001. NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J Mol Biol 312: 167–175. 10.1006/jmbi.2001.4924 [DOI] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6: 801–814. 10.1101/gad.6.5.801 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. 2006. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77. 10.1016/j.cell.2006.04.044 [DOI] [PubMed] [Google Scholar]
- Huet J, Sentenac A. 1987. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci 84: 3648–3652. 10.1073/pnas.84.11.3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J, Cottrelle P, Cool M, Vignais ML, Thiele D, Marck C, Buhler JM, Sentenac A, Fromageot P. 1985. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J 4: 3539–3547. 10.1002/j.1460-2075.1985.tb04114.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Bošković A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla ME. 2015. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 22: 662–671. 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert JP, Roques C, Pandita RK, Paquet E, Herst P, Gingras AC, Pandita TK, et al. 2016. The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol Cell 62: 409–421. 10.1016/j.molcel.2016.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir S, Hockemeyer D, de Lange T. 2014. TALEN gene knockouts reveal no requirement for the conserved human shelterin protein Rap1 in telomere protection and length regulation. Cell Rep 9: 1273–1280. 10.1016/j.celrep.2014.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. 2001. Sprap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630. 10.1016/S0960-9822(01)00503-6 [DOI] [PubMed] [Google Scholar]
- Karlseder J, Kachatrian L, Takai H, Mercer K, Hingorani S, Jacks T, de Lange T. 2003. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol Cell Biol 23: 6533–6541. 10.1128/MCB.23.18.6533-6541.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibe T, Osawa GA, Keegan CE, de Lange T. 2010. Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol Cell Biol 30: 1059–1066. 10.1128/MCB.01498-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G, Liu K, Liu C, Lustig AJ. 1993. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev 7: 1146–1159. 10.1101/gad.7.7a.1146 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini-Denchi E, Sfeir A. 2016. Stop pulling my strings—what telomeres taught us about the DNA damage response. Nat Rev Mol Cell Biol 17: 364–378. 10.1038/nrm.2016.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101: 471–483. 10.1016/S0092-8674(00)80858-2 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2019. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47: e47. 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Wilson NM, Petracek ME, Berman J. 1989. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet 16: 225–239. 10.1007/BF00422108 [DOI] [PubMed] [Google Scholar]
- Lototska L, Yue JX, Li J, Giraud-Panis MJ, Songyang Z, Royle NJ, Liti G, Ye J, Gilson E, Mendez-Bermudez A. 2020. Human RAP1 specifically protects telomeres of senescent cells from DNA damage. EMBO Rep 21: e49076. 10.15252/embr.201949076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ, Kurtz S, Shore D. 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250: 549–553. 10.1126/science.2237406 [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marión RM, Montero JJ, López de Silanes I, Graña-Castro O, Martínez P, Schoeftner S, Palacios-Fábrega JA, Blasco MA. 2019. TERRA regulate the transcriptional landscape of pluripotent cells through TRF1-dependent recruitment of PRC2. Elife 8: e44656. 10.7554/eLife.44656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz-Potoczny M, Lobanova A, Loeb AM, Kirak O, Olbrich T, Ruiz S, Lazzerini Denchi E. 2021. TRF2-mediated telomere protection is dispensable in pluripotent stem cells. Nature 589: 110–115. 10.1038/s41586-020-2959-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, Thanasoula M, Carlos AR, Gómez-López G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. 2010. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol 12: 768–780. 10.1038/ncb2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, Gómez-López G, García F, Mercken E, Mitchell S, Flores JM, de Cabo R, Blasco MA. 2013. RAP1 protects from obesity through its extratelomeric role regulating gene expression. Cell Rep 3: 2059–2074. 10.1016/j.celrep.2013.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, Gómez-López G, Pisano DG, Flores JM, Blasco MA. 2016. A genetic interaction between RAP1 and telomerase reveals an unanticipated role for RAP1 in telomere maintenance. Aging Cell 15: 1113–1125. 10.1111/acel.12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Shore D. 2001. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol 21: 8082–8094. 10.1128/MCB.21.23.8082-8094.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8: 2257–2269. 10.1101/gad.8.19.2257 [DOI] [PubMed] [Google Scholar]
- Nanavaty V, Sandhu R, Jehi SE, Pandya UM, Li B. 2017. Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA:DNA hybrids. Nucleic Acids Res 45: 5785–5796. 10.1093/nar/gkx184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora GJ, Buncher NA, Opresko PL. 2010. Telomeric protein TRF2 protects Holliday junctions with telomeric arms from displacement by the Werner syndrome helicase. Nucleic Acids Res 38: 3984–3998. 10.1093/nar/gkq144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Marcand S. 2005. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J 24: 3117–3127. 10.1038/sj.emboj.7600778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul NK, Baksh KA, Arias JF, Zamble DB. 2020. The impact of a His-tag on DNA binding by RNA polymerase α-C-terminal domain from Helicobacter pylori. Protein Expr Purif 167: 105541. 10.1016/j.pep.2019.105541 [DOI] [PubMed] [Google Scholar]
- Pinzaru AM, Kareh M, Lamm N, Lazzerini-Denchi E, Cesare AJ, Sfeir A. 2020. Replication stress conferred by POT1 dysfunction promotes telomere relocalization to the nuclear pore. Genes Dev 34: 1619–1636. 10.1101/gad.337287.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt JM, Ryvkin P, Wanat JJ, Donahue G, Ricketts MD, Barrett SP, Waters HJ, Song S, Chavez A, Abdallah KO, et al. 2013. Rap1 relocalization contributes to the chromatin-mediated gene expression profile and pace of cell senescence. Genes Dev 27: 1406–1420. 10.1101/gad.218776.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan SK, Su T, Yen L, Jacquet K, Huang C, Côté J, Kurdistani SK, Carey MF. 2016. EP400 deposits H3.3 into promoters and enhancers during gene activation. Mol Cell 61: 27–38. 10.1016/j.molcel.2015.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. Deeptools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Terrones D, Gaume X, Ishiuchi T, Weiss A, Kopp A, Kruse K, Penning A, Vaquerizas JM, Brino L, Torres-Padilla ME. 2018. A molecular roadmap for the emergence of early-embryonic-like cells in culture. Nat Genet 50: 106–119. 10.1038/s41588-017-0016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196: 801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy J, Bae NS, Scrafford J, Baumann P. 2009. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J 28: 3390–3399. 10.1038/emboj.2009.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck W, Fortin I, Sermwittayawong D, Côté J, Tan S. 2005. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol Cell Biol 25: 5535–5542. 10.1128/MCB.25.13.5535-5542.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiaputra D, Ahmad S, Dalwadi U, Steunou AL, Lu S, Ross JD, Dong MQ, Côté J, Yip CK. 2018. Molecular architecture of the essential yeast histone acetyltransferase complex NuA4 redefines its multimodularity. Mol Cell Biol 38: e00570-17. 10.1128/MCB.00570-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. 2010. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327: 1657–1661. 10.1126/science.1185100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh BN, Akhtar A. 2019. The many lives of KATs—detectors, integrators and modulators of the cellular environment. Nat Rev Genet 20: 7–23. 10.1038/s41576-018-0072-4 [DOI] [PubMed] [Google Scholar]
- Shore D, Nasmyth K. 1987. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51: 721–732. 10.1016/0092-8674(87)90095-X [DOI] [PubMed] [Google Scholar]
- Shore D, Stillman DJ, Brand AH, Nasmyth KA. 1987. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J 6: 461–467. 10.1002/j.1460-2075.1987.tb04776.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Perez JV, Svitko W, Ricketts MD, Dean E, Schultz D, Marmorstein R, Johnson FB. 2020. Rap1-mediated nucleosome displacement can regulate gene expression in senescent cells without impacting the pace of senescence. Aging Cell 19: e13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steunou AL, Rossetto D, Cȏté J. 2014. Regulating chromatin by histone acetylation. In Fundamentals of chromatin (ed. Workman JL, Abmayr SM), pp. 147–212. Springer, New York. [Google Scholar]
- Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, et al. 2010. Telomere-independent Rap1 is an IKK adaptor and regulates NF-κB-dependent gene expression. Nat Cell Biol 12: 758–767. 10.1038/ncb2080 [DOI] [PubMed] [Google Scholar]
- Vignais ML, Woudt LP, Wassenaar GM, Mager WH, Sentenac A, Planta RJ. 1987. Specific binding of TUF factor to upstream activation sites of yeast ribosomal protein genes. EMBO J 6: 1451–1457. 10.1002/j.1460-2075.1987.tb02386.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizlin-Hodzic D, Ryme J, Simonsson S, Simonsson T. 2009. Developmental studies of Xenopus shelterin complexes: the message to reset telomere length is already present in the egg. FASEB J 23: 2587–2594. 10.1096/fj.09-129619 [DOI] [PubMed] [Google Scholar]
- Wotton D, Shore D. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11: 748–760. 10.1101/gad.11.6.748 [DOI] [PubMed] [Google Scholar]
- Xu P, Li C, Chen Z, Jiang S, Fan S, Wang J, Dai J, Zhu P, Chen Z. 2016. The NuA4 core complex acetylates nucleosomal histone H4 through a double recognition mechanism. Mol Cell 63: 965–975. 10.1016/j.molcel.2016.07.024 [DOI] [PubMed] [Google Scholar]
- Yanez GH, Khan SJ, Locovei AM, Pedroso IM, Fletcher TM. 2005. DNA structure-dependent recruitment of telomeric proteins to single-stranded/double-stranded DNA junctions. Biochem Biophys Res Commun 328: 49–56. 10.1016/j.bbrc.2004.12.134 [DOI] [PubMed] [Google Scholar]
- Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. 2009. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell 137: 99–109. 10.1016/j.cell.2009.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. 2011. Human telomeric proteins occupy selective interstitial sites. Cell Res 21: 1013–1027. 10.1038/cr.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Ramírez CM, Mateos-Gomez PA, Pinzaru A, Ceccarini G, Kabir S, Fernández-Hernando C, Sfeir A. 2013. Nontelomeric role for Rap1 in regulating metabolism and protecting against obesity. Cell Rep 3: 1847–1856. 10.1016/j.celrep.2013.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chen F, Chen R, Xie D, Yang J, Zhao X, Guo R, Zhang Y, Shen Y, Goke J, et al. 2019. Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Res 47: 8485–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.