Here, Huth et al. investigated the role of components of the nonsense-mediated mRNA decay (NMD) pathway in regulating embryonic stem cell (ESC) differentiation, and show that NMD controls expression levels of the translation initiation factor Eif4a2 and its premature termination codon-encoding isoform (Eif4a2PTC). Their findings expose an intricate link between mRNA homeostasis and mTORC1 activity that must be maintained for normal dynamics of cell state transitions.
Keywords: embryonic stem cells, RNA biology, cell fate regulation, exit from naïve pluripotency, nonsense-mediated mRNA decay, pluripotency, translation initiation
Abstract
Cell fate transitions depend on balanced rewiring of transcription and translation programs to mediate ordered developmental progression. Components of the nonsense-mediated mRNA decay (NMD) pathway have been implicated in regulating embryonic stem cell (ESC) differentiation, but the exact mechanism is unclear. Here we show that NMD controls expression levels of the translation initiation factor Eif4a2 and its premature termination codon-encoding isoform (Eif4a2PTC). NMD deficiency leads to translation of the truncated eIF4A2PTC protein. eIF4A2PTC elicits increased mTORC1 activity and translation rates and causes differentiation delays. This establishes a previously unknown feedback loop between NMD and translation initiation. Furthermore, our results show a clear hierarchy in the severity of target deregulation and differentiation phenotypes between NMD effector KOs (Smg5 KO > Smg6 KO > Smg7 KO), which highlights heterodimer-independent functions for SMG5 and SMG7. Together, our findings expose an intricate link between mRNA homeostasis and mTORC1 activity that must be maintained for normal dynamics of cell state transitions.
Mouse embryonic stem cells (ESCs) capture the developmentally transient naïve pluripotent state in vitro. ESC self-renewal can be maintained in a naïve pluripotent state using inhibitors against Mek1/2 (PD0325901) and Gsk3 (CHIR990201; collectively termed “2i”). ESC self-renewal is sustained by an interactive transcription factor (TF) network, which has been extensively characterized (Young 2011; Dunn et al. 2014). Within 24–36 h after withdrawal of 2i, ESCs shut down naïve identity and initiate differentiation by transiting into formative pluripotency (Wray et al. 2011; Kalkan et al. 2017; Lackner et al. 2021). During this transition, the naïve gene regulatory network (GRN) is extinguished and expression of formative factors such as Otx2, Pou3f1 (Oct6), Dnmt3a/b, and Fgf5 is initiated. A similar transition is evident during peri-implantation development, where the TF network maintaining naïve pluripotency dissolves between embryonic day (E) 4.5 and E5.5 (Boroviak et al. 2014; Acampora et al. 2016; Mohammed et al. 2017).
High-resolution dissection of the exit from the naïve state is facilitated by the availability of Rex1 promoter-driven destabilized GFP reporter ESC lines (Rex1::GFPd2) (Kalkan et al. 2017; Mulas et al. 2017). Rex1 is a known marker of naïve pluripotency, and Rex1-GFPd2 down-regulation is initiated within 24 h after 2i withdrawal (N24) and completed after 48 h (N48). Extinction of Rex1-GFPd2 expression coincides with functional commitment to differentiation.
Several genome-wide screens have uncovered drivers of the exit from naïve pluripotency (Betschinger et al. 2013; Leeb et al. 2014; Li et al. 2020; Lackner et al. 2021), many of which are involved in transcriptional regulation and epigenetic modification. These screens also identified a large cohort of post-transcriptional regulators. RNA modifiers such as m6A (Batista et al. 2014; Geula et al. 2015), negative regulators of mRNA stability such as Pum1 (Leeb et al. 2014), and components of the nonsense-mediated mRNA decay (NMD) pathway (Leeb et al. 2014; Li et al. 2015; Lou et al. 2016; Lackner et al. 2021) have been shown to play a role in ESC differentiation. Nonetheless, how post-transcriptional regulatory mechanisms contribute to cell fate changes remains poorly understood.
NMD is a translation-coupled mechanism that is crucial to maintain cellular RNA homeostasis by promoting degradation of potentially deleterious mRNAs containing a premature termination codon (PTC) (Kurosaki et al. 2019). However, PTC-independent NMD activity has also been shown (Mendell et al. 2004; Wittmann et al. 2006; Tani et al. 2012). NMD is triggered by phosphorylation of the RNA helicase UPF1. Phosphorylated UPF1 (p-UPF1) is essential for NMD and recruits NMD downstream effectors of two distinct RNA degradation pathways (Medghalchi et al. 2001). NMD-mediated mRNA degradation can occur either by the SMG6-mediated endonucleolytic cleavage pathway or via the exonucleolytic cleavage pathway, which is mediated by a SMG5–SMG7 heterodimer (Kurosaki et al. 2019). How division of labor between the two pathways is arranged remains elusive, but transcriptome-wide analysis demonstrated that both decay pathways have highly overlapping mRNA targets (Colombo et al. 2017). SMG5, SMG6, and SMG7 have also been shown to participate in recycling of NMD components by interacting with PP2A to mediate dephosphorylation of p-UPF1 (Anders et al. 2003; Chiu et al. 2003). Notably, recent data suggest that the endonucleolytic and exonucleolytic decay pathways are not acting in a strictly independent manner, and the SMG5–SMG7 heterodimer has been shown to play an important role in the SMG6-mediated endonucleolytic decay pathway (Boehm et al. 2021).
Although NMD components constitute some of the top hits in genome-wide screens studying exit from naïve pluripotency, neither the contribution of individual NMD effector proteins (SMG5, SMG6, and SMG7) nor the key mRNA targets of NMD that lead to a delayed cell fate transition are known.
Here we identify a role for NMD in ensuring normal differentiation kinetics by facilitating establishment of proper cell state-specific gene expression programs and by controlling expression of Eif4a2, a key translation initiation factor. We identify an Eif4a2-dependent increased mTORC1 activity in NMD-deficient ESCs as a factor contributing to their inability to properly commit to differentiation.
Results
Graded defects in exit from naïve pluripotency in NMD-deficient ESCs
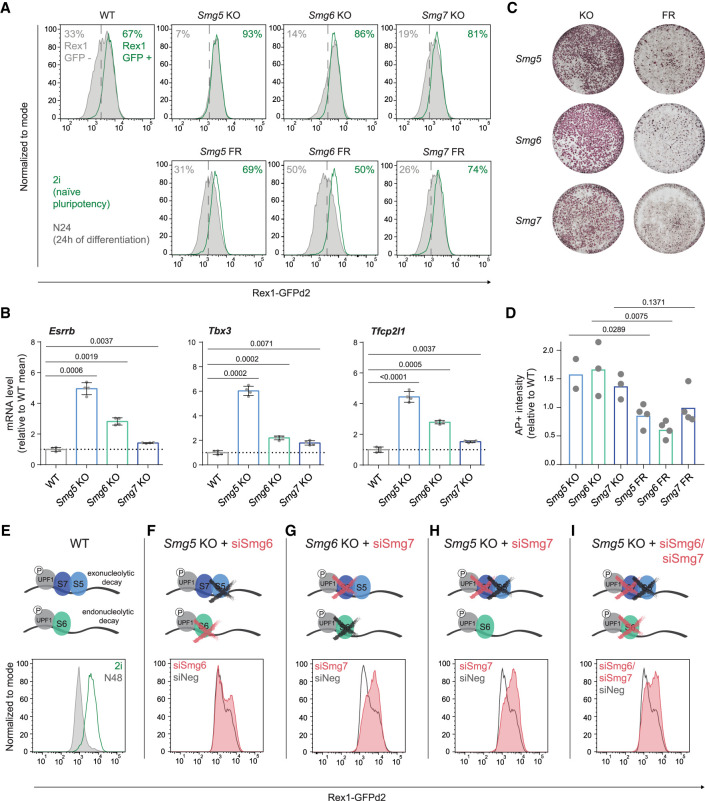
To delineate the molecular function of NMD in the exit from naïve pluripotency, we generated Rex1-GFPd2 reporter ESC lines (Wray et al. 2011; Li et al. 2018) deficient for the three NMD downstream effectors Smg5, Smg6, or Smg7 (NMD KO ESCs), as well as corresponding rescue cell lines in which the missing NMD factor was re-expressed (NMD Flag rescue ESCs [FR]) (Supplemental Fig. S1A–C). All Smg factor KO ESCs showed normal cell cycle and growth profiles (Supplemental Fig. S1D,E). However, they all exhibited pronounced delays in the exit from naïve pluripotency. This manifested in delayed down-regulation of the Rex1-GFPd2 reporter 24 h after the onset of differentiation (N24), and significant delays in the down-regulation of components of the core naïve transcription factor network (such as Esrrb, Tbx3, and Tfcp2l1) (Fig. 1A,B; Supplemental Fig. S1F). Notably, the three Smg factor KOs displayed variable degrees of impairment in exit from naïve pluripotency: The strongest effect was reproducibly observed in the absence of Smg5 and the weakest was observed in the absence of Smg7.
Figure 1.
Hierarchy of defects in the exit from naïve pluripotency in NMD-deficient ESCs. (A) Rex1-GFPd2 analysis by flow cytometry in naïve pluripotency-supporting conditions (2i, green profiles) and 24 h after induction of differentiation by withdrawing 2i in N2B27 medium (N24, gray profiles) in WT, NMD KO, and NMD rescue ESCs (Flag rescue [FR]). The plots shown correspond to one experiment, representative for n = 5 independent replicates. (B) RT-qPCR analysis of the indicated naïve transcription factors (TFs) at N24 in WT and Smg factor KO ESCs. Mean and standard deviation (SD) are plotted for each cell line; n = 2 biological replicates. Expression was normalized to β-actin; results are shown as expression level relative to WT. Two-tailed t-tests were performed to compare WT with Smg factor KOs, and resulting P-values are indicated in the plot. (C) Commitment assay in NMD KO and NMD rescue ESCs. Alkaline phosphatase (AP) staining was used to visualize ESC colonies. (D) Quantification of commitment assays. Mean and SD are shown. Unpaired, two-tailed t-tests were performed to compare each Smg factor KO with its respective Flag rescue (FR) cell line. P-values are indicated in the plot. (E–I) Rex1-GFPd2 flow cytometry analysis of WT cells (E) and Smg factor KO cells (F–I) at N48, after siRNA-mediated knockdown (KD) of Smg6 or Smg7. Plots show one out of two independent experiments with consistent results.
Smg factors are known to regulate telomere maintenance (Reichenbach et al. 2003; Azzalin et al. 2007; Chawla and Azzalin 2008). However, no telomere shortening was observed in NMD KO ESCs, which exhibited either similar (Smg6 KO) or even longer (Smg5 and Smg7 KOs) telomeres compared with wild-type (WT) ESCs (Supplemental Fig. S1G).
To examine later differentiation events, we assayed the ability of Smg factor KO ESCs to irreversibly exit the naïve state and commit to differentiation. We exposed them to differentiation conditions for 72 h and then reapplied 2i medium. Only cells that retain naïve identity are able to grow in 2i and form alkaline phosphatase (AP)-positive colonies. We observed a clear Smg factor KO-dependent delayed entry into commitment (Fig. 1C,D).
To monitor the effect of NMD deficiency on long-term differentiation potential, we performed 3D aggregate embryoid body (EB) differentiation throughout a 10-d time course (Keller et al. 1993). Down-regulation of naïve (Esrrb) and general (Pou5f1, also known as Oct4) pluripotency markers was severely impaired in NMD KO EBs (Supplemental Fig. S1H,I). Although Smg factor KO EBs up-regulated the formative marker genes Fgf5 and Oct6 with kinetics similar to those of WT, the subsequent shutdown of the formative program, initiated in WT cells around day 4 (d4), was impaired in NMD mutant cells. Furthermore, expression of the endo–mesoderm defining TF Brachyury (T) was not properly down-regulated after a peak of expression between d4 and d6, suggesting a general function of NMD in early cell fate decisions. In addition, teratomas derived from Smg6 KO ESCs showed a lower degree of differentiation than WT and rescue controls (Supplemental Fig. S1J).
Together, these data show a global differentiation defect of Smg factor-deficient cells, with an unexpectedly variable and graded degree of phenotypic strengths (Smg5 KO > Smg6 KO > Smg7 KO) during the exit from naïve pluripotency. This does not align with a proposed heterodimer-dependent function of SMG5 and SMG7, which would predict similar phenotypes for Smg5 and Smg7 KOs (Ohnishi et al. 2003; Okada-Katsuhata et al. 2012; Jonas et al. 2013). The results therefore suggest additional functions for SMG5 and SMG7 that are heterodimer-independent.
Cooperativity between Smg factors regulates NMD function and the exit from naïve pluripotency
To investigate potential cooperativity between NMD effectors in regulating the exit from naïve pluripotency, we performed siRNA-mediated knockdowns of Smg6 and Smg7 in Smg5 and Smg6 KO ESCs (Fig. 1E–I; Supplemental Fig. S1K). Thereby, we generated cells double- and triple-depleted for the three NMD downstream effectors. WT cells showed a near-complete loss of Rex1-GFPd2 expression at 48 h of differentiation (N48) (Fig. 1E). siRNA-mediated depletion of Smg6 in Smg5 KO cells resulted in only a minor, nonsynergistic increase in the differentiation delay assessed in Rex1-GFPd2 reporter assays (Fig. 1F) despite the strong defects observed in both single KOs. In contrast, the depletion of Smg7 in a Smg6 KO background yielded a strong synergistic differentiation phenotype (Fig. 1G). Depletion of Smg7 in Smg5 KO ESCs also resulted in a clear synergistic effect (Fig. 1H), further highlighting heterodimer-independent functions for SMG5 and SMG7. Combined depletion of all the NMD effectors by double knockdown of Smg6 and Smg7 in Smg5 KO ESCs resulted in differentiation delays on par with Smg6/Smg7 or Smg5/Smg7 double depletion (Fig. 1I). To assess the impact of NMD effector depletion and codepletion on NMD-specific mRNA target gene expression, we assessed expression levels of the known NMD target gene Gadd45b by RT-qPCR (Mendell et al. 2004). Consistent with observed differentiation delays, we observed the highest transcript levels of Gadd45b in Smg6/Smg7 and Smg5/Smg7 double-deficient cells. Double depletion of Smg5 and Smg6 resulted in a significantly weaker phenotype (Supplemental Fig. S1L). Together, this suggests that differentiation delays scale with the extent of NMD impairment.
To further study the cooperativity of NMD factors, we sought to generate stable NMD double-KO (dKO) ESCs. However, attempts using CRISPR–Cas9 to generate all possible dKO ESC lines yielded only Smg5/Smg6 dKO ESCs. Neither Smg5/Smg7 nor Smg6/Smg7 dKOs could be established despite multiple attempts using efficient gRNAs (Supplemental Table S1). Although our inability to obtain Smg5/Smg7 or Smg6/Smg7 dKO ESCs does not rule out the feasibility of generating such dKO ESCs, our experiments indicate a strong counterselection against these double-mutant cell lines. This suggests that Smg7 takes on a more important role for ESC self-renewal in the absence of its heterodimerization partner Smg5 or in the absence of Smg6. This is also consistent with the exacerbated differentiation defects observed in Smg5/Smg7 or Smg6/Smg7 double-depletion experiments discussed above. Smg5/Smg6 dKO cells showed a deficiency in down-regulating naïve TF mRNAs similar to that observed in Smg5 single-KO cells, indicating an epistatic role of Smg5 in regulating differentiation-relevant functions (Supplemental Fig. S1M).
Taken together, our results show that NMD effectors act synergistically during the exit from pluripotency. Smg7 can be depleted without impacting the ESC state or strongly affecting early differentiation, but codepletion with either Smg5 or Smg6 results in apparently nonviable phenotypes. This suggests an important and potentially essential role for Smg7 that is only revealed in the absence of either Smg5 or Smg6.
SMG7 alone is sufficient for p-UPF1 binding and is required for mediating the SMG5–p-UPF1 interaction
To better understand the interplay between SMG5, SMG6, and SMG7 and to identify the molecular basis for an apparent heterodimer-independent function of SMG5 and SMG7, we performed coimmunoprecipitation (co-IP) experiments. We precipitated SMG5 or SMG7 in WT and NMD KO ESCs in the presence or absence of RNA. SMG5 and p-UPF1, but not SMG6, coimmunoprecipitated with SMG7 in an RNA-independent manner in WT ESCs, suggesting protein–protein complex formation (Fig. 2A; Supplemental Fig. S2A). Interestingly, SMG7 efficiently bound p-UPF1 in the absence of its heterodimerization partner SMG5. This SMG7–p-UPF1 interaction was also maintained in Smg5/Smg6 dKO ESCs, suggesting that SMG7 can interact with p-UPF1 independently of other downstream NMD effectors. Conversely, however, SMG5 failed to bind p-UPF1 in the absence of SMG7 (Fig. 2B; Supplemental Fig. S2B), indicating that SMG7 provides the critical docking site for p-UPF1 during NMD and that p-UPF1 binding to SMG7 is independent of Smg5. When we performed the same co-IP experiments in the presence of RNA, we observed an interaction between SMG5 or SMG7 with SMG6 in the absence of the respective heterodimerization partner (Supplemental Fig. S2C,D). This RNA-dependent interaction was not observed in WT cells, suggesting aberrant cobinding of components of the endonucleolytic (SMG6) and exonucleolytic (SMG5 or SMG7) branches of NMD to the same RNA molecules in Smg5 or Smg7 KO ESCs, albeit without SMG6–SMG7 or SMG6–SMG5 protein–protein complex formation.
Figure 2.
SMG7 alone is sufficient for p-UPF1 binding and is required for mediating the SMG5–p-UPF1 interaction. (A) Western blot analysis showing results of SMG7 co-IP in the indicated cell lines after cyanase-mediated nucleic acid removal. The antibodies used are indicated. (B) Western blot analysis of SMG5 co-IP (Flag antibody co-IP) in the indicated WT and Smg5 Flag rescue (FR)-based cell lines after cyanase treatment. The antibodies used are indicated. A and B show one representative of two experiments with consistent results. (C) Western blot analysis for UPF1 and p-UPF1 expression in WT and NMD KO ESCs. Numbers in the plot are relative intensities compared with WT, normalized to total UPF1 levels.
Continuous NMD activity relies on cyclic phosphorylation and dephosphorylation of UPF1. We observed increased p-UPF1 levels in all NMD KO cells (Fig. 2C), in line with previous reports (Okada-Katsuhata et al. 2012). Notably, p-UPF1 levels were elevated in all three Smg factor KOs and followed the same gradient as the differentiation phenotype and deregulation of the NMD target Gadd45b: Their increase was strongest in Smg5 KOs and least pronounced in Smg7 KOs. UPF1 is phosphorylated by the kinase SMG1 upon NMD target recognition (Huang et al. 2011; Yepiskoposyan et al. 2011). Smg1 mRNA is itself an NMD target, but in contrast to p-UPF1, Smg1 is up-regulated to a similar extent in all three NMD KO ESCs (Supplemental Fig. S2E; Colombo et al. 2017). Therefore, the graded increase of p-UPF1 levels in NMD KOs is unlikely to be a direct effect of increased Smg1 expression, and impaired UPF1 dephosphorylation might underlie the elevated p-UPF1 levels in NMD mutant cells.
Taken together, this confirms that SMG5 and SMG7 form a heterodimer but suggests that they have additional, biochemically separable, and heterodimer-independent roles that result in distinct phenotypes upon Smg5 or Smg7 depletion.
Integrating transcriptome-wide gene expression with mRNA half-life analysis identifies relevant NMD targets during the exit from naïve pluripotency
To uncover the molecular mode of action by which NMD regulates the exit from naïve pluripotency, we applied a combinatorial approach based on the following logic: An NMD target transcript causally relevant in the context of differentiation must show elevated transcript levels, following the same graded pattern as the differentiation phenotype (fold change [FC] vs. WT: Smg5 KO > Smg6 KO > Smg7 KO) and must show a concomitant increased mRNA half-life.
To identify primary phenotypically relevant NMD targets, we first performed RNA-seq of Smg5 KO, Smg6 KO, Smg7 KO, and WT cells in 2i and at N24 to assess the impact of NMD factor depletion on global gene expression (Supplemental Fig. S3A; Supplemental Table S2). In 2i, we identified 881 transcripts significantly deregulated in either of the Smg factor KO ESCs compared with WT (Padj ≤ 0.01), 516 of which were up-regulated and 252 of which were down-regulated in all three NMD KO ESCs (Fig. 3A). This shows a strong overlap in deregulated genes between all three Smg factor KO cell lines, further supporting the notion of largely overlapping target transcripts between SMG5, SMG6, and SMG7 (Colombo et al. 2017). Of the 516 transcripts up-regulated in all three KOs, 256 showed a graded increase in deregulation (FC vs. WT: Smg5 KO > Smg6 KO > Smg7 KO) (Fig. 3B; Supplemental Table S2) without necessarily exhibiting significance in all pairwise comparisons between Smg factor KO ESCs. Gene ontology (GO) analysis of these 256 graded up-regulated transcripts in 2i revealed an enrichment for, e.g., methyltransferase and helicase activity (Supplemental Fig. S3B; Supplemental Table S3). The remaining up-regulated transcripts in 2i without a graded expression pattern in the KOs showed enrichment for, e.g., telomere maintenance and helicase activity. Within genes down-regulated in 2i, we detected a GO term enrichment for transcripts involved in neural development, somite development, and other differentiation-related processes, suggesting that NMD acts to destabilize naïve pluripotent cell identity already in the ESC state.
Figure 3.
Integrating transcriptome-wide gene expression with mRNA half-life analysis identifies relevant NMD targets during the exit from naïve pluripotency. (A) Differentially expressed genes between WT and Smg5, Smg6, and Smg7 KO ESCs in self-renewal (2i) (Padj ≤ 0.01; null hypothesis: |fold change (FC)| < 1.5). All genes fulfilling this cutoff in any of the three Smg factor KOs are shown. Clusters are indicated at the left. (B) Log2 fold change of deregulated transcripts in 2i belonging to clusters 2 and 4 (564 transcripts). Mean and SD are indicated in the graph. Paired, two-tailed t-test was used to calculate the indicated P-values. (C) Differentially expressed genes between WT and NMD KO ESCs at N24 (Padj ≤ 0.01; null hypothesis: |FC| < 1.5). All genes fulfilling this cutoff in any of the three Smg factor KOs are shown. (D) Log2 fold change of deregulated transcripts at N24 belonging to clusters 2 and 3 (808 transcripts). Mean and SD are indicated in the graph. Paired, two-tailed t-test was used to calculate P-values. (E) Half-life analysis in WT, Smg5 KO, and Smg6 KO ESCs in 2i and within the first 6 h after 2i withdrawal (N0–N6). Half-lives were calculated in minutes based on SLAM-seq experiments. Mean and SD are indicated in the graph. Paired t-test was used to calculate the indicated P-values. (F) Overlap between genes with a half-life (HL) increase of >10% in 2i and at the onset of differentiation (N0–N6) in both Smg5 and Smg6 KO cells. One-thousand-three-hundred-fourteen genes showed an increase in HL in all conditions. (G) Venn diagrams showing the overlap between the set of 1314 transcripts with increased HLs (blue), genes up-regulated in RNA-seq (gray), and genes up-regulated following the expression gradient (FC vs. WT: Smg5 KO > Smg6 KO > Smg7 KO; orange) in 2i (top panel) or at N24 (bottom panel). (H) FACS plots showing the effects of siRNA-mediated depletion of Esrrb (top) or Nanog (bottom) on the differentiation delay exhibited by Smg5, Smg6, or Smg7 KOs, measured by Rex1-GFPd2 flow cytometry analysis. WT profiles are shown in light gray (target siRNA) and in dashed black lines (negative control siRNA). Indicated mutant profiles are shown in blue (negative control siRNA) or red (target siRNA). One of three experiments with consistent results is shown.
At N24, we identified 1174 significantly deregulated transcripts in Smg factor KO cells compared with WT cells (deregulated in either Smg factor KO; Padj ≤ 0.01) (Fig. 3C). Out of the 727 transcripts up-regulated in all NMD KO ESCs, 433 were up-regulated following a graded pattern as described above (FC vs. WT: Smg5 KO > Smg6 KO > Smg7 KO) (Fig. 3D; Supplemental Table S2). GO analysis of these 433 transcripts revealed enrichment for pluripotency-related terms, such as LIF response and stem cell maintenance, which was not found within the nongraded up-regulated genes (Supplemental Fig. S3C; Supplemental Table S3). Within the graded down-regulated transcripts, we detected a specific enrichment of differentiation-related terms, such as neural tube development and pattern specification. Together, this suggests that genes showing graded expression at N24 are involved in and reflect the different levels of differentiation delays observed in NMD KO cells.
Previous reports showed increased c-Myc levels in NMD-deficient ESCs and proposed a causative involvement of c-Myc in NMD KO-induced differentiation delays (Li et al. 2015). In our data sets, we did not detect increased c-Myc transcript levels or observe increased protein levels in any of the three Smg factor KO ESCs (Supplemental Fig. S3D,E). On the contrary, we reproducibly detected reduced levels of c-MYC upon NMD disruption in ESCs. Furthermore, genetic depletion of c-Myc in Smg factor KO ESCs did not rescue their differentiation delays (Supplemental Fig. S3F,G), indicating that c-Myc is not causally involved in the naïve-to-formative transition defects observed in Smg5, Smg6, or Smg7 KO ESCs. We attribute this discrepancy to the different, 2i-based culture conditions in our study that facilitated the identification of differentiation-relevant NMD targets, which are obscured in more heterogeneous FCS/LIF-based culture conditions.
Elevated transcript levels of direct NMD targets are expected to result from increased half-lives, based on impairment of mRNA degradation. We presumed that the NMD-responsive part of the machinery driving the extinction of the naïve network must show sensitivity to NMD depletion in 2i and/or at the onset of differentiation. We therefore chose to measure half-lives in 2i to identify factors potentially upstream of the naïve-to-formative differentiation cascade. We further performed half-life measurements between 0 and 6 h after 2i withdrawal (N0–N6) to detect the potential impact on the down-regulation of the naïve network because, during this time window, the sharpest drop of naïve TF mRNAs can be observed (Leeb et al. 2014; Lackner et al. 2021). Using SLAM-seq (Herzog et al. 2017), we were able to calculate half-lives for 4342 transcripts in 2i and 5250 transcripts at the onset of differentiation (N0–N6) in WT and Smg5 and Smg6 KOs (Supplemental Fig. S3H; Supplemental Table S4). We detected a global increase of transcript half-lives in Smg5 and Smg6 KO ESCs compared with WT ESCs at both time points. On average, mRNA half-life was significantly longer in NMD KOs than in WT, increasing from 141 to 167 and 188 min in 2i and from 149 to 160 and 182 min between N0 and N6 in WT and Smg5 and Smg6 KOs, respectively (P < 10−4 for all comparisons between KO and WT) (Fig. 3E). Overall, both the identity of transcripts and amplitude of half-life changes showed a strong overlap between Smg5 and Smg6 KO ESCs (Supplemental Fig. S3I,J). Transcriptome-wide comparisons showed that the vast majority of genes exhibiting elevated transcript levels showed a concomitant increase in half-lives (Supplemental Fig. S3K,L). Despite a relatively low sequencing depth, this allowed us to follow a step-wise filtering approach to stringently define a set of genes that showed a consistent increase in half-lives of >10% in four direct comparisons (2i: WT vs. Smg5 KO, WT vs. Smg6 KO; N0–N6: WT vs. Smg5 KO, WT vs. Smg6 KO) (Fig. 3F; Supplemental Table S4), and within this set of genes to identify those that are of potential phenotypic relevance (graded expression pattern: FC vs. WT: Smg5 KO > Smg6 KO > Smg7 KO). Thereby, we identified a set of 33 candidate genes in 2i and 54 candidate genes at N24. Twenty-four of those genes fulfil all criteria in both 2i and at N24 (Fig. 3G). We detected Esrrb as the only naïve transcription factor encoding mRNA within the group of 54 candidate NMD target transcripts with a graded expression at N24. This prompted us to probe whether the phenotype in Smg factor KO ESCs is dependent on increased levels of Esrrb. Indeed, siRNA-mediated depletion of Esrrb but not Nanog resulted in a partial rescue of Rex1-GFPd2 down-regulation kinetics (Fig. 3H), suggesting that increased half-life of Esrrb results in increased transcript levels at N24, which in turn causes a delay in the exit from naïve pluripotency. Bona fide NMD targets show elevated transcript levels after NMD inhibition achieved through translational inhibition by cycloheximide (CHX). Esrrb mRNA did not exhibit this hallmark feature of an NMD target and did not react to CHX treatment (Supplemental Fig. S4A), suggesting that although Esrrb up-regulation is causative for the Smg factor KO phenotype, other primary targets are directly regulated by NMD to ensure proper exit from the naïve pluripotent state. We propose that the increased half-life of Esrrb might be due to a potential deregulation of other, NMD-independent mRNA-degradation processes in Smg factor KO ESCs.
Eif4a2 is a bona fide NMD target in ESCs, and its NMD sensitivity is evolutionarily conserved
We reasoned that genes with functional relevance for the exit from naïve pluripotency defects of Smg factor KO ESCs will be found within the set of 24 candidate genes showing increased half-lives and graded expression in 2i and at the onset of differentiation (Supplemental Fig. S4B). We tested how many of these were bona fide NMD targets and displayed increased transcript levels upon treatment with CHX. Indeed, 14 out of 18 evaluated transcripts within this group showed significant increases in transcript levels after CHX treatment (Supplemental Fig. S4C). Among these, our attention was drawn to the RNA helicase Eif4a2, a component of the eIF4F translation initiation complex, which mediates 5′ cap recognition by the small ribosomal subunit (Yoder-Hill et al. 1993; Li et al. 2001). Several layers of evidence suggest Eif4a2 as a bona fide and phenotypically relevant NMD target: First, Eif4a2 is a highly expressed gene (FPKM in WT in 2i = 150), and its graded increase in both 2i and at N24 combined with a consistent increase in half-life indicates that it can be of primary relevance for the exit from naïve pluripotency delay in NMD-deficient ESCs (Supplemental Figs. S3M, S4D). Second, the Eif4a2 locus encodes for two splice isoforms: one full-length protein (eIF4A2FL) and one predicted PTC-containing isoform (eIF4A2PTC) (Fig. 4A). Third, Eif4a2 mRNA levels were significantly increased after translation inhibition by CHX treatment (Fig. 4B), similar to the known NMD target Gadd45b. Eif4a2PTC showed an even more pronounced sensitivity to CHX treatment, indicating that both Eif4a2FL and Eif4a2PTC mRNAs are NMD targets. Graded Eif4a2PTC up-regulation in NMD mutant cells in 2i and at N24 was confirmed by RT-qPCR validation (Supplemental Fig. S4E). Furthermore, Eif4a2PTC up-regulation is directly caused by the absence of Smg factors and hence is rescued by re-expression of the missing NMD effector in the respective KO cells.
Figure 4.
Eif4a2 is a bona fide NMD target in ESCs. (A) Genome browser view of the Eif4a2 locus visualizing RNA-seq results in WT, Smg5 KO, and Smg6 KO in 2i and at N24. (*) RT-qPCR primers amplifying all Eif4a2 isoforms; (#) RT-qPCR primers amplifying the Eif4a2PTC isoform. (B) Expression levels of the indicated genes after treatment with cycloheximide (CHX) or DMSO in WT ESCs, measured by RT-qPCR. Data shown are normalized to untreated WT ESCs. Mean and SD are plotted; n = 4 biological replicates. Unpaired, two-tailed t-test was performed between the DMSO and CHX comparison for each gene, and the resulting P-values are indicated in the plot. (C) Plots showing expression levels of Eif4a2PTC as a fraction of total Eif4a2 expression in WT and Smg factor KO ESCs, measured by RT-qPCR. Data from n = 4 independent experiments are shown. Unpaired, two-tailed t-test was performed to compare WT with Smg factor KOs. P-values are shown in the plot. (D) Western blot analysis for eIF4A2 protein levels in WT and NMD KO ESCs in self-renewal (2i) and at N24. Vinculin was used as a loading control. (E) Schematic representation of the Eif4a2 gene structure in mice, humans, and dogs. (F) The percentage of transcriptome-wide sequence identity of the indicated genomic features (coding exon, intron, and untranslated region [UTR]) between mice, dogs, humans, and rats. Pink indicates the relative position of the Eif4a2 PTC-containing exon, and black lines indicate the mean identity.
Eif4a2PTC transcript levels in WT cells are ∼20% of total Eif4a2 transcript (Fig. 4C). In Smg5 and Smg6 KO ESCs, levels rise to 50%60%. eIF4A2FL protein levels were increased in Smg factor KO cells in 2i and at N24 (Fig. 4D; Supplemental Fig. S4F). The Eif4a2PTC isoform produced a protein that was weakly detected by Western blot analysis in WT and NMD rescue cells (Fig. 4D; Supplemental Fig. S4H) and strongly increased in all three Smg factor KO ESCs, reaching up to 50% of full-length protein levels in Smg5 and Smg6 KO cells (Fig. 4D; Supplemental Fig. S4G). The up-regulation of Eif4a2 was unique among eIF4F complex members, since the others, including the close homolog Eif4a1, showed neither increased expression (Supplemental Fig. S4D,I) nor significantly longer half-lives in Smg factor KOs (Supplemental Table S4), and did not react to CHX treatment (Supplemental Fig. S4J). Taken together, this identifies Eif4a2 as a bona fide NMD target in ESCs and shows that NMD disruption leads to the production of a specific eIF4A2PTC protein.
The linkage of NMD to Eif4a2 expression appears to be conserved in evolution. The gene structure of Eif4a2 is strikingly similar and the exact position of the PTC in the PTC-containing exon is identical between mice, dogs, and humans (Fig. 4E). In a transcriptome-wide comparison, the Eif4a2 PTC exon showed high conservation at the nucleotide level despite being under no apparent selective pressure to maintain protein-coding potential (only two amino acids are encoded in this exon before the PTC) (Fig. 4F): Less than 5% of all exons and <3% of all untranslated regions (UTRs) showed higher conservation rates between mice and rats than the Eif4a2PTC exon (Supplemental Fig. S4K). In the comparisons with humans and dogs, these percentages were even lower (3.4% and 1.1% in humans, 1.3% and 0.7% in dogs, respectively). Hence, the sequence identity of the Eif4a2 PTC-containing exon and therefore the potential of Eif4a2 to be regulated by NMD are features that appear to be conserved in mammalian evolution.
Eif4a2PTC up-regulation is causative for defects in exit from naïve pluripotency in Smg5-deficient ESCs
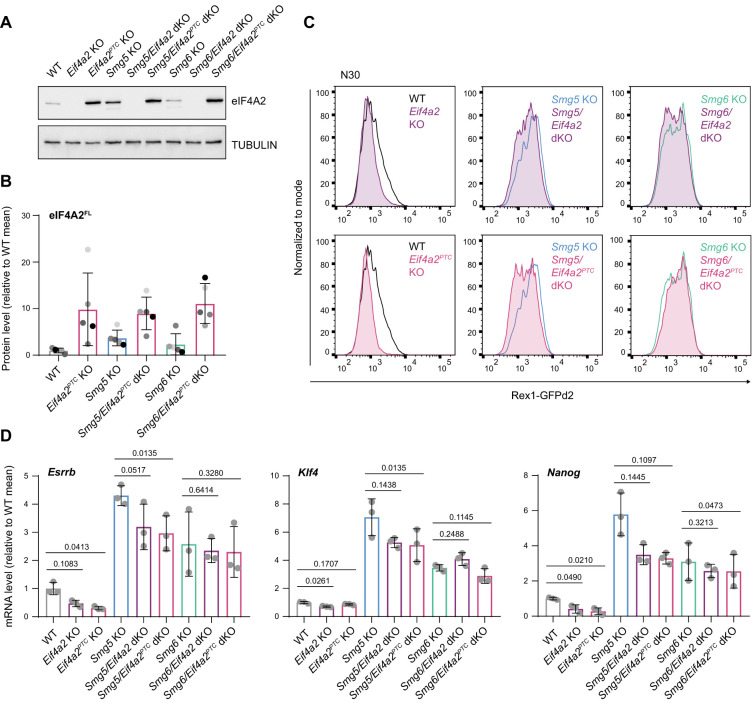
To delineate a potential causative relationship between increased Eif4a2 levels and the differentiation defect observed in NMD-deficient cells, we generated Eif4a2 KO cells and Smg5/Eif4a2 or Smg6/Eif4a2 double-deficient cells by deletion of either all potential Eif4a2 isoforms (Eif4a2 KO) or, specifically, Eif4a2PTC (Eif4a2PTC KO) (Fig. 5A; Supplemental Fig. S5A,B). Notably, deletion of the PTC exon resulted in a clear increase in full-length eIF4A2 protein levels in WT and NMD KO ESCs (Fig. 5B). Both the complete absence of Eif4a2 and specific deletion of Eif4a2PTC accelerated Rex1-GFPd2 down-regulation kinetics and down-regulation of naïve marker transcripts at N24 in WT cells (Fig. 5C,D). In Smg5 mutants, deletion of Eif4a2 or Eif4a2PTC partially restored normal Rex1-GFPd2 down-regulation (Fig. 5C). Furthermore, transcript levels of Esrrb, Klf4, and Nanog were significantly reduced compared with Smg5 single mutants at N30 (Fig. 5D), without, however, reaching WT levels, suggesting an incomplete rescue in Smg5/Eif4a2 and Smg5/Eif4a2PTC dKO cells. The differentiation defect observed in Smg6 KO ESCs did not show a clear dependence on Eif4a2, suggesting that other mechanisms deregulated in Smg6 KO ESCs are contributing to the differentiation phenotype to a larger extent than the up-regulation of Eif4a2PTC.
Figure 5.
Eif4a2 is causative for defects in exit from naïve pluripotency in Smg5-deficient ESCs. (A) Western blot analysis for eIF4A2 expression in the indicated cell lines. Tubulin was used as a loading control. (B) Quantification of eIF4A2FL protein expression upon Eif4a2PTC deletion in WT and NMD KO ESCs, measured by quantitative Western blot analysis. Expression levels were normalized to GAPDH; results are shown as expression levels relative to the WT mean. Mean and SD for n = 5 independent experiments are shown. (C) Rex1-GFPd2 flow cytometry analysis at N30 in WT, Smg5 KO, and Smg6 KO ESCs deleted for Eif4a2 (purple) or Eif4a2PTC (pink). Parental cell lines are shown for comparison. (D) RT-qPCR analysis of Esrrb, Nanog, and Klf4 in the indicated cell lines. Expression was normalized to Rpl32; results are shown as expression levels relative to the WT mean. Mean and SD for n = 3 independent experiments are shown. A paired, two-tailed t-test was performed to compare each parental cell line with its respective Eif4a2 or Eif4a2PTC KO. P-values are indicated in the plot.
To further confirm the specificity of Eif4a2 depletion in restoring differentiation potential in a Smg5-deficient ESC background, we compared the differentiation potential of Tcf7l1 KO and Eif4a2/Tcf7l1 dKO ESCs. Tcf7l1 encodes for an HMG-box transcription factor and is a strong positive regulator of the exit from naïve pluripotency (Hoffman et al. 2013; Lackner et al. 2021). TCF7L1 is a transcriptional corepressor and acts downstream from the Wnt signaling cascade. Tcf7l1 KO ESCs showed defects in exit from naïve pluripotency on par with NMD KOs but exhibited an NMD-independent, Gsk3 inhibition-like phenotype (Lackner et al. 2021). Tcf7l1 KO cells were insensitive to codepletion of Eif4a2 (Supplemental Fig. S5C,D), suggesting that Eif4a2 is a specific genetic interactor of NMD in regulating differentiation.
In sum, the phenotypic rescue in Smg5/Eif4a2PTC double-KO cells, in which eIF4A2FL protein levels are even further increased, supports the proposition that Eif4a2PTC, rather than the full-length isoform, is a major mediator of the observed exit from naïve pluripotency defect, at least in Smg5-deficient ESCs.
To test the sufficiency of Eif4a2 up-regulation to cause a differentiation delay, we overexpressed Flag-tagged Eif4a2FL or Eif4a2PTC in WT ESCs (Supplemental Fig. S5E). Indeed, at N24, Eif4a2PTC-overexpressing cells showed increased transcript levels for the naïve TFs Klf4, Esrrb, Tfcp2l1, and Nanog and increased protein levels for NANOG (Supplemental Fig. S5F–H). Cells overexpressing Eif4a2FL exhibited a weaker effect and no up-regulation of NANOG protein levels (Supplemental Fig. S5H). This is in accord with an increase of Eif4a2PTC rather than Eif4a2FL having a major impact on the exit from naïve pluripotency. Notably, the differentiation delays elicited by increasing expression levels of Eif4a2 isoforms did not reach the intensity observed in NMD KO ESCs, suggesting that factors in addition to Eif4a2 contribute to the failure to properly shut down naïve pluripotency in NMD-deficient ESCs.
Eif4a2-mediated differentiation delay is functionally linked to Eif4a2PTC-dependent regulation of mTORC1 activity and translation
To identify potential direct eIF4A2FL and eIF4A2PTC targets, we performed RIP-seq (Keene et al. 2006) using Eif4a2 KO ESCs engineered to only express either Flag-tagged eIF4A2PTC or eIF4A2FL (Eif4a2PTC FR and Eif4a2FL FR) (Supplemental Fig. S6A). Expression levels of eIF4A2FL closely matched endogenous levels and the PTC isoform was weakly detectable, similar to the levels in WT. As expected for a translation initiation factor, we identified non-protein-coding transcripts such as Meg3, Rian, and Malat1 as depleted in Flag-eIF4A2FL RIP samples, indicating the specificity of the assay (Supplemental Table S5). Out of 9812 expressed transcripts (CPM > 1 in all input samples), we detected 2601 transcripts bound by eIF4A2FL significantly stronger than in empty vector (EV) control cells (Padj ≤ 0.05, log2FC > 0), which was confirmed by RT-qPCR on a selected set of candidates (Supplemental Fig. S6B). The vast majority of mRNAs showed a close correlation between expression level and eIF4A2FL binding, as expected for a general translation initiation factor. Only 31 transcripts showed eIF4A2FL enrichment significantly higher than expected based on transcript abundance. Among those was Mtor, the core component of both the mTORC1 and mTORC2 complexes (Supplemental Table S5). In contrast, RIP-seq of Flag-eIF4A2PTC resulted in only minimal enrichments on RNA, possibly owing to the low protein levels of eIF4A2PTC protein.
To understand the protein–protein interaction landscape of both eIF4A2 isoforms in ESCs, we performed co-IP followed by mass spectrometry. To compensate for the highly unstable nature of eIF4A2PTC, which is suggested by the discrepancy between Eif4a2PTC transcript and eIF4A2PTC protein levels relative to the respective full-length counterpart, we performed sample preparation after short-term treatment with the irreversible proteasome inhibitor epoxomicin (Supplemental Fig. S6C; Meng et al. 1999). Thereby, we identified 254 and 306 proteins specifically bound by eIF4A2FL or eIF4A2PTC, respectively, over Flag-only control immunoprecipitation (IP) in EV cells (Fig. 6A; Supplemental Table S6).
Figure 6.
Eif4a2-mediated differentiation delay is caused by PTC isoform-dependent regulation of translation. (A) Venn diagram showing the overlap between eIF4A2FL- and eIF4A2PTC-bound proteins. Seventy-five proteins coprecipitate with both isoforms. (B) Volcano plot showing proteins detected by mass spectrometry after eIF4A2FL coimmunoprecipitation. Black dots represent significantly enriched or depleted proteins (compared with empty vector [EV] co-IP), and purple dots represent proteins specifically enriched only in eIF4A2FL co-IP (178 proteins; see A). (C) Volcano plot showing proteins detected by mass spectrometry after eIF4A2PTC coimmunoprecipitation. Black dots represent significantly enriched or depleted proteins, and pink dots represent proteins specifically enriched only in eIF4A2PTC co-IP (231 proteins; see A). (D) Translation initiation measured in ES-DMEM-2i medium by S35 incorporation into WT, Smg factor KO, Tsc2 KO, and Smg factor KO ESCs deficient for Eif4a2 or Eif4a2PTC. Results are shown as incorporation levels relative to the WT mean. Mean and SD are plotted; n = 4 independent experiments with three biological replicates each. A two-tailed, paired t-test was used to calculate P-values. (E) Quantification of p-4E-BP1 protein expression in 2i in WT and Smg factor KO ESCs, measured by Western blot analysis. Expression was normalized to GAPDH; results are shown as expression levels relative to WT (dotted line). Mean and SD for n = 9 independent experiments are shown. Paired, two-tailed t-tests were performed to compare WT with Smg factor KO ESCs. P-values are indicated in the plot. (F) Expression levels of Nanog, Esrrb, and Tfcp2l1 measured in WT, Smg factor KO, Tsc2 KO, and Tcf7l1 KO ESCs at N30 after treatment with DMSO or 20 nM rapamycin, measured by RT-qPCR. Expression levels are normalized to Rpl32. Mean and standard error of the mean (SEM) are shown; n = 2 independent experiments. (G) Rex1-GFPd2 analysis by flow cytometry at N30 in WT, Smg5 KO, and Smg6 KO ESCs transfected with control (siNeg) or siRNAs targeting Esrrb (siEsrrb) and treated with DMSO or 20 nM rapamycin (Rapa). (H) Rex1-GFPd2 profiles of WT, Smg5 KO, and Smg6 KO upon concomitant depletion of Eif4a2PTC (Eif4a2PTC KO) and siRNA-mediated Esrrb down-regulation (siEsrrb). Parental cell lines and control siRNA transfections (siNeg) are shown for comparison.
Among the 178 proteins specifically bound to eIF4A2FL and not to the PTC isoform were all eIF4G isoforms and several components of the eIF3 complex (Fig. 6B). This highlights the integration of eIF4A2FL but not eIF4A2PTC into a functional translation initiation complex.
Among the 75 proteins that coprecipitated with both isoforms, we identified several negative regulators of mRNA stability and regulation, such as AGO2, IGFBP1, IGFBP3, PUM1, TRIM71, CNOT, and the NMD components SMG6, SMG7, and UPF1 (Supplemental Fig. S6D). The interaction of eIF4A2 with SMG6 and SMG7 is consistent with previous reports (Schweingruber et al. 2016; Wilczynska et al. 2019). Together, this supports an intricate link between eIF4A2 and the mRNA destabilization machinery, which is potentially maintained in its PTC isoform. In addition, the GATOR complex member and mTOR regulator WDR59 precipitated with both isoforms (Supplemental Fig. S6D). eIF4A1 did not coprecipitate with either eIF4A2 isoform, suggesting that eIF4A1 and eIF4A2 participate in distinct, biochemically separable complexes.
Among the 231 proteins specifically associated with the PTC isoform and not the full-length protein, we detected a large cohort of chaperones, illustrating the unstable nature of eIF4A2PTC. However, we also detected interactions with the FGF/ERK pathway component RAF1, the GATOR complex member MIOS, and the key mTORC1 regulator TSC2 (Fig. 6C).
Taken together, these results show that eIF4A2FL is mainly involved in translation initiation but also interacts with mRNA-destabilizing proteins, including NMD factors, CCR4–NOT components, and the RISC member AGO2. In contrast, eIF4A2PTC protein showed little association with translation initiation complex members but still binds mRNA binding proteins and shows interaction with the negative mTORC1 pathway regulator TSC2.
eIF4A2 is part of the eIF4F complex, which integrates signals from the mTORC1 pathway to regulate translation levels (Sonenberg and Hinnebusch 2009). Together, the identification of several components of the mTORC1 complex and TSC2 as eIF4A2FL and eIF4A2PTC interactors and the detection of Mtor as a transcript enriched for eIF4A2 binding prompted us to ask whether increased levels of Eif4a2 and Eif4a2PTC in Smg factor KO cells impact on translational homeostasis in ESCs. To this end, we performed radioactive S35-based translation assays in Smg5 or Smg6 KO ESCs and corresponding Eif4a2 and Eif4a2PTC dKOs (Fig. 6D). We detected a significant Eif4a2PTC-dependent increase in translation rates in Smg5 KO ESCs, similar in magnitude to that observed in Tsc2 KOs, in which deregulated mTORC1 activity leads to an increase in translation (Grabole et al. 2016). Although increased translation rates in Smg6 KO ESCs were not observed at a statistically significant level, we detected significant reductions of translation levels upon Eif4a2PTC or Eif4a2 depletion in Smg6 KO ESCs. Because full-length eIF4A2 levels increase upon PTC isoform KO (Fig. 5B), these results permit the conclusion that increased translation rates in Smg5 KO cells are mainly dependent on the PTC isoform of Eif4a2.
In line with a possible intersection of Eif4a2 function with the mTOR pathway, increased mTORC1 activity was evident by significantly increased p-4E-BP1 levels in Smg5 (P = 0.0005) and in Smg6 (P = 0.0303) KO ESCs (Fig. 6E). Increased levels of p-4E-BP1 are dependent on eIF4A2PTC, as shown by a reduction of p-4E-BP1 levels upon codeletion of Smg5 or Smg6 with Eif4a2PTC (Supplemental Fig. S6E). To determine the extent to which increased mTORC1 activity is causative for the observed differentiation defects in Smg5, Smg6, and Smg7 KO ESCs, we treated differentiating cells with the mTORC1 inhibitor rapamycin (Betschinger et al. 2013; Villegas et al. 2019). As expected, in Tsc2 KO ESCs, which exhibit an mTORC1-dependent phenotype, we detected a complete restoration of the down-regulation kinetics of the naïve markers Nanog, Esrrb, and Tfcp2l1 as well as Rex1-GFPd2 levels upon rapamycin treatment, whereas the mTORC1-unrelated Tcf7l1 mutants exhibited strong differentiation delays in both the presence and absence of rapamycin (Fig. 6F; Supplemental Fig. S6F–H). In contrast, rapamycin rescued differentiation defects of Smg5, Smg6, and Smg7 KO ESCs, and mRNA levels of naïve TFs were clearly reduced upon rapamycin treatment in all Smg factor KO cells. Together, this shows that elevated mTORC1 activity is a key factor determining the differentiation delays of Smg factor-deficient ESCs. In Smg5 KO ESCs treated with rapamycin, naïve TF expression was still clearly elevated compared with WT, indicating that deregulation of mTORC1-independent mechanisms also contributes to the delayed differentiation phenotype.
We therefore tested whether treatment of Smg5 or Smg6 KO cells with rapamycin together with concomitant destabilization of the naïve TF network is sufficient to restore normal differentiation kinetics. Indeed, simultaneous siRNA-mediated depletion of Esrrb with inhibition of mTORC1 activity resulted in WT-like Rex1-GFPd2 differentiation kinetics in both Smg5 and Smg6 KO cells (Fig. 6G). Similarly, deletion of Eif4a2PTC combined with siRNA-mediated depletion of Esrrb could restore near-WT-like Rex1-GFPd2 down-regulation kinetics in Smg factor KO ESCs (Fig. 6H).
In summary, deregulation of a feedback circuit between NMD and translation initiation, encoded in a PTC-containing isoform of Eif4a2, results in elevated mTORC1 activity and increased translation rates in NMD-deficient ESCs. Although the relative contribution of Eif4a2 to the translation state might differ between Smg5 and Smg6 KO ESCs, increased mTORC1 activity appears to be a crucial contributor to both KO phenotypes. We conclude that, in combination, the perturbance in RNA homeostasis, including increased levels of naïve pluripotency-specific transcripts, and the deregulation of translation through eIF4A2PTC are the major contributors to NMD-associated differentiation defects in ESCs.
Discussion
Depletion of Smg5, Smg6, or Smg7 results in a graded impairment of NMD activity and causes corresponding delays in the exit from naïve pluripotency. Differentiation defects of Smg5, Smg6, and Smg7 KOs scale with deregulation of known NMD target genes and levels of p-UPF1 (Smg5 KO > Smg6 KO > Smg7 KO), indicating that NMD defects directly translate to differentiation delays at the exit from naïve pluripotency.
The striking difference in NMD target regulation and extent of differentiation delays between Smg5 and Smg7 KOs (Smg5 KO ≫ Smg7 KO) together with the strong synergistic effects after codepletion are difficult to reconcile with the proposed obligatory heterodimer dependence of these two factors (Ohnishi et al. 2003; Jonas et al. 2013). Our data suggest a model in which a division of labor between SMG5 and SMG7 is causative for the different effects observed in the respective KOs (Fig. 7). Smg factors have a dual role in triggering RNA degradation and in mediating UPF1 dephosphorylation; p-UPF1 increase in all three Smg factor KOs is in accord with such a role of Smg factors in PP2A recruitment (Ohnishi et al. 2003; Okada-Katsuhata et al. 2012). Our data suggest that SMG7 acts as the main adapter or recruiter for p-UPF1, at least in naïve pluripotent ESCs. However, by itself it is unable to efficiently dephosphorylate UPF1, as evidenced by high p-UPF1 levels in Smg5 KO ESCs, where SMG7 binding to p-UPF1 is unaffected. We propose that SMG7 binding to p-UPF1 without later UPF1 dephosphorylation by SMG5 results in jamming of the dephosphorylation cycle and the ensuing stalling of the mRNA degradation circuit in Smg5 KO ESCs. Aberrant recruitment of SMG6 to mRNA targets already bound by SMG7 and p-UPF1 cannot properly restore NMD function in Smg5 KOs. SMG5 alone in Smg7 KOs is unable to bind p-UPF1. This suggests that the main role of SMG5 lies in its dephosphorylation activity, whereas recruitment to p-UPF1 is SMG7-dependent. This is consistent with the strongest increase of p-UPF1 levels and consequently the strongest differentiation delays in Smg5 KO ESCs.
Figure 7.
Schematic model of Smg factor activity resulting in a graded impairment of NMD and differentiation in Smg5 KO, Smg6 KO, and Smg7 KO ESCs. Schematic illustration of the proposed interactions between NMD effectors and p-UPF1 dephosphorylation in WT, Smg5 KO, Smg6 KO, and Smg7 KO ESCs that lead to graded defects in NMD and in the exit from naïve pluripotency.
The relatively weaker phenotype in Smg7 KOs is consistent with recent reports that SMG5 can compensate for the loss of SMG7 and that the presence of either SMG5 or SMG7 is sufficient to support SMG6-mediated endonucleolytic decay of NMD targets (Boehm et al. 2021). Notably, our observations of a weak Smg7 KO phenotype without impact on cell proliferation and cell fitness are distinct from those recently made in HEK293 cells (Boehm et al. 2021) but are in accordance with the observation that Smg6 depletion results in stronger NMD defects than Smg7 deficiency (Colombo et al. 2017). It remains unclear whether the differences in cell fitness are due to distinct roles of Smg7 between mice and humans or between ESCs, somatic cells, and established immortalized human cell lines.
Our data support a model in which SMG5 recruitment to p-UPF1-bound mRNAs is dependent on SMG7. Hence, in Smg7 KO ESCs, the exonucleolytic decay pathway is completely inactive, but Smg6 and the endonucleolytic pathway can largely compensate for loss of Smg7 activity, resulting in only mild NMD defects in Smg7 KO cells. However, SMG6 is unable to compensate for the complete absence of the SMG5–SMG7 heterodimer, suggested by the apparent loss of viability in a Smg5/Smg7 dKO situation. This is consistent with the recent finding that the SMG5–SMG7 heterodimer is critical for NMD, even for SMG6-mediated endonucleolytic decay (Boehm et al. 2021), and our data further support an essential and partially redundant function of either SMG5 or SMG7 in the endonucleolytic decay pathway. Our results are consistent with this function being UPF1 dephosphorylation, which is also required in the SMG6-mediated endonucleolytic mRNA decay axis.
It is a conundrum that despite the weak defects in Smg7 KO cells, facultative essentiality for Smg7 is revealed in the absence of either Smg5 or Smg6, and that Smg7 alone, in the simultaneous absence of Smg5 and Smg6, is sufficient to sustain minimal NMD activity required for survival. This suggests that SMG7 can either interact with potentially novel interactors in the absence of its normal interaction partners or act as a jack of all trades, fulfilling minimal roles in target recognition, recruitment of the degradation machinery, and UPF1 dephosphorylation. Taken together, the mode of action described above explains both the graded p-UPF1 levels and the graded impact on NMD of Smg5, Smg6, and Smg7 KOs (Fig. 7). This model remains, in part, speculative but is fully in line with genetic and biochemical data.
We propose that the exit from naïve pluripotency defect in NMD mutant ESCs cannot be attributed to a single NMD target but results from a combination of strong impact on transcript levels with a major, not previously recognized influence of NMD on mTORC1 activity and the translation state of ESCs. Our data link the NMD-controlled Eif4a2PTC genetically and biochemically to translation and mTORC1 activity. Consistent with our co-IP mass spectrometry results, a direct protein–protein interaction between eIF4A2 and TSC2 has been reported in Drosophila (Tsokanos et al. 2016); our data indicate that in mammalian cells, this interaction occurs between TSC2 and the eIF4A2PTC isoform. Together, this supports the conclusion that NMD deficiency results in a dysbalanced translational state that is dependent on increased levels of eIF4A2PTC protein and the likely resulting elevated mTORC1 activity. Control of protein synthesis rates is known to be fundamental for maintenance of self-renewal and differentiation (Bornelöv et al. 2019; Di Stefano et al. 2019) and for maintaining an ESC-specific chromatin state (Bulut-Karslioglu et al. 2018; Corsini et al. 2018). ESCs require down-regulation of translation rates and ribosome biogenesis to successfully differentiate (You et al. 2015; Corsini et al. 2018).
Although we can assign the main responsibility for the differentiation defect observed in NMD-deficient ESCs to increased mTORC1 activity and the NMD-responsive Eif4a2PTC isoform, our data reveal a complex phenotype of NMD-deficient ESCs. The relative contribution of elevated Eif4a2PTC to the overall differentiation defect differs between Smg5 and Smg6 KO cells. Whereas the differentiation defect of Smg5 KO ESCs depends, to a large extent, on elevated Eif4a2PTC expression, such dependence could not be detected in Smg6 KOs. However, the deregulation of the naïve pluripotency circuitry in Smg factor KO ESCs was dependent on their significantly increased mTORC1 activity. This indicates that NMD controls mTORC1 activity and translation at various levels, not exclusively through regulation of Eif4a2PTC expression.
We note that our mass spectrometry analysis detected the core pluripotency transcription factor proteins SOX2 and OCT4 as high-confidence interactors of eIF4A2FL and eIF4A2PTC, respectively (Supplemental Table S6). However, without further experimentation, it cannot be ruled out that an interaction of cytoplasmic eIF4A2 with known nuclear factors is an artefact introduced after cell lysis before co-IP, although both OCT4 and SOX2 have been reported as RNA binding proteins before (Castello et al. 2012; He et al. 2016; Holmes et al. 2020).
NMD is triggered during translation. NMD-mediated regulation of Eif4a2, and specifically its PTC-containing isoform, provides a link back from NMD to translation initiation, thereby establishing a feedback circuit. We speculatively suggest that such a mechanism embedded in an NMD-responsive PTC-containing Eif4a2 isoform can be used as a rheostat to balance NMD with translational activity. Eif4a2 up-regulation upon NMD dysfunction increases translation initiation activity, which in turn directly increases the chances of triggering NMD in successive rounds of eIF4A2-initiated translation. Because Eif4a1 is not subject to regulation by NMD, such a mechanism could also be used to shift the balance between levels and activity of eIF4A1 and eIF4A2. This is interesting because eIF4A1 and eIF4A2 have been proposed to have distinct cellular functions: Both are involved in cap-dependent translation initiation, but eIF4A2 appears also to play a role in translation inhibition through interaction with the CCR4–NOT complex (Meijer et al. 2013; Wilczynska et al. 2019). In line with the latter observation, our mass spectrometry data show interaction of both eIF4A2FL and eIF4A2PTC with several components of the CCR4–NOT complex and with AGO2.
These models remain speculation without further knowledge of the specific role of eIF4A2PTC and its potential biochemical interaction with the translation initiation machinery and also with its full-length twin eIF4A2FL. Underscoring the importance of the Eif4a2PTC isoform, it shows an unexpectedly high level of evolutionary conservation in various mammalian species, including humans. The exact function of eIF4A2PTC and how it integrates into the translation machinery remain interesting questions for future research.
In conclusion, we show that maintenance of proper NMD activity is essential for restructuring of gene regulatory networks during the naïve-to-formative transition. In NMD-deficient cells, multiple transcripts, including naïve transcription factor mRNAs and most prominently those of the translation initiation factor Eif4a2, show increased transcription levels. Our results place NMD as a central player in shaping transcriptomes to maintain proper cellular identity. We provide evidence for an intricate feedback circuit between NMD and translation. NMD susceptibility is hardwired in an evolutionarily conserved fashion into the Eif4a2 gene structure and sequence. This direct link between NMD and a translation initiation factor will need to be considered when studying NMD-associated phenotypes. Our results are also relevant for improving therapeutic interventions and provide a rationale for choosing the appropriate Smg factor in pharmacological approaches to inhibiting NMD activity, depending on the desired strength and extent of NMD inactivation (Kurosaki et al. 2019; Pawlicka et al. 2020).
Materials and methods
Cloning
gRNAs were designed using http://crisprscan.org. Annealed oligos were cloned in a gRNA-expressing vector (Addgene 41824) using a BsaI site (Mali et al. 2013; Lackner et al. 2021). gRNA sequences are indicated in Supplemental Table S7. For generating rescue cell lines, the coding sequence of the gene of interest was amplified by PCR and cloned into a pCAG-3xFlag-empty-pgk-hph vector (Betschinger et al. 2013).
Teratoma assay
Paraffin-embedded teratoma tissue blocks were cut on a rotary microtome RM2255 (Leica). The 3-µm sections were then stained for hematoxylin and eosin in the automated slide stainer Gemini AS (Histocom) and mounted in Eukitt. Slides were scanned on a VS120 (Olympus) slide scanner.
Cell culture
Mouse embryonic stem cells (ESCs) containing a Rex1-GFPd2-IRES-BSD reporter construct carrying a Cas9 transgene (RC9) (Wray et al. 2011; Li et al. 2018) were used as a parental cell line for all of the knockout cells generated in this study. A list of the primers used to perform genotyping PCRs in the derived KO cell lines is in Supplemental Table S7. ESCs were cultured on 0.2% gelatin-coated (Sigma-Aldrich G1890) plates in DMEM high-glucose (Sigma-Aldrich D5671) supplemented with 15% FBS (Gibco 10270-106), 2 mM L-glutamine (Sigma-Aldrich G7513), 0.1 mM NEAA (Sigma-Aldrich M7145), 1 mM sodium pyruvate (Sigma-Aldrich S8636), 10 µg/mL penicillin–streptomycin (Sigma-Aldrich P4333), 55 µM β-mercaptoethanol (Fisher Scientific 21985-023), 2i (1.5 μM CHIR99021 and 0.5 μM PD0325901), and 10 ng/mL LIF (batch tested, in-house; ES-DMEM + 2i medium). All cell lines were regularly negatively tested for mycoplasma infection.
Monolayer differentiation
For differentiation, ESCs were plated on gelatin-coated plates at a density of 1 × 104 cells/cm2 in basal medium (N2B27) composed of a 1:1 mix of DMEM/F12 (Gibco 21331020) and neurobasal medium (Gibco 21103049) supplemented with N2 (homemade), B27 serum-free supplement (Gibco 17504-044), 2 mM L-glutamine (Sigma-Aldrich G7513), 0.1 mM NEAA (Sigma-Aldrich M7145), 10 µg/mL penicillin–streptomycin (Sigma-Aldrich P4333), 55 µM β-mercaptoethanol (Fisher Scientific 21985-023), and 2i (3 μM CHIR99021 and 1 μM PD0325901; N2B27 + 2i medium). The following day, N2B27 + 2i medium was withdrawn, and cells were washed once with PBS and then differentiated for the indicated time in N2B27.
Rapamycin treatment
ESCs were plated on gelatin-coated plates in N2B27 + 2i + DMSO (Sigma-Aldrich D2650) or N2B27 + 2i + 20 nM rapamycin (Enzo Life Sciences BML-A275-0005). The following day, N2B27 + 2i medium was removed and cells were differentiated in N2B27 + DMSO or N2B27 + 20 nM rapamycin for the indicated time after one wash with PBS.
Commitment assay
ESCs were plated on gelatin-coated plates in N2B27 + 2i medium at a density of 2 × 103 cells/cm2. The following day, N2B27 + 2i medium was withdrawn and cells were differentiated for 3 d in N2B27. After 72 h, medium was changed to N2B27 + 2i + blasticidin (BSD, Invivogen), and medium was refreshed every 2 d. Alkaline phosphatase staining was performed after 4 d in N2B27 + 2i + BSD (Sigma-Aldrich 86R). Plates were then imaged using an Olympus Cell-Sense microscope (Olympus).
EB differentiation assay
EBs were formed as hanging drops in ESC culture medium without LIF and 2i (200 cells/20 µL drop). After 2 d, EBs were collected in a 10-cm Petri dish; medium was changed every 2 d. Cells were harvested every 2 d and RNA was extracted using the ExtractMe kit (LabConsulting EM09).
Generation of knockout cell lines
Cells (2 × 105) were reverse-transfected in a gelatin-coated six-well plate in ES-DMEM + 2i using Lipofectamine 2000 (Fisher Scientific 11668-027). Two gRNAs were used for each gene (1 µg of each) together with 0.5 µg of pCAG-dsRed. dsRED/GFP-double-positive cells were sorted using a BD FACS Aria III flow cytometer. Sorted cells were plated at clonal density in ES-DMEM + 2i and single-cell-derived colonies were manually picked 1 wk after sorting. Colonies were then trypsinized, and half of the cell suspension was plated in a 96-well plate for expansion. The remaining cells were lysed and DNA was extracted for PCR genotyping (Lackner et al. 2021). Clonal rescue cell lines were generated using piggyBac-based transgenesis. Clones with stable expression of rescue transgenes were isolated and used for further analysis. A correct karyotype (modal average of 40 chromosomes) was confirmed for all ESC lines, and cultures were regularly tested negative for mycoplasma infection.
RNAi assay
In a gelatin-coated 12-well plate, 1.5 × 104 cells/cm2 were transfected in N2B27 + 2i using DharmaFECT 1 (Fisher Scientific T-2001). For RNAi assays, esiRNA for Esrrb and Nanog (Sigma-Aldrich) and FlexiTube siRNAs for Smg6 and Smg7 (Qiagen) were used at a final concentration of 20 nM. The following day, medium was changed to N2B27 after two PBS washes and cells were differentiated for the indicated time. For the Esrrb RNAi assay combined with rapamycin treatment, 1 × 104 cells/cm were transfected with 200 ng of esiRNA targeting Esrrb (Sigma-Aldrich EMU015811) in N2B27 + 2i medium. The day after, cells were treated for 4 h with either DMSO or 20 nM rapamycin in N2B27 + 2i medium before being differentiated in N2B27 + DMSO or N2B27 + 20 nM rapamycin for the indicated time. A PBS wash was performed after the removal of 2i, before switching to differentiation medium.
Cell cycle analysis
ESCs were plated in gelatin-coated plates in ES-DMEM + 2i at a density of 1 × 104 cells/cm2. After 2 d, cells were harvested and fixed in 70% ethanol. Cells were then incubated with 100 μg/mL RNase A solution (Qiagen 19101). Propidium iodide solution (50 mg/L; Sigma-Aldrich P4170) was added to the cells. Cell cycle profiles were recorded on a LSRFortessa flow cytometer (BD bioscience).
Flow cytometry
Cells were harvested using 0.25% trypsin/EDTA, and trypsin was neutralized using ES-DMEM. Rex1-GFPd2 expression levels were measured with a LSRFortessa flow cytometer (BD bioscience). High-throughput measurements were acquired using a 96-well plate HTS unit. Data analysis was performed using FlowJo software (BD Bioscience).
Translation inhibition assay
ESCs were plated in gelatin-coated plates in ES-DMEM-2i at a density of 2 × 104 cells/cm2. The following day, cells were treated for 8 h with 100 µg/mL either DMSO or cycloheximide (CHX; Sigma-Aldrich 01810) and then harvested for RNA extraction.
Telomere quantification
For telomere quantification, genomic DNA was extracted from cells using the Puregene core kit A (Qiagen). DNA was quantified using PicoGreen assay for dsDNA (Fisher Scientific P11496) on a NanoDrop 3300 Fluorospectrometer (Fisher Scientific). PCR reactions were performed on a CFX384 Touch real-time PCR detection system (Bio-Rad) using telomere-specific primers and primers for the single-copy gene 36B4. For each primer pair, a standard curve was created with known amounts of DNA to determine primer efficiency.
RNA analysis
RNA was extracted using the ExtractMe kit (LabConsulting EM09) according to the manufacturer's protocol. cDNA was generated from 0.3–1 µg of RNA using the SensiFAST cDNA synthesis kit (Bioline BIO-65054). Real-time PCR was performed on the CFX384 Touch real-time PCR detection system (Bio-Rad) using the Sensifast SYBR no Rox kit (Bioline BIO-98020). All RT-qPCR primers used in this study are listed in Supplemental Table S7.
Protein analysis
Proteins were extracted using RIPA buffer (Sigma-Aldrich 20-188) supplemented with Complete Mini EDTA-free protease inhibitor cocktail (Roche 04693159001) and PhosSTOP (phosphatase inhibitor cocktail [Roche 04906845001]). Five percent BSA was used for blocking for phospho antibodies, and 5% milk was used for all other antibodies. Primary antibodies were incubated overnight at 4°C or for 1 h at room temperature. Washes were performed using PBS-T (Sigma-Aldrich P4417). Secondary antibodies were incubated for 1 h at room temperature. Primary antibodies were used at a dilution of 1:250 for anti-SMG5 (rabbit; Abcam ab33033), 1:1000 for anti-SMG6 (rabbit; Abcam ab87539), 1:2000 for anti SMG7 (rabbit; NovusBio NBP1-22967), 1:1000 for anti-p-UPF1 (rabbit; Millipore 07-1016), 1:1000 for anti-UPF1 (D15G6; rabbit; Cell Signaling 12040S), 1:1000 for anti-Flag M2 (mouse; Sigma-Aldrich F1804), 1:1000 for anti-c-MYC (D84C12; rabbit; Cell Signaling 5605T), 1:1000 for anti-TCF7L1 (rabbit; Fisher Scientific PA5-40327), 1:1000 for anti-eIF4A2 (rabbit; Abcam ab31218), 1:1000 for anti-eIF4A1 (rabbit; Cell Signaling 2490T), 1:1000 for anti-NANOG (rabbit; NovusBio NB100-58842), 1:1000 for anti-p-4E-BP1(Ser65; rabbit; Cell Signaling 9451T), 1:5000 for anti-Tubulin (mouse; Sigma-Aldrich T8203), 1:5000 for anti-GAPDH (mouse; Sigma-Aldrich G8795), and 1:5000 for anti-Vinculin (E1E9V; rabbit; Cell Signaling 13901T). Secondary antibodies were used at a dilution of 1:10,000 for antirabbit IgG (Amersham NA934) and 1:15,000 for goat antimouse IgG (Santa Cruz Biotechnology sc-2064). Chemiluminescence signal from antibody binding was detected using ECL Select detection kit (GE Healthcare GERPN2235) with a ChemiDoc system (Bio-Rad). Western quantification was performed using ImageJ or ImageLab.
Immunoprecipitation
Cells were plated in ES-DMEM + 2i, and after 2 d were harvested in IP lysis buffer (10 mM Tris Base, 10 mM NaCl, 2 mM EDTA, 0.5% Triton X-100) supplemented with Complete Mini EDTA-free protease inhibitor cocktail (Roche 04693159001) and PhosSTOP (phosphatase inhibitor cocktail [Roche 04906845001]). One milligram of lysate was used for the immunoprecipitation. Briefly, Dynabeads (Fisher Scientific 10004D) were coated with 5 µg of anti-Flag M2 antibody (mouse; Sigma-Aldrich F1804) for 1 h. For RNA-free IP, lysates were treated with 50 U/mL cyanase (Süd-Laborbedarf SLG CY1000) and 0.5 µL/mL 2 M MnSO4 and incubated for 10 min on ice. Lysates were cleared by centrifugation at 16,000g for 15 min at 4°C, and 10% of each lysate was kept as input. NaCl was added to a final concentration of 150 mM. Dynabeads were washed three times with wash buffer (137 mM NaCl, 20 mM Tris base, 0.5% [v/v] Tergitol-type NP-40) and one time with lysis buffer before incubation with the lysates for 3 h at 4°C. Dynabeads were washed three times with wash buffer supplemented with Complete Mini EDTA-free protease inhibitor cocktail (Roche 04693159001) and PhosSTOP (phosphatase inhibitor cocktail [Roche 04906845001]). Samples were eluted in 2× NuPAGE sample buffer.
For the immunoprecipitation coupled with mass spectrometry, cells were plated in ES-DMEM + 2i. The following day, they were treated with 1 µM epoxomicin (Gentaur 607-A2606) for 3 h. Cells were harvested and lysed in IP lysis buffer. One milligram of lysate was used for the immunoprecipitation. Immunoprecipitation protocol was followed (see above) with the addition of the cross-linking of the anti-Flag M2 antibody to the Dynabeads using dimethyl pimelimidate (DMP) (Sigma-Aldrich D-8388). Briefly, after antibody coupling, Dynabeads were washed three times with 200 mM sodium borate (pH 9) and then incubated for 30 min with DMP–sodium borate solution. They were washed with the following buffers: three times with 250 mM Tris (pH 8.0), twice with 100 mM glycine (pH 2), three times with TBS-T, and once with lysis buffer. Beads were then incubated with the lysates for 2 h at 4°C. Five washes were performed with wash buffer (137 mM NaCl, 20 mM Tris base).
Sample preparation for mass spectrometry analysis
Beads with cross-linked antibodies were transferred to new tubes and resuspended in 30 µL of 2 M urea in 50 mM ammonium bicarbonate (ABC). Disulfide bonds were reduced with 10 mM dithiothreitol for 30 min at room temperature before adding 25 mM iodoacetamide and incubating for 15 min at room temperature in the dark. The remaining iodoacetamide was quenched by adding 5 mM DTT, and the proteins were digested with 150 ng of trypsin (Trypsin Gold, Promega) for 90 min at room temperature. The supernatant was transferred to a new tube, the beads were washed with another 30 µL of 2 M urea in 50 mM ABC, and the wash was combined with the supernatant. After diluting to 1 M urea with 50 mM ABC, an additional 150 ng of trypsin was added and incubated overnight at 37°C in the dark. The digest was stopped by addition of trifluoroacetic acid (TFA) to a final concentration of 0.5%, and the peptides were desalted using C18 Stagetips (Rappsilber et al. 2007). Peptides were separated on an Ultimate 3000 RSLC nano-flow chromatography system (Thermo Fisher) using a precolumn for sample loading (Acclaim PepMap C18, 2 cm long × 0.1-mm diameter, 5-μm particle size, Thermo Fisher) and a C18 analytical column (Acclaim PepMap C18, 50 cm long × 0.75-mm diameter, 2-μm particle size, Thermo Fisher), applying a segmented linear gradient from 2% to 35% and finally 80% solvent B (80% acetonitrile, 0.1% formic acid; solvent A: 0.1% formic acid) at a flow rate of 230 nL/min over 120 min. Eluting peptides were analyzed on a Q Exactive HF-X Orbitrap mass spectrometer (Thermo Fisher), which was coupled to the column with a customized nano-spray Easy-Spray ion source (Thermo Fisher) using coated emitter tips (New Objective).
mRNA half-life measurement
ESCs were plated in N2B27 + 2i at a density of 2 × 104 cells/cm2. The day after, the medium was changed to N2B27 + 2i + 100 µM 4SU (Carbosynth NT06186) and cells were cultured in this condition for 12 h. After a PBS wash, cells were incubated with N2B27 or N2B27 + 2i medium + 10 mM uridine (Sigma-Aldrich U6381) for 3 and 6 h. RNA extraction was carried out using Trizol (Fisher Scientific 10296-010) following the manufacturer's instruction with the addition of 0.1 mM DTT during the isopropanol precipitation. RNA was resuspended in 1 mM DTT. Five micrograms of RNA was treated with 10 mM iodoacetamide (Sigma-Aldrich I1149) followed by ethanol precipitation (Herzog et al. 2017). Two nanograms of RNA was used for library preparation. Libraries were prepared using the QuantSeq 3′mRNA-seq library preparation kit for Illumina FWD (Lexogen R3142) and were analyzed on a HiSeqV4 SR100.
RNA immunoprecipitation
Cells (3 × 106) were plated in ES-DMEM + 2i. After 2 d, cells were harvested and lysed using the Magna-RIP kit (Millipore 17-700) according to the manufacturer's protocol. Briefly, cells were lysed by a freeze and thaw cycle and lysates were stored at −80°C. Immunoprecipitations were performed for 3 h at 4°C using 5 µg of an anti-Flag M2 antibody (mouse; Sigma-Aldrich F1804). For RIP-seq, libraries were prepared using the QuantSeq 3′mRNA-seq library preparation kit for Illumina FWD (Lexogen R3142) and analyzed on a HiSeqV4 SR50. Relative binding to input and empty vector control was calculated.
Translation rate measurement using S35 incorporation
ESCs were cultured in ES-DMEM + 2i and 100 µCi S35 (Hartmann IS-103) was added to the culture medium for 30 min (Yamasaki et al. 2009). Cells were washed twice in PBS and lysed in RIPA buffer (Sigma-Aldrich 20-188) supplemented with Complete Mini EDTA-free protease inhibitor cocktail (Roche 04693159001) as described above. Proteins were acetone-precipitated and resuspended in RIPA buffer after two rounds of acetone washes. Protein measurements were performed using a micro BCA protein assay kit (Thermo Fisher 23235). Seven microliters of protein extracts was then spotted on a nitrocellulose membrane and exposed to a BAS storage phosphor screen (GE Healthcare). After 6–12 h, the signal was acquired using a Typhoon scanner (GE Healthcare). Radioactive signal was quantified using the ImageQuant software.
RNA-seq differential analysis
RNA-seq data and analysis used in this study were obtained from a previous study (Lackner et al. 2021). In brief, quality control was performed using FastQC (version 0.11.5). Transcripts were mapped to the mm10 mouse reference genome and counts were determined using STAR (version 2.5.3). RPKM values were calculated using DESeq2 (version 1.20.0). Differential expression analysis was carried out using limma (version 3.30.13). P-values were corrected for multiple testing using the Benjamini–Hochberg method (FDR). Clustering was performed using all genes significantly regulated (Padj < 0.01) in at least one of the Smg factor mutant cells using hclust. Cluster numbers are indicated in Figure 3, A and C. For RNA-seq, we used two independently derived KO cell lines for each of the Smg factors.
GO enrichment analysis
GO annotations were obtained from the R package org.Mm.eg.db (version 3.4.0). For the enrichment of GO terms in the different gene lists, we only considered terms with 5–500 genes assigned to them. Significance of the enrichment was determined using Fisher's exact test with all expressed genes as background (Lackner et al. 2021). GO terms that did not differ in more than five genes were clustered, and one representative term for each cluster was defined (using the R base function hclust on the L1 distance of the binary membership matrix). As a representative GO term for each cluster, we chose the term with the lowest number of annotated genes. Multiple hypothesis testing was executed using the Benjamini–Hochberg method on all representative terms to calculate adjusted P-values.
Quantification of mRNA half-lives
QuantSeq data were analyzed using SLAM-DUNK (version 0.2.4). For calculation of RNA half-lives, transcripts with CPM ≥ 2 were considered. Briefly, background (no 4SU treatment) was subtracted to calculate T > C conversion rates of transcripts of 4SU and 3- and 6-h uridine time points. Half-lives were calculated by plotting T > C conversion rates of 4SU and 3- and 6-h uridine as a single exponential fit to a decay model (Herzog et al. 2017).
Exon conservation
Pairwise whole-genome alignments of mouse GRCm38/mm10 against dog canFam3, rat RGSC6.0/rn6, and human GRCh38/hg38 from UCSC (http://hgdownload.cse.ucsc.edu/downloads.html) were used for the analysis. With the Ensembl annotation of the mouse genome, we extracted the respective regions of the genes (introns, exons, and UTRs) and calculated their sequence identities compared with the other organism in the pairwise alignment (number of matching nucleotide pairs in the alignment) × 100/length). As gene region annotation can be ambiguous due to variations in transcript splicing, we used the following definitions: Everything that was annotated as coding sequence in any protein-coding transcript we counted as coding exon. UTR was everything that was annotated as UTR in any protein-coding transcript except if it was also annotated as coding sequence, in which case it was counted as coding exon. Regions not covered by the above definitions were counted as introns.
RIP-seq analysis
Quality control of the transcripts from input and RIP samples was performed using FastQC. Reads were trimmed using bbduk (version 38.57). Reads were mapped to the mm10 mouse reference genome with STAR (version 2.5.3a). Afterward, indexing was performed using SAMtool (version 1.5), and reads in transcripts were counted with HTSeq-count (version 0.11.2). Transcripts that had CPM ≥ 1 in all the input samples were considered for the differential analysis. Differential analysis was performed using DESeq2 (version 1.24.0). To identify significantly enriched transcripts in eIF4A2FL RIP, we considered only the 1231 transcripts with CPM ≥ 30 in eIF4A2FL RIP and either log fold change eIF4A2FL input/eIF42FL RIP ≥ 1, RIP ≤ −1 (Padj ≤ 0.05), log fold change eIF4A2FL RIP/EV RIP ≥ 1, or RIP/EV RIP ≤ −1 (Padj ≤ 0.05).
Mass spectrometry data acquisition and analysis
The mass spectrometer was operated in data-dependent acquisition mode (DDA), and survey scans were obtained in a mass range of 375–1500 m/z with lock mass activated, at a resolution of 1.2 × 105 at 200 m/z and an AGC target value of 3 × 106. The eight most intense ions were selected with an isolation width of 1.6 m/z and fragmented in the HCD cell at 28% collision energy, and the spectra were recorded for a maximum of 250 msec at a target value of 1 × 105 and a resolution of 3 × 104. Peptides with a charge of +1 or more than +7 were excluded from fragmentation, the peptide match feature was set to preferred, the exclude isotope feature was enabled, and selected precursors were dynamically excluded from repeated sampling for 30 sec.
Raw data were processed using the MaxQuant software package (version 1.6.0.16) (Tyanova et al. 2016), the UniProt mouse reference proteome (January 2019, http://www.uniprot.org) and target sequences, and a database of the most common contaminants. The search was performed with full trypsin specificity and a maximum of two missed cleavages at a protein and peptide spectrum match false discovery rate of 1%. Carbamidomethylation of cysteine residues was set as fixed, and oxidation of methionine and N-terminal acetylation were set as variable modifications. For label-free quantification the “match between runs” feature and the LFQ function were activated; all other parameters were left at default.
MaxQuant output tables were further processed in R (https://www.R-project.org). Reverse database identifications, contaminant proteins, protein groups identified only by a modified peptide, protein groups with less than three quantitative values in one experimental group, and protein groups with less than two razor peptides were removed from further analysis. Due to differences in overall contaminant levels between samples, LFQ values were renormalized using the sample median of the “background” protein subset (as identified in controls). Missing values were replaced by randomly drawing data points from a normal distribution modeled on the whole data set (data mean shifted by −1.8 standard deviations, width of distribution of 0.3 standard deviations). Differences between groups were statistically evaluated using the limma package (Ritchie et al. 2015) at 5% FDR (Benjamini–Hochberg). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Pérez-Riverol et al. 2019) under the accession number PXD019588. NGS data have been deposited on GEO (GSE153457).
Supplementary Material
Acknowledgments
We thank Thomas Sauer and Johanna Stranner at the Max Perutz Laboratories Biooptics-FACS Facility for expert support, Markus Hartl and the Max Perutz Laboratories Mass Spectrometry Facility for mass spectrometry analysis, and Irmgard Fischer for help with microscopy. We thank Stefan Weitzer and Irene Gösler for help with S35-based experiments, and Oliver Mühlemann and Javier Caceres for valuable discussions and feedback. NGS was performed at the Vienna Biocenter Core Facilities (VBCF). This work was supported by the Austrian Science Fund (FWF; P31334) and by a joint grant by the FWF and the German Research Foundation (DFG) to A.B. and M.L. (FWF: I3786; DFG: 398882498). Martin Leeb is a Wiener Wissenschafts Forschungs und Technologiefunds Vienna Research Group leader (VRG14-006) and was supported by an FWF Erwin Schroedinger return fellowship.
Author contributions: M.H., L.S., and E.G. performed experiments and analyzed data. R.S., M.R., F.T.-T., L.H., A.L., and M.G. performed bioinformatic analysis, supervised by A.B., A.v.H., C.B., and M.L. J.R., S.S., and M.O. performed experiments. K.W. and C.S. analyzed teratoma samples. A.C., K.F.L., and A.P. supported translation measurements. V.H. and S.L.A. performed half-life measurements. M.H., L.S., E.G., A.S., and M.L. designed experiments. M.L. designed and supervised the study and wrote the manuscript, together with M.H., L.S., and E.G., with feedback from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.347690.120.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Acampora D, Omodei D, Petrosino G, Garofalo A, Savarese M, Nigro V, Di Giovannantonio LG, Mercadante V, Simeone A. 2016. Loss of the Otx2-binding site in the Nanog promoter affects the integrity of embryonic stem cell subtypes and specification of inner cell mass-derived epiblast. Cell Rep 15: 2651–2664. 10.1016/j.celrep.2016.05.041 [DOI] [PubMed] [Google Scholar]
- Anders KR, Grimson A, Anderson P. 2003. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J 22: 641–650. 10.1093/emboj/cdg056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. 2007. Telomeric repeat-containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801. 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- Batista Pedro J, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley Donna M, Lujan E, Haddad B, Daneshvar K, et al. 2014. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15: 707–719. 10.1016/j.stem.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. 2013. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153: 335–347. 10.1016/j.cell.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm V, Kueckelmann S, Gerbracht JV, Kallabis S, Britto-Borges T, Altmüller J, Krüger M, Dieterich C, Gehring NH. 2021. SMG5–SMG7 authorize nonsense-mediated mRNA decay by enabling SMG6 endonucleolytic activity. Nat Commun 12: 3965. 10.1038/s41467-021-24046-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornelöv S, Selmi T, Flad S, Dietmann S, Frye M. 2019. Codon usage optimization in pluripotent embryonic stem cells. Genome Biol 20: 119. 10.1186/s13059-019-1726-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, Nichols J. 2014. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol 16: 513–525. 10.1038/ncb2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, Macrae TA, Oses-Prieto JA, Covarrubias S, Percharde M, Ku G, Diaz A, McManus MT, Burlingame AL, Ramalho-Santos M. 2018. The transcriptionally permissive chromatin state of embryonic stem cells is acutely tuned to translational output. Cell Stem Cell 22: 369–383.e8. 10.1016/j.stem.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann Benedikt M, Strein C, Davey Norman E, Humphreys David T, Preiss T, Steinmetz Lars M, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406. 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Chawla R, Azzalin CM. 2008. The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet Genome Res 122: 194–201. 10.1159/000167804 [DOI] [PubMed] [Google Scholar]
- Chiu SY, Serin G, Ohara O, Maquat LE. 2003. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87. 10.1261/rna.2137903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Karousis ED, Bourquin J, Bruggmann R, Mühlemann O. 2017. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 23: 189–201. 10.1261/rna.059055.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini NS, Peer AM, Moeseneder P, Roiuk M, Burkard TR, Theussl H-C, Moll I, Knoblich JA. 2018. Coordinated control of mRNA and rRNA processing controls embryonic stem cell pluripotency and differentiation. Cell Stem Cell 22: 543–558.e12. 10.1016/j.stem.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Di Stefano B, Luo E-C, Haggerty C, Aigner S, Charlton J, Brumbaugh J, Ji F, Rabano Jiménez I, Clowers KJ, Huebner AJ, et al. 2019. The RNA helicase DDX6 controls cellular plasticity by modulating P-body homeostasis. Cell Stem Cell 25: 622–638.e13. 10.1016/j.stem.2019.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. 2014. Defining an essential transcription factor program for naïve pluripotency. Science 344: 1156–1160. 10.1126/science.1248882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. 2015. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347: 1002–1006. 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- Grabole N, Zhang JD, Aigner S, Ruderisch N, Costa V, Weber FC, Theron M, Berntenis N, Spleiss O, Ebeling M, et al. 2016. Genomic analysis of the molecular neuropathology of tuberous sclerosis using a human stem cell model. Genome Med 8: 94. 10.1186/s13073-016-0347-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Sidoli S, Warneford-Thomson R, Tatomer Deirdre C, Wilusz Jeremy E, Garcia Benjamin A, Bonasio R. 2016. High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol Cell 64: 416–430. 10.1016/j.molcel.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, von Haeseler A, Zuber J, Ameres SL. 2017. Thiol-linked alkylation of RNA to assess expression dynamics. Nat Methods 14: 1198–1204. 10.1038/nmeth.4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JA, Wu C-I, Merrill BJ. 2013. Tcf7l1 prepares epiblast cells in the gastrulating mouse embryo for lineage specification. Development 140: 1665–1675. 10.1242/dev.087387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes ZE, Hamilton DJ, Hwang T, Parsonnet NV, Rinn JL, Wuttke DS, Batey RT. 2020. The Sox2 transcription factor binds RNA. Nat Commun 11: 1805. 10.1038/s41467-020-15571-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Lou C-H, Chan W, Shum EY, Shao A, Stone E, Karam R, Song H-W, Wilkinson MF. 2011. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell 43: 950–961. 10.1016/j.molcel.2011.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Weichenrieder O, Izaurralde E. 2013. An unusual arrangement of two 14-3-3-like domains in the SMG5–SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev 27: 211–225. 10.1101/gad.206672.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T, Olova N, Roode M, Mulas C, Lee HJ, Nett I, Marks H, Walker R, Stunnenberg HG, Lilley KS, et al. 2017. Tracking the embryonic stem cell transition from ground state pluripotency. Development 144: 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. 2006. RIP-ChIP: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1: 302–307. 10.1038/nprot.2006.47 [DOI] [PubMed] [Google Scholar]
- Keller G, Kennedy M, Papayannopoulou T, Wiles MV. 1993. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol 13: 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Popp MW, Maquat LE. 2019. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nature Reviews Molecular Cell Biology 20: 406–420. 10.1038/s41580-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner A, Sehlke R, Garmhausen M, Giuseppe Stirparo G, Huth M, Titz-Teixeira F, van der Lelij P, Ramesmayer J, Thomas HF, Ralser M, et al. 2021. Cooperative genetic networks drive embryonic stem cell transition from naïve to formative pluripotency. EMBO J 40: e105776. 10.15252/embj.2020105776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Dietmann S, Paramor M, Niwa H, Smith A. 2014. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell 14: 385–393. 10.1016/j.stem.2013.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Belsham GJ, Proud CG. 2001. Eukaryotic initiation factors 4A (eIF4A) and 4G (eIF4G) mutually interact in a 1:1 ratio in vivo. Journal of Biological Chemistry 276: 29111–29115. 10.1074/jbc.C100284200 [DOI] [PubMed] [Google Scholar]
- Li T, Shi Y, Wang P, Guachalla LM, Sun B, Joerss T, Chen YS, Groth M, Krueger A, Platzer M et al. 2015. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J 34: 1630–1647. 10.15252/embj.201489947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yu JSL, Tilgner K, Ong SH, Koike-Yusa H, Yusa K. 2018. Genome-wide CRISPR-KO screen uncovers mTORC1-mediated Gsk3 regulation in naive pluripotency maintenance and dissolution. Cell Rep 24: 489–502. 10.1016/j.celrep.2018.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QV, Rosen BP, Huangfu D. 2020. Decoding pluripotency: genetic screens to interrogate the acquisition, maintenance, and exit of pluripotency. Wiley Interdiscip Rev Syst Biol Med 12: e1464. 10.1002/wsbm.1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou C-H, Dumdie J, Goetz A, Shum EY, Brafman D, Liao X, Mora-Castilla S, Ramaiah M, Cook-Andersen H, Laurent L, et al. 2016. Nonsense-mediated RNA decay influences human embryonic stem cell fate. Stem Cell Reports 6: 844–857. 10.1016/j.stemcr.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. 2001. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 10: 99–105. 10.1093/hmg/10.2.99 [DOI] [PubMed] [Google Scholar]
- Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. 2013. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 340: 82–85. 10.1126/science.1231197 [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078. 10.1038/ng1429 [DOI] [PubMed] [Google Scholar]
- Meng L, Mohan R, Kwok BHB, Elofsson M, Sin N, Crews CM. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci 96: 10403–10408. 10.1073/pnas.96.18.10403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H, Hernando-Herraez I, Savino A, Scialdone A, Macaulay I, Mulas C, Chandra T, Voet T, Dean W, Nichols J, et al. 2017. Single-cell landscape of transcriptional heterogeneity and cell fate decisions during mouse early gastrulation. Cell Rep 20: 1215–1228. 10.1016/j.celrep.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulas C, Kalkan T, Smith A. 2017. NODAL secures pluripotency upon embryonic stem cell progression from the ground state. Stem Cell Reports 9: 77–91. 10.1016/j.stemcr.2017.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200. 10.1016/S1097-2765(03)00443-X [DOI] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. 2012. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 40: 1251–1266. 10.1093/nar/gkr791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicka K, Kalathiya U, Alfaro J. 2020. Nonsense-mediated mRNA decay: pathologies and the potential for novel therapeutics. Cancers 12: 765. 10.3390/cancers12030765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Reichenbach P, Höss M, Azzalin CM, Nabholz M, Bucher P, Lingner J. 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13: 568–574. 10.1016/S0960-9822(03)00173-8 [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber C, Soffientini P, Ruepp MD, Bachi A, Mühlemann O. 2016. Identification of interactions in the NMD complex using proximity-dependent biotinylation (BioID). PLoS One 11: e0150239. 10.1371/journal.pone.0150239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N. 2012. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 9: 1370–1379. 10.4161/rna.22360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokanos FF, Albert MA, Demetriades C, Spirohn K, Boutros M, Teleman AA. 2016. eIF4A inactivates TORC1 in response to amino acid starvation. EMBO J 35: 1058–1076. 10.15252/embj.201593118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11: 2301–2319. 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- Villegas F, Lehalle D, Mayer D, Rittirsch M, Stadler MB, Zinner M, Olivieri D, Vabres P, Duplomb-Jego L, De Bont E, et al. 2019. Lysosomal signaling licenses embryonic stem cell differentiation via inactivation of Tfe3. Cell Stem Cell 24: 257–270.e8. 10.1016/j.stem.2018.11.021 [DOI] [PubMed] [Google Scholar]
- Wilczynska A, Gillen SL, Schmidt T, Meijer HA, Jukes-Jones R, Langlais C, Kopra K, Lu WT, Godfrey JD, Hawley BR, et al. 2019. eIF4A2 drives repression of translation at initiation by Ccr4-Not through purine-rich motifs in the 5′UTR. Genome Biol 20: 262. 10.1186/s13059-019-1857-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jäck H-M. 2006. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol 26: 1272–1287. 10.1128/MCB.26.4.1272-1287.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. 2011. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 13: 838–845. 10.1038/ncb2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42. 10.1083/jcb.200811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Mühlemann O. 2011. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17: 2108–2118. 10.1261/rna.030247.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder-Hill J, Pause A, Sonenberg N, Merrick WC. 1993. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J Biol Chem 268: 5566–5573. 10.1016/S0021-9258(18)53358-5 [DOI] [PubMed] [Google Scholar]
- You KT, Park J, Kim VN. 2015. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev 29: 2004–2009. 10.1101/gad.267112.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. 2011. Control of the embryonic stem cell state. Cell 144: 940–954. 10.1016/j.cell.2011.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.