Abstract
Background
Biological nitrogen fixation (BNF) is an important nitrogen source for legume plants, and highly efficient nitrogen fixation requires sufficient phosphorus (P). However, the mechanism of maintaining nitrogen fixation of the legume nodules under low P concentration remains largely unknown.
Results
A nodule-localized SPX protein, GmSPX8, was discovered by transcriptome and functional analysis of its role in N2 fixation was characterized in soybean nodules. GmSPX8 was preferentially expressed in nodules and its expression was gradually increased during nodule development. And also the expression pattern was investigated using reporter gene β-glucuronidase (GUS) driven by the promoter of GmSPX8. GmSPX8 was greatly induced and the GUS activity was increased by 12.2% under P deficiency. Overexpression of GmSPX8 in transgenic plants resulted in increased nodule number, nodule fresh weight and nitrogenase activity by 15.0%, 16.0%, 42.5%, subsequently leading to increased N and P content by 17.0% and 19.0%, while suppression of GmSPX8 showed significantly impaired nodule development and nitrogen fixation efficiency under low P stress. These data indicated that GmSPX8 conferred nodule development and nitrogen fixation under low P condition. By yeast two-hybrid screening, GmPTF1 was identified as a potential interacting protein of GmSPX8, which was further confirmed by BiFC, Y2H and pull down assay. Transcript accumulation of GmPTF1 and its downstream genes such as GmEXLB1 and EXPB2 were increased in GmSPX8 overexpressed transgenic nodules, and in the presence of GmSPX8, the transcriptional activity of GmPTF1 in yeast cells and tobacco leaves was greatly enhanced.
Conclusions
In summary, these findings contribute novel insights towards the role of GmSPX8 in nodule development and nitrogen fixation partly through interacting with GmPTF1 in soybean under low P condition.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-022-03556-2.
Keywords: Legume plants, Nodules, Biological nitrogen fixation, Phosphorus, SPX proteins, BiFC, Y2H, Transcriptional activity
Background
Legume plants, such as Medicago truncatula and soybean (Glycine max (L.) Merr.), could establish symbiotic associations with rhizobia to form nodules for biological nitrogen fixation (BNF). BNF has the capacity to fix atmospheric nitrogen (N2) into ammonia, which is essential to meet the requirement for N nutrient during plant growth and development processes [1–4]. The amount of N source, fixed by BNF, is about 50 million tons per year, contributing nearly half of the N provided by manufactured fertilizer [5, 6]. Simultaneously, excessive application of chemical fertilizer in agricultural production results in the deterioration of environmental quality and soil systems. Therefore, BNF is a very valuable alternative to N fertilizer [7].
BNF is a really complex process. To improve BNF ability and efficiency in legumes, and to realize BNF in non-leguminous by bio-engineering measure, numerous studies have been focused on the genetics of BNF, especially in the recent years [8]. In Medicago truncatula, SHR-SCR module determined the fate of cortical cell to enable de novo nodule organogenesis [9]. In soybean, Nodule Number Locus 1 (GmNNL1) interacted with NopP effector from Bradyrhizobium USDA110 to inhibit nodulation [10]. Under salinity stress, glycogen synthase kinase 3 (GSK3)-like kinase inhibited legume-rhizobia symbiosis through phosphorylating GmNSP1 [11]. All these significant findings would promote our understanding on the legume-rhizobia interactions for better BNF.
Nodule formation was an energy-consuming process, which required a large amount of phosphorus (P) [12–15]. Phosphorus deficiency directly impaired nodule initiation, development and N2 fixation. Sufficient P supply significantly promoted soybean nodulation with 63% and 85% increases in nodule number and nodule size. Moreover, under P starvation condition, P content in nodules was much higher than other organs, e.g. shoot, root, and leaves [15–21]. Thus, maintaining a relative level of P in nodules is important to plant growth and BNF [22]. Recently, several P homeostasis related genes have proven to be involved in soybean nodulation [22]. Overexpression of GmPAP12 increased nodule number, nodule fresh weight, nitrogenase activity and the resultant higher N content under low P condition, while its RNAi transgenic lines displayed impaired nodule development and nitrogen fixation ability [23]. GmPT5, a high-affinity P transporter, controls P transport from roots to nodules, essential for maintaining Pi homeostasis in nodules [22]. Therefore, mining functional genes involved in nodulation would greatly improve our understanding on soybean nodulation, and further promote BNF in agricultural production [6, 24, 25].
Proteins containing SPX (Syg1, Pho81 and Xpr1) domain are vital components in P signaling pathway and P homeostasis in the cell. The SPX domain is named after the conserved domain in the N-terminal of yeast gpa1 (Syg1), yeast phosphatase (Pho81) and the human xenotropic and polytropic retrovirus receptor 1 (Xpr1) [26]. In plants, SPX-containing-proteins could be divided into four subfamilies according to the presence of additional domains: SPX-EXS, SPX-MFS, SPX-RING and SPX [27–31]. Among them, SPX family proteins refer to proteins only containing the SPX domain, which have important role in P signaling pathway in plants [26]. In Arabidopsis, there are four members of SPX proteins, AtSPX1-AtSPX4, among which AtSPX1-AtSPX3 are responsive to P starvation in roots and shoots. AtSPX1 interacts with PHR1 and has a cellular P-dependent inhibitory effect on PHR1 [32, 33]. In rice, six SPX proteins (OsSPX1-OsSPX6) were identified responding to P starvation. OsSPX1, an ortholog of AtSPX1, interacts with the OsPHR2 in the nucleus to inhibit phosphate starvation responses [34]. OsSPX4, a cytoplasmic SPX protein, negatively regulate P Signaling by interacting with OsPHR2 and preventing translocation of OsPHR2 to the nucleus [35]. The interaction of OsSPX4 and OsPHR2 is competitively inhibited by transcription factor bHLH6, which regulates P homeostasis by antagonizing SPX4 [36]. In soybean, nine SPX members (GmSPX1-GmSPX9) are characterized, among which GmSPX1 is a negative regulator and GmSPX3 is a positive one in the P signaling network [27, 31]. Although great efforts have been made on the function of SPX proteins responding to P deficiency in plants, their roles and molecular mechanisms of SPX proteins in soybean nodulation have not been well addressed.
In this study, a nucleus-localized SPX protein, GmSPX8, in soybean nodulation was characterized under low P condition. GmSPX8 was preferentially expressed in nodules. The overexpression and suppression analysis demonstrated that GmSPX8 was responsible for nodule development and nitrogen fixation under P deficiency in soybean. Interacting proteins of GmSPX8 was hunted and verified.
Results
Plant growth and nodule development were impaired under P-deficient treatment in soybean
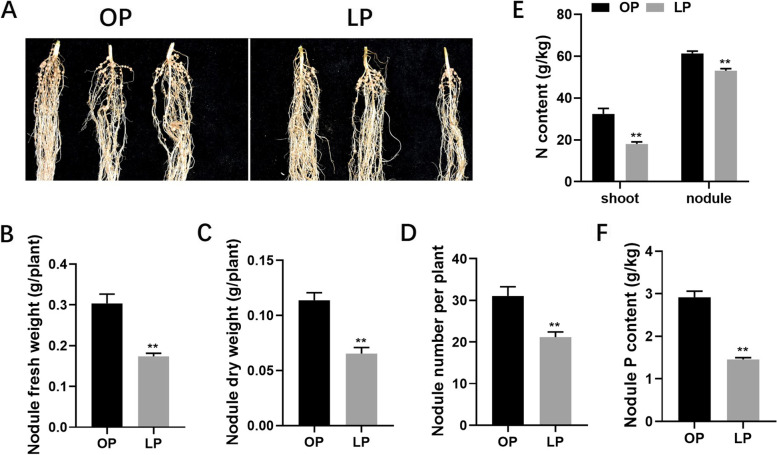
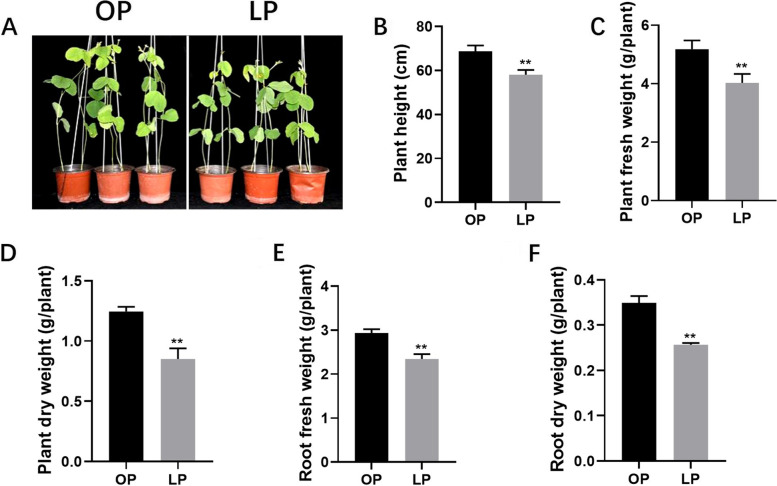
Zhonghuang15 (ZH15) was grown under P-sufficient and P-deficient conditions, inoculated with rhizobium Bradyrhizobium diazoefficiens USDA110. The phenotype data was collected at 28dpi and analyzed accordingly (Fig. 1). Comparing the data under P-sufficient treatment, the nodule number, nodule fresh-, dry- weight and total P content under low P condition were decreased by 30.1%, 46.6%, 51.6% and 39.8%, respectively. Consequently, the nitrogen fixation was impaired as total N content was reduced by 44.2% and 13.4% in shoot and nodule, respectively (Fig. 1). In addition, plant height, plant fresh- and dry- weight have reductions of 13.0%, 16.0% and 32.0%, respectively, and the root fresh- and dry- weight were simultaneously reduced by 16.9% and 26.5% (Fig. 2). Thus, the nodulation, nitrogen fixation ability and plant growth were significantly impaired in low P condition in soybean.
Fig. 1.
Analysis of soybean nodulation under P-sufficient and P-deficient conditions. A Photographs of soybean nodules. B Nodule fresh weight. C Nodule dry weight. D Nodule number. E N content of shoot and nodules. F P content of nodules. Soybean plants and nodules were harvested at 28 dpi in different P conditions (P-sufficient: OP, 500 μM KH2PO4 and P-deficient: LP, 5 μM KH2PO4). Data were presented as the average of three different biological replicates and 20 plants for each replicate. Bars showed the means ± SE values. Asterisks indicate significant difference within a P level in t tests. * p < 0.05, ** p < 0.01
Fig. 2.
Effects of P deficiency on soybean growth. A Phenotype of plant performance. B Plant height. C Plant fresh weight. D Plant dry weight. E Root fresh weight. F Root dry weight. Soybean seedlings were grown in P sufficient conditions, respectively, with Bradyrhizobium diazoefficiens USDA110 inoculation and plants were harvested at 28 dpi. Data are means of four replicates, and error bars show the SE values. Asterisks in B-F indicate significant difference between different P level in t test. * p < 0.05, ** p < 0.01
Transcriptome analysis of soybean nodules in response to low P stress using RNA-seq
To explore genes responsible for nodule development in response to low P stress, transcriptome dataset of nodules was collected under P-sufficient and P-deficient conditions. Comparing to the treatment of P-sufficient, P deficiency triggered a total of 7723 differentially expressed genes (DEGs) in nodules, with 4382 up-regulated DEGs and 3341 repressed ones (Table S1 and Figure S1A). All the DEGs could be classified into eight biological processes, two cellular components and 20 molecular function terms through Gene ontology (GO) category analysis, and the categories of molecular function contained most DEGs (Figure S1B). These findings suggested that specific regulatory signaling was induced in nodules to maintain nodule formation and development under P deficient condition, and resulted in the differential symbiotic phenotypes of nodules observed under P-sufficient and P-deficient conditions.
GmSPX8 was preferentially expressed in soybean nodules under low P stress
Given that the SPX domain-containing proteins were reported responsive to the fungus infection in low P stress in soybean [27], eight SPX domain-containing genes among the up-regulated DEGs, GmSPX1, GmSPX3, GmSPX4, GmSPX7, GmSPX8, GmSPX9, Glyma.03G032400 and Glyma.10G261900, were selected for further analysis. Their expression in nodules under P deficient condition was confirmed by qRT-PCR, which was in accordance with their RNA-seq data (Figure S2 and Table S2). Of the eight SPX domain-containing genes, GmSPX8 presented a highest expression level, implying its role in nodule formation and development.
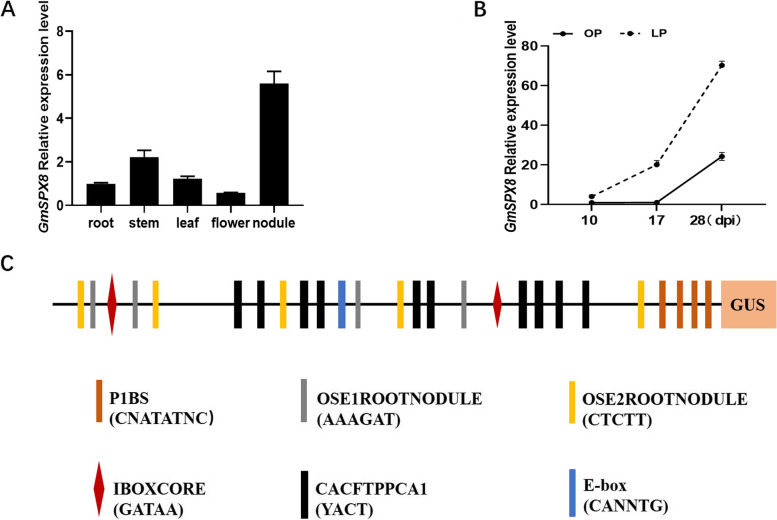
The spatial–temporal expression patterns of GmSPX8 in different soybean organs were assayed via qRT-PCR. This gene mainly expressed in nodules, and its expression was increased gradually during nodule development, while relative low level expressed in root, stem, leaf and flower (Fig. 3A-B).
Fig. 3.
Expression profiles of GmSPX8 in various tissues. A Expression patterns of GmSPX8 in different organs. B Transcript abundance of GmSPX8 in nodules at different time points determined by qRT-PCR. C Schematic representation of the Cis-elements in the promoter of GmSPX8 analyzed by PlantCARE
To understand the expression pattern of GmSPX8, its promoter was investigated. Several nodulation and P signaling-related motifs were characterized in its promoter sequence (1500 bp), such as P1BS (PHR1 binding site) element (CNATATNC), E-box (CANNTG), OSE1ROOTNODULE (AAAGAT), and OSE2ROOTNODULE (CTCTT) (Fig. 3C). Visual GUS expression driven by GmSPX8 promoter was detected in nodules of transgenic composite soybean plants under both P conditions, and the GUS intensity was increased by 12.2% in low P stress than that in P sufficient condition (Figure S3). These data suggested that the GmSPX8 promoter tends to have a higher activity under P deficient condition.
GmSPX8 conferred nodule development and nitrogen fixation under low P stress
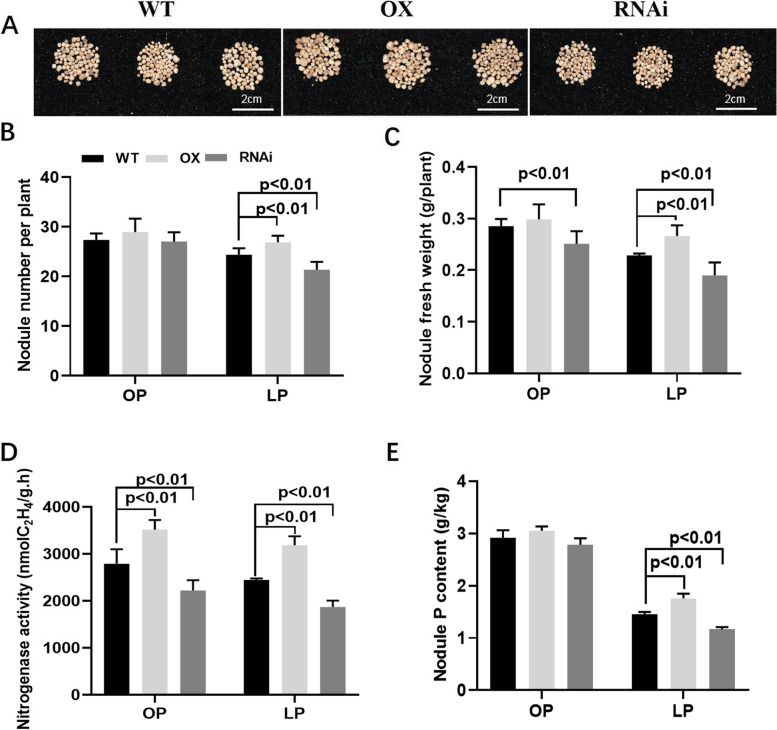
To further dissect the function of GmSPX8 in nodule development and nitrogen fixation, transgenic composite soybean plants overexpressing or suppressing (RNAi) of GmSPX8 were generated and evaluated under P deficient condition (Fig. 4A). Comparing to the non-transgenic wild type (WT), GmSPX8 overexpressed transgenic plants under low P stress preserved 10.4% and 16.5% more of nodule number and nodule fresh weight, respectively, while GmSPX8 RNAi plants inversely have fewer nodule number and fresh weight with reductions of 12.3% and 21.9%, respectively. Simultaneously, nitrogenase activity and P contents in GmSPX8 overexpressed transgenic plants were significantly increased by 37.8%and 35.0% under low P stress, while both were decreased in RNAi plants by 23.6%and 19.8%, respectively (Fig. 4B-E). All these data demonstrated that GmSPX8 involved in both nodule development and nitrogen fixation under P deficient condition.
Fig. 4.
Nodulation analysis of transgenic composite soybean plants either overexpressing (OX) or suppressing (RNAi) of GmSPX8 under P-sufficient and P-deficient conditions. A Growth performance of nodules. B Nodule number. C Nodule total fresh weight per plant. D Nitrogenase activities of nodules measured by acetylene reduction assay. E Nodule P content. Transgenic composite soybean plants were harvested at 28 dpi under P-sufficient and P-deficient conditions. Data were presented as the average of three different biological replicates and 20 plants for each replicate. Bars showed the means ± SD values
GmSPX8 interacting with GmPTF1 to regulate nodule development and nitrogen fixation under low P stress
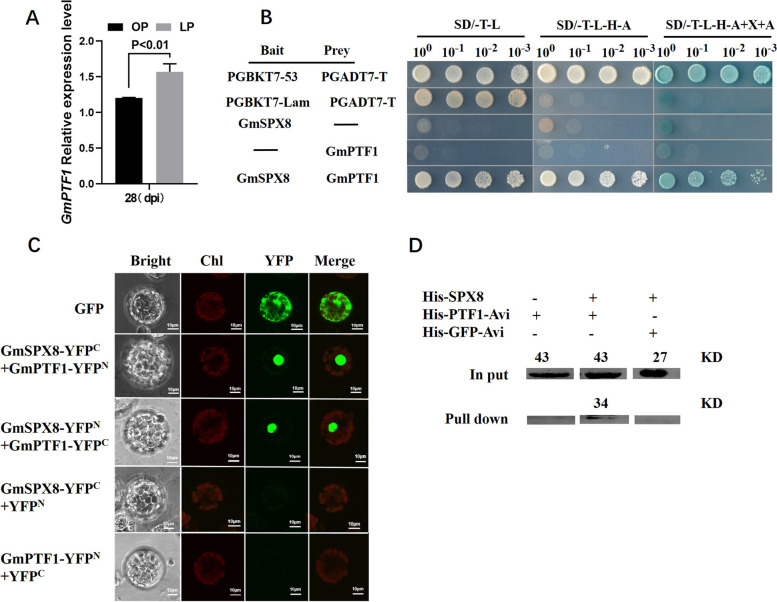
In order to characterize the molecular mechanism of GmSPX8 involving in soybean nodulation in P-deficient condition, we isolated six potential partners of GmSPX8 from cDNA library of soybean nodules through yeast two-hybrid (Y2H) screening analysis, and GmPTF1 (XM_006588849) was selected for further analysis, due to its nuclear localization and its response to low P stress [37] (Table 1). Under low P condition, GmPTF1 had a higher abundance in nodules under low P condition than under P-sufficient condition (Fig. 5A). In order to further confirm the physical interaction between GmSPX8 and GmPTF1 in vivo, Y2H assay was performed. Co-transformed with BD-GmSPX8 and AD-GmPTF1, yeast cells grew well on SD/-Trp-Leu-His-Ade + X-α-gal + AbA selective medium, but those co-transformed with BD/AD-GmPTF1 or BD-GmSPX8/AD did not grow on the same medium (Fig. 5B). Bimolecular fluorescence complementation (BiFC) assay in Arabidopsis protoplasts also showed that strong yellow fluorescence (YFP) signal was detected in the nucleus when GmSPX8-YFPN and GmPTF1-YFPC or GmSPX8-YFPC and GmPTF1-YFPN were co-expressed in Arabidopsis protoplasts (Fig. 5C).The interaction between GmSPX8 and GmPTF1 was also confirmed in vitro through the pull-down assay using recombinant purified proteins in E.coli (Fig. 5D). Thus, GmPTF1 interacted with GmSPX8 in soybean nodules under low P stress.
Table 1.
Interacting partners of GmSPX8 screened by yeast two-hybrid in soybean nodules
| ACCESSION | ANNOTATION |
|---|---|
| XM_003543662 | Glycine max outer plastidial membrane protein |
| XM_003526765 | Glycine max superoxide dismutase |
| XM_003531923 | Glycine max transcription factor bHLH93 |
| XM_006588849 | Glycine max transcription factor bHLH48 |
| XM_003552460 | Glycine max outer plastidial membrane protein |
| XM_003554268 | Glycine max outer mitochondria membrane protein |
Fig. 5.
GmSPX8 interacted with GmPTF1 in soybean nodules. A Relative expression level of GmPTF1 in nodules. B Interaction between GmSPX8 and Gm PTF1 was shown by Y2H assay. Positive yeast cells containing GmSPX8 and GmPTF1 were selected on SD/-T-L–H-A medium containing 125 ng/ml AbA and 40 ug/ml X-α-Gal. C BiFC analysis of the interaction between GmSPX8 and GmPTF1 in Arabidopsis protoplasts. GmSPX8-YFPN and GmPTF1-YFPC or GmSPX8-YFPC and GmPTF1-YFPN were cotransformed into Arabidopsis protoplasts. The fluorescent signal of YFP was detected by fluorescence microscope. D Analysis of interaction between GmSPX8 and GmPTF1 by Pull-down assay using recombinant proteins purified from E. coli. All these experiments were repeated three times with similar results
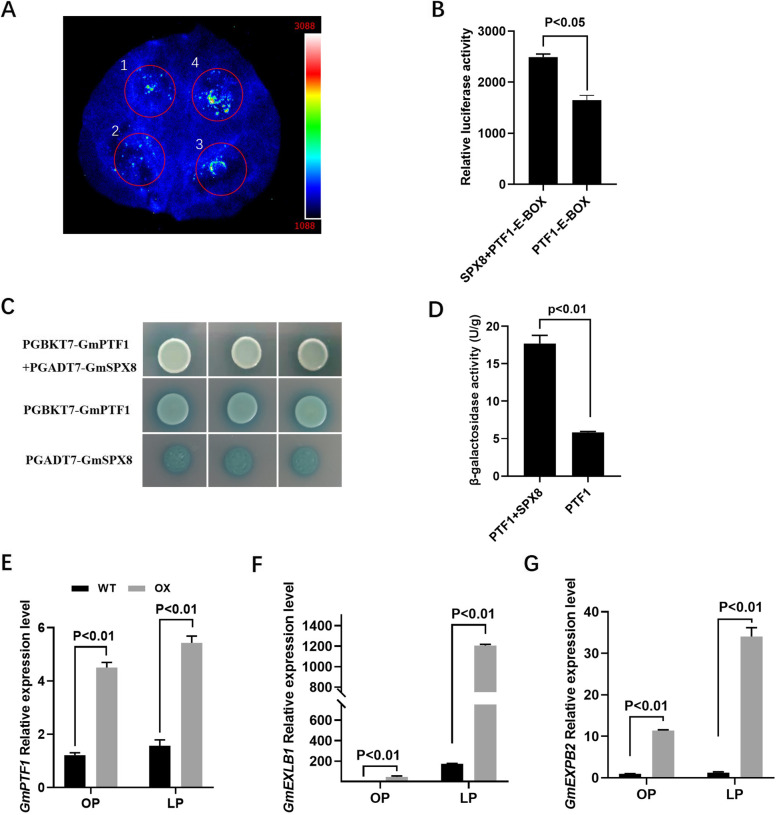
To determine GmSPX8 on GmPTF1 transcription activity, transcriptional activation assay was performed using N. benthamiana leaves. The transcription activity of GmPTF1 was monitored through the fluorescence intensity of luciferase (LUC). In the presence of GmPTF1, intense LUC signals were observed, and with GmSPX8, the intensity of LUC signals was increased around 50%, which demonstrated that GmSPX8 could enhance the promoter activation activity of GmPTF1 (Fig. 6A-B). In addition, the activation analysis of GmPTF1 was done in yeast cells AH109, containing a β-galactosidase gene as a selection marker (Fig. 6C). In the presence of GmSPX8, the activity of β-galactosidase in yeast cells was two times higher than that of GmPTF1 alone (Fig. 6D). Furthermore, the transcript accumulation of GmPTF1 was increased more higher in GmSPX8 overexpressed nodules than in wild type in P-deficient conditions (Fig. 6E). GmEXPB2 (β-expansin gene), a Cell Wall β-Expansin, was reported to influence soybean nodulation and development, and GmEXLB1 (expansin-like B1) and GmEXPB2 were directly regulated by GmPTF1 [38, 39]. To explore this regulation in nodules, the transcript abundances of GmEXLB1 and GmEXPB2 were surveyed in transgenic composite plants with overexpressed GmSPX8. GmEXLB1 and GmEXPB2 were greatly increased in GmSPX8 overexpressed nodules in P deficient condition (Fig. 6F-G). All these findings suggested that GmSPX8 was involved in soybean nodulation via directly interacting with GmPTF1.
Fig.6.
GmSPX8 positively regulated GmPTF1 and its downstream genes. A GmSPX8 regulated the transcriptional activity of GmPTF1. 1. PGreenII62-SK empty vector and E-box-LUC, 2. PSK-GmSPX8 and E-box-LUC, 3. PSK-GmPTF1 and E-box-LUC, 4. PSK-GmPTF1 and PSK-GmSPX8 and E-box-LUC. B Relative luciferase activity of tobacco leaves expressing the indicated constructs. C Effect of GmSPX8 on the transcription activity of GmPTF1 in yeast cells. D β-galactosidase activity of yeast cells expressing both GmSPX8 and GmPTF1 or GmPTF1 alone. Expression abundance of GmPTF1 (E), GmEXBL1 (F) and GmEXPB2 (G) in nodules of wild type and GmSPX8 overexpressed nodules under P-sufficient and P-deficient conditions
Discussion
BNF could provide essential N source for legume growth and development, and phosphorus concentration significantly influenced nodule growth and development. Therefore, numerous studies have focused on nitrogen fixation and plant growth under P deficient condition [15–17, 40]. In this study, nodule development was significantly retarded under P deficient condition and subsequently leading to reduced nitrogen fixation ability and N content and P content. Accordingly, plant morphogenesis, e.g. plant height, fresh weight and dry weight, was influenced (Fig. 1–2).
Previous studies have mainly focused on isolating PSI (phosphorus starvation induced) genes and unraveling phosphorus signaling networks in nodules of legume plants through transcriptome and metabolome profiling, e.g. common bean (Phaseolus vulgaris), Medicago truncatula and soybean. Purple acid phosphatases (PAPs), phosphate transporters (PTs), MYB (v-myb avian myeloblastosis viral oncogene homolog) and bHLH (basic/helix-loop-helix) proteins transcription factors had been reported to involve in Pi homeostasis in nodules [17, 25, 41]. In Arabidopsis, soybean and rice, SPX proteins were considered as an important P sensor [31, 33, 34]. GmSPX8 in soybean responded to P starvation in leaves and roots, and was up-regulated by P starvation in mycorrhizal nodules of soybean, implicating its involvement in P homeostasis in soybean nodules [27, 31].In this study, to isolate PSI genes in soybean, transcriptome profiling was conducted with nodules under different P conditions, and eight SPX domain-containing proteins were identified (TableS1 and Figure S2). Of the eight proteins, GmSPX8 was preferentially expressed in nodules inoculated with Bradyrhizobium diazoefficiens USDA110, and its expression level was gradually increased during nodule development (Fig. 3A-B), which was observed for the first time. The promoter analysis revealed that several nodulation- and P deficiency-related motifs presented in GmSPX8 promoter region, and higher promoter activity was observed in transgenic nodules (Fig. 3C and Figure S3). Furthermore, overexpression of GmSPX8 in transgenic composite nodules increased nodule number, nodule fresh weight, nitrogenase activity and increased P content in nodules under low P condition, whereas suppression of GmSPX8 inversely decrease nodule number, nodule fresh weight, nitrogenase activity and decrease P content in nodules (Fig. 4). All these data provided that GmSPX8 played a vital role in nodulation and P homeostasis of nodules in soybean.
There are several studies on the molecular mechanism of SPX proteins involving in P signaling in plants [34]. OsSPX1 and OsSPX2 interacted with OsPHR2 to inhibit P starvation response in a P-dependent manner in planta [34]. OsbHLH6 interacted with OsSPX4 for Pi signaling and homeostasis in rice [36]. OsPTF1, belonging to bHLH family, was induced under low P condition in roots and its overexpression increased the tolerance to Pi starvation in rice [42]. Moreover, GmPTF1 responded to P starvation primarily through regulating the expression of GmEXPB2 in soybean [43]. In our study, GmPTF1 was isolated as a potential target of GmSPX8 from soybean nodules and overexpression of GmSPX8 increased the expression of GmPTF1 and its downstream PSI genes, such as GmEXPB2and GmEXLB1 (Table 1 and Fig. 6E-G). In addition, GmSPX8 could enhance the transcriptional activation activity of GmPTF1 in yeast cells and tobacco leaves (Fig. 6A-D). Taken together, these findings indicate that GmSPX8 responds to P starvation by regulating GmPTF1 and its downstream PSI genes to maintain P content for nodule development and BNF in soybean.
legumes require high energy such as ATP during nodulation and biological N2 fixation processes, thus P requirement was high, and also these processes were inhibited through a feedback loop formed by the release of free P in BNF, especially under excessive P conditions [12, 15, 44]. PTF1 could bind to the E-box (CANNTG) element in the promoter regions of regulated downstream genes in Arabidopsis, maize and soybean [43, 45, 46]. Here, we found that the E-box element presented in the promoter region of GmSPX8, and transcriptional activation assay confirmed the binding of GmPTF1 to the E-box of GmSPX8 (Fig. 6 A-D). All these data implied that GmPTF1 might regulate the expression of GmSPX8 to maintain the stabilization of P through a feedback regulation in soybean nodules under P starvation.
Conclusions
Our data showed that GmSPX8 is preferentially expressed in soybean nodules under P deficiency. The functional analysis of GmSPX8 in transgenic composite soybean plants demonstrated that GmSPX8 conferred to nodule development and nitrogen fixation under low P condition. GmSPX8 interacts with GmPTF1 in nodules and overexpression of GmSPX8 increased transcription accumulation of GmPTF1 and its downstream genes. All these findings show that GmSPX8 regulates nodule development and nitrogen fixation through its interaction with GmPTF1 in soybean under low P condition.
Materials and methods
Plant materials and growth conditions
Soybean (Glycine max (L.) Merr.) seeds used in this study are originally obtained from State Key Laboratory for North China Crop Improvement and Regulation, Hebei Agricultural University. A genotype of soybean Zhonghuang 15(ZH15) was used in this study for phenotypic and functional analysis. Soybean seeds were surface sterilized and germinated in Petri dishes with wet and sterile filter papers for three days under dark conditions in a growth chamber (28℃, 16/8 h light/dark photoperiod). One-week seedlings were inoculated with rhizobia strain Bradyrhizobium diazoefficiens USDA110, and planted into vermiculite watered with nitrogen-free nutrient solution containing 5 μM (P-deficient condition: LP) or 500 μM (P-sufficient condition: OP) of KH2PO4. Nodules at 28 days post inoculation (dpi) with rhizobia are mature and have high nitrogen fixation ability [47, 48]. Soybean plants and nodules were separately harvested at 28 dpi for measuring fresh weight, dry weight, height of shoot, total P and N content, nodule number, and nitrogenase activity.
For spatial expression analysis of selected genes responding to P supply and rhizobia inoculation, soybean seedlings were inoculated with Bradyrhizobium diazoefficiens USDA110, and then transplanted into vermiculite watered with nitrogen-free nutrient solution, which contained 5 μM (P-deficient condition: LP) or 500 μM (P-sufficient condition: OP) of KH2PO4. Shoots, leaves, roots and mature nodules were harvested separately at 28 days after inoculation. Nodules at different developmental stages were separately harvested at 10, 17, 28 day post inoculation. All tissues were frozen in liquid nitrogen and stored at -80℃ for further RNA extraction and qRT-PCR analysis.
RNA isolation and RNA-seq analysis
Nodule samples were collected from three independent biological replicates for different Pi treatment. Samples were ground in liquid nitrogen and subjected to total RNA extraction using Trizol reagent (Invitrogen, USA). mRNA was purified from total RNA using poly-T oligo-attached magnetic beads (TIANGEN BIOTECH). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-) (TIANGEN BIOTECH). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H (TIANGEN BIOTECH). These cDNA libraries were sequenced on an Illumina Novaseq6000 platform and 150 bp paired-end reads were generated. Feature Counts v1.5.0-p3 was used to count the read numbers mapped to each gene, and FPKM [49] of each gene was calculated based on the length of the gene and read counts mapped to this gene. Differential expression analysis was performed using the DESeq2 R package (1.16.1), which provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 were assigned as differentially expressed [49–51].
Quantitative real-time PCR (qRT-PCR)
For qRT-PCR, total mRNA was isolated and cDNA was synthesized accordingly. The specific primers for qRT-PCR are shown in Table S3. The qRT-PCR condition was: 30 s at 95℃, followed by 40 cycles of 5 s at 95℃, 15 s at 60℃ and 12 s at 72℃, and a final 5 s at 72℃. qRT-PCR was performed using SYBR Premix EX Tag™ (TaKaRa) on a CFX96™ real-time system (Bio-rad). The cycle threshold (CT) values of each sample were standardized using GmActin11 and the relative fold change (FC) of gene expression was calculated based on the 2−ΔΔCT method [52].
Construction of GmSPX8 overexpression and RNAi cassettes and soybean hairy root transformation
For overexpression construct, full-length ORF of GmSPX8 was cloned into pCAMBIA1390 under CaMV 35S promoter between BamHI and PstI enzyme sites. For RNAi constructs, about 200 bp fragment specific to GmSPX8 was cloned into pTCK-303 vector between BamHI and KpnI, SpeI and SacI, respectively, under CaMV 35S promoter [49]. Hairy root transformation through Agrobacterium rhizogenes strain K599 and determination of GUS positive roots were performed as previously described [50]. Transgenic composite plants were inoculated with Bradyrhizobium diazoefficiens USDA 110 and grown in different P conditions. Nodule samples were harvested at 28 day after rhizobium inoculation and analyzed [50, 51].
Acetylene reduction assay
Nitrogenase activity was measured by Acetylene Reduction Assay with the available protocol [53].
Measurement of N and P contents
Dried samples were ground and digested with HNO3 in a microwave oven. The resulting samples were subjected to themeasurement of N and P content. P content was measured by the color reaction of P-molybdate blue at the obsorbance of 700 nm, and N content was determined using semimicro-kjeldahl determination method in a nitrogen analyzer [23, 54].
Yeast two-hybrid assay
The ORF of GmSPX8 as bait was cloned into pGBKT7-BD vector and then transformed into the yeast strain Y2H Gold. Interacting proteins of GmSPX8 was screened from yeast cDNA library of soybean nodules using Matchmaker Gold Yeast Two-Hybrid System (Clontech, 630,489, USA) following the manufacturer’s instruction. pGBKT7-53 and pGADT7-T were used as positive control, pGBKT7-Lam and pGADT7-T were used as negative control. The Y2H assay was biologically repeated three times.
Bimolecular fluorescence complementation (BiFC) analysis
Full length CDS of GmSPX8 and GmPTF1 were cloned into vector p326YFPN and p326YFPC, respectively, to generate GmSPX8-YFPN, GmPTF1-YFPC, GmSPX8-YFPC and GmPTF1-YFPN [55]. The resulting constructs were then co-transformed into Arabidopsis protoplasts by polyethylene glycol (PEG)-mediated transformation as described previously [56]. YFP fluorescence was imaged using a confocal microscope.
Expression and purification of fusion proteins and in vitro pull-down assays
Full length CDS of GmPTF1 was cloned into pET-28a ( +) vector containing an His-tag in the amino-terminus, and GmSPX8 was cloned into a modified pET-28a ( +) vector with an additional Avi-tag at the C-terminal end (His-GmSPX8-Avi) [57]. The resulting constructs were introduced into Escherichia coli strain BL21 (DE3) (EMD Chemicals, Gibbstown, NJ) with or without birA (encoding biotin protein ligase) for biotinylation [58]. Recombinant proteins were induced by 0.5 mM isopropyl-β-d-thiogalactoside for 4 h at 28 °C, and GmSPX8 was purified by affinity chromatography using streptavidin agarose resin (Thermo Fisher Scientific, Waltham, MA). The pull-down assays were performed as described previously [35].
Construction of pGmSPX8-GUScassette, histochemical GUS staining and activity assay
The 1500-bp promoter fragment of GmSPX8 was cloned into the PcamG vector between SacI and SalI restriction enzyme sites to make pGmSPX8-GUS construct [59]. For GUS staining, transgenic soybean root nodules were incubated at 37 °C for 12 h in 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-Gluc) solution [60], and then the root tissues were washed with ethanol (70% v/v) before photographing. For GUS activity, total nodule proteins were extracted and incubated in a mixture containing 10 mM 4-methylumbelliferyl β-D-glucuronide (MUG; Sigma, USA) for 1 h at 37 °C. The fluorescence product of 4- methylumbelliferone (4-MU) was monitored using a Versa Fluor Fluorometer (Bio-Rad) with excitation at 365 nm and emission at 455 nm. The assay was repeated at least three times, and the data was calculated as the mean of independent experiments with the respective standard deviation.
Transcription activation assay in yeast cells
The Matchmaker Gold Yeast Two-Hybrid System (Clontech, Mountain View, CA, USA) was used to test GmPTF1 transcriptional activation activity. GmPTF1 and GmSPX8 were cloned into pGBKT7-BD and pGADT7-AD vector, respectively to generate pGBKT7-BD-GmPTF1 and pGADT7-AD-GmSPX8 constructs. pGBKT7-BD-GmPTF1 and pGADT7-AD-GmSPX8 or pGBKT7-BD-GmPTF1 alone were transformed into yeast strain AH109, and the yeast cells were grown in SD/-Trp-His + X-α-gal plates to assay the activation ability of GmPTF1 in yeast.
Transcriptional activity assays in tobacco
Full length ORF of GmSPX8 and GmPTF1 were cloned into the entry vector PGreenII62-SK (PSK-GmSPX8 and PSK-GmPTF1). A 186 bp length of promoter of GmSPX8 containing E-box (CAAATG) was fused to the LUC reporter gene on the PGreenII0800-LUC vector (E-box-LUC) [61]. The resulting constructs were introduced into A. tumefaciens bacteria GV3101 (Psoup-P19) and transiently transformed into the abaxial side of leaves of 4-week-old N. benthamiana plants. After 2 d of inoculation, the infiltrated leaves were harvested and sprayed with 1 mM D-luciferin. The fluorescence was detected after 10 min using a plant imaging system (Tanon-5200Multi; Tanon, Shanghai, China). The images were analyzed by imagej software (National institutes of Health, China).
Statistical methods
Statistical analyses were performed using SPSS 17.0 software (IBM, United States).
Supplementary Information
Additional file 1: Figure S1. (A) The volcano plot showing DEGs. (B) GOanalysis.
Additional file 2: Figure S2. Heatmap presentation of expression of candidategenes obtained from RNA-seq data under P-sufficient and P-deficient conditions.(A) Expression of selected genesfrom RNA-seq data analysis. (B) Validationof the expression from RNA-seq data by quantitative real-time PCR (qRT-PCR).OP: P sufficient condition. LP: P deficient condition.
Additional file 3: Figure S3. (A) Expression pattern of GmSPX8 intransgenic nodules harboring PGmSPX8-GUS construct. Transgeniccomposite soybean plants were grown in different P conditions and nodules wereharvested at 28 dpi for GUS staining. (B)GUS activity of transgenic nodules driven by the promoter of GmSPX8. Values aremeans of 10 independent lines for each P treatment. Bars showed the means ± SDvalues
Additional file 4: Table S1. Transcriptome data showing all the DEGs.
Additional file 5: Table S2. Cycle threshold (CT) Value of RT-PCR.
Additional file 6: Table S3. Primers used in this study.
Acknowledgements
We thank Prof. Dongcheng Liu (Hebei Agricultural University, China) for the constructive suggestions and helpful comments on the manuscript.
Abbreviations
- BNF
Biological Nitrogen Fixation
- ORF
Open Reading Frame
- cDNA
Complementary DNA
- GUS
β-Glucuronidase
- N
Nitrogen
- P
Phosphorus
- qPCR
Real-time quantitative PCR
- Trp
Trptophan; Leu: Leucine
- His
Histidine
- Ade
Adenine
- ABA
Aureobasidin A
- WT
Wild Type
- OX
Overexpression
- DPI
Days Post Inoculation
Authors’ contributions
CZ and HD conceived the project; XX conducted the experiments and analyzed the data; ZY carried out the BiFC assays; HD and XX wrote the manuscript; CZ revised the manuscript; WL, YK and XL provided suggestions during all the process of experiments. All authors have read and approved the manuscript.
Funding
This research was funded by the Project of Hebei Province Science and Technology Support Program (17927670H, 16227516D-1) and the Hebei Hundred-Talent Program (E2011100003). The funding bodies did not play any roles in experiment design, data analysis, and manuscript preparation.
Availability of data and materials
All the data used in this study are included in this published article and its additional files. The RNA-seq data can be found in the NCBI SRA database with the accession PRJNA739502. Our SRA record, https://www.ncbi.nlm.nih.gov/sra/PRJNA739502 could be accessible upon this publication in BMC Plant Biology.
Declarations
Ethics approval and consent to participate
All necessary permissions for planting and investigating this cultivar were obtained from Hebei Agricultural University, and the collection and research of this cultivar have complied with the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xinzhu Xing and Hui Du contributed equally to this work.
References
- 1.Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM. Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol. 2010;52(1):61–76. doi: 10.1111/j.1744-7909.2010.00899.x. [DOI] [PubMed] [Google Scholar]
- 3.Oldroyd GE. Plant science. Nodules and hormones Science. 2007;315(5808):52–53. doi: 10.1126/science.1137588. [DOI] [PubMed] [Google Scholar]
- 4.Marx H, Minogue CE, Jayaraman D, Richards AL, Kwiecien NW, Siahpirani AF, Rajasekar S, Maeda J, Garcia K, Del Valle-Echevarria AR, et al. A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat Biotechnol. 2016;34(11):1198–1205. doi: 10.1038/nbt.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan SL, Li R, Chen HF, Zhang CJ, Chen LM, Hao QN, Chen SL, Shan ZH, Yang ZL, Zhang XJ, et al. RNA-Seq analysis of nodule development at five different developmental stages of soybean (Glycine max) inoculated with Bradyrhizobium japonicum strain 113–2. Sci Rep. 2017;7:42248. doi: 10.1038/srep42248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udvardi M, Poole PS. Transport and Metabolism in Legume-Rhizobia Symbioses. Annu Rev Plant Biol. 2013;64(64):781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 7.Sutton MA, Oenema O, Erisman JW, Leip A, van Grinsven H, Winiwarter W. Too much of a good thing. Nature. 2011;472(7342):159–161. doi: 10.1038/472159a. [DOI] [PubMed] [Google Scholar]
- 8.Charpentier M, Oldroyd G. How close are we to nitrogen-fixing cereals? Curr Opin Plant Biol. 2010;13(5):556–564. doi: 10.1016/j.pbi.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Dong W, Zhu Y, Chang H, Wang C, Yang J, Shi J, Gao J, Yang W, Lan L, Wang Y, et al. An SHR-SCR module specifies legume cortical cell fate to enable nodulation. Nature . 2021;589(7843):586–90. [DOI] [PubMed]
- 10.Zhang B, Wang M, Sun Y, Zhao P, Liu C, Qing K, Hu X, Zhong Z, Cheng J, Wang H, et al. Glycine max NNL1 restricts symbiotic compatibility with widely distributed bradyrhizobia via root hair infection. Nat Plants. 2021;7(1):73–86. doi: 10.1038/s41477-020-00832-7. [DOI] [PubMed] [Google Scholar]
- 11.He C, Gao H, Wang H, Guo Y, He M, Peng Y, Wang X. GSK3-mediated stress signaling inhibits legume-rhizobium symbiosis by phosphorylating GmNSP1 in soybean. Mol Plant. 2021;14(3):488–502. doi: 10.1016/j.molp.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 12.Makoudi B, Kabbadj A, Mouradi M, Amenc L, Domergue O, Blair M, Drevon JJ, Ghoulam C. Phosphorus deficiency increases nodule phytase activity of faba bean–rhizobia symbiosis. Acta Physiol Plant. 2018;40(3):63. [Google Scholar]
- 13.Ferguson BJ, Mens C, Hastwell AH, Zhang MB, Su HA, Jones CH, Chu XT, Gresshoff PM. Legume nodulation: The host controls the party. Plant, Cell Environ. 2019;42(1):41–51. doi: 10.1111/pce.13348. [DOI] [PubMed] [Google Scholar]
- 14.Schulze J, Temple G, Temple SJ, Beschow H, Vance CP. Nitrogen fixation by white lupin under phosphorus deficiency. Ann Bot-London. 2006;98(4):731–740. doi: 10.1093/aob/mcl154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabeza RA, Liese R, Lingner A, von Stieglitz I, Neumann J, Salinas-Riester G, Pommerenke C, Dittert K, Schulze J. RNA-seq transcriptome profiling reveals that Medicago truncatula nodules acclimate N-2 fixation before emerging P deficiency reaches the nodules. J Exp Bot. 2014;65(20):6035–6048. doi: 10.1093/jxb/eru341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valentine AJ, Kleinert A, Benedito VA. Adaptive strategies for nitrogen metabolism in phosphate deficient legume nodules. Plant Sci. 2017;256:46–52. doi: 10.1016/j.plantsci.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez G, Valdes-Lopez O, Ramirez M, Goffard N, Weiller G, Aparicio-Fabre R, Fuentes SI, Erban A, Kopka J, Udvardi MK, et al. Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 2009;151(3):1221–1238. doi: 10.1104/pp.109.143842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv L, Yu K, Lu H, Zhang X, Liu X, Sun C, Xu H, Zhang J, He X, Zhang D. Transcriptome-wide identification of novel circular RNAs in soybean in response to low-phosphorus stress. PloS one. 2020;15(1):e0227243. doi: 10.1371/journal.pone.0227243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li CC, Zhou J, Wang XR, Liao H. A purple acid phosphatase, GmPAP33, participates in arbuscule degeneration during arbuscular mycorrhizal symbiosis in soybean. Plant, Cell Environ. 2019;42(6):2015–2027. doi: 10.1111/pce.13530. [DOI] [PubMed] [Google Scholar]
- 20.Tang C, Hinsinger P, Drevon JJ, Jaillard B. Phosphorus deficiency impairs early nodule functioning and enhances proton release in roots of Medicago truncatula L. Ann Bot-London. 2001;88(1):131–138. [Google Scholar]
- 21.Hogh-Jensen H, Schjoerring JK, Soussana JF. The influence of phosphorus deficiency on growth and nitrogen fixation of white clover plants. Ann Bot-London. 2002;90(6):745–753. doi: 10.1093/aob/mcf260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin L, Zhao J, Tian J, Chen L, Sun Z, Guo Y, Lu X, Gu M, Xu G, Liao H. The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 2012;159(4):1634–1643. doi: 10.1104/pp.112.199786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Yang Z, Kong Y, Li X, Li W, Du H, Zhang C. GmPAP12 Is Required for Nodule Development and Nitrogen Fixation Under Phosphorus Starvation in Soybean. Front Plant Sci. 2020;11:450. doi: 10.3389/fpls.2020.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiou TJ, Lin SI. Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol. 2011;62:185–206. doi: 10.1146/annurev-arplant-042110-103849. [DOI] [PubMed] [Google Scholar]
- 25.Xue Y, Zhuang Q, Zhu S, Xiao B, Liang C, Liao H, Tian J. Genome Wide Transcriptome Analysis Reveals Complex Regulatory Mechanisms Underlying Phosphate Homeostasis in Soybean Nodules. Int J Mol Sci. 2018;19(10):2924. doi: 10.3390/ijms19102924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 2012;193(4):842–851. doi: 10.1111/j.1469-8137.2011.04002.x. [DOI] [PubMed] [Google Scholar]
- 27.Yao Z, Tian J, Liao H. Comparative characterization of GmSPX members reveals that GmSPX3 is involved in phosphate homeostasis in soybean. Ann Bot. 2014;114(3):477–488. doi: 10.1093/aob/mcu147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamburger D, Rezzonico E, MacDonald-Comber Petetot J, Somerville C, Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 2002;14(4):889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Secco D, Baumann A, Poirier Y. Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol. 2010;152(3):1693–1704. doi: 10.1104/pp.109.149872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Huang W, Ying Y, Li S, Secco D, Tyerman S, Whelan J, Shou H. Functional characterization of the rice SPX-MFS family reveals a key role of OsSPX-MFS1 in controlling phosphate homeostasis in leaves. New Phytol. 2012;196(1):139–148. doi: 10.1111/j.1469-8137.2012.04227.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Zhou X, Xu Y, Yao M, Xie F, Gai J, Li Y, Yang S. Soybean SPX1 is an important component of the response to phosphate deficiency for phosphorus homeostasis. Plant Sci. 2016;248:82–91. doi: 10.1016/j.plantsci.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008;54(6):965–975. doi: 10.1111/j.1365-313X.2008.03460.x. [DOI] [PubMed] [Google Scholar]
- 33.Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodriguez J, et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA. 2014;111(41):14947–14952. doi: 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Ruan W, Shi J, Zhang L, Xiang D, Yang C, Li C, Wu Z, Liu Y, Yu Y, et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci USA. 2014;111(41):14953–14958. doi: 10.1073/pnas.1404680111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lv Q, Zhong Y, Wang Y, Wang Z, Zhang L, Shi J, Wu Z, Liu Y, Mao C, Yi K, et al. SPX4 Negatively Regulates Phosphate Signaling and Homeostasis through Its Interaction with PHR2 in Rice. Plant Cell. 2014;26(4):1586–1597. doi: 10.1105/tpc.114.123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Q, Lu H, Guo H, Wang Y, Zhao P, Li Y, Wang F, Xu J, Mo X, Mao C. OsbHLH6 interacts with OsSPX4 and regulates the phosphate starvation response in rice. Plant J. 2021;105(3):649–667. doi: 10.1111/tpj.15061. [DOI] [PubMed] [Google Scholar]
- 37.Li XH, Wu B, Kong YB, Zhang CY. GmPTF1, a novel transcription factor gene, is involved in conferring soybean tolerance to phosphate starvation. Genet Mol Res. 2014;13(1):926–937. doi: 10.4238/2014.February.19.3. [DOI] [PubMed] [Google Scholar]
- 38.Yang ZJ, Gao Z, Zhou HW, He Y, Liu YX, Lai YL, Zheng JK, Li XX, Liao H. GmPTF1 modifies root architecture responses to phosphate starvation primarily through regulating GmEXPB2 expression in soybean. Plant J. 2021;107(2):525–543. doi: 10.1111/tpj.15307. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Zhao J, Tan Z, Zeng R, Liao H. GmEXPB2, a Cell Wall beta-Expansin, Affects Soybean Nodulation through Modifying Root Architecture and Promoting Nodule Formation and Development. Plant Physiol. 2015;169(4):2640–2653. doi: 10.1104/pp.15.01029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Udvardi M, Poole PS. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol. 2013;64:781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 41.Nasr Esfahani M, Inoue K, Chu HD, Nguyen KH, Van Ha C, Watanabe Y, Burritt DJ, Herrera-Estrella L, Mochida K, Tran LP. Comparative transcriptome analysis of nodules of two Mesorhizobium-chickpea associations with differential symbiotic efficiency under phosphate deficiency. The Plant journal : for cell and molecular biology. 2017;91(5):911–26. [DOI] [PubMed]
- 42.Yi KK, Wu ZC, Zhou J, Du LM, Guo LB, Wu YR, Wu P. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 2005;138(4):2087–2096. doi: 10.1104/pp.105.063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, Gao Z, Zhou H, He Y, Liu Y, Lai Y, Zheng J, Li X, Liao H. GmPTF1 modifies root architecture responses to phosphate starvation primarily through regulating GmEXPB2 expression in soybean. Plant J. 2021;107(2):525–543. doi: 10.1111/tpj.15307. [DOI] [PubMed] [Google Scholar]
- 44.Vengavasi K, Pandey R, Abraham G, Yadav RK. Comparative Analysis of Soybean Root Proteome Reveals Molecular Basis of Differential Carboxylate Efflux under Low Phosphorus Stress. Genes. 2017;8(12):341. doi: 10.3390/genes8120341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z, Liu C, Zhang Y, Wang B, Ran Q, Zhang J. The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J Exp Bot. 2019;70(19):5471–5486. doi: 10.1093/jxb/erz307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(9):3582–3587. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng XG, Wang L, Wang H, Yu GH, Ba YL, Liu MM. Specific Expression of a Novel Nodulin GmN479 Gene in the Infected Cells of Soybean (Glycine max) Nodules. Agr Sci China. 2011;10(10):1512–1524. [Google Scholar]
- 48.Zhang S, Wang Y, Li K, Zou Y, Chen L, Li X. Identification of Cold-Responsive miRNAs and Their Target Genes in Nitrogen-Fixing Nodules of Soybean. Int J Mol Sci. 2014;15(8):13596–13614. doi: 10.3390/ijms150813596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du Y, He W, Deng C, Chen X, Gou L, Zhu F, Guo W, Zhang J, Wang T. Flowering-Related RING Protein 1 (FRRP1) Regulates Flowering Time and Yield Potential by Affecting Histone H2B Monoubiquitination in Rice (Oryza Sativa) PloS one. 2016;11(3):e0150458. doi: 10.1371/journal.pone.0150458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim YK, Kim S, Um JH, Kim K, Choi SK, Um BH, Kang SW, Kim JW, Takaichi S, Song SB, et al. Functional implication of beta-carotene hydroxylases in soybean nodulation. Plant Physiol. 2013;162(3):1420–1433. doi: 10.1104/pp.113.215020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li C, Li C, Zhang H, Liao H, Wang X. The purple acid phosphatase GmPAP21 enhances internal phosphorus utilization and possibly plays a role in symbiosis with rhizobia in soybean. Physiol Plant. 2017;159(2):215–227. doi: 10.1111/ppl.12524. [DOI] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Oh HS, Son O, Chun JY, Stacey G, Lee MS, Min KH, Song ES, Cheon CI. The Bradyrhizobium japonicum hsfA gene exhibits a unique developmental expression pattern in cowpea nodules. Molecular plant-microbe interactions : MPMI. 2001;14(11):1286–92. [DOI] [PubMed]
- 54.Grunwald U, Guo W, Fischer K, Isayenkov S, Ludwig-Muller J, Hause B, Yan X, Kuster H, Franken P. Overlapping expression patterns and differential transcript levels of phosphate transporter genes in arbuscular mycorrhizal, Pi-fertilised and phytohormone-treated Medicago truncatula roots. Planta. 2009;229(5):1023–1034. doi: 10.1007/s00425-008-0877-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du H, Kim S, Hur YS, Lee MS, Lee SH, Cheon CI. A Cytosolic Thioredoxin Acts as a Molecular Chaperone for Peroxisome Matrix Proteins as Well as Antioxidant in Peroxisome. Mol Cells. 2015;38(2):187–194. doi: 10.14348/molcells.2015.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 57.Du H, Kim S, Nam KH, Lee MS, Son O, Lee SH, Cheon CI. Identification of uricase as a potential target of plant thioredoxin: Implication in the regulation of nodule development. Biochem Biophys Res Commun. 2010;397(1):22–26. doi: 10.1016/j.bbrc.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 58.Tirat A, Freuler F, Stettler T, Mayr LM, Leder L. Evaluation of two novel tag-based labelling technologies for site-specific modification of proteins. Int J Biol Macromol. 2006;39(1–3):66–76. doi: 10.1016/j.ijbiomac.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 59.Kong Y, Wang B, Du H, Li W, Li X, Zhang C. GmEXLB1, a Soybean Expansin-Like B Gene, Alters Root Architecture to Improve Phosphorus Acquisition in Arabidopsis. Front Plant Sci. 2019;10:808. doi: 10.3389/fpls.2019.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong L, Zhou W, Wang H, Ding S, Lu Q, Wen X, Peng L, Zhang L, Lu C. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell. 2013;25(8):2925–2943. doi: 10.1105/tpc.113.111229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhai K, Deng Y, Liang D, Tang J, Liu J, Yan B, Yin X, Lin H, Chen F, Yang D, et al. RRM Transcription Factors Interact with NLRs and Regulate Broad-Spectrum Blast Resistance in Rice. Molecular cell. 2019;74(5):996–1009. doi: 10.1016/j.molcel.2019.03.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. (A) The volcano plot showing DEGs. (B) GOanalysis.
Additional file 2: Figure S2. Heatmap presentation of expression of candidategenes obtained from RNA-seq data under P-sufficient and P-deficient conditions.(A) Expression of selected genesfrom RNA-seq data analysis. (B) Validationof the expression from RNA-seq data by quantitative real-time PCR (qRT-PCR).OP: P sufficient condition. LP: P deficient condition.
Additional file 3: Figure S3. (A) Expression pattern of GmSPX8 intransgenic nodules harboring PGmSPX8-GUS construct. Transgeniccomposite soybean plants were grown in different P conditions and nodules wereharvested at 28 dpi for GUS staining. (B)GUS activity of transgenic nodules driven by the promoter of GmSPX8. Values aremeans of 10 independent lines for each P treatment. Bars showed the means ± SDvalues
Additional file 4: Table S1. Transcriptome data showing all the DEGs.
Additional file 5: Table S2. Cycle threshold (CT) Value of RT-PCR.
Additional file 6: Table S3. Primers used in this study.
Data Availability Statement
All the data used in this study are included in this published article and its additional files. The RNA-seq data can be found in the NCBI SRA database with the accession PRJNA739502. Our SRA record, https://www.ncbi.nlm.nih.gov/sra/PRJNA739502 could be accessible upon this publication in BMC Plant Biology.