Abstract
Bacterial biofilms show enormous levels of antibiotic resistance, but little is known about the underlying molecular mechanisms. Multidrug resistance pumps (MDRs) are responsible for the extrusion of chemically unrelated antimicrobials from the bacterial cell. Contribution of the MDR-mediated efflux to antibiotic resistance of Pseudomonas aeruginosa biofilms was examined by using strains overexpressing and lacking the MexAB-OprM pump. Resistance of P. aeruginosa biofilms to ofloxacin was dependent on the expression of MexAB-OprM but only in the low concentration range. Unexpectedly, biofilm resistance to ciprofloxacin, another substrate of MexAB-OprM, did not depend on the presence of this pump. Dose-dependent killing indicated the presence of a small “superresistant” cell fraction. This fraction was primarily responsible for very high resistance of P. aeruginosa biofilms to quinolones. Bacterial cells recovered from a biofilm and tested under nongrowing conditions with tobramycin exhibited higher resistance levels than planktonic cells but lower levels than cells of an intact biofilm.
Biofilms, communities of cells adhering to a substratum, likely represent the prevalent form of microorganisms in nature (7). Resistance to antimicrobials is a general feature of all biofilms that are the major cause of recalcitrant infections (8, 13, 26). Biofilms might be responsible for 65% of all bacterial infections (39). Life-threatening infection caused by Pseudomonas aeruginosa biofilms in cystic fibrosis patients is a well-known example (15). One obvious difference between planktonic cells and biofilm is the presence of a polysaccharide matrix enveloping the community that retards diffusion of antimicrobials into the biofilm. However, direct measurements of diffusion rates show that at least some antibiotics equilibrate throughout the biofilm within minutes or hours (9, 29, 41). P. aeruginosa expresses a β-lactamase, and a combination of retarded diffusion and an enzyme that destroys the antibiotic at the rate at which it arrives at the cell surface could explain resistance to ampicillin (41). However, this would not explain resistance to quinolones, for example. Retarded diffusion alone can postpone but not prevent the death of biofilm cells from quinolones. This realization has left the mechanism of biofilm resistance largely unexplained (6).
Do biofilm cells express mechanisms that contribute to biofilm resistance? Not much is known apart from an example of increased β-lactamase production in P. aeruginosa biofilms (14). The genetic analysis of biofilms has just recently begun, and its focus has been on genes required for biofilm formation. It has been found that pili and flagella are important for the early stages of biofilm formation in Pseudomonas (32, 33) and Escherichia coli (40). It was also reported that a quorum-sensing factor of P. aeruginosa is required for the formation of the biofilm architecture; in the absence of the factor, biofilms were thin and dense (10). This altered biofilm was solubilized by sodium dodecyl sulfate (SDS), unlike a wild-type biofilm. It remained unclear whether the quorum-sensing factor induced an increase in the intrinsic antimicrobial resistance of biofilm cells.
All known organisms have multidrug resistance pumps (MDRs) that can extrude chemically unrelated antimicrobials from the cell (20, 30, 34). In P. aeruginosa, the MexAB-OprM MDR is expressed in wild-type planktonic cells and is largely responsible for high levels of “intrinsic resistance” of this microorganism to antibiotics (21, 30, 37). Mutations leading to overexpression of the pump have been identified in clinical isolates of multidrug-resistant strains (6). The substrates of MexAB-OprM include quinolones, tetracycline, β-lactams, trimethoprim, and many other compounds. The pump extrudes antimicrobials across the outer membrane, which explains its ability to confer resistance to β-lactams that target the cell wall synthesis. The pump is composed of three different peptides: a MexB translocase belonging to the resistance-nodulation-division (RND) family of solute/proton antiporters, an outer membrane porin, OprM; and a “membrane fusion protein,” MexA, that apparently docks MexB to OprM. The broad specificity of MDRs seems to qualitatively match the broad resistance of biofilms to antimicrobials. However, biofilms can show very high levels of resistance (for example, a minimal bactericidal concentration of 1 mg/ml in the case of P. aeruginosa and tobramycin), and it is unclear whether mechanisms operating in planktonic cells that confer significantly lower levels of resistance play a role in biofilms. Nothing is currently known with regard to the role of MDRs in biofilm resistance to antibiotics.
The contribution of MDR-mediated efflux to biofilm antibiotic resistance was examined in this study. A detailed examination of dose-response killing of biofilm cells expressing or lacking MexAB-OprM led to a number of unexpected findings.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains used in this study were K767 PAO1 prototroph (25), K1119 PAO1 ΔmexAB-oprM (22), K766 PAO1 nalB mexAB-oprM++, PAO-JP1 PAO1 ΔlasI Tcr (35), and PA14 pilB (31). Mueller-Hinton broth (MHB) (Difco, Detroit, Mich.) was used to culture P. aeruginosa unless otherwise noted.
Antibiotics.
Ciprofloxacin and ofloxacin were obtained from Bayer and the R.W. Johnson Research Institute, respectively. Tetracycline and tobramycin were from Sigma, St. Louis, Mo.
Susceptibility testing of planktonic and biofilm cells.
Planktonic stationary-phase cells were used in dose-response killing experiments, for a better comparison to biofilms. The minimal bactericidal concentration for planktonic cells was defined as an antimicrobial concentration required for complete eradication of the cells during a 6-h incubation in MHB. Complete eradication was recorded if one or no colonies were present on triplicate plates inoculated with undiluted culture. This point was plotted as 100/ml on the dose-response killing curves. Both modifications were implemented to allow a better comparison with bacterial biofilms. Phosphate-buffered saline (PBS), pH 7.4 (Sigma), was used to determine planktonic cell resistance under nongrowing conditions.
The disposable MBEC device was used to form biofilms (MBEC Biofilm Technologies Ltd., Calgary, Canada) (5). The device is a platform carrying 96 polystyrene pegs that fit in a microtiter plate. For biofilm formation, the device was placed in a tray filled with MHB and cells (104/ml) and was incubated on a tilting shaker, which provides a shearing force, for 16 h at 37°C. After biofilms formed on the pegs, the pegs were washed in PBS, and the device was placed in a microtiter plate with MHB for drug susceptibility testing. Following 6 h of incubation in the presence of an antimicrobial agent, the pegs were washed twice in PBS, and the device was placed in a microtiter plate with PBS and sonicated for 5 min in a water bath sonicator (Branson ultrasonic cleaner; Branson Ultrasonics Corporation, Danbury, Conn.). For each antimicrobial concentration tested, cells were collected from three parallel pegs, plated with appropriate dilutions, and counted separately, and the mean values are presented.
Dose-dependent killing of cells recovered from a biofilm.
Biofilm formation was carried out as described above, and cells were dislodged from pegs by sonication into MHB containing a given amount of antibiotic. Following a 6-h incubation with antibiotic, the number of live cells was determined by plate counts.
RESULTS
In this study, we used the recently developed Calgary device for biofilm susceptibility testing. The device allows us to test a fairly large number of samples simultaneously, enabling us for the first time to collect detailed data on the dose response of P. aeruginosa biofilms to several antibiotics. We have intentionally chosen two strains of P. aeruginosa for this study that have a fixed status of MexAB-OprM expression, an overproducing mutant and a deletion strain. This would limit the number of uncontrollable variants and clearly show a potential role for an MDR in biofilm resistance. A set of representative antibiotics was selected for this study. Of these, ofloxacin, ciprofloxacin, and tetracycline are substrates of the MexAB-OprM pump. MDRs primarily extrude amphipathic substances (20), but recently RND-type MDRs extruding hydrophilic aminoglycosides were found in Burkholderia pseudomallei (27) and P. aeruginosa (2).
MexAB-OprM and biofilm resistance.
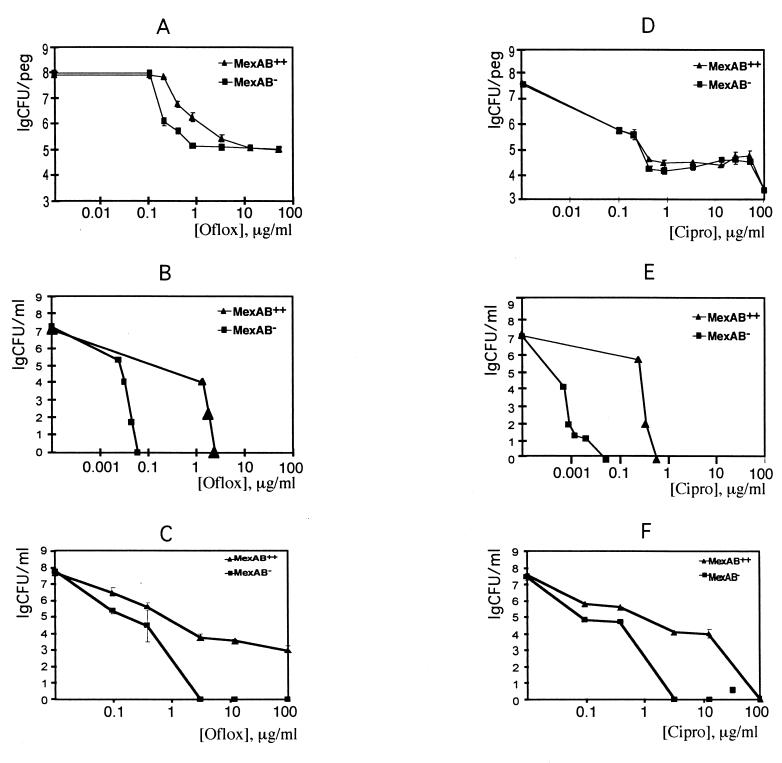
P. aeruginosa biofilms were formed overnight and treated with antimicrobials for 6 h. Cells were then dislodged from the plastic pegs by mild sonication, and the number of live cells was determined by plate counts. MexAB-OprM conferred a distinct protective effect on biofilms exposed to ofloxacin (Fig. 1). At low concentrations, ofloxacin was 50- to 100-fold more effective in killing biofilm cells of the deletion mutant than the strain overexpressing the pump. Unexpectedly, as the concentration of ofloxacin increased, the difference in resistance between the two strains gradually diminished. After the number of live cells dropped by 3 orders of magnitude, the remaining fraction, consisting of about 105 cells, did not decline upon further increase in antibiotic concentration. There was therefore no difference in resistance to ofloxacin between these “superresistant” cells of the two strains. By contrast, the rate of killing of planktonic cells in growth medium by ofloxacin showed a rapid increase upon an increase in the antibiotic concentration, and a large difference in resistance between the MexAB-OprM expressing and deletion strains was observed throughout the entire concentration range. Interestingly, the majority of biofilm cells (∼99.9%) were eradicated with a clinically achievable antibiotic concentration (1 to 5 μg/ml), and the MexAB-OprM pump contributed to resistance at this concentration range.
FIG. 1.
Role of the MexAB-OprM pump in resistance of biofilms and planktonic cells to killing by antibiotics. (A through C) Biofilms, stationary-phase cells, and nongrowing cells, respectively, treated with ofloxacin (Oflox). (D through F) Biofilms, stationary-phase cells, and nongrowing cells, respectively, treated with ciprofloxacin (Cipro). (G through I) Biofilms, stationary-phase cells, and nongrowing cells, respectively, treated with tetracycline (Tet). (J through L) Biofilms, stationary-phase cells, and nongrowing cells, respectively, treated with tobramycin (Tobra).
In the case of ciprofloxacin, there was no difference in biofilm resistance between cells expressing and lacking MexAB-OprM (Fig. 1). A large difference in resistance between these strains was observed as expected with planktonic cells (Fig. 1). The ability of MexAB-OprM to confer resistance to ofloxacin shows that the pump is indeed active in the biofilm. It appears that other drug resistance mechanisms operating in the biofilm mask the contribution of the MexAB-OprM pump to ciprofloxacin resistance. As in the case of ofloxacin, a distinct plateau in the dose-response curve was observed with ciprofloxacin, suggesting the presence of superresistant cells.
We found that tetracycline, a poorly lethal antibiotic, showed good killing activity against planktonic cells of the MexAB-OprM deletion strain (about a 3-log decrease at 0.5 μg/ml) and poor killing of the overexpression strain (Fig. 1). It was interesting to learn whether the very prominent difference between strain susceptibility to tetracycline would be observed in the biofilm. This was not the case; the results showed a dramatic increase in the MexAB-OprM deletion strain's resistance to tetracycline in a biofilm. A difference in strain susceptibility was observed only at the very high range of concentrations (Fig. 1).
Tobramycin is apparently not a substrate of the MexAB-OprM pump (or a very poor one), and in planktonic cells the efficient killing with this antibiotic was similar for both strains. Complete eradication was achieved at around 2 μg/ml (Fig. 1). The biofilms showed very high resistance to tobramycin, and the strains behaved similarly. At a very high concentration (1 mg/ml), the deletion mutant was more sensitive than the overproducing strain.
Contribution of slow growth to antibiotic resistance.
The dramatic increase in resistance of the deletion MexAB-OprM strain to tetracycline upon transition from the planktonic to the biofilm state could be due largely to slow growth. Bactericidal action generally decreases with lower growth rate, and slow growth is considered to be one of the components of biofilm resistance (3, 8). We wanted to learn how slow growth contributed to resistance of P. aeruginosa to antibiotics used in this study.
For these model experiments, we chose conditions that would provide minimal or no growth. Dose-response killing of stationary-phase planktonic cells was studied in simple phosphate buffer (Fig. 1). Almost identical killing dynamics were observed for planktonic cells in phosphate buffer and for biofilm cells in growth medium in the case of tetracycline. Thus, a decrease in growth rate could largely explain biofilm resistance to tetracycline, a poorly lethal antibiotic. By contrast, cells in phosphate buffer were very sensitive to tobramycin. Biofilm cells exhibited approximately 100-fold-higher resistance than planktonic cells in phosphate buffer. Thus, another unknown mechanism(s) is responsible for high resistance of P. aeruginosa to aminoglycosides. Slow growth evidently increased resistance of planktonic cells to quinolones. Very high resistance to quinolones in phosphate buffer was observed with the strain overexpressing MexAB-OprM. This suggests that slow growth and MDRs might contribute to ofloxacin resistance of biofilms.
Antibiotic resistance of bacterial cells recovered from a biofilm.
Whether increased resistance derives primarily from the biofilm structure (decreased diffusion or binding of antibiotics) or is an intrinsic property of biofilm cells remains an important question. What has been firmly established is that cells derived from the biofilm are not mutants. After regrowth, planktonic cells from a biofilm are similar to cells of the parent strain. We confirmed this by examining the sensitivity of cells taken from biofilms after antibiotic treatment and regrown in rich medium (data not shown). What has also been established is that MICs for cells taken from the biofilm and immediately exposed to antibiotic are the same as MICs for the wild type (42). This is to be expected: once a potentially resistant cell starts growing in the presence of an antibiotic, resistant mechanisms will be “diluted” after several divisions, and the sample will be scored as having no growth. There seemed to be a simple way to probe the issue of intrinsic resistance of biofilm cells. The rationale is to test the ability of cells to resist killing rather than to study their ability to grow. Biofilm cells can be dislodged, and their dose-response killing can be studied. In this format, there is no growth due to the presence of high concentrations of antibiotic, and resistance mechanisms would not be “diluted.” However, disruption of a biofilm and transfer of cells into fresh growth medium could change the cell properties (activate them from possible dormancy, for example) and lead to susceptibility. This suggests that only a positive result (preservation of increased resistance of liberated cells) would provide useful information. According to our results with planktonic cells in buffer, only tobramycin produced effective killing. Using this antibiotic would therefore allow us to observe possible resistance of liberated planktonic cells that was not due to slow growth alone.
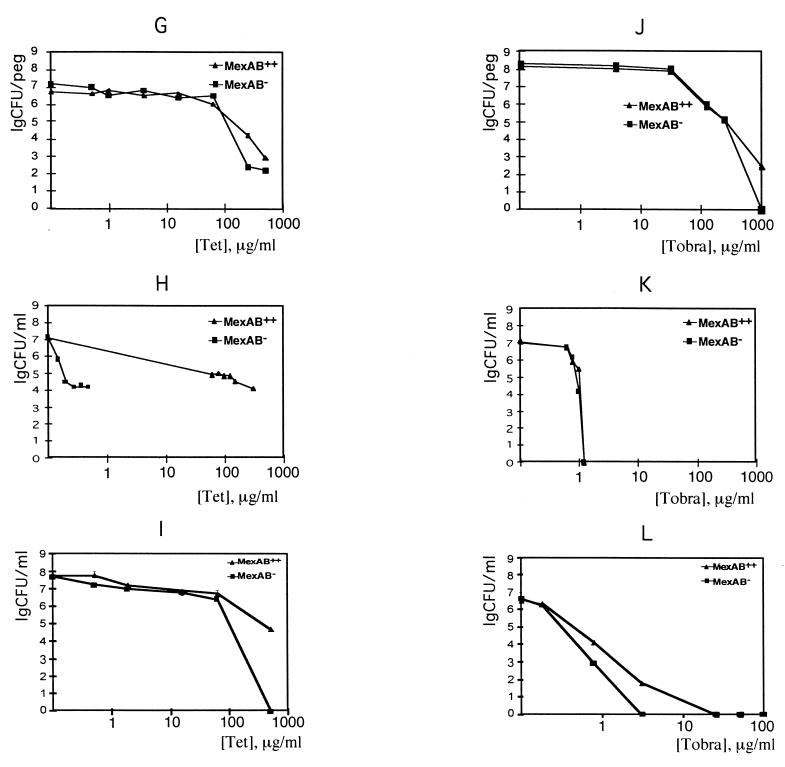
Resistance of liberated cells to tobramycin was substantially greater than that of planktonic cells in PBS and unexpectedly was similar to resistance of biofilms in growth medium (Fig. 2). When cells are liberated from the biofilm by sonication, one would expect the biofilm matrix to be liberated as well. This matrix could bind some of the tobramycin, contributing to higher resistance of liberated cells. We examined this possibility by studying dose-response killing of liberated cells compared to killing in a 10-fold- and a 150-fold-diluted suspension of liberated cells. The decline of the cell number with an increase in tobramycin was more pronounced in the suspension diluted 10-fold than in undiluted cells, suggesting that the undiluted suspension might contain a factor protecting cells from the antibiotic. The patterns of decline in cell counts in a 10-fold- and 150-fold-diluted suspensions were comparable. This means that in diluted cells, only intrinsic resistance of cells determines the pattern of killing. Even the diluted biofilm cells were substantially more resistant than stationary-phase planktonic cells, as judged both by population decline (Fig. 1K) and by the concentrations required for eradication. This experiment suggests that resistance to tobramycin is due in part to intrinsic resistance that is acquired by cells growing in a biofilm.
FIG. 2.
Dose-response killing of P. aeruginosa wild-type cells liberated from a biofilm. Cells were dislodged from a biofilm by sonication, placed in Mueller-Hinton growth medium, and treated with tobramycin (Tobra) (diamonds). Similarly prepared cells were diluted prior to tobramycin treatment 10-fold (squares) and 150-fold (triangles).
Antibiotic resistance of a biofilm deficient in quorum-sensing factor.
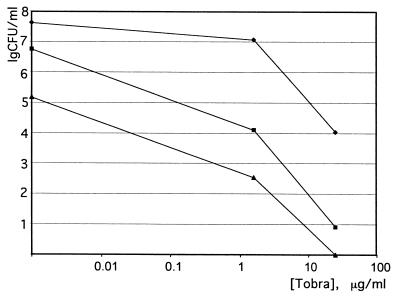
Recently, it was shown that a P. aeruginosa strain deficient in the production of N-(3-oxododecanoyl)-l-homoserine lactone (HSL) quorum-sensing factor forms a dense biofilm that is dislodged from the surface by 0.2% SDS, unlike the wild-type biofilm (10). It was also reported that MexAB-OprM extrudes HSL (11, 36), raising the question of the interrelation between HSL, MexAB-OprM, and antibiotic resistance. In order to probe directly for a possible role of HSL in biofilm resistance to antibiotics, we used the lasI mutant strain defective in HSL production. We found no difference in susceptibilities of the biofilms formed by the lasI strain and the wild type in resistance to ofloxacin and tobramycin (Fig. 3) or in resistance to SDS (not shown). In order to verify that the method we are using allows us to distinguish between cells with different capabilities to form biofilms, a pilB strain defective in pilus synthesis and biofilm formation (32) was tested. Biofilms formed by the pilB strain had 100-fold fewer cells than the wild-type PAO1 or the K766 and K1119 strains (data not shown). The apparent discrepancy between our results and the above-cited study is probably due to the difference in the substratum used. The surface material has a strong effect on biofilm adherence (26). We employed polystyrene, which the MBEC device is made of, while glass was used in the other study (10).
FIG. 3.
Killing of P. aeruginosa biofilms by ofloxacin (Oflx) and tobramycin (Tobra). Conditions were as described for Fig. 1. PAO-JP1 is a lasI mutant defective in the production of HSL, and PAO1 is the wild-type parent strain.
DISCUSSION
Role of MDRs.
Since MDRs play a significant role in multidrug resistance of planktonic cells, it might seem obvious that they should have a role in biofilm resistance as well. However, our results show that this is not necessarily the case. There was no difference in biofilm resistance to ciprofloxacin between cells overexpressing the MexAB-OprM pump and those lacking it. On the other hand, a notable (50- to 100-fold) difference in susceptibility between the strains was observed in the case of ofloxacin at a relatively low concentration range. These differences do not follow from the very similar and large differences between the susceptibilities of two strains to these antibiotics in planktonic cells. One interesting possibility that would account for the unexpected lack of a role for MexAB-OprM in biofilm resistance to ciprofloxacin is that another MDR(s), perhaps with preferential selectivity for ciprofloxacin, is being expressed in the biofilm. Many MDRs are regulated by environmental factors: EmrAB of E. coli is induced by its drug substrates (4, 23), as are BMR of B. subtilis (1) and QacA of S. aureus (16). In E. coli, the RND pump AcrAB is induced by stress (24, 31). Two of these RND pumps, MexCD-OprJ (38) and MexEF-OprN (19), have substrate spectra similar to that of MexAB-OprM, and mutants overexpressing these pumps have been identified in resistant clinical isolates (12, 18, 43). MexCD-OprJ and MexEF-OprN are not expressed in planktonic cells. There are 10 additional putative RND pumps in the genome of P. aeruginosa (Pseudomonas Genome Project, http://www.pseudomonas.com). Examination of mutants with disruptions in the MDRs will provide a definitive answer as to whether and which MDRs have an important role in biofilm resistance.
Intrinsic resistance of biofilm cells.
Cells liberated from a biofilm into growth medium were considerably more resistant to tobramycin than planktonic cells. This experiment suggests that cells become intrinsically more resistant when growing in the biofilm and retain part of this resistance even outside the biofilm. This is the first observation that shows increased resistance of cells liberated from the biofilm and provides the bases for searches for genes that might be specifically induced in and responsible for biofilm resistance to antibiotics.
Superresistant cells.
The detailed dose-response killing examination of biofilms reported in this study allowed us to detect the presence of a subpopulation of cells that show little sensitivity to quinolones and are responsible for a characteristic “plateau” on the killing plot. Not much has been published on dose-response killing of microbial biofilms due to the lack of a convenient susceptibility test, but the few existing studies report a pattern of killing that is very similar to our observations. In E. coli, increasing concentrations of ciprofloxacin or imipenem caused an initial decrease in live cells of a biofilm by 2 to 3 orders of magnitude, while the remaining small population was essentially insensitive to a further increase in drug concentration (3). This pattern was also observed with amoxicillin and clindamycin in Lactobacillus acidophilus, and with erythromycin and metronidazole in the case of Gardnerella vaginalis biofilms, where initial rapid killing was followed by a plateau of resistant cells (28). To our knowledge, the phenomenon of superresistant cells in the biofilm has not been discussed. With a number of other antibiotics, the initial killing was more gradual due to high resistance of the bulk of the cells, and no plateau was observed. This is to be expected—a superresistant fraction will show up only if the majority of cells are distinctly more sensitive. It appears that four different microorganisms tested with a number of unrelated drugs behave very similarly, showing a rapid decline of the bulk of the population in response to an increase in a potent antibiotic and a resistant subpopulation. It thus seems that the biofilm employs a dual strategy for survival—expression of energy-dependent resistance mechanisms like MDRs in the active cells that make up the majority of the population, and (possibly) passive resistance of a superresistant subpopulation. Superresistant cells are ultimately responsible for the very high levels of biofilm resistance, at least in the cases discussed above. Unlike planktonic cells, where a small remaining population would be eliminated by the immune system, biofilm cells are protected by the matrix (17), and remaining superresistant cells will be responsible for biofilm regrowth after treatment with an antibiotic. Elucidating the nature of these superresistant cells is a formidable challenge, since they make up only a small part of the total population. However, their importance in biofilm survival will undoubtedly drive the effort to understand the mechanisms of their remarkable resistance.
ACKNOWLEDGMENTS
We thank M. Mittelman for helpful discussions and H. Ceri for advice on biofilm cultivation. K. Poole, B. H. Iglewski, R. Kolter, and G. A. O'Toole are gratefully acknowledged for the kind gift of bacterial strains.
This research was supported by NIH grant GM54412-01.
ADDENDUM
After this paper was submitted, a study by Aires and coauthors (2) reported that OprM is part of another MDR, MexXY, that extrudes aminoglycosides. We did not find significant differences in killing by tobramycin between cells expressing and lacking OprM; one substantial difference between our conditions and those of Aires et al. was that large amounts of divalent metals were included in the media in the latter study, which could have affected OprM and/or MexXY.
REFERENCES
- 1.Ahmed M, Borsch C M, Taylor S S, Vazquez-Laslop N, Neyfakh A A. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J Biol Chem. 1994;269:28506–28513. [PubMed] [Google Scholar]
- 2.Aires J R, Kohler T, Nikaido H, Plesiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashby M J, Neale J E, Knott S J, Critchley I A. Effect of antibiotics on non-growing planktonic cells and biofilms of Escherichia coli. J Antimicrob Chemother. 1994;33:443–452. doi: 10.1093/jac/33.3.443. [DOI] [PubMed] [Google Scholar]
- 4.Brooun A, Tomashek J J, Lewis K. Purification and ligand binding of EmrR, a regulator of a multidrug transporter. J Bacteriol. 1999;181:5131–5133. doi: 10.1128/jb.181.16.5131-5133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceri H, Olson M E, Stremick C, Read R R, Morck D, Buret A. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H Y, Yuan M, Livermore D M. Mechanisms of resistance to beta-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J Med Microbiol. 1995;43:300–309. doi: 10.1099/00222615-43-4-300. [DOI] [PubMed] [Google Scholar]
- 7.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 8.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 9.Darouiche R O, Dhir A, Miller A J, Landon G C, Raad I I, Musher D M. Vancomycin penetration into biofilm covering infected prostheses and effect on bacteria. J Infect Dis. 1994;170:720–723. doi: 10.1093/infdis/170.3.720. [DOI] [PubMed] [Google Scholar]
- 10.Davies G D, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 11.Evans K, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukuda H, Hosaka M, Iyobe S, Gotoh N, Nishino T, Hirai K. nfsC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:790–792. doi: 10.1128/AAC.39.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gander S. Bacterial biofilms: resistance to antimicrobial agents. J Antimicrob Chemother. 1996;37:1047–1050. doi: 10.1093/jac/37.6.1047. [DOI] [PubMed] [Google Scholar]
- 14.Giwercman B, Jensen E T T, Hoiby N, Kharazmi A, Costerton J W. Induction of β-lactamase production in Pseudomonas aeruginosa biofilm. Antimicrob Agents Chemother. 1991;35:1008–1010. doi: 10.1128/aac.35.5.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grkovic S, Brown M H, Roberts N J, Paulsen I T, Skurray R A. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J Biol Chem. 1998;273:18665–18673. doi: 10.1074/jbc.273.29.18665. [DOI] [PubMed] [Google Scholar]
- 17.Hoyle B D, Jass J, Costerton J W. The biofilm glycocalyx as a resistance factor. J Antimicrob Chemother. 1990;26:1–5. doi: 10.1093/jac/26.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Jakics E B, Iyobe S, Hirai K, Fukuda H, Hashimoto H. Occurrence of the nfxB type mutation in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:2562–2565. doi: 10.1128/aac.36.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 20.Lewis K. Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem Sci. 1994;19:119–123. doi: 10.1016/0968-0004(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 21.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X Z, Zhang L, Srikumar R, Poole K. Beta-lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:399–403. doi: 10.1128/aac.42.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomovskaya O, Lewis K, Matin A. EmrR is a negative upstream regulator of the E. coli multidrug resistance pump EmrA. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma D, Alberti M, Lynch C, Nikaido H, Hearst J E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 25.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mittelman M W. Adhesion to biomaterials. In: Fletcher M, editor. The molecular and ecological diversity of bacterial adhesion. New York, N.Y: Wiley; 1997. pp. 89–127. [Google Scholar]
- 27.Moore R A, DeShazer D, Reckseidler S, Weissman A, Woods D E. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muli F W, Struthers J K. The growth of Gardnerella vaginalis and Lactobacillus acidophilus in Sorbarod biofilms. J Med Microbiol. 1998;47:401–405. doi: 10.1099/00222615-47-5-401. [DOI] [PubMed] [Google Scholar]
- 29.Nichols W W, Evans M J, Slack M P E, Walmsley H L. The penetration of antibiotics into aggregates of mucoid and non-mucoid Pseudomonas aeruginosa. J Gen Microbiol. 1989;135:1291–1303. doi: 10.1099/00221287-135-5-1291. [DOI] [PubMed] [Google Scholar]
- 30.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 34.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson J P, Pesci E C, Iglewski B H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson J P, Van Delden C, Iglewski B H. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poole K. Bacterial multidrug resistance—emphasis on efflux mechanisms and Pseudomonas aeruginosa. J Antimicrob Chemother. 1994;34:453–456. doi: 10.1093/jac/34.4.453. [DOI] [PubMed] [Google Scholar]
- 38.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 39.Potera C. Forging a link between biofilms and disease. Science. 1999;283:1837–1839. doi: 10.1126/science.283.5409.1837. [DOI] [PubMed] [Google Scholar]
- 40.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 41.Stewart P S. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother. 1996;40:2517–2522. doi: 10.1128/aac.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright T L, Ellen R P, Lacroix J M, Sinnadurai S, Mittelman M W. Effects of metronidazole on Porphyromonas gingivalis biofilm. J Periodont Res. 1997;32:473–477. doi: 10.1111/j.1600-0765.1997.tb00560.x. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida T, Muratani T, Iyobe S, Mitsuhashi S. Mechanisms of high-level resistance to quinolones in urinary tract isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1994;38:1466–1469. doi: 10.1128/aac.38.7.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]