Abstract
The tumor suppressive role of CYLD lysine 63 deubiquitinase (CYLD) is known in melanoma. To the best of our knowledge, however, the precise mechanism underlying the tumor suppressive function of CYLD has yet to be clarified. In the present study, a novel melanoma mouse model was generated, which revealed accelerated tumor growth in Cyld-knockout (Cyld−/−) compared with Cyld-wild-type (Cyld+/+) mice. To determine the underlying molecular mechanism, mutation analysis of primary tumor-derived cell lines from Cyld+/+ and Cyld−/− mice was performed using RNA sequencing data. Variant calling revealed no common mutations in Cyld−/− compared with Cyld+/+ cells. Thus, the epigenetic processes influencing development and progression of melanoma were investigated. Initial analysis of expression pattern of known hypermethylated genes in melanoma (suppressor of cytokine signalling, methylthioadenosine phosphorylase, cadherin 1) in the presence or absence of 5′-Aza-deoxyctidine treatment revealed that CYLD does not play a key role in DNA methylation. Chromatin accessibility and histone H3 modification assay uncovered a role of CYLD in the formation of chromatin structure. Subsequent inhibitor experiments confirmed the effect of CYLD on H3K9me2 level associated with heterochromatin. Furthermore, enhanced H3K9 dimethylation in Cyld−/− melanoma cells was associated with upregulation of euchromatic histone lysine methyltransferase 2 (EHMT2). Moreover, the specific inhibitor of EHMT2, CM272, resulted in decreased proliferation and relaxation of compact chromatin in Cyld-deficient melanoma cells. These results reveal a novel role of CYLD in histone methylation and chromatin packaging.
Keywords: melanoma, CYLD lysine 63 deubiquitinase, epigenetic, chromatin accessibility, H3K9me2, euchromatic histone lysine methyltransferase 2
Introduction
The deubiquitinase CYLD lysine 63 deubiquitinase (CYLD) was first described in cylindromatosis (1) and its tumor suppressive properties have been investigated in various types of cancer, such as pancreas, breast and liver cancer (2-4). In malignant melanoma, the expression of CYLD is downregulated by elevated expression of the transcription factor SNAIL1, which results in increased proliferative and migratory potential of melanoma cells (5,6). Recently, our group generated a novel transgenic (Tg) melanoma mouse model, Tg(Grm1) Cyld, showing enhanced tumor development and growth in Cyld-knockdown (Cyld−/−) mice compared with Cyld-wild type (Cyld+/+) mice (7). Our previous study described a novel regulatory role of CYLD in vasculogenic mimicry and lymph- and angiogenesis (7). To the best of our knowledge, however, the underlying mechanisms resulting in accelerated tumor growth in Cyld−/− mice have not yet been determined.
Besides transcriptomic changes, epigenetic dysregulation, including DNA methylation and post-translational modification of histones, has been shown to be associated with cancer in a number of studies (8,9). These changes lead to organization of DNA into chromatin and ensure genomic integrity (10). Since epigenetic processes are reversible, they are currently the focus of research for novel therapeutic approaches to circumvent drug resistance in cancer (11,12). The present study aimed to determine the underlying mechanism of accelerated melanoma development and the role of CYLD in epigenetic processes of chromatin formation and histone methylation.
Materials and methods
Murine melanoma cell lines
No animals were sacrificed as murine melanoma cell lines generated in 2019 were used (7). In brief, tissue samples from primary tumors (ear and tail) from two Tg(Grm1)Cyld−/− and two Tg(Grm1)Cyld+/+ male mice (age, 153 and 217 days) were collected and washed with Braunol® (7.5%; B Braun Meisungen AG), followed by 1X PBS, 70% ethanol and 1X PBS. Tumor tissue was added to a mixture of DMEM (Sigma-Aldrich; Merck KGaA) and collagenase. Following incubation for 3 h at 37°C, the cell suspension was centrifuged (4 min at 600 g, room temperature) and the cells were seeded in T25 flasks, as previously described (7,13).
Cell culture
Murine melanoma cell lines (mCyld+/+: EPv24 and EPv40 ear; mCyld−/−: EC36 and EC111 ear) derived from tumor tissue of Tg(Grm1) model animals (13) were cultivated in DMEM (4,500 mg glucose/l, 110 mg sodium pyruvate/l and l-glutamine) with 10% fetal bovine serum, amphotericin B (2.5 μg/ml) and 5% penicillin/streptomycin (all Sigma-Aldrich; Merck KGaA) in a humidified atmosphere containing 8% CO2 at 37°C. Cells were reseeded at a ratio 1:3 to 1:5 twice weekly.
Human primary melanoma cell line Mel Juso [provided by Dr Judith Johnson (Ludwig Maximilian University of Munich, Munich, Germany), stably transfected with control green fluorescent protein [human (h)CYLD−], CYLD (hCYLD+) or CYLDC/S (catalytically inactive mutant of CYLD) vector (5,6) were cultivated (8% CO2 at 37°C) in RPMI-1640 (Sigma-Aldrich; Merck KGaA) with NaHCO3 and the aforementioned supplements. For cell counting the light microscope AE2000 from Motic Incorporation Ltd. was used (×4 or ×20 magnification).
For inhibitor studies, murine and human melanoma cells were treated with Chaetocin (inhibitor of histone methyltransferase SUV39H1; 10 nM; Absource Diagnostics GmbH), the Jumonji histone demethylase inhibitor JIB04 (500 nM; Sigma-Aldrich; Merck KGaA) and CM272 [dual EHMT2/DNA methyltransferase (DNMTs) inhibitor; 200 nM; kindly provided by Matías A Ávila (14)] for 24 h at 37°C.
For simplification, the following nomenclature has been used in the rest of the text: murine Cyld-expressing (mCyld+/+) and Cyld-deficient (mCyld−/−) cells; human CYLD-expressing (hCYLD+) and CYLD-deficient (hCYLD−) and CYLD mutant (hCYLDC/S) cells.
RNA isolation, reverse transcription-quantitative (RT-q) PCR
Total RNA and cDNA of the murine and human melanoma cells were generated as previously described (7). RT-qPCR was performed with LightCycler 480 system (Roche Diagnostics GmbH) to analyze mRNA expression, as previously described (15). For each reaction, 1.0 cDNA template, 0.5 forward and reversed primers each (20 μM each) and 10.0 μl SYBR Green I (Roche Diagnostics GmbH) were used in a total volume of 20 μl. The primers were as follows: β-actin forward, 5′-TGG AAT CCT GTG GCA TCC ATG AAA C-3′ and reverse, 5′-TAA AAC GCA GCT CAG TAA CAG TCC G-3′; cadherin (CDH)1 forward, 5′-ACG TAT CAG GGT CAA GTG CC-3′ and reverse, 5′-CCT GAC CCA CAC CAA AGT CT-3′; suppressor of cytokine signaling (SOCS)1 forward, 5′-TAA CCC GGT ACT CCG TGA CT-3′ and reverse, 5′-CTC ACC CTC CAC AAC CAC TC-3′; methylthioadenosine phosphorylase (MTAP) forward, 5′-ATC GTG ACC ACA GCT TGC GGG-3′ and reverse, 5′-TCT GCC CGG GAG CTG AA-3′ and euchromatic histone lysine methyltransferase 2 (EHMT2) forward, 5′-TAC CCA TCC CCT GTG TCA AT-3′ and reverse, 5′-TCC TTG TCA TAC CAG CAT CG-3′. All samples were analyzed in duplicate and normalized to β-actin.
Protein isolation and western blotting
Murine and human melanoma cells were lysed in RIPA buffer (Roche Diagnostics GmbH) for 15 min at 4°C and cell debris was separated via centrifugation at 600 x g and 4°C for 10 min. Protein concentration was determined using the Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). A total of 35 μg total lysate per lane was separated on 10.00-12.75% SDS-PAGE gels and subsequently transferred onto a PVDF membrane. Following blocking for 1 h at room temperature with 5% BSA/0.1% TBS-T (Sigma-Aldrich; Merck KGaA) or 5% non-fat dry milk/TBS-T (0.1% Tween-20), the membrane was incubated overnight at 4°C with the following antibodies: β-actin (1:3,000; cat. no. A5441; Sigma-Aldrich; Merck KGaA), histone 3 (H3) K9me2 (1:1,000; cat. no. ab3251; Abcam), H3K9me3 (1:1,000; cat. no. 07-442; Merck KGaA), H3 (1:1,000; cat. no. 4499; Cell Signaling, Technology, Inc.), EHMT2 (1:1,000; cat. no. 3306; Cell Signaling Technology, Inc.) and CYLD [1:1,000; cat. nos. 12797 (mCYLD) and 4495 (hCYLD), Cell Signaling Technology, Inc.].
After washing three times with TBS-T (0.1% Tween-20), membranes were incubated for 1 h with horseradish peroxidase-conjugated secondary anti-rabbit (cat. no. 7074) or anti-mouse (7076) antibody (both 1:3,000; both Cell Signaling Technology, Inc.). Finally, the membrane was washed three times in TBS-T and the immunoreaction was visualized using Clarity™ Western ECL Substrate (Bio-Rad Laboratories, Inc.) and Chemostar chemiluminescence imager (Intas Pharmaceuticals, Ltd.). Signal intensity was quantified using LabImage software Version 4.2.1 (Kapelan Bio-Imaging GmbH).
Histone extraction
For isolation of histone extracts, Histone Extraction kit was used (cat. no. ab113476; Abcam). A total of 1x107 melanoma cells (murine or human) were seeded in T75 flasks and harvested after 24 h incubation (8% CO2 at 37°C) with a cell scraper. Following isolation according to the manufacturer's instructions, protein concentration was quantified using the Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). For western blot analysis, 10 μg purified histone was used. Aliquots were stored at -80°C.
5-Aza-deoxycytidine (AZA) treatment
Murine melanoma cells were seeded in a T75 flask (750,000 cells/flask) 24 h (8% CO2 at 37°C) before treatment. Cells were treated with 5 μM AZA (Sigma-Aldrich; Merck KGaA) for 72 h (8% CO2 at 37°C) and harvested for qPCR and western blot analysis.
Clonogenic assay
To investigate proliferation, colony formation ability was assessed. A total of 300 cells (murine and human melanoma cell lines) was seeded in a 6-well plate and cultivated for 10 days (8% CO2 at 37°C). After washing twice with PBS, cells were fixed and stained with 6.0 glutaraldehyde and 0.5% crystal violet (Sigma-Aldrich; Merck KGaA), respectively, for 1 h at room temperature). The colony (>50 cells) size was analyzed using cellSens dimension-software (V1.12) and an IX83 fluorescence microscope (both Olympus Corporation).
Chromatin accessibility assay
The EpiQuik™ Chromatin Accessibility Assay kit was used according to the manufacturer's instructions (BioCat GmbH). Briefly, 1×106 cells (murine or human melanoma cells) were collected 24 h after seeding. Following cell lysis and chromatin isolation, one half of the lysed cell was digested with a nuclease mix (Nse) and the other was left untreated (No-Nse). Subsequently, DNA was purified and analyzed via qPCR as aforementioned. Fold enrichment (FE) was calculated as follows: FE=2(Nse Ct-No-Nse Ct)/-2(Nse Ct-No-Nse Ct) ×100%. FE>400% indicated closed chromatin and FE<1,600% indicated open chromatin (values between 400 and 1,600% represent partially open chromatin). FE was normalized to Cyld-deficient cells and the control.
H3 modification multiplex assay
Histone H3 Modification Multiplex Assay kit (cat. no. ab185910, Abcam) was used to quantify 21 H3 modifications according to the manufacturer's instructions. A total of 50 ng/well histone extract from Cyld-expressing (Cyld+/+) and -deficient (Cyld−/−) murine cell lines, 50 ng/well histone extract was used. H3 lysine mono-di- and trimethylation was measured at sites K4, K9, K27, K36 and K79.
DNA sequencing (seq) and copy number variation (CNV) analysis
DNA-seq was performed using input samples of corresponding chromatin immunoprecipitation (ChIP) experiments. Chromatin from 1×107 cells (mCyld−/− and mCyld+/+) of each sample was crosslinked in 1:10 volume of fixation buffer (50 mM HEPES/KOH, pH 7.9, 11% formaldehyde) for 10 min at room temperature and quenched by 0.125 M glycine. Following two washes with PBS and PMSF, cells were scraped and centrifuged at 4°C and 3,500 x g for 10 min. The supernatant was discarded and the pellet was resuspended in 15 ml lysis buffer 1 (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40, 1X Roche Complete, EDTA-free protease inhibitor) and incubated for 10 min on ice. Following centrifugation of 5 min at 4°C and 3,500 x g, the supernatant was discarded and the pellet resuspended in 15 ml lysis buffer 2 (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, 1X Roche Complete, EDTA-free protease inhibitor). The suspension was incubated on ice for an additional 10 min and examined under the light microscope (magnification: x200) for quality assessment of isolated nuclei from cytoplasmic fractions. The nuclei were pelleted at 4°C and 3,500 x g for 5 min. The supernatant was discarded and the pellet was resuspended in sonication buffer (1x107 cells/450 μl sonication buffer: 16.7 mM Tris-HCl, pH 8.0, 1.2 mM EDTA, 16,7 mM NaCl, 0.01% SDS, 1.10% Triton X-100, 1X Roche Complete, EDTA-free protease inhibitor). Cross-linked chromatin was sheared to an average DNA fragment size of 200-700 bp using the Covaris ME220 Focused-ultrasonicator (Covaris, Inc.). Then, 20 μl supernatant was used for DNA purification using the QIAquick PCR purification kit (Qiagen GmbH). Library preparation was performed with purified DNA using the TruSeq® ChIP Sample Preparation kit according to the manufacturer's instructions (Illumina, Inc.). Quality and quantity of the final libraries were checked for size (200-500 bp) by TapeStation 4200 using the High-Sensitivity DNA kit (Agilent Technologies, Inc.) and concentration was determined by Qubit 4 Fluorometer (Thermo Fisher Scientific, Inc.). Each library was diluted to 4 nM and pooled to a final concentration of 5 nM. DNA libraries were sequenced for 50 cycles on an Illumina HiSeq4000 with single-end module (HiSeq 3000/4000 SBS kit, 50 cycles; cat. no. FC-410-1001; llumina, Inc.). Sequence tags of all experiments were mapped to the current mouse reference sequence (GRCm38/mm10) using Bowtie 2 (v 2.2.7) (16,17). Only uniquely mapped tags were used for CNV determination via Control-FREEC (v11.6) (18).
RNA-seq and mutational analysis
Total RNA samples were isolated from mCyld−/− and mCyld+/+ cell lines using Total RNA kit I (Omega Bio-Tek, Inc.) according to manufacturer's instructions. All RNA samples were examined for integrity and purity by TapeStation 4200 (Agilent Technologies, Inc.).
Library preparation was performed with two biological replicates using the TruSeq® Stranded Total RNA Library Prep Human/Mouse/Rat kit (cat. no. 20020596, Illumina, Inc.) according to the manufacturer's instructions. The resulting libraries were checked for size (200-500 bp) by TapeStation 4200 using the High-Sensitivity DNA kit (Agilent Technologies, Inc.) and concentration was determined by Qubit 4 Fluorometer (Thermo Fisher Scientific, Inc.). Each library was diluted to 4 nM and pooled to a final concentration of 5 nM. Paired-end seq was performed using a HiSeq4000 with paired-end module (Illumina, Inc.) according to the manufacturer's instructions. The samples were sequenced from each side of a fragment ~75 bp long with an average number 20 million reads per sample. Following quality check using FastQC (Babraham Bioinformatics; bioinformatics.babraham. ac.uk/projects/fastqc/) paired-end reads were aligned to the mouse reference genome (GRCm38, mm10) using the STAR alignment software (v 2.5.2a) (17,19). Following mapping, only reads that mapped to a single unique location were selected for further analysis. The mapped reads were used for variant calling by Genome Analysis Toolkit (GATK; v4.1; gatk. broadinstitute.org) (20). Single nucleotide variants (SNVs) were identified with a cut-off of 1% minor allele frequency and a minimum of 10 variant reads. Variants were annotated using SnpEff 5.0e (21). Intronic variants were discarded and all exon variants were analyzed to determine their effect on the resulting coding sequence (non-synonymous or synonymous). Plots of contingency tables were produced using variant calling output and R script Genotype-Variants (v1.1; github. com/cfarkas/Genotype-variants) of the genotype-variant pipeline as described by Farkas et al (22).
Schematic illustrations
Schematic illustrations were abstracted and modified from 'Les Laboratoires Servier-smart' Medical Art (smart.servier.com/). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License.
Statistical analysis
Data are presented as the mean ± SEM of ≥3 independent experiments, unless otherwise stated. Statistical significance was determined using Student's unpaired t-test or one-way ANOVA followed by Bonferroni's post hoc test. Data were analyzed using GraphPad Prism Version 9 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Absence of CNVs and common mutations in Cyld−/− and Cyld+/+ cells
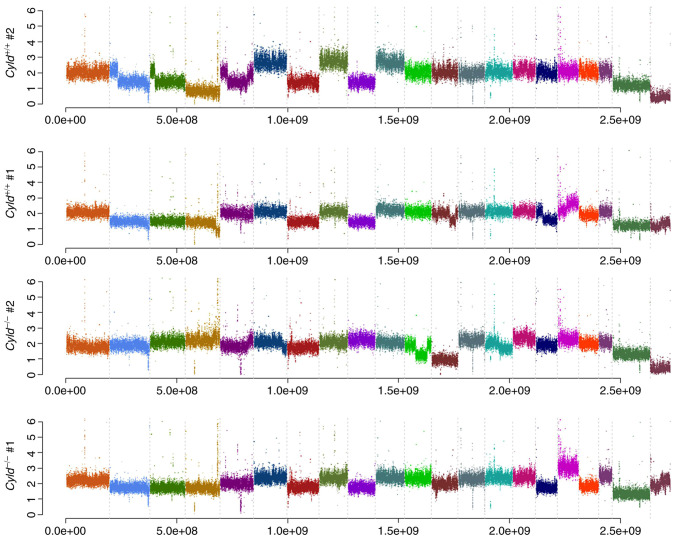
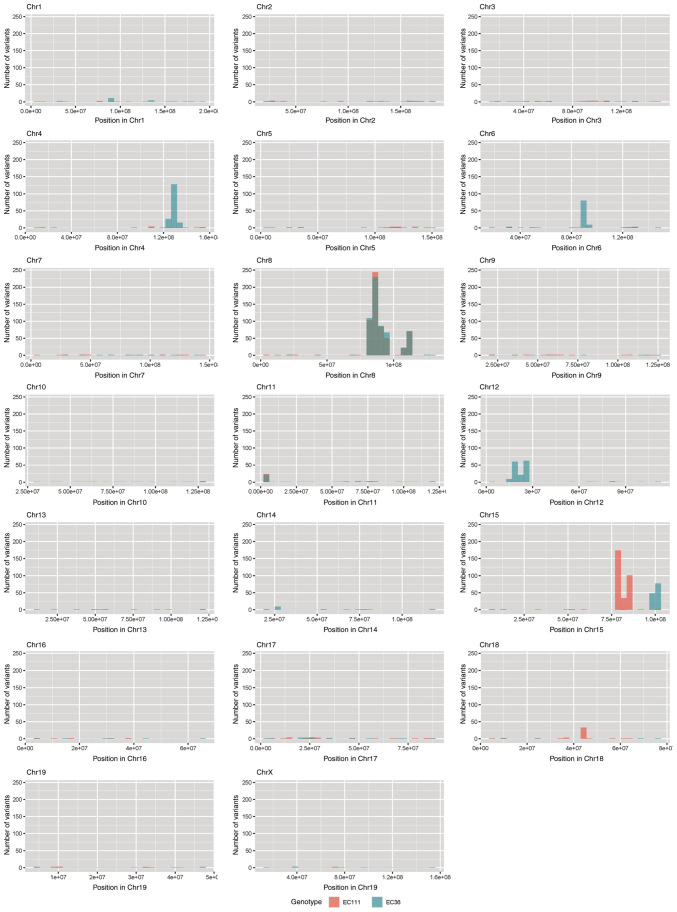
Melanoma is one of the tumors with the highest mutation burden among solid tumors (23). To investigate whether CYLD affects melanoma-associated mutations, sequencing analyses were performed. No common CNVs were detected in mCyld−/− cell and mCyld+/+ cells (Fig. 1). Therefore, SNVs were investigated to identify potential mutations underlying CYLD-dependent changes in tumor development of Tg(Grm1)Cyld mice. Variant calling was performed using Mutect2 of GATK (v4.2) to discover somatic variants in the RNA-seq dataset mCyld−/− vs. mCyld+/+ cells. Few common somatic mutations were determined and all but one variants (pre-mRNA processing factor 39 on chromosome 12) in mCyld−/− were located on chromosome 8 near the CYLD genomic location (Fig. 2). In summary, no Cyld-dependent significant copy number gains or losses and just a few somatic mutations were detected.
Figure 1.
Copy number variation analysis. Frequency plots illustrate no Cyld-dependent significant copy number gains or losses in mCyld−/− compared with mCyld+/+. Each color indicates one chromosome. CYLD, CYLD lysine 63 deubiquitinase.
Figure 2.
Somatic mutation analysis. Bar plots showing the number of nucleotide variants found in both mCyld−/− cell lines but not in mCyld+/+ cell lines. Each bar represents the number of variants per 10 million base pairs along the chromosomal location in mCyld−/− cell line #1 (red) and #2 (blue). Common variants found in both mCyld−/− cell lines are only present on Chr 8 (gray) due to knockout of Cyld. Chr, chromosome; CYLD, CYLD lysine 63 deubiquitinase.
CYLD does not affect DNA methylation
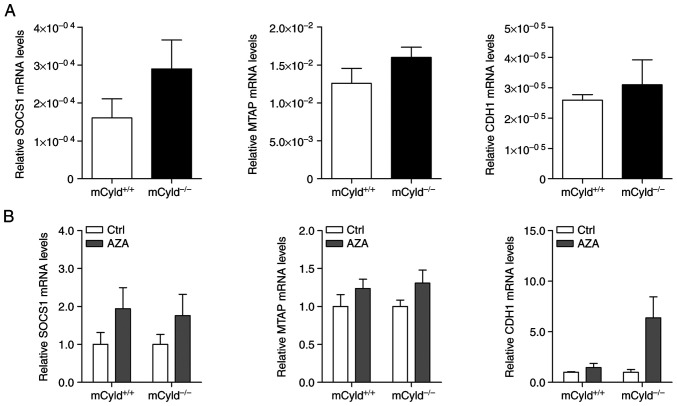
Besides genomic mutations, epigenetic dysregulation has also been linked to tumor development and progression in a large number of studies (9-12). To investigate the role of CYLD in controlling methylation of CpG islands, expression of known hypermethylated genes (MTAP, CDH1 and SOCS1) in human melanoma was investigated (24,25). No significant differences in MTAP, CDH1 and SOCS1 mRNA expression were observed between mCyld+/+ and mCyld−/− cell lines (Fig. 3A), indicating that CYLD was not involved in DNA methylation. DNA demethylation was induced by treatment with AZA to investigate CYLD-dependent alterations in gene expression. AZA treatment was associated with a notable (but not significant) increase in CYLD protein expression (Fig. S1). mRNA expression of hypermethylated genes was not significantly altered in mCyld+/+ and mCyld−/− cells following AZA treatment (Fig. 3B), confirming the aforementioned results.
Figure 3.
CYLD and DNA methylation. Quantitative PCR analysis of known hypermethylated genes (SOCS1, MTAP and CDH1) in mCyld+/+ and mCyld−/− cell lines (A) before and (B) after treatment with AZA for 72 h (normalized to untreated Ctrl). CYLD, CYLD lysine 63 deubiquitinase; SOCS1, suppressor of cytokine signaling; MTAP, methylthioadenosine phosphorylase; CDH1, cadherin 1; Ctrl, control; AZA, 5-Aza-deoxycytidine.
In summary, these results indicated that CYLD had no effect on DNA methylation.
CYLD deficiency results in more compact chromatin structure and affects H3 modification associated with heterochromatin
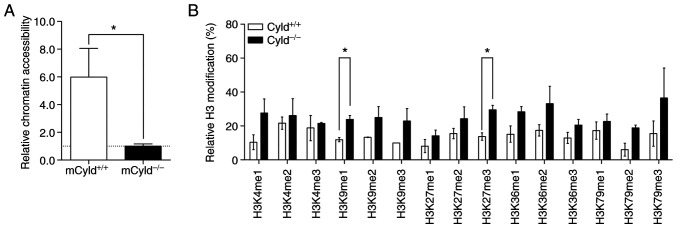
Epigenetic events affect the accessibility of chromatin via formation of heterochromatin and euchromatin (26). To determine whether CYLD affected chromatin structure, chromatin accessibility was assessed using the EpiQuik chromatin accessibility kit. This assay showed that mCyld+/+ cells exhibited a more open chromatin structure compared with mCyld−/− cells (Fig. 4A).
Figure 4.
Effect of CYLD on chromatin accessibility and H3 modification. (A) Chromatin accessibility assay of mCyld+/+ and mCyld−/− cell lines. Quantification of relative chromatin accessibility to mCyld−/− cells. (B) H3 modification assay of mCyld+/+ and mCyld−/− cell lines (normalized to total amount of H3). n=2. *P<0.05. CYLD, CYLD lysine 63 deubiquitinase; H3, histone 3.
Heterochromatin is associated with certain H3 modifications (27); to the best of our knowledge, however, there are no studies regarding H3 modification and CYLD. Therefore, H3 modification assay was performed. Although modifications were notably higher in mCyld−/− compared with mCyld+/+ cells, this was only significant for H3K9me1- and H3K27me3- modifications (Fig. 4B). Taken together, these data suggested that CYLD was involved in chromatin structure and histone methylation.
CYLD-dependent effects of inhibitors Chaetocin, JIB04 and CM272 on H3 methylation and cell proliferation
The aforementioned results suggested an association between CYLD, chromatin structure and histone methylation. Because aberrant histone modification disrupts epigenetic balance and contributes to melanoma progression (28), the effect of histone demethylase/methyltransferase inhibitors on CYLD-dependent proliferation were analyzed.
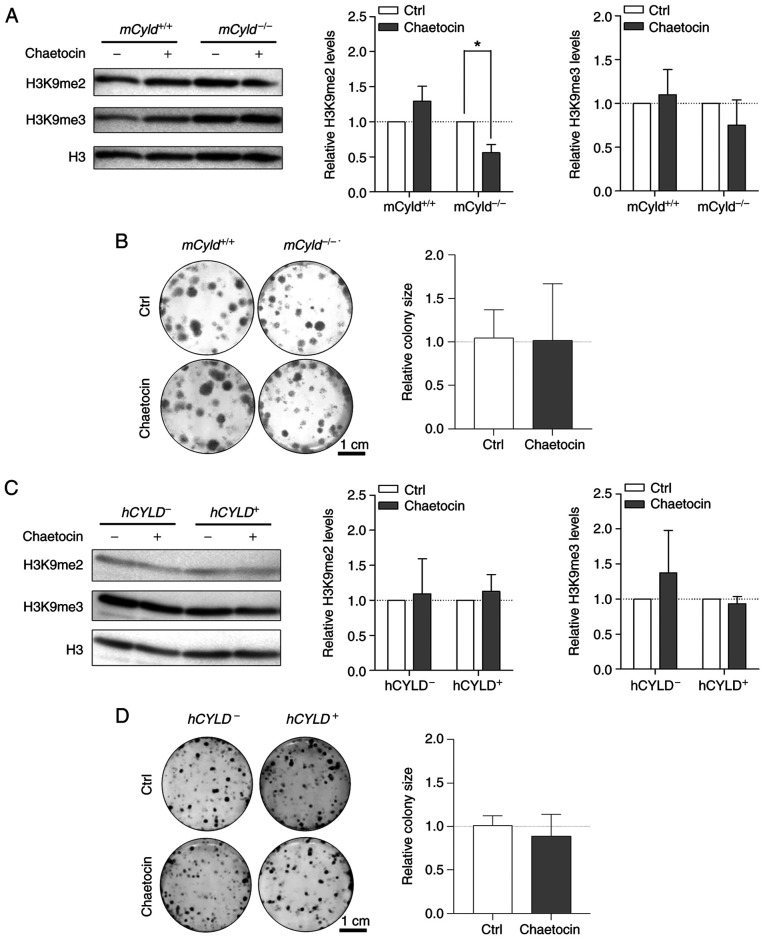
Treatment with Chaetocin, a lysine-specific histone methyltransferase, leads to inhibition of the histone methyltransferase suppressor of variegation 3-9 homolog 1 (SUV39H1, also known as KMT1A), which is involved in formation of hetero-chromatin by di-and trimethylating H3K9 (29,30). Here, treatment resulted in significantly decreased H3K9 dimethylation only in mCyld−/− cells; trimethylation of H3K9 was not significantly affected (Fig. 5A). A previous study has demonstrated that Chaetocin exerts anti-proliferative effects in vivo in melanoma cells (31). However, clonogenic assay revealed no significant change in colony size between mCyld−/− and mCyld+/+ cells (Fig. 5B).
Figure 5.
Effect of histone inhibitor Chaetocin on Cyld+/+ and Cyld−/− melanoma cells. Representative (A) western blot of H3K9me2 and H3K9me3 levels following 24 h treatment with Chaetocin (10 nM) relative to H3 and (B) anchorage-dependent clonogenic assay of murine cell lines. Representative (C) western blot of H3K9me2 and H3K9me3 levels following 24 h treatment with Chaetocin (10 nM) relative to H3 and (D) anchorage-dependent clonogenic assay (hCyld+/+:hCyld−/− cell colony size). *P<0.05. CYLD, CYLD lysine 63 deubiquitinase; H3, histone 3; ctrl, control; m, mouse; h, human.
Using the established human CYLD melanoma system, no significant changes in H3K9 di- and trimethylation levels were observed via western blot analysis after treatment with Chaetocin (Fig. 5C). Consistent with the results in the murine cell lines, proliferation was not significantly altered by Chaetocin in human cell lines (Fig. 5D). Treatment with Chaetocin had no significant effect on CYLD protein expression in murine and human melanoma cells (Fig. S2A and B).
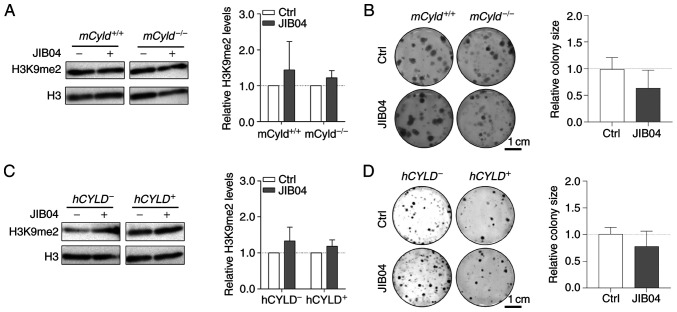
The counterpart of histone lysine methyltransferases are histone lysine demethylases (KDMs), including family members KDM3 [Jumonji Domain-Containing Protein 1A (JMJD1)], KDM4 (JMJD2) and KDM2 (JmjC domain-containing histone demethylation protein 1) (32). To the best of our knowledge, there are no inhibitors targeting specific KDMs. Nevertheless, there are broad-spectrum KDM inhibitors, such as the small molecule JIB04, which increases H3K9me2 levels (33). Since the aforementioned results excluded an effect of CYLD on H3K9 trimethylation on proliferation, cells were treated with JIB04; this treatment had no significant effect on levels of H3K9me2 in mCyld−/− or mCyld+/+ cells (Fig. 6A). Clonogenic assay revealed no significant changes in colony size following JIB04 treatment in mCyld−/− compared with mCyld+/+ cells (Fig. 6B). There was no significant change in H3K9me2 levels (Fig. 6C) or colony size (Fig. 6D) in hCYLD− compared with hCYLD+ cells. CYLD protein expression was not significantly affected by treatment with JIB04 (Fig. S2C and D).
Figure 6.
Effect of histone inhibitor JIB04 on Cyld+/+ and Cyld−/− melanoma cells. Representative (A) western blot of H3K9me2 levels (relative to H3) following 24 h treatment with JIB04 (500 nM) and (B) anchorage-dependent clonogenic assay in murine cell lines. Representative (C) western blot of H3K9me2 levels (relative to H3) following 24 h treatment with JIB04 (500 nM) and (D) anchorage-dependent clonogenic assays of h cell lines (Cyld+/+:Cyld−/− colony size). JIB04, Jumonji histone demethylase inhibitor; Ctrl, control; h, human; m, mouse; CYLD, CYLD lysine 63 deubiquitinase; H3, histone 3.
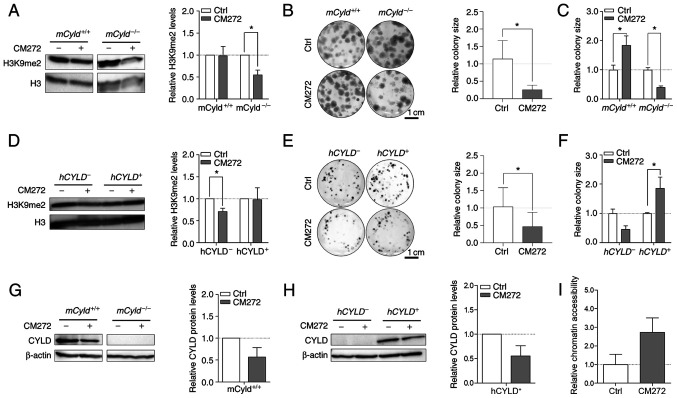
Selective inhibitor, CM272 suppresses activity of EHMT2 (also known as G9a) (14). This inhibitor contributes to decreased dimethylation of H3K9 and may hinder melanoma growth (14). To the best of our knowledge, however, no data concerning the role of CYLD in this context are available. Western blot analysis confirmed a significant decrease H3K9me2 levels following CM272 treatment only in mCyld−/− cell lines (Fig. 7A). Clonogenic assay revealed a significant decrease in colony size in mCyld−/− cells, which was consistent with H3K9me2 western blot results (Fig. 7B and C). mCyld+/+ cells exhibited a significantly increased colony size (Fig. 7C), as well as significantly decreased CYLD expression following CM272 treatment (Fig. 7D). In conclusion, CM272 treatment of CYLD-expressing cells led to decreased expression and tumor suppressive function of CYLD. These results were confirmed in human cells: hCYLD− cells exhibited significantly decreased H3K9me2 levels (Fig. 7E) and proliferation following CM272 treatment (Fig. 7F and G). hCYLD+ cells showed decreased CYLD expression following CM272 treatment (Fig. 7H) but this was not significant.
Figure 7.
Effect of histone inhibitor CM272 on Cyld+/+ and Cyld−/− melanoma cells. Representative western blot of H3K9me2 levels following 24 h treatment with CM272 relative to H3 of (A) murine cell lines. Representative images of anchorage-dependent clonogenic assays of (B and C) murine cell lines (mCyld+/+:mCyld−/− colony size). Representative western blot of H3K9me2 levels following 24 h treatment with CM272 relative to H3 of (D) human cell lines and representative images of anchorage-dependent clonogenic assays of (E and F) human cell lines (hCYLD+:hCYLD− colony size). Representative western blot of CYLD protein expression following 24 h treatment with CM272 relative to β-actin of (G) murine and (H) human cells. (I) Analysis of the chromatin accessibility of the mCyld−/− cell lines after 24 h treatment with CM272 (200 nM) (quantification of relative chromatin accessibility to untreated mCyld−/− cells). *P<0.05. Ctrl, control; h, human; m, mouse; CYLD, CYLD lysine 63 deubiquitinase; H3, histone 3.
It was hypothesized that treatment of Cyld-deficient cells with CM272 would lead to looser chromatin structure. Chromatin accessibility analysis of mCyld−/− cells showed more open chromatin structure following CM272 treatment compared with control cells (Fig. 7I) but this was not significant.
Catalytically inactive CYLD mutant cells (hCYLDC/S) exhibited similar trends to hCYLD− cells. There was a significant decrease in H3K9me2 levels (Fig. S3A) and colony size (Fig. S3B), whereas CYLD expression was not significantly altered following CM272 treatment (Fig. S3C). Moreover, hCYLDC/S cells exhibited more compact chromatin structure, similar to hCYLD− cells, whereas hCYLD+ cells exhibited a more open chromatin structure (Fig. S3D).
Together, these data showed that the inhibitors Chaetocin and JIB04 exerted no CYLD-dependent effect on cell proliferation. The inhibitor CM272 had anti-proliferative effects on Cyld-deficient cells but pro-proliferative effects on Cyld-expressing cells. The pro-proliferative effects were dependent on enzymatic function of CYLD.
High levels of EHMT2 in Cyld-deficient cells increase H3K9 dimethylation
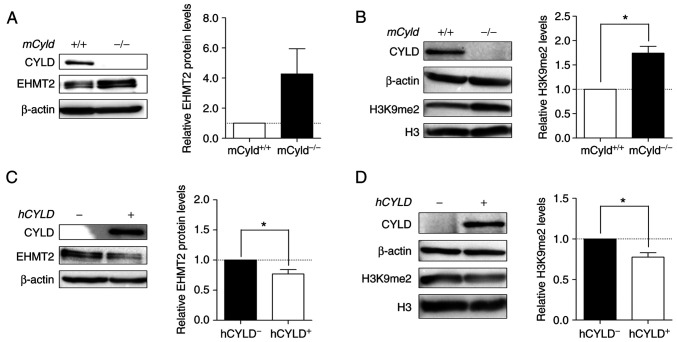
Histone lysine methyltransferase EHMT2 specifically mono- and dimethylates H3K9 and catalyzes trimethylation of H3K27 (34). Thus, the present study investigated expression levels of this methyltransferase and its primary methylation site H3K9me2. Western blot analysis demonstrated notably enhanced EHMT2 expression in mCyld−/− cells (Fig. 8A). mCyld−/− cells showed a significant increase in H3K9me2 compared with Cyld+/+ cells (Fig. 8B). Same investigations in our human cell system verified the negative association between CYLD and EHMT2 expression as well as a positive association between H3K9me2 and EHMT2 (Fig. 8C and D). hCYLDC/S cells showed similar H3K9me2 and EHMT2 expression to hCYLD− cells (Fig. S3E and F).
Figure 8.
Aberrant EHMT2 and H3K9me2 expression. Representative western blot analysis of (A) EHMT2 protein expression (relative to β-actin) and (B) validation of H3K9me2 level of mCyld+/+ and mCyld−/− cell lines (relative to H3). Western blot analysis of (C) EHMT2 protein expression (relative to β-actin) and (D) quantification of H3K9me2 level of hCYLD+ and hCYLD− cell lines (relative to H3). *P<0.05. EHMT2, euchromatic histone lysine methyltransferase 2; H3, histone 3; m, mouse; h, human; CYLD, CYLD lysine 63 deubiquitinase.
Discussion
The deubiquitinase CYLD is downregulated in different types of cancer, including hepatocellular carcinoma, breast cancer and malignant melanoma (6,35,36). Using the melanoma mouse model Tg(Grm1) and murine cell lines generated from tumors, our previous study demonstrated the tumor suppressive function of CYLD in proliferation, migration and lymph- and angiogenesis (7). Nevertheless, the underlying molecular mechanism of early tumor onset in Cyld−/− mice has not yet been uncovered.
Chromosomal aberrations or nucleotide polymorphisms affect pathways involved in tumor formation and progression in breast cancer or cutaneous melanoma (37,38). However, the present study was unable to identify chromosomal alterations and detected only a few nucleotide variations in mCyld−/− compared with mCyld+/+ cells. Most mutations occurred in proximity to CYLD. To the best of our knowledge, clustering of mutations near knockout sites has not previously been investigated but it is hypothesized that the sequence in which the CYLD gene locus is located is genetically unstable or particularly susceptible to single nucleotide polymorphisms (39). CYLD interacts with and deubiquitinates p53 in response to DNA damage (40) and therefore affects genomic instability. To investigate this, mutation analysis of normal mouse tissue is required. The present study identified few mutations but did not determine a link with melanoma-associated mutations or early tumor onset in CYLD knockout mice.
In addition to genomic instability and abnormal gene expression, dysregulation of epigenetic mechanisms, such as DNA methylation and histone modification are hallmarks of cancer (8). Certain studies have shown that CYLD expression is suppressed by epigenetic regulatory mechanisms, such as DNA methylation or histone deacetylation (41,42). The present western blot analysis demonstrated enhanced CYLD expression following AZA treatment. This was consistent with CYLD regulation in gastric adenocarcinoma, where hypermethylation of the CYLD promoter results in decreased expression of CYLD (41). Colon and hepatocellular carcinoma show no expression changes of CYLD following AZA treatment (35). Although, we describe a transcriptional control of the CYLD promoter via overexpression of the transcriptional factor SNAIL1 (6), epigenetic regulatory mechanisms may also contribute to downregulation of CYLD. However, the present analysis suggested that CYLD serves no key role in DNA methylation in a mouse model.
In addition to DNA methylation, histone tails undergo post-translational modifications that alter chromatin structure and dynamics (26). CYLD interacts with certain histone deacetylases (HDACs); in hepatic stellate cells, CYLD regulates hepatocyte growth factor expression via interaction with HDAC7, leading to lower hepatocellular damage and liver fibrosis (43). Moreover, Wickström et al (44) showed negative regulation of cell cycle by via inhibitory interaction of CYLD and HDAC6 in keratinocytes and melanoma cells. To the best of our knowledge, there is no evidence of a direct effect of the tumor suppressor CYLD on other types of histone modification, such as methylation. The present data revealed that the overall accessibility of chromatin is decreased in Cyld−/− melanoma cells, suggesting that CYLD deficiency favors the formation of more condensed heterochromatin and associated histone modifications, such as H3 methylation. H3 modification assay revealed higher levels of heterochromatin-specific histone modifications H3K9me and H3K27me in Cyld−/− compared with Cyld+/+ cells.
Histone-modifying enzymes affect the structure of chromatin and are deregulated in cancer, including prostate, breast, colon, skin, and lung cancers (45,46). Cancer drug discovery has focused on development of competitive analogs of co-factors, such as JIB04 and CM272, which modulate activity of epigenetic enzymes upregulated in tumors (47,48). Although inhibitors Chaetocin and JIB04 had no influence on proliferation of melanoma cells, treatment with dual-inhibitor CM272 showed CYLD-dependent effects. Previously, anti-proliferative effects of CM272 were observed in hematological tumor (14). Moreover, CM272 inhibits fibrogenesis and proliferation in cholangiocarcinoma and hepatocellular carcinoma (49-51). Furthermore, decreased cell viability and induction of type I interferon response have been observed in murine melanoma cells (52). The present study revealed decreased proliferation of Cyld−/− melanoma cells following CM272 treatment, suggesting that this inhibitor may be a potential therapy for melanoma. Notably, Cyld+/+ cells showed enhanced proliferation following treatment, potentially due to decreased CYLD protein expression. Therefore, CM272 should be used in clinical trials to investigate its effect on CYLD expression. Downregulation of CYLD in patients with melanoma has been described in previous studies (6,53). Nevertheless, a recent study reported heterogeneous expression between melanoma cell lines and melanoma in vivo (54). This shows the importance of defining CYLD status when using CM272 inhibitor in melanoma therapy, as treatment of Cyld-expressing cells results in an increase in proliferation. Moreover, Rodriguez-Madoz et al (55) showed that CM272 treatment leads to heterochromatin relaxation. This was also observed in Cyld−/− melanoma cells in the present study, suggesting that this inhibitor reverses silencing of tumor suppressors, which decreases proliferation when CYLD is lost. In summary, in vitro analysis of the effect of inhibitors revealed that CYLD is involved in histone methylation modification associated with chromatin structure.
There was an inverse association between CYLD and expression of the potential target of CM272, EHMT2. The methyltransferase EHMT2 is deregulated in many tumor entities and is associated with increased migration, invasion and poor prognosis for patients, demonstrating its important role in tumorigenesis (33,56,57). Additionally, this lysine methyltransferase contributes to H3K9 and H3K27 methylation, which are associated with gene silencing and chromatin condensation (58). In a zebrafish melanoma model, histone methyltransferase (HKMT) SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) promotes melanoma formation (59). This enzyme also methylates H3K9 and serves as an oncogene in melanoma (59). Moreover, a multimeric complex of four H3K9 HKMTs, including EHMT2/G9a/KMT1C, GLP/KMT1D, SETDB1/KMT1E and Suv39h1/KMT1A, is involved in regulating gene expression and heterochromatin assembly (60). Thus, the role of these enzymes and their association with CYLD in development of melanoma should be further investigated. However, the Suv39h1 inhibitor Chaetocin did not affect proliferation of Cyld+/+ or Cyld−/− cells, suggesting no effect of this HKMT on melanoma cells. Nevertheless, it is unclear whether there is a direct or indirect connection between CYLD and EHMT2. The present results indicated direct interaction due to the negative association between expression of CYLD and EHMT2 and effect of the inhibitor CM272 on proliferation and chromatin structure. To demonstrate this potential association between CYLD and EHMT2, rescue experiments should be performed in follow-up studies.
In the present study, analysis of human melanoma cells showed that hCYLDC/S behaved like hCYLD-deficient cells (Fig. S3). This suggested involvement of the deubiquitinase function of CYLD in epigenetic processes. Certain studies have demonstrated the role of deubiquitinases in crosstalk between different histone post-translational modifications (61). For example, ubiquitin specific peptidase (USP28) enhances tumorigenicity of breast cancer cells via stabilization of lysine-specific demethylase 1 (62). Moreover, knockdown of USP24 increases Suv39h1 expression, which results in enhanced H3K9me levels and prevents the progression of malignant lung cancer (63). Furthermore, H3 polyubiquitination suppresses transcription of fetal and cell cycle genes in postnatal mouse liver by crosstalk with H3K9 methylation and loss of ubiquitin ligase CRL4 inhibit H3K9 methylation (64). The present data and existing literature suggest that CYLD may inhibit H3K9 dimethylation via deubiquitination of upstream histone modifiers, such as EHMT2 or H3K9, and therefore suppress cell proliferation due to activation of silenced tumor suppressors.
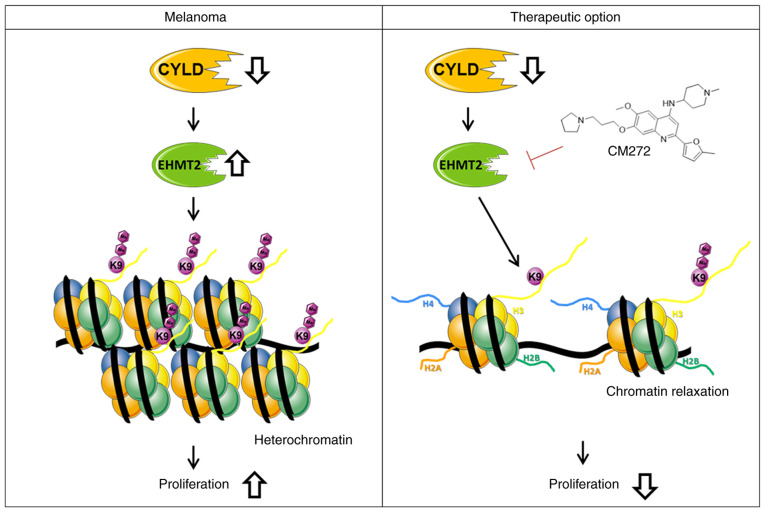
The present study revealed modulation of chromatin structure in a CYLD-dependent manner in melanoma. The present results indicated that increased expression of EHMT2 led to increased H3K9me2, subsequently promoting a more compact heterochromatin structure when CYLD is lost (Fig. 9). These results may explain enhanced tumor onset of CYLD knockout mice. Further studies should investigate the role of CYLD in epigenetic processes as the present results indicated other H3 modifications (such as H3K27me) might be CYLD-dependent. To the best of our knowledge, the present study is the first to demonstrate an association between tumor suppressor CYLD, chromatin structure and histone methylation. Moreover, suppressing proliferation of the higher H3K9me2 expressing Cyld−/− cells via CM272 may be a promising strategy for melanoma treatment (Fig. 9). The high similarity of mouse and human melanoma cells illustrates the relevance of our generated cells for the study of molecular mechanisms in melanoma.
Figure 9.
Model of CYLD deficiency in melanoma cells. The upregulation of EHMT2 and associated di-methylation of H3K9 leads to heterochromatin and the induction of proliferation of proliferation. CM272, which inhibits the activity of EHMT2. The associated decrease in H3K9me2 levels results in loosening of chromatin and decreased proliferation of CYLD-deficient melanoma cells (abstracted and modified from smart.servier.com/). EHMT2, euchromatic histone lysine methyltransferase 2; CYLD, CYLD lysine 63 deubiquitinase.
Supplementary Data
Acknowledgments
The authors would like thank Professor Suzie Chen (Rutgers University, Chemical Biology, Piscataway, USA) for providing Tg(Grm1) animals and Dr Ramin Massoumi (Department of Laboratory Medicine, Lund University, Sweden) for providing C57Bl/6 Cyld−/− mice. The authors would like to thank Mr. Alexander-Oliver Matthies (Friedrich-Alexander-University; FAU) for preparation of RNA samples and RNA-Seq libraries and Professor Christoph Bock (Biomedical Sequencing Facility, Research Center for Molecular Medicine of the Austrian Academy of Sciences), Vienna, Austria) for sequencing samples. The authors would also like to thank Mr. Ingmar Henz for technical assistance and Mrs. Valerie Fritz for critical discussion (both FAU).
Funding Statement
The present study was supported by the German Research Foundation (grant nos. DFG FOR2127 and TRR305, B12), FEDER/Ministerio de Ciencia, Innovación y Universidades-Agencia Estatal de Investigación, Spain (grant nos. PID2019-104878RB-100/AEI/10.13039/501100011033 and PID2020-PID2020-120387RB-I00), Fundación Fuentes Dutor and the Ramón y Cajal Program (grant no. RYC-2018-024475-1).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the NCBI BioProject database (accession no. PRJNA796721; (dataview.ncbi.nlm.nih.gov/object/PRJNA796721?reviewer=q05a8apg3kla63kggpoh5gi6dt).
Authors' contributions
AKB, SK and MS conceived the project, analyzed the data and wrote the manuscript. MS designed and performed the experiments. AKB, SK, MGFB, APL, MAA, SF and MKF provided materials, analyzed the data and wrote the manuscript. All authors have read and approved the final manuscript. MS and AKB confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 2.Hellerbrand C, Massoumi R. Cylindromatosis-a protective molecule against liver diseases. Med Res Rev. 2016;36:342–359. doi: 10.1002/med.21381. [DOI] [PubMed] [Google Scholar]
- 3.Orfanidou T, Xanthopoulos K, Dafou D, Pseftogas A, Hadweh P, Psyllaki C, Hatzivassiliou E, Mosialos G. Down-regulation of the tumor suppressor CYLD enhances the transformed phenotype of human breast cancer cells. Anticancer Res. 2017;37:3493–3503. doi: 10.21873/anticanres.11717. [DOI] [PubMed] [Google Scholar]
- 4.Xie S, Wu Y, Hao H, Li J, Guo S, Xie W, Li D, Zhou J, Gao J, Liu M. CYLD deficiency promotes pancreatic cancer development by causing mitotic defects. J Cell Physiol. 2019;234:9723–9732. doi: 10.1002/jcp.27658. [DOI] [PubMed] [Google Scholar]
- 5.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 6.Massoumi R, Kuphal S, Hellerbrand C, Haas B, Wild P, Spruss T, Pfeifer A, Fässler R, Bosserhoff AK. Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J Exp Med. 2009;206:221–232. doi: 10.1084/jem.20082044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jel MM, Schott M, Lamm S, Neuhuber W, Kuphal S, Bosserhoff AK. Loss of CYLD accelerates melanoma development and progression in the Tg(Grm1) melanoma mouse model. Oncogenesis. 2019;8:56. doi: 10.1038/s41389-019-0169-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strub T, Ballotti R, Bertolotto C. The 'ART' of epigenetics in melanoma: From histone 'alterations, to resistance and therapies'. Theranostics. 2020;10:1777–1797. doi: 10.7150/thno.36218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nebbioso A, Tambaro FP, Dell'Aversana C, Altucci L. Cancer epigenetics: Moving forward. PLoS Genet. 2018;14:e1007362. doi: 10.1371/journal.pgen.1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Putiri EL, Robertson KD. Epigenetic mechanisms and genome stability. Clin Epigenetics. 2011;2:299–314. doi: 10.1007/s13148-010-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, Körbel C, Laschke MW, Gimotty PA, Philipp SE, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strub T, Ghiraldini FG, Carcamo S, Li M, Wroblewska A, Singh R, Goldberg MS, Hasson D, Wang Z, Gallagher SJ, et al. SIRT6 haploinsufficiency induces BRAFV600E melanoma cell resistance to MAPK inhibitors via IGF signalling. Nat Commun. 2018;9:3440. doi: 10.1038/s41467-018-05966-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiffner S, Braunger BM, de Jel MM, Coupland SE, Tamm ER, Bosserhoff AK. Tg(Grm1) transgenic mice: A murine model that mimics spontaneous uveal melanoma in humans? Exp Eye Res. 2014;127:59–68. doi: 10.1016/j.exer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 14.San José-Enériz E, Agirre X, Rabal O, Vilas-Zornoza A, Sanchez-Arias JA, Miranda E, Ugarte A, Roa S, Paiva B, Estella-Hermoso de Mendoza A, et al. Discovery of first-in-class reversible dual small molecule inhibitors against G9a and DNMTs in hematological malignancies. Nat Commun. 2017;8:15424. doi: 10.1038/ncomms15424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich P, Koch A, Fritz V, Hartmann A, Bosserhoff AK, Hellerbrand C. Wild type Kirsten rat sarcoma is a novel microRNA-622-regulated therapeutic target for hepatocellular carcinoma and contributes to sorafenib resistance. Gut. 2018;67:1328–1341. doi: 10.1136/gutjnl-2017-315402. [DOI] [PubMed] [Google Scholar]
- 16.Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11. 2010 doi: 10.1002/0471250953.bi1107s32. Unit 11.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O, Barillot E. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farkas C, Fuentes-Villalobos F, Rebolledo-Jaramillo B, Benavides F, Castro AF, Pincheira R. Streamlined computational pipeline for genetic background characterization of genetically engineered mice based on next generation sequencing data. BMC Genomics. 2019;20:131. doi: 10.1186/s12864-019-5504-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schinke C, Mo Y, Yu Y, Amiri K, Sosman J, Greally J, Verma A. Aberrant DNA methylation in malignant melanoma. Melanoma Res. 2010;20:253–265. doi: 10.1097/CMR.0b013e328338a35a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Micevic G, Theodosakis N, Bosenberg M. Aberrant DNA methylation in melanoma: Biomarker and therapeutic opportunities. Clin Epigenetics. 2017;9:34. doi: 10.1186/s13148-017-0332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan MA, Shilatifard A. Chromatin signatures of cancer. Genes Dev. 2015;29:238–249. doi: 10.1101/gad.255182.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharakhov IV, Sharakhova MV. Heterochromatin, histone modifications, and nuclear architecture in disease vectors. Curr Opin Insect Sci. 2015;10:110–117. doi: 10.1016/j.cois.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vardabasso C, Hake SB, Bernstein E. Histone variant H2A.Z.2: A novel driver of melanoma progression. Mol Cell Oncol. 2016;3:e1073417. doi: 10.1080/23723556.2015.1073417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR) 3-9. Nat Chem Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 30.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 31.Han X, Han Y, Zheng Y, Sun Q, Ma T, Zhang J, Xu L. Chaetocin induces apoptosis in human melanoma cells through the generation of reactive oxygen species and the intrinsic mitochondrial pathway, and exerts its anti-tumor activity in vivo. PLoS One. 2017;12:e0175950. doi: 10.1371/journal.pone.0175950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MS, Cho HI, Yoon HJ, Ahn YH, Park EJ, Jin YH, Jang YK. JIB-04, a small molecule histone demethylase inhibitor, selectively targets colorectal cancer stem cells by inhibiting the Wnt/β-catenin signaling pathway. Sci Rep. 2018;8:6611. doi: 10.1038/s41598-018-24903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zylicz JJ, Dietmann S, Gunesdogan U, Hackett JA, Cougot D, Lee C, Surani MA. Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. Elife. 2015;4:e09571. doi: 10.7554/eLife.09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hellerbrand C, Bumes E, Bataille F, Dietmaier W, Massoumi R, Bosserhoff AK. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis. 2007;28:21–27. doi: 10.1093/carcin/bgl081. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi M, Jono H, Shinriki S, Nakamura T, Guo J, Sueta A, Tomiguchi M, Fujiwara S, Yamamoto-Ibusuki M, Murakami K, et al. Clinical significance of CYLD downregulation in breast cancer. Breast Cancer Res Treat. 2014;143:447–457. doi: 10.1007/s10549-013-2824-3. [DOI] [PubMed] [Google Scholar]
- 37.Bauer M, Kantelhardt EJ, Stiewe T, Nist A, Mernberger M, Politt K, Hanf V, Lantzsch T, Uleer C, Peschel S, et al. Specific allelic variants of SNPs in the MDM2 and MDMX genes are associated with earlier tumor onset and progression in Caucasian breast cancer patients. Oncotarget. 2019;10:1975–1992. doi: 10.18632/oncotarget.26768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elefanti L, Sacco G, Stagni C, Rastrelli M, Menin C, Russo I, Alaibac M. TLR7 Gln11Leu single nucleotide polymorphism and susceptibility to cutaneous melanoma. Oncol Lett. 2016;12:275–280. doi: 10.3892/ol.2016.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenner MW, Leone PE, Walker BA, Ross FM, Johnson DC, Gonzalez D, Chiecchio L, Dachs Cabanas E, Dagrada GP, Nightingale M, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110:3291–3300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- 40.Fernández-Majada V, Welz PS, Ermolaeva MA, Schell M, Adam A, Dietlein F, Komander D, Büttner R, Thomas RK, Schumacher B, Pasparakis M. The tumour suppressor CYLD regulates the p53 DNA damage response. Nat Commun. 2016;7:12508. doi: 10.1038/ncomms12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghadami E, Nikbakhsh N, Fattahi S, Kosari-Monfared M, Ranaee M, Taheri H, Amjadi-Moheb F, Godazandeh G, Shafaei S, Nosrati A, et al. Epigenetic alterations of CYLD promoter modulate its expression in gastric adenocarcinoma: A footprint of infections. J Cell Physiol. 2019;234:4115–4124. doi: 10.1002/jcp.27220. [DOI] [PubMed] [Google Scholar]
- 42.Zhong S, Fields CR, Su N, Pan YX, Robertson KD. Pharmacologic inhibition of epigenetic modifications, coupled with gene expression profiling, reveals novel targets of aberrant DNA methylation and histone deacetylation in lung cancer. Oncogene. 2007;26:2621–2634. doi: 10.1038/sj.onc.1210041. [DOI] [PubMed] [Google Scholar]
- 43.Pannem RR, Dorn C, Hellerbrand C, Massoumi R. Cylindromatosis gene CYLD regulates hepatocyte growth factor expression in hepatic stellate cells through interaction with histone deacetylase 7. Hepatology. 2014;60:1066–1081. doi: 10.1002/hep.27209. [DOI] [PubMed] [Google Scholar]
- 44.Wickström SA, Masoumi KC, Khochbin S, Fässler R, Massoumi R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010;29:131–144. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chi P, Allis CD, Wang GG. Covalent histone modifications-miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kampranis SC, Tsichlis PN. Histone demethylases and cancer. Adv Cancer Res. 2009;102:103–169. doi: 10.1016/S0065-230X(09)02004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Chang J, Varghese D, Dellinger M, Kumar S, Best AM, Ruiz J, Bruick R, Peña-Llopis S, Xu J, et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat Commun. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabal O, San José-Enériz E, Agirre X, Sánchez-Arias JA, Vilas-Zornoza A, Ugarte A, de Miguel I, Miranda E, Garate L, Fraga M, et al. Discovery of reversible DNA methyltransferase and lysine methyltransferase G9a inhibitors with antitumoral in vivo efficacy. J Med Chem. 2018;61:6518–6545. doi: 10.1021/acs.jmedchem.7b01926. [DOI] [PubMed] [Google Scholar]
- 49.Fernández-Barrena MG, Arechederra M, Colyn L, Berasain C, Avila MA. Epigenetics in hepatocellular carcinoma development and therapy: The tip of the iceberg. JHEP Rep. 2020;2:100167. doi: 10.1016/j.jhepr.2020.100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barcena-Varela M, Caruso S, Llerena S, Aacute;lvarez-Sola G, Uriarte I, Latasa MU, Urtasun R, Rebouissou S, Alvarez L, Jimenez M, et al. Dual targeting of histone methyltransferase G9a and DNA-methyltransferase 1 for the treatment of experimental hepatocellular carcinoma. Hepatology. 2019;69:587–603. doi: 10.1002/hep.30168. [DOI] [PubMed] [Google Scholar]
- 51.Colyn L, Bárcena-Varela M, Aacute;lvarez-Sola G, Latasa MU, Uriarte I, Santamaría E, Herranz JM, Santos-Laso A, Arechederra M, Ruiz de Gauna M, et al. Dual targeting of G9a and DNA methyltransferase-1 for the treatment of experimental cholangiocarcinoma. Hepatology. 2021;73:2380–2396. doi: 10.1002/hep.31642. [DOI] [PubMed] [Google Scholar]
- 52.De Beck L, Prosper F, Maes K, Vanderkerken K, Breckpot K. Can CM272, a dual G9a/DNMT1 inhibitor, be used as an immunomodulating agent to enhance the efficacy of existing immunotherapies in melanoma?. Presented at BACR: Novel combination strategies for cancer treatment (poster session); Antwerp. 2019. [Google Scholar]
- 53.Ke H, Augustine CK, Gandham VD, Jin JY, Tyler DS, Akiyama SK, Hall RP, Zhang JY. CYLD inhibits melanoma growth and progression through suppression of the JNK/AP-1 and β1-integrin signaling pathways. J Invest Dermatol. 2013;133:221–229. doi: 10.1038/jid.2012.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.La T, Jin L, Liu XY, Song ZH, Farrelly M, Feng YC, Yan XG, Zhang YY, Thorne RF, Zhang XD, Teng L. Cylindromatosis is required for survival of a subset of melanoma cells. Oncol Res. 2020;28:385–398. doi: 10.3727/096504020X15861709922491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez-Madoz JR, San Jose-Eneriz E, Rabal O, Zapata-Linares N, Miranda E, Rodriguez S, Porciuncula A, Vilas-Zornoza A, Garate L, Segura V, et al. Reversible dual inhibitor against G9a and DNMT1 improves human iPSC derivation enhancing MET and facilitating transcription factor engagement to the genome. PLoS One. 2017;12:e0190275. doi: 10.1371/journal.pone.0190275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dang NN, Jiao J, Meng X, An Y, Han C, Huang S. Abnormal overexpression of G9a in melanoma cells promotes cancer progression via upregulation of the Notch1 signaling pathway. Aging (Albany NY) 2020;12:2393–2407. doi: 10.18632/aging.102750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato S, Weng QY, Insco ML, Chen KY, Muralidhar S, Pozniak J, Diaz JMS, Drier Y, Nguyen N, Lo JA, et al. Gain-of-function genetic alterations of G9a drive oncogenesis. Cancer Discov. 2020;10:980–997. doi: 10.1158/2159-8290.CD-19-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan MR, Hsu MC, Chen LT, Hung WC. G9a orchestrates PCL3 and KDM7A to promote histone H3K27 methylation. Sci Rep. 2015;5:18709. doi: 10.1038/srep18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferré F, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M, Ait-Si-Ali S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 61.Pinto-Fernandez A, Kessler BM. DUBbing cancer: Deubiquitylating enzymes involved in epigenetics, DNA damage and the cell cycle as therapeutic targets. Front Genet. 2016;7:133. doi: 10.3389/fgene.2016.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y, Wang Y, Yang XH, Kang T, Zhao Y, Wang C, Evers BM, Zhou BP. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep. 2013;5:224–236. doi: 10.1016/j.celrep.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang YC, Wang SA, Chen PH, Hsu TI, Yang WB, Chuang YP, Su WC, Liaw HJ, Chang WC, Hung JJ. Variants of ubiquitin-specific peptidase 24 play a crucial role in lung cancer malignancy. Oncogene. 2016;35:3669–3680. doi: 10.1038/onc.2015.432. [DOI] [PubMed] [Google Scholar]
- 64.Li G, Ji T, Chen J, Fu Y, Hou L, Feng Y, Zhang T, Song T, Zhao J, Endo Y, et al. CRL4DCAF8 ubiquitin ligase targets histone H3K79 and promotes H3K9 methylation in the liver. Cell Rep. 2017;18:1499–1511. doi: 10.1016/j.celrep.2017.01.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the NCBI BioProject database (accession no. PRJNA796721; (dataview.ncbi.nlm.nih.gov/object/PRJNA796721?reviewer=q05a8apg3kla63kggpoh5gi6dt).