ABSTRACT
Dysregulated long non-coding RNAs (lncRNAs) play an important role in cancer progression. However, there have been limited reports to date of the involvement of ubiquitin-binding protein domain protein 10 antisense RNA 1 (UBXN10-AS1) in cancer. Our aim was to explore the role and underlying mechanism of UBXN10-AS1 in the occurrence of colon adenocarcinoma (COAD). Real-time quantitative PCR and Western blotting were performed to determine the expression of UBXN10-AS1, miR-515-5p, and Slit guidance ligand 3 (SLIT3). Cell Counting Kit-8 and wound healing scratch assays were performed to measure COAD cell proliferation and migration. A xenograft assay was performed to examine tumor growth in vivo. Luciferase reporter and RNA immunoprecipitation (RIP) assays were used to determine the binding interaction among miR-515-5p, UBXN10-AS1, and SLIT3. The results showed that UBXN10-AS1 and SLIT3 were expressed at low levels in COAD tissues, while miR-515-5p was expressed at high levels. UBXN10-AS1 overexpression suppressed tumor growth in vitro and in vivo. The luciferase reporter and RNA RIP assays demonstrated that UBXN10-AS1 targeted miR-515-5p, which in turn targeted SLIT3. Functionally, miR-515-5p overexpression reversed the inhibition of COAD cell proliferation and migration by UBXN10-AS1 overexpression, and SLIT3 overexpression counteracted the oncogenicity of miR-515-5p. Our study shows that UBXN10-AS1 modulates the miR-515-5p/SLIT3 axis, thereby resulting in the inhibition of COAD cell proliferation and migration.
KEYWORDS: UBXN10-AS1, in vivo, proliferation, migration, colon adenocarcinoma
Introduction
Colon cancer is a frequently diagnosed gastrointestinal malignancy of the bowel. According to the GLOBOCAN database, approximately 1,148,515 new cases and 576,858 deaths were recorded in 2020 globally, ranking fifth in terms of incidence and mortality among human neoplastic disorders [1]. Tobacco use, environmental pollution, excessive alcohol consumption, and obesity increase the risk of colon cancer and the economic burden [2]. Tremendous progress has been made in the diagnostic and therapeutic modalities for colon cancer [3]; however, the prognosis of metastatic colon cancer remains poor. Importantly, colon adenocarcinoma (COAD) accounts for 95% of colon cancer cases, and the 5-year overall survival is approximately 65%. Therefore, elucidation of the mechanisms underlying COAD progression is needed to explore how disease interventions may be improved.
Long non-coding RNAs (lncRNAs) are a major subclass of non-coding RNAs that are longer than 200 nucleotides. Increasing evidence has demonstrated that non-coding transcriptomes are critical for the complex regulation that occurs during physiological and pathological processes [4–6]. For decades, many lncRNAs have been reported, and their cellular functionalities have been characterized in association with COAD [7,8]. For example, Zhou et al. explored 10 prognostic autophagy-related lncRNAs by analyzing RNA sequencing data of a COAD cohort from TCGA [7]. The lncRNA SNHG17 is unregulated in COAD tissues and promotes the formation of malignant phenotypes of COAD cells [9]. An oncogenic function for lncRNA ZEB1-AS1 has also been reported in COAD [10]. In addition, highly expressed lncRNA USP2-AS1 displays positive association with negative prognostic factors exhibited by patients with COAD. By contrast, there have been few reports to date regarding the 623-bp lncRNA UBXN10-AS1 [11]. Shao et al. reported that UBXN10-AS1 is expressed at low levels in prostate cancer tissues and is related to patient prognosis [12]. However, the role of UBXN10-AS1 in COAD remains unknown.
The Slit guidance ligand 3 (SLIT3) gene is located on 5q34-q35.1 and consists of 37 exons. SLIT3 is broadly expressed in normal tissues and encodes a 37-kDa protein that interacts with roundabout homolog receptors to affect cell migration. Originally, SLIT3 was found to play a role in the central nervous system [13]. Increasing evidence indicates that it also has a role in cancer. For example, SLIT3 deficiency inhibits hepatocellular carcinoma progression both in vitro and in vivo [14]. Its anti-tumorigenic role has also been reported in studies on thyroid cancer [15]. However, little is known about its function in the progression of COAD.
In the present study, we found that the lncRNA UBXN10-AS1 is downregulated in COAD tissues and is predominantly localized in the cytoplasmic fraction of COAD cells. After performing bioinformatics analysis, it was predicted that SLIT3 may be downstream of the UBXN10-AS1-mediated ceRNA network, exerting effects on apoptosis and cell proliferation. Therefore, SLIT3 was considered as a gene of interest in COAD. Bioinformatics analysis revealed that miR-515-5p is the only miRNA that can connect UBXN10-AS1 and SLIT3. Moreover, miR-515-5p is of interest because of its role in different cancers, similar to that of other miRNAs [16–18]. MiR-515-5p promotes breast cancer progression by regulating the Hippo signaling pathway [19]; it also regulates prostate cancer cell growth and metabolism [20]. MiR-515-5p is downregulated in metastasized tissues in in vivo lung xenografts, and its exogenous expression can suppress cancer metastasis [21]. However, the function of miR-515-5p in COAD remains unexplored.
In our study, we hypothesized that UBXN10-AS1, miR-515-5p, and SLIT3 play a role in COAD progression. Therefore, the purpose of our study was to explore the expression and functionality of UBXN10-AS1 in COAD and determine the UBXN10-AS1-mediated mechanism in COAD progression.
Methods
Patient tissues and cell lines
Surgical COAD tissues and their corresponding uncancerous tissues were collected from COAD patients diagnosed at the Clinical Medical College & Affiliated Hospital of Chengdu University Hospital. Informed consent was obtained from the patients included. Ethical approval was obtained from the Ethics Committee of the Clinical Medical College & Affiliated Hospital of Chengdu University.
Normal colon (FHC) and colorectal cancer (HCT116, SW480, and SW620) cell lines were purchased from the American Tissue Collection Center (ATCC, USA) and maintained in DMEM containing 10% FBS and 1% streptomycin/penicillin at 37°C and 5% CO2.
Cell transfection
All transfections were carried out using Lipo3000 (Invitrogen, USA), as specified by the manufacturer. The packaged lentivirus vectors for UBXN10-AS1-overexpression (LV-UBXN10-AS1) and empty lentivirus vectors (LV-NC), pcDNA 3.1-SLIT3 for SLIT3 overexpression and empty pcDNA 3.1 vectors (NC), si-UBXN10-AS1 for UBXN10-AS1 silencing, si-SLIT3 for SLIT3 silencing, and si-NC were obtained from GenePharma (Shanghai, China) along with miR-515-5p mimic or mimic NC. For UBXN10-AS1 overexpression, the LV-UBXN10-AS1 or LV-NC vectors were introduced in 5 × 106 SW480 and SW620 cells (70% confluence) at an MOI of 20–30. At 72 h post-transfection, its efficiency was verified using RT-qPCR. For transfection of pcDNA 3.1-SLIT3, miR-515-5p mimic, or their matching NCs, the indicated vectors or mimics were transfected in 4 × 105 SW480 and SW620 cells (70% confluence) for 48 h. The efficiency of transfection was determined by RT-qPCR or Western blotting.
Real-time quantitative PCR (RT-qPCR)
Total RNA extraction was performed using the TRIzol reagent. RNA reverse transcription into cDNA was performed with a TonkBio™ RT reagent Kit (TonkBio, USA) or a One Step miRNA cDNA Synthesis Kit (HaiGene, Biotech Co. Ltd, China) according to the manufacturer’s instructions. qPCR was performed using Universal SYBR Green qPCR Supermix (US EVERBRIGHT® INC., China). The qPCR data were analyzed by the 2−ΔΔCt method [22] with normalization relative to GAPDH or U6. The primers used are listed in Table 1.
Table 1.
PCR primers used in this study
| Gene | Primer type | Sequence |
|---|---|---|
| UBXN10-AS1 | Forward | 5ʹ-GTTGCATAGGTCCCTCGGTT-3ʹ |
| Reverse | 5ʹ-AGACGAGCAGAAACACCACC-3ʹ | |
| miR-515-5p | Forward | 5ʹ-TTCTCCAAAAGAAAGCACTTTCTG-3ʹ |
| Reverse | 5ʹ-TGGTGTCGTGGAGTCG-3ʹ | |
| SLIT3 | Forward | 5′- AGCGCCTTGACCTGGACA −3 |
| Reverse | 5′- TCGGCGTGCTCTGGAAAA −3′ | |
| miR-515-5p | Forward | 5ʹ-CTCGCTTCGGCAGCACA-3ʹ |
| Reverse | 5ʹ-ACGCTTCACGAATTTGCGT-3ʹ | |
| GAPDH | Forward | 5ʹ-GAGAAACCTGCCAAGTATGATGAC-3ʹ |
| Reverse: | 5ʹ-GGAGTTGCTGTTGAAGTCAC-3ʹ |
Western blot analysis
Cell pellets were collected and sonicated in pre-cooled RIPA lysis buffer before successive centrifugations. The supernatants were collected and their protein concentrations were determined using a DC protein assay. Total lysate (30 µg) was separated by 10% SDS-PAGE. A semi-dry transfer system (Bio-Rad, USA) was used to transfer the proteins to PVDF membranes. After treatment with 10 mL of blocking buffer at room temperature, the membranes were successively treated with 10 mL of blocking buffer containing 1:1,000 anti-SLIT3 or anti-GAPDH. The next day, bands were detected following incubation with a secondary antibody for 1 h at room temperature. ECL reagents were used to visualize the bands.
Cell proliferation assay
Cell Counting Kit-8 (CCK8, MCE, China) was used to assess cell proliferation in vitro, as described previously [23]. Cells (104–105 cells/well) were seeded into 96-well plates and cultured in a CO2 incubator at 37°C for 24, 48, 72, and 96 h. Subsequently, 10 μL of CCK8 reagent was added to the wells, and the plate was incubated for 1 h. The absorbance was measured at 450 nm using a microplate reader.
Cell migration assay
Wound healing assay was performed to assess cell migration, as reported previously [24]. Cells (104–105 cells/well) were cultured in 24-well tissue culture plates until they reached 70–80% confluence. A wound-like gap was created in the wells using a 200 µL micropipette tip. Cells were grown for an additional 24 h and then fixed with 3.7% paraformaldehyde for 30 min. The fixed cells were stained with 1% crystal violet for 30 min. Microscope images of the stained monolayers were captured at 0 h and 24 h.
Luciferase reporter assay
Luciferase reporter assays were performed to confirm the direct interaction among miR-515-5p, UBXN10-AS1, and SLIT3. The luciferase reporter vectors were pmirGLO luciferase constructs fused with the wild-type (WT) UBXN10 fragment that binds miR-515-5p or the 3ʹ UTR of SLIT3 (UBXN10-AS1-WT, SLIT3-WT) or the corresponding mutant (MUT) fragment (UBXN10-AS1-MUT, SLIT3-MUT) obtained from Cobioer Biosciences Co. Ltd. Cells at 70–80% confluence were transfected with miR-515-5p mimic or NC coupled with UBXN10-AS1-WT/UBXN10-AS1-MUT or SLIT3-WT/SLIT3-MUT vectors. After 48 h, the cell luciferase signals were quantified using a Luciferase Reporter Assay Substrate Kit (Abcam, USA) as previously described [25].
Luciferase activity was calculated as follows:
Luciferase activity = Firefly luciferase/Renilla luciferase.
RNA immunoprecipitation (RIP) assay
Cells (5 × 106) were sonicated using a Bioruptor. After removal of cell debris, the remaining lysate was added to antibody-coupled beads containing AGO2 or IgG. The immunoprecipitated RNA complex was collected and uncrosslinked. Finally, RNA extraction was performed as previously described for RT-qPCR [26].
Subcellular distribution
UBXN10-AS1 nucleus/cytoplasm localization in SW480 and SW620 cells was determined using a Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions [27].
Tumor formation in nude mice
Ten BALB/c-nu/nu male mice (body weights 18–20 g) were purchased from the Animal Resource Center at the Wuhan Institute of Virology, Chinese Academy of Sciences, China. The mice were housed in cages with appropriate temperature and humidity and a 12-h light/dark cycle. SW480 cells containing LV-UBXN10-AS1 or LV-NC were injected subcutaneously in the left armpit of nude mice. Starting at four days post-implantation, tumor volumes were recorded every four days. Tumor volume = (longest diameter of the tumor) × (shortest diameter)2 × 0.5 [28]. The tumor weights were recorded on day 28 after the mice were euthanized.
Statistical analyses
Statistical analyses were performed using the GraphPad Prism 9.0 software. The results are presented as mean ± SD (n = 3). Comparisons between groups were performed using the unpaired Student’s t-test and those among groups using one-way ANOVA followed by Tukey’s post hoc test. Statistical significance was set at P < 0.05.
Results
In this study, we aimed to explore the expression and functionality of UBXN10-AS1 in COAD in vitro and in vivo. The UBXN10-AS1-mediated mechanism involved in COAD progression was further investigated. Our findings revealed that UBXN10-AS1 inhibits the progression of COAD by sponging miR-515-5p to upregulate SLIT3.
UBXN10-AS1 may regulate the miR-515-5p/SLIT3 axis in COAD
The expression of UBXN10-AS1 was downregulated in COAD samples based on the GEPIA database (Figure 1a), suggesting that its negative effect on COAD warranted further exploration. The key gene (SLIT3) involved in apoptosis and cell proliferation was identified using STRING analysis from the mRNA microarray (GSE126095) with adjusted P < 0.05 and logFC < −2 (Figure 1b). Next, to identify the miRNA connecting UBXN10-AS1 and SLIT3, miRDB was used to predict miRNAs that could bind to UBXN10-AS1, while starBase was used to predict miRNAs that could bind to SLIT3. Finally, miR-515-3p was confirmed to be the common miRNA between miRDB and starBase (Figure 1c).
Figure 1.
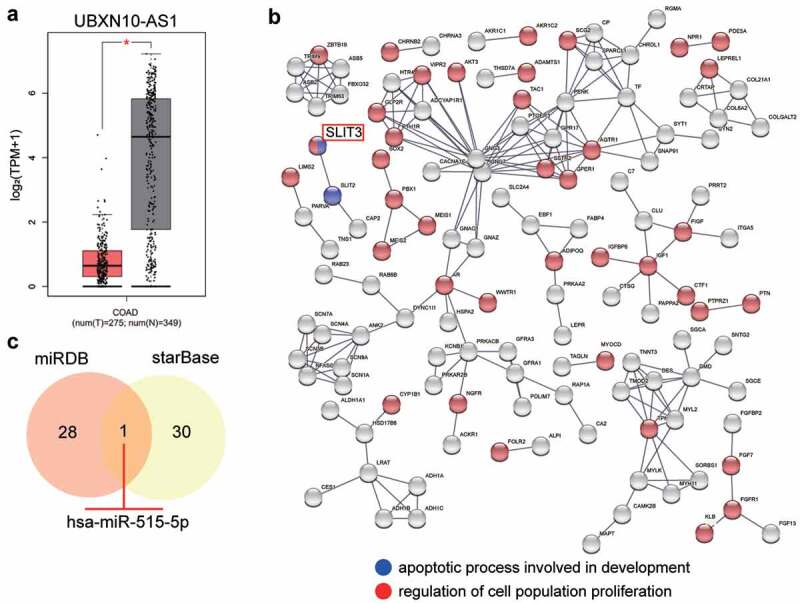
UBXN10-AS1 might be the upstream of miR-515-5p/SLIT3 axis in COAD. (A) The expression of UBXN10-AS1 in COAD samples and normal samples from GEPIA database. B The GO enrichment of upregulated genes in GSE126095 was performed by STRING. GSE126095, mRNA expression microarray. **P < 0.001. C The miR-515-5p might bind to UBXN10-AS1 and SLIT3 by the prediction of miRDB and starBase. miRDB, an online tool for prediction of miRNAs sponged by UBXN10-AS1. starBase, an online tool for prediction of miRNAs targeting SLIT3.
UBXN10-AS1 overexpression inhibits COAD tumorigenesis
To analyze the effect of UBXN10-AS1 expression in COAD, RT-qPCR analysis was performed to detect its expression in COAD cells and tissues. As depicted in Figure 2(a,b), UBXN10-AS1 expression was low in COAD cells and tissues; moreover, in SW480 and SW620 cells, its expression was reduced by approximately 0.5-fold. Next, we determined the subcellular localization of UBXN10-AS1 in SW480 and SW620 cells. The results showed that UBXN10-AS1 was primarily enriched in the cytoplasm (Figure 2c), suggesting that it may carry out its functions via translational and posttranslational modifications in the cytoplasm. Therefore, we performed in vivo and in vitro assays to assess the impact of UBXN10-AS1 overexpression on COAD progression. Both SW480 and SW620 cells were successfully transfected with the vectors to overexpress UBXN10-AS1, as evidenced by the high expression of UBXN10-AS1 in cells from both the cell lines (Figure 2d). CCK8 assays showed that UBXN10-AS1 overexpression caused an impaired proliferation in COAD cells (Figure 2e). Meanwhile, the wound healing assay proved that UBXN10-AS1 overexpression impaired the COAD cell migration (figure 2f). Subsequently, the anti-tumorigenic role of UBXN10-AS1 overexpression was assessed in SW480 xenografted nude mice. We subcutaneously injected SW480 cells infected with LV-UBXN10-AS1 vectors or LV-NC vectors into BALB/c mice and then monitored the tumor growth. As shown in Figure 2g, LV-UBXN10-AS1 cell-derived xenograft tumors were smaller than those in the NC groups. Moreover, LV-UBXN10-AS1 cell-derived xenograft tumors grew more slowly than those in the NC groups. Our findings show that UBXN10-AS1 plays an anti-tumorigenic role in COAD in vitro and in vivo.
Figure 2.
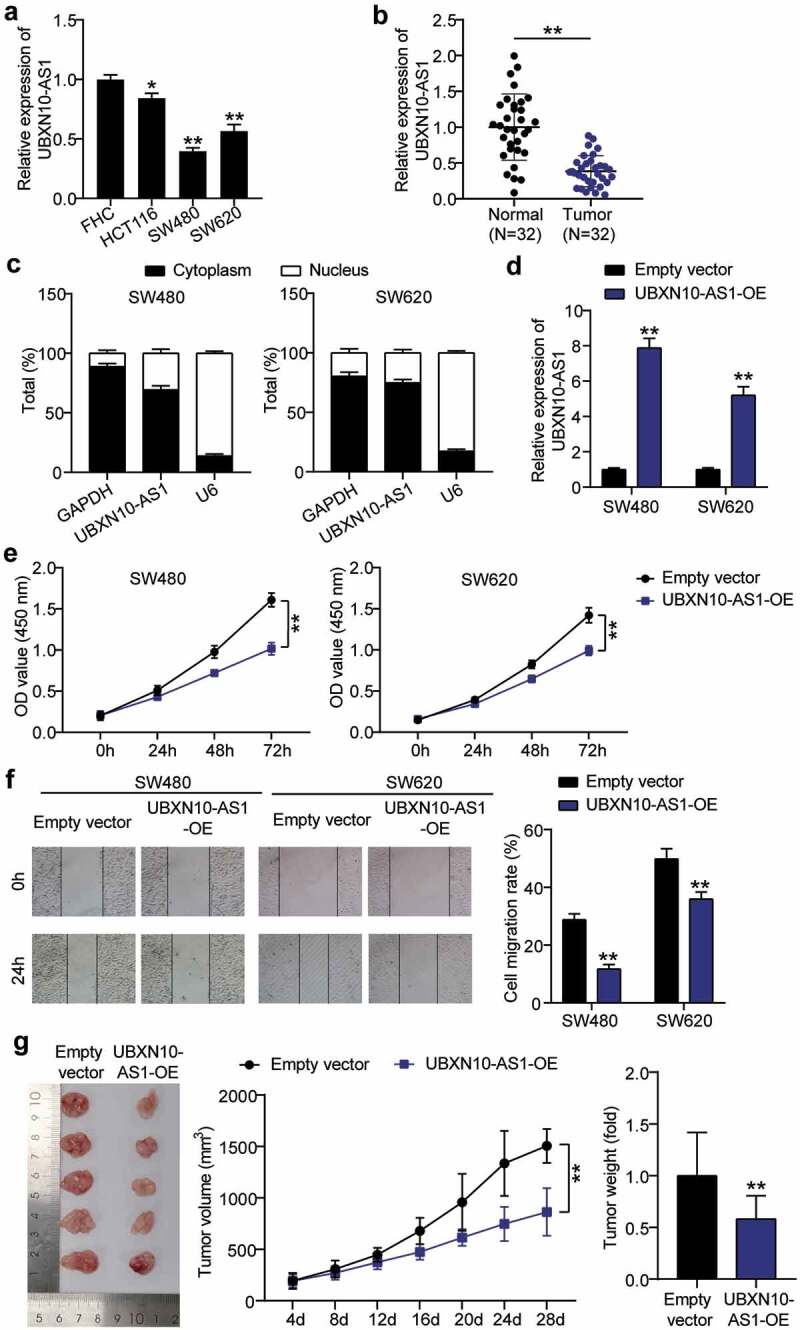
TRIM28 promotes prostate cancer tumorgenesis in vitro and in vivo. A. RT-qPCR analysis assessing UBXN10-AS expression was detected in COAD cells and FHC cells. *P < 0.05,**P < 0.001 vs. FHC. B. RT-qPCR analysis assessing UBXN10-AS expression was detected in COAD tissue and normal tissues. **P < 0.001. C. Subcellular localization of UBXN10-AS1 in SW480 and SW620 cells. D. UBXN10-AS expression in SW480 and SW620 cells infected with Lv-UBXN10-AS and Lv-NC by RT-qPCR. E. CCK8 assay determining the proliferation of cells in (D). F. Wound healing assay assessing the migration of cells in (D). G. UBXN10-AS1 overexpression inhibits in vivo tumor growth. The tumor volume and weight were significantly lower in the UBXN10-AS-overepxression group than those in the NC group. (D-G) Empty vector, empty lentivirus vectors. UBXN10-AS1-OE, UBXN10-AS1-overexpressing lentivirus vectors. **P < 0.001 vs. empty vectors.
MiR-515-5p is a target of UBXN10-AS1 in COAD
Considering the enrichment of UBXN10-AS1 in the cytoplasm, bioinformatic target prediction conducted in the present study revealed that UBXN10-AS1 shared complementary base pairs with miR-515-5p (Figure 3a). The interaction between miR-515-5p and UBXN10-AS1 was verified using luciferase reporter assays. As shown in Figure 3b, miR-515-5p mimic transfection reduced the UBXN10-AS1-WT-mediated luciferase activity but had no impact on UBXN10-AS1-MUT-mediated luciferase activities. Moreover, the results of the RNA RIP experiment demonstrated abundant precipitation of miR-515-5p and UBXN10-AS1 in the AGO2 pellets (Figure 3c), hinting toward the coexistence of UBXN10-AS1 in the same RNA-induced silencing complex. We also noted the abundant expression of miR-515-5p in the SW480 and SW620 cells as well as in the COAD tissues (Figure 3d,e). Furthermore, Pearson analysis showed a prominent correlation between miR-515-5p and UBXN10-AS1 in the COAD tissues (figure 3f). Thus, we concluded that UBXN10-AS1 targets miR-515-5p.
Figure 3.
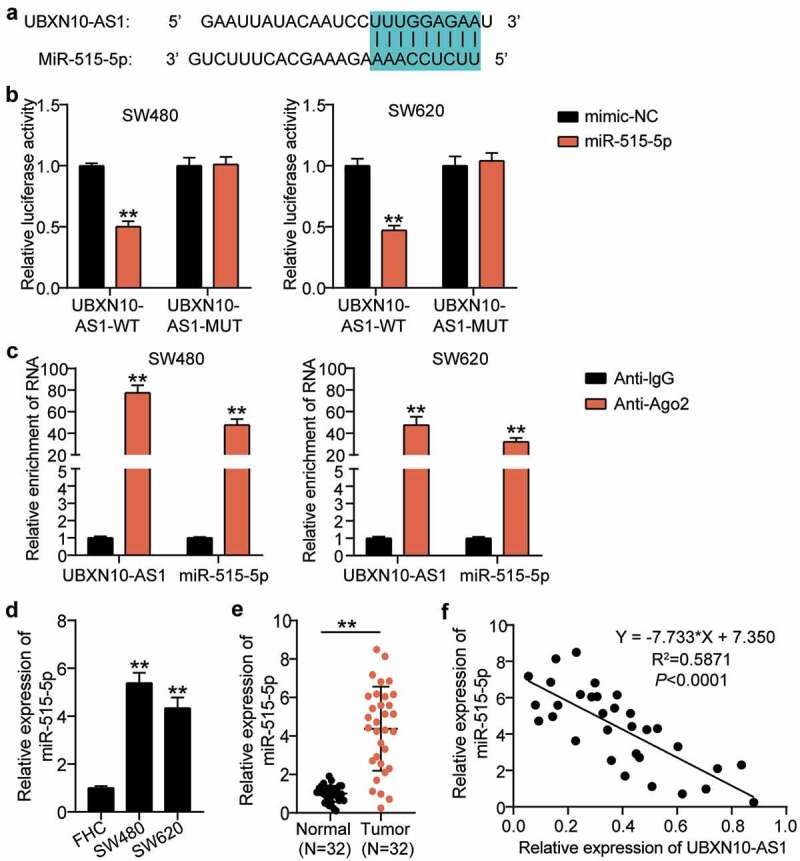
UBXN10-AS1 targeted miR-515-5p directly in COAD. A. By means of searching on starBase, miR-515-5p was predicted to have the potentials to bind with UBXN10-AS1. B. Luciferase reporter assay in SW480 and SW620 cells after co-transfection with miR-515-5p mimic or NC and UBXN10-AS1-WT or UBXN10-AS1-MUT. **P < 0.001 vs. mimic-NC. C. RIP assay was implemented, and relative enrichment of miR-515-5p and UBXN10-AS1 was detected by RT-qPCR. **P < 0.001 vs. Anti-IgG. D. The relative expression of miR-515-5p in SW480 and SW620 cells. **P < 0.001 vs. FHC. E. The relative expression of miR-515-5p in COAD tissues. **P < 0.001. F. Correlation between the expression of miR-515-5p and UBXN10-AS1 in COAD tissues was analyzed by Pearson’s correlation analysis.
UBXN10-AS1 regulates COAD cell proliferation and migration by sponging miR-515-5p
Rescue assays were performed to elucidate whether the interplay between UBXN10-AS1 and miR-515-5p influences COAD progression. We first tested miR-515-5p expression in the SW480 and SW620 cells transfected with miR-515-5p mimic or LV-UBXN10-AS1. As shown in Figure 4a, RT-qPCR results revealed a drastic reduction of miR-515-5p expression in COAD cells transfected with LV-UBXN10-AS1, which was completely reversed by the simultaneous transfection of the cells with LV-UBXN10-AS1 and miR-515-5p mimic, indicating that UBXN10-AS1 can downregulate miR-515-5p expression in COAD cells. The CCK8 assays demonstrated that the increased proliferation caused by the miR-515-5p mimic could be offset by concomitant transfection with LV-UBXN10-AS1 (Figure 4b). Meanwhile, the pro-migratory effect of the miR-515-5p mimic on the SW480 and SW620 cells was counteracted by LV-UBXN10-AS1 transfection (Figure 4c). As described above, LV-UBXN10-AS1 reduced miR-515-5p expression via ceRNA activity and influenced COAD cell proliferation and migration.
Figure 4.
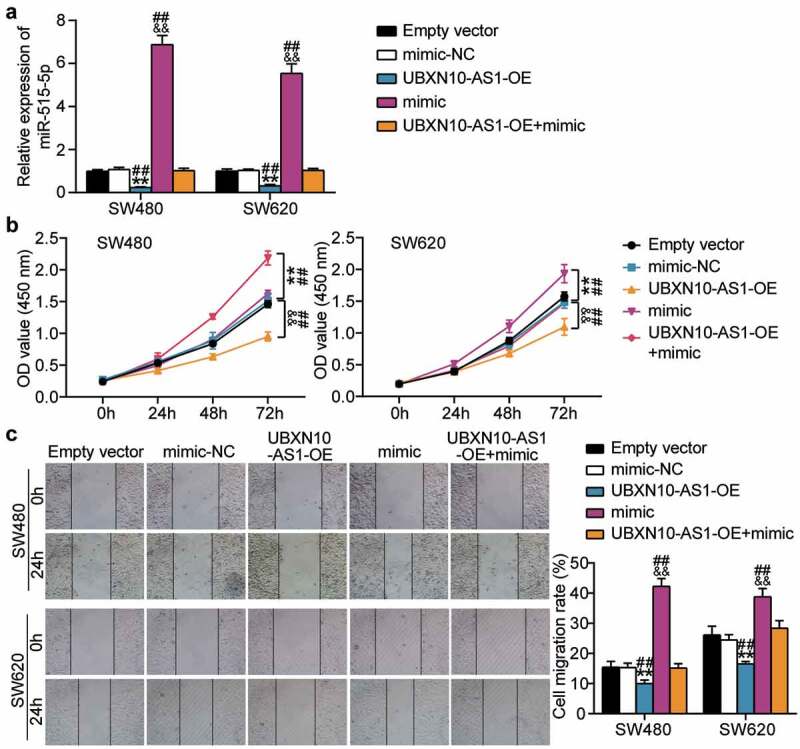
Upregulation of miR-515-5p abrogates the anti-tumorigenic effects of UBXN10-AS1 overexpression in COAD cells. A. RT-qPCR examining miR-515-5p expression in SW480 and SW620 cells. B. CCK8 assays detected the proliferation of SW480 and SW620 cells. C. Scratch wound-healing migration assay examined the migration of SW480 and SW620 cells. Empty vector, empty lentivirus vectors. UBXN10-AS1-OE, UBXN10-AS1-overexpressing lentivirus vectors. mimic, miR-515-5p mimic. **P < 0.001 vs. Empty vector; &&P < 0.001 vs. mimic-NC; ##P < 0.001 vs. UBXN10-AS1-OE+mimic.
SLIT3 is a target of miR-515-5p
After determining the effect of the ceRNA activity of UBXN10-AS1 on miR-515-5p, we further sought to identify the mRNA controlled by the UBXN10-AS1/miR-515-5p axis. Prediction with the starBase software revealed a highly matched sequence between miR-515-5p and 3ʹ UTR of SLIT3 (Figure 5a). This prediction was further validated by luciferase reporter assays, which demonstrated that miR-515-5p dramatically reduced the luciferase activity of 3ʹ UTR SLIT3-WT (Figure 5b). In contrast to the high expression of miR-515-5p in COAD cells and tissues, we found clear evidence of the downregulation of SLIT3 (Figure 5c,d). Further correlation analysis showed that SLIT3 was inversely correlated with miR-515-5p expression (Figure 5e). More importantly, UBXN10-AS1 overexpression induced increased expression of SLIT3, while its silencing drastically reduced SLIT3 expression in the SW480 and SW620 cells (figure 5f). All these data determined the ceRNA activity of UBXN10-AS1, miR-515-5p, and SLIT3.
Figure 5.
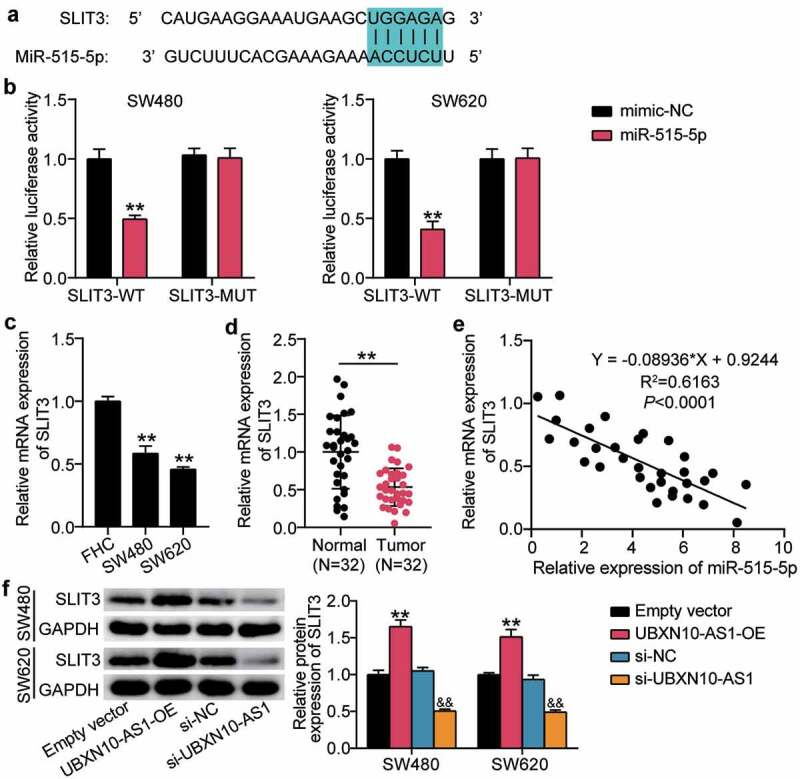
miR-515-5p targets SLIT3 directly in COAD. A. The wild-type putative binding sequences of miR-515-5p in the sequence of the SLIT3 3ʹ-UTR. B. SLIT3 3ʹ-UTR (WT and MUT) luciferase vectors were co-transfected with miR-515-5p mimic or mimic NC, the activity of SLIT3 3ʹ-UTR was measured by luciferase. Renilla was used as control. **P < 0.001 vs. mimic-NC. C. Detection of SLIT3 expression in SW480 and SW620 cells by RT-qPCR, **P < 0.001 vs. FHC. D. Detection of SLIT3 expression in COAD tissues by RT-qPCR. **P < 0.001. E. The relationship between SLIT3 mRNA and miR-515-5p in COAD tissues was determined by Pearson’s correlation analysis. F. Western blotting analysis of SLIT3 expression in SW480 and SW620 cells transfected with UBXN10-AS1 overexpression or silence. Empty vector, empty lentivirus vectors. UBXN10-AS1-OE, UBXN10-AS1-overexpressing lentivirus vectors. **P < 0.001 vs. empty vector, &&P < 0.001 vs. si-NC.
MiR-515-5p regulates COAD cell proliferation and migration by downregulation of SLIT3
After validating the direct binding and regulation of miR-515-5p on SLIT3 mRNA, we further explored whether SLIT3 is required for miR-515-5p activity during COAD malignancy. We transfected the SW480 and SW620 cells with miR-515-5p mimic, pcDNA-SLIT3 vector (OE), and OE+miR-515-5p mimic. Western blotting showed that overexpression of miR-515-5p markedly reduced SLIT3 expression, which was restored by additional pcDNA-SLIT3 transfection (Figure 6a). In vitro assays demonstrated that ectopic SLIT3 expression reduced proliferation and migration, while the anti-proliferation and anti-migration effects of SLIT3 overexpression were nullified by the miR-515-5p mimic (Figure 6b,c). Collectively, miR-515-5p exerts its anti-tumorigenic effect by downregulating SLIT3.
Figure 6.
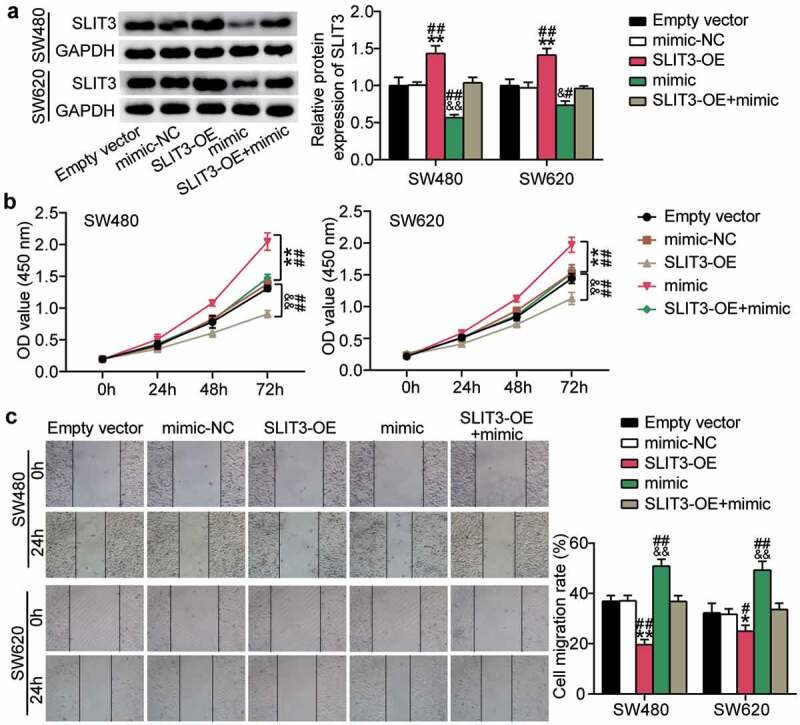
SLIT3 overexpression offsets the pro-tumorigenic role of miR-515-5p overexpression in COAD. A. The expression changes of SLIT3 protein were assessed by Western blots. B. CCK8 assays detected the proliferation capacity of indicated SW480 and SW620 cells. C. scratch wound-healing migration assays detected the migration capacity of indicated SW480 and SW620 cells. Empty vector, pcDNA 3.1 empty vectors. SLIT3-OE, SLIT3 overexpression vectors. mimic, miR-515-5p mimic. *P < 0.001, **P < 0.001 vs. Empty vector; &P < 0.05, &&P < 0.001 vs. mimic-NC; #P < 0.05, ##P < 0.001 vs. SLIT3-OE+mimic.
Upregulation of SLIT3 neutralizes UBXN10-AS1-mediated tumor-suppression in vitro
To further elucidate the role of SLIT3 in UBXN10-AS1-mediated tumor repression, we introduced the UBXN10-AS1-OE vectors into the SW620 and SW480 cells coupled with si-SLIT3. Western blots showed significant si-SLIT3-mediated nullification of increased SLIT3 expression by UBXN10-AS1-OE vectors (Figure 7a). Functionally, upon SLIT3 silencing, the proliferative and migratory rates of the SW620 and SW480 cells were enhanced; however, these effects were almost fully mitigated by UBXN10-AS1 overexpression (Figure 7b,c). In summary, SLIT3 silencing almost fully eliminated the tumor-suppressing role of UBXN10-AS1 in COAD.
Figure 7.
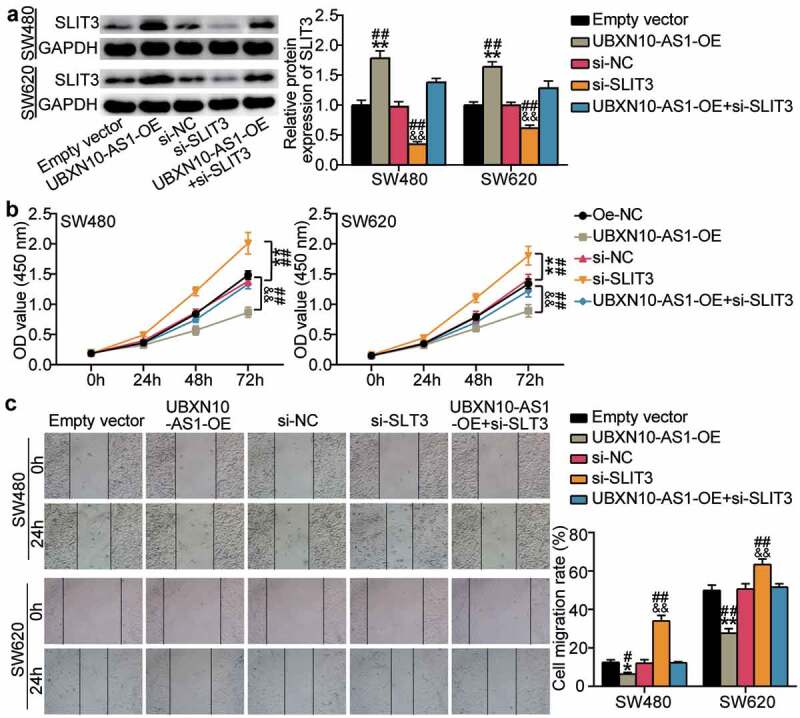
Upregulation of SLIT3 neutralizes UBXN10-AS1’s tumor-suppressionin vitro. A. The expression of SLIT3 protein were determined by Western blots. B. CCK8 assays detected the proliferation capacity of indicated SW480 and SW620 cells. C. wound healing assay detected the migration capacity of indicated SW480 and SW620 cells. Empty vector, empty lentivirus vectors. UBXN10-AS1-OE, UBXN10-AS1-overexpressing lentivirus vectors. si-NC, siRNA negative control. *P < 0.05, **P < 0.001 vs. Empty vector; &&P < 0.001 vs. si-NC; #P < 0.05, ##P < 0.001 vs. si-SLIT3+ UBXN10-AS1-OE.
Discussion
In the present study, we found that UBXN10-AS1 is expressed at low levels in COAD tissues and that its overexpression suppressed COAD cell proliferation in vitro and in vivo. More importantly, we identified that UBXN10-AS1 acts as a tumor suppressor partially due to its ceRNA activity to regulate the miR-515-5p/SLIT3 axis.
LncRNAs have been reported to be aberrantly expressed in various malignancies, and they can reshape oncogene or anti-oncogene expression epigenetically, transcriptionally, and post-transcriptionally, which is why they can serve as oncogenes or tumor suppressors [29]. In the past several decades, an increasing number of lncRNAs have been found to have such functions [30]. In keeping with this trend, we found that UBXN10-AS1 was downregulated in COAD tissues and cells. To date, the unregulated expression of UBXN10-AS1 has only been reported in studies on prostate cancer and thyroid carcinoma [12,31], and its function has remained understudied. Our gain-of-function analysis demonstrated that UBXN10-AS1 overexpression reduced COAD cell proliferation and migration in vitro and slowed tumor growth in vivo. To the best of our knowledge, this is the first study demonstrating the suppressive effect of UBXN10-AS1 on COAD progression in vitro and in vivo.
To identify the UBXN10-AS1-mediated mechanism of regulation of COAD progression, we compared UBXN10-AS1 expression in the cytoplasm and nucleus of SW480 and SW620 cells. Our subcellular distribution analysis demonstrated that UBXN10-AS1 was mainly located in the cytoplasm, indicating that it can perform cytoplasmic functions, such as miRNA sponging. Indeed, as evidenced by luciferase reporter and RNA RIP assays, UBXN10-AS1 could exert its effects via sponging miR-515-5p. MiR-515-5p has been reported to be downregulated in several cancers, such as prostate and breast cancer, and it can function as a tumor suppressor during cancer progression. For example, prostate cancer patients with high-miR-515-5p expression display favorable clinical outcomes [20]. Furthermore, miR-515-5p overexpression can reduce the malignant phenotype of hepatocellular carcinoma cells [32]. However, our data show that miR-515-5p was abundantly expressed in COAD tissues and cells. Here, the functional study of cells also demonstrated that miR-515-5p can increase COAD cell proliferation and migration. Our findings suggest that miR-515-5p is an oncomiR in COAD. These opposing effects of miR-515-5p suggested that miR-515-5p may function as an oncomiR or an anti-oncogene in different tumor models. Reports of such functions have increased considerably [33]. Interestingly, a negative correlation between miR-515-5p and UBXN10-AS1 also indicated that UBXN10-AS1 may at least partially downregulate miR-515-5p expression, thus offsetting the its oncogenicity in COAD. Our rescue assays showed that UBXN10-AS1 overexpression can nullify miR-515-5p overexpression-mediated increased COAD cell proliferation and migration. These results showed that UBXN10-AS1 can sponge miR-515-5p and suppress COAD cell proliferation and migration.
MiRNA/mRNA interactions enable miRNAs to downregulate mRNA expression via targeted binding. The regulatory mechanism of miR-515-5p in COAD progression has not been previously reported. In the current study, we identified SLIT3 as a downstream target of miR-515-5p in COAD cells. The tumor suppressor role of SLIT3 in thyroid cancer and hepatocellular carcinoma has been reported earlier [14,15]. In the present study, we found that SLIT3 was expressed at low levels in COAD tissues and was inversely related to miR-515-5p expression. More importantly, SLIT3 overexpression enhanced the effect of UBXN10-AS1 overexpression and inhibited COAD cell proliferation and migration. Thus, SLIT3 may have a dual role in cancer progression. Meanwhile, miR-515-5p can restore the suppressed COAD cell proliferation and migration regulated by SLIT3 overexpression, confirming that miR-515-5p suppresses COAD cell activities by targeting SLIT3. This relationship was further validated by our subsequent results showing that UBXN10-AS1 promoted SLIT3 expression. Thus, UBXN10-AS1 competitively binds miR-515-5p and stepwise abolishes the inhibition of SLIT3 expression, resulting in tumor inhibition.
However, our study has some limitations. First, no in vivo assays were performed. Second, the UBXN10-AS1-mediated cell functions are complex, and further studies are warranted to fully understand its function in COAD progression. Third, the sample size was small.
Conclusions
UBXN10-AS1 was expressed at low levels in COAD cells, and it may suppress COAD cell proliferation and migration via the miR-515-5p/SLIT3 axis. Our findings provide further insights into lncRNA-associated targets for COAD treatment.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Availability of Data and Material
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
YT and JXC conducted the study and collected and analyzed the data. JXC and BL designed the study and experimental layout and procured the funding. YT analyzed and interpreted the data. BL carried out the literature analysis and drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval
The present study was approved by the Ethics Committee of the Clinical Medical College & Affiliated Hospital of Chengdu University (Chengdu, China). The processing of clinical tissue samples was in strict compliance with the ethical standards of the Declaration of Helsinki.
Consent to participate
All patients provided attested written informed consent.
Consent for publication
Consent for publication was obtained from the participants.
References
- [1].Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- [2].Otani K, Kawai K, Hata K, et al. Colon cancer with perforation. Surg Today. 2019;49(1):15–20. [DOI] [PubMed] [Google Scholar]
- [3].Wu C. Systemic therapy for colon cancer. Surg Oncol Clin N Am. 2018;27(2):235–242. [DOI] [PubMed] [Google Scholar]
- [4].Li Z, Sun Y, He M, et al. Differentially-expressed mRNAs, microRNAs and long noncoding RNAs in intervertebral disc degeneration identified by RNA-sequencing. Bioengineered. 2021;12(1):1026–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Abolghasemi M, Tehrani SS, Yousefi T, et al. Critical roles of long noncoding RNAs in breast cancer. J Cell Physiol. 2020;235(6):5059–5071. [DOI] [PubMed] [Google Scholar]
- [6].Safa A, Gholipour M, Dinger ME, et al. The critical roles of lncRNAs in the pathogenesis of melanoma. Exp Mol Pathol. 2020;117:104558. [DOI] [PubMed] [Google Scholar]
- [7].Zhou W, Zhang S, Li HB, et al. Development of prognostic indicator based on autophagy-related lncRNA analysis in colon adenocarcinoma. Biomed Res Int. 2020;2020:9807918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang Z, Qian W, Wang S, et al. Analysis of lncRNA-associated ceRNA network reveals potential lncRNA biomarkers in human colon adenocarcinoma. Cell Physiol Biochem. 2018;49(5):1778–1791. [DOI] [PubMed] [Google Scholar]
- [9].Liu J, Zhan Y, Wang J, et al. lncRNA-SNHG17 promotes colon adenocarcinoma progression and serves as a sponge for miR-375 to regulate CBX3 expression. Am J Transl Res. 2020;12(9):5283–5295. [PMC free article] [PubMed] [Google Scholar]
- [10].Ni X, Ding Y, Yuan H, et al. Long non-coding RNA ZEB1-AS1 promotes colon adenocarcinoma malignant progression via miR-455-3p/PAK2 axis. Cell Prolif. 2020;53(1):e12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li D, Bao J, Yao J, et al. lncRNA USP2-AS1 promotes colon cancer progression by modulating Hippo/YAP1 signaling. Am J Transl Res. 2020;12(9):5670–5682. [PMC free article] [PubMed] [Google Scholar]
- [12].Shao N, Zhu Y, Wan FN, et al. Identification of seven long noncoding RNAs signature for prediction of biochemical recurrence in prostate cancer. Asian J Androl. 2019;21(6):618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen L, Yao JH, Zhang SH, et al. Slit-like 2, a novel zebrafish slit homologue that might involve in zebrafish central neural and vascular morphogenesis. Biochem Biophys Res Commun. 2005;336(1):364–371. [DOI] [PubMed] [Google Scholar]
- [14].Ng L, Chow AKM, Man JHW, et al. Suppression of Slit3 induces tumor proliferation and chemoresistance in hepatocellular carcinoma through activation of GSK3β/β-catenin pathway. BMC Cancer. 2018;18(1):621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Guan H, Wei G, Wu J, et al. Down-regulation of miR-218-2 and its host gene SLIT3 cooperate to promote invasion and progression of thyroid cancer. J Clin Endocrinol Metab. 2013;98(8):E1334–44. [DOI] [PubMed] [Google Scholar]
- [16].Abolghasemi M, Tehrani SS, Yousefi T, et al. MicroRNAs in breast cancer: roles, functions, and mechanism of actions. J Cell Physiol. 2020;235(6):5008–5029. [DOI] [PubMed] [Google Scholar]
- [17].Vaghari-Tabari M, Majidinia M, Moein S, et al. MicroRNAs and colorectal cancer chemoresistance: new solution for old problem. Life Sci. 2020;259:118255. [DOI] [PubMed] [Google Scholar]
- [18].Salamati A, Majidinia M, Asemi Z, et al. Modulation of telomerase expression and function by miRNAs: anti-cancer potential. Life Sci. 2020;259:118387. [DOI] [PubMed] [Google Scholar]
- [19].Qiao K, Ning S, Wan L, et al. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38(1):418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang X, Zhou J, Xue D, et al. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int J Biol Macromol. 2019;129:227–232. [DOI] [PubMed] [Google Scholar]
- [21].Pardo OE, Castellano L, Munro CE, et al. miR-515-5p controls cancer cell migration through MARK4 regulation. EMBO Rep. 2016;17(4):570–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. [DOI] [PubMed] [Google Scholar]
- [23].Yan S, Xu J, Liu B, et al. Long non-coding RNA BCAR4 aggravated proliferation and migration in esophageal squamous cell carcinoma by negatively regulating p53/p21 signaling pathway. Bioengineered. 2021;12(1):682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Martinotti S, Ranzato E. Scratch wound healing assay. Methods Mol Biol. 2020;2109:225–229. [DOI] [PubMed] [Google Scholar]
- [25].Barriscale KA, O’Sullivan SA, McCarthy TV. A single secreted luciferase-based gene reporter assay. Anal Biochem. 2014;453:44–49. [DOI] [PubMed] [Google Scholar]
- [26].Gagliardi M, Matarazzo MR. RIP: RNA immunoprecipitation. Methods Mol Biol. 2016;1480:73–86. [DOI] [PubMed] [Google Scholar]
- [27].Mitra S, Muralidharan SV, Di Marco M, et al. Subcellular distribution of p53 by the p53-responsive lncRNA NBAT1 determines chemotherapeutic response in neuroblastoma. Cancer Res. 2021;81(6):1457–1471. [DOI] [PubMed] [Google Scholar]
- [28].Chio CC, Wang YS, Chen YL, et al. Down-regulation of Fas-L in glioma cells by ribozyme reduces cell apoptosis, tumour-infiltrating cells, and liver damage but accelerates tumour formation in nude mice. Br J Cancer. 2001;85(8):1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang Y, Jin T, Shen H, et al. Identification of long non-coding RNA expression profiles and co-expression genes in thyroid carcinoma based on The Cancer Genome Atlas (TCGA) database. Med Sci Monit. 2019;25:9752–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ni JS, Zheng H, Ou YL, et al. miR-515-5p suppresses HCC migration and invasion via targeting IL6/JAK/STAT3 pathway. Surg Oncol. 2020;34:113–120. [DOI] [PubMed] [Google Scholar]
- [33].Lee YS, Dutta A. MicroRNAs in cancer. Annual Review of Pathology: Mechanisms of Disease. 2009;4(1):199–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.


