ABSTRACT
To explore a new marker which can detect bacterial vaginosis (BV) with high sensitivity and specificity quantitatively. According to the Nugent Score, vaginal secretions from study participants were divided into BV, healthy, and BV-intermediate groups. First, we compared the obvious differences and high abundance of bacteria in the three groups using 16S rRNA-sequencing, and screened out candidate markers. Then, quantitative detection of these candidate markers from the three groups was done using real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR), followed by evaluation of the sensitivity and specificity. Finally, we verified the new markers using clinical cases. Gardnerella vaginalis, Atopobium vaginae, Lactobacillus, Megasphaera were screened out by 16S rRNA-sequencing. RT-qPCR data were transformed and analyzed through ROC curves. PCR results for these bacteria were log-transformed using Lactobacillus crispatus as the numerator and other BV-related bacteria as the denominator. Four new indicators were found. Of these, log L. crispatus/G. vaginalis (L/G) <0 was the best indicator. The sensitivity, specificity, positive predictive value, and negative predictive value of our system were 93.5%, 97.2%, 96.6 and 94.6%, respectively. Combination of data for 16S rRNA-sequencing and RT-qPCR revealed four indicators for BV detection. Of these, log L/G < 0 was the best indicator. Creating a molecular-diagnostic system independent of the Nugent Score for BV could have an important impact on the clinical management of BV.
Abbreviation: log L. crispatus/G. vaginalis (logL/G); Bacterial vaginosis (BV); vaginal secretions (VSs); polymerase chain reaction (PCR); rRNA-sequencing (rRNA-seq); real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR); operational taxonomic unit (OTU); non-metric multidimensional scaling (NMDS); receiver operating characteristic (ROC).
KEYWORDS: Bacterial vaginosis, indicator, lactobacillus crispatus, gardnerella vaginalis
Introduction
Bacterial vaginosis (BV) is a clinical manifestation caused by a decrease in the abundance of Lactobacillus species and microbial imbalance in the vagina. An increase in thin vaginal secretions (VSs) is the main clinical manifestation. The prevalence of BV in women of child-bearing age is 8–40% [1]. Studies have shown that BV is related to premature birth, miscarriage, infertility, premature rupture of membranes, neonatal infection and pelvic inflammation [2,3]. Also, BV may increase the risk of the trichomonad vaginitis, vulvovaginal candidiasis, vulnerability to infection by the human immunodeficiency virus, and various sexually transmitted diseases [4–8], and a high reproductive rate [9,10]. About 10–40% of women with BV may be asymptomatic. Some studies have reported that >50% of pregnant women manifest no symptoms [11,12]. Therefore, the clinical diagnosis of BV may be reliant mainly on laboratory indicators.
The Amsel criteria are used widely to diagnose BV. The presence of three out of four positive criteria indicates that the cause of vaginal complaints is BV: (i) thin, white, yellow, homogeneous discharge; (ii) clue cells on microscopy; (iii) pH of vaginal fluid >4.5; (iv) release of a ‘fishy’ odor upon addition of 10% potassium hydroxide solution. However, application of the Amsel criteria can be subjective (e.g., judging a fishy smell) and is, therefore, limited.
Another way to diagnose BV is to use the Nugent Score: a Gram stain scoring system for vaginal swabs first proposed in 1991 [13]. A score from 0 to 10 is obtained from combining three other scores. A score of 0–3 is considered ‘negative’ for BV, 4–6 is considered ‘indeterminate’ for BV, and a score of 7+ is considered to indicate BV. However, use of the Nugent Score requires highly trained and experienced staff. Also, reliance on morphology for identifying bacteria is subjective, and can lead to mis-identification of bacteria. For example, Lactobacillus species with indeterminate staining can be mistaken for Gardnerella vaginalis or other Gram-variable bacilli [14,15]. During the past decade, molecular diagnostic tools have increased in importance to aid diagnoses [16–18], and have led to various methods to diagnose BV [19].
Major studies using polymerase chain reaction (PCR) methods have been employed to directly detect bacteria in VSs in relation to BV. About 35 bacterial species have been implicated in BV, including G. vaginalis, Atopobium vaginae, Porphyromonas asaccharolytica, various genera (Megasphaera, Sneathia, Prevotella, Peptostreptococcus) and bacterial vaginosis-associated bacterium (BVAB)1–3 [20–25]. Some commercial reagent kits can be used to diagnose BV through combined detection of A. vaginae, G. vaginalis, Megasphaera species (types 1 and 2), BVAB (type 1 and/or type 2) and Lactobacillus species. Other some reagent kits use single quantitative detection of G. vaginalis to diagnose BV. However, which may make the detecting indicator more complicated or simpler, especially, single bacterial species detection may ignore imbalanced vaginal flora. Moreover, the quantitative detection of bacteria will also be affected by secretion appearance, volume, and other uncontrollable factors [26–28].
Compared with PCR methods, although 16S rRNA-sequencing (16S rRNA-seq) technology cannot detect vaginal flora quantitatively, it reflects the relative abundance and different varieties of vaginal microflora in BV patients. In this study, we propose to use 16S rRNA sequencing technology to screen out the differential bacteria in vaginal secretions of healthy people and BV patients, and to diagnose BV by comparing the ratio of beneficial bacteria (e.g. Lactobacillus) to harmful bacteria (e.g. Gardnerella vaginalis), which can better reflect the thinking of flora balance, while avoiding the problem of inaccurate absolute quantification of bacteria in vaginal secretions; and to find out the sensitivity and specificity from the differential bacteria The most optimal index to reduce the number of molecular targets needed to diagnose BV in practice, and finally to achieve the purpose of saving test cost and accurately diagnosing BV.
Materials and methods
Ethical approval of the study protocol
The study protocol was approved (2021056) by the Ethics Committee of Tongde Hospital of Zhejiang Province, China.
Grouping the screening step and establishing inclusion and exclusion criteria: the inclusion criteria were women: (i) with a regular menstrual cycle: the menstrual cycle lasts from 21 to 35 days, lasts from 2 to 8 days, and the menstrual volume is 20 to 60 ml; (ii) aged 18–46 years; (iii) who were not pregnant; (iv) not using vaginal contraceptives; (v) without Candida species, Neisseria gonorrhoeae, Trichomonas species or other pathogens in their VSs; (vi) who had not used antibiotics in the previous 3 months; (vii) No sexual intercourse within 24 hours. (viii) To rule out ovarian and uterine related tumors.
Women were divided into the healthy-volunteer (N) group, BV intermediate (M) group and BV (B) group.
For group N, VSs were collected from healthy women. These women had undergone physical examination in our hospital previously. Their Nugent Score was 0–3, the pH of the VS was 3.8–4.5, and Candida species and Trichomonas species were absent. The cleanliness degree was I–II. The VS had a normal appearance, without a fishy odor, and vaginal itching/burning sensations were absent.
For group B, vaginal secretions were collected from outpatients in the gynecology department of our hospital. Candida species and Trichomonas species were not detected. The Nugent Score was 7–10. The VS met two items of the Amsel criteria (clue cells were present and pH >4.5).
For group M, VSs were collected from outpatients in the gynecology department of our hospital. Candida species and Trichomonas species were absent, and the Nugent Score was 4–6.
The enrollment criteria for clinical verification differed according to the group. For group N, the criteria were identical to the upper inclusion standard. For group B, the Nugent Score was 7–10, clue cells were present, and pH >4.5. Candida and Trichomonas species and other pathogens were not eliminated.
Clinical validation phase case enrollment criteria
Group N: pregnancy was not excluded, other criteria were the same as in the screening phase. Group B: Nugent score 7–10 and positive clue cells or vaginal discharge PH > 4.5, age 18–46 years, no history of sexual intercourse for 24 hours. To verify the accuracy of the index in a complex setting, no other exclusion criteria were set.
Clinical specimens
A gynecologist used a sterile swab to wipe-off secretions outside the vulva. Then, two sterile nylon vaginal swabs were placed into the lower one-third of the vagina. and sent for analyses within 1 h. In one sample, 0.5 mL of physiologic (0.9%) saline was added to measure the degree of cleanliness, presence of Candida species or Trichomonas species, other routine tests, and Gram staining for the Nugent Score. Another sample was immediately stored at − 80°C for DNA extraction, 16S rRNA sequence and RT qPCR.
DNA extraction
DNA was extracted using a commercial reagent kit from Hunan Shengxiang Biological Technical (Hunan, China) according to manufacturer instructions, The vaginal secretion specimens containing the nucleic acids to be extracted were lysed by the lysis solution to lyse the cells, and the DNA was obtained by washing, elution and purification processes using magnetic beads that specifically recognize and efficiently bind to the DNA molecules and adsorb the beads to the tube walls using a magnetic separator. Then nuclear purity was measured using Qubit™ 2.0 (Thermo Fisher, Waltham, MA, USA). Extracted DNA was analyzed by a spectrophotometer at wavelengths of 260 nm (excitation) and 280 nm (emission). DNA was stored at −40°C.
16S rRNA-seq
The V4 variable area of bacteria (primer: 515 F-806 R) was amplified and sequenced through a HiSeq™ platform (Illumina, San Diego, CA, USA). First, the purity and concentration of the DNA in the sample was measured by agarose gel electrophoresis. Samples were placed in a centrifuge tube and diluted to 1 ng/μL in sterile water. Using diluted DNA as a template and specific primers with barcodes, amplification was carried out by Phusion® High-Fidelity PCR Master Mix with GC Buffer (New England Biolabs, Ipswich, MA, USA) and high-efficiency enzymes.
PCR outcomes were detected with 2% agarose gel by electrophoresis. Strips of size 400–450 bp were recycled by gel kits (Qiagen, Hilden, Germany). A TruSeq® DNA PCR-Free Sample Preparation kit (Illumina) was used to construct DNA libraries. The constructed libraries were detected quantitatively by Qubit and RT-qPCR, and the relevant libraries were sequenced on the HiSeq platform.
Data manipulation
Data were obtained through splicing and filtering of primary data gained by sequencing. These data were used to analyze operational taxonomic unit (OTU) clusters and species classification. We obtained information on the species, abundance, and distribution of bacteria. Multiple sequence comparisons were performed on OTUs and phylogenetic trees were constructed, and further differences in community structure were obtained for different samples and subgroups, which were presented by downscaling plots such as PCoA, PCA and NMDS.
Fluorescence RT-qPCR
DNA extracted from VSs was used to detect 14 common pathogens. We used a 7500 Real-time PCR system (Applied Biosystems, Foster City, CA, USA) and the TaqMan™ Gene Expression Assays kit (Thermo Fisher). For the latter, the total reaction volume was 20 µL (20 × TaqMan Gene Expression Assay (1.0 μL), 2 × TaqMan Gene Expression Master Mix (10.0 μL), DNA template (4.0 μL), double-distilled H2O (5.0 μL). The PCR parameters were: 50°C for 2 min, 95°C for 10 min in pre-denaturation, 95°C for 15 s in denaturation, 60°C for 1 min in annealing extension, in a total of 40 cycles.
Results
Basic information of patients
Many doctors utilize the Amsel criteria for diagnosing BV. However, application of the Amsel criteria can be subjective and is, therefore, limited. The Nugent Score, a Gram stain grading system for vaginal swabs initially introduced in 1991, is another method for BV diagnosis. A well-versed team is required to employ the Nugent Score, though. Furthermore, relying just on morphology to identify bacteria is prone to error due to the subjectivity involved. This research aims to discover a new bacterial vaginosis quantitative marker with excellent sensitivity and specificity. From April to September 2020, a total of 109 women were enrolled. Group N comprised 39 cases, and their mean age was 39.6 ± 7.15 years. Group B was composed of 37 cases of mean age 38.9 ± 8.03 years. Group M comprised 33 cases of mean age 37.9 ± 6.03 years. There was no significant difference according to age (ANOVA) among the three groups (P = 0.623). Clue cells were detected in group B (23/37, 62.16%) but not in the other two groups.
16S rRNA-seq
Heatmaps were drawn for different groups to show the genus level (Figure 1(a)). There were obvious differences in the genus level among the three groups. Lactobacillus species were in high abundance in group N. Bacteria of genera Gardnerella, Mobiluncus, Megasphaera, Sneathia, Peptostreptococcus, Veillonella, Prevotella, Gemella and Mycoplasma showed high abundance in group B. Group M showed high abundance of bacteria of genera Atopobium, Klebsiella, Enterobacteriaceae, Streptococcus,Ureaplamsa and Sphingomonas. Bacteria of genera Lactobacillus, Gardnerella, Atopobium, Prevotella, Shuttleworthia and Streptococcus were the most abundant among the three groups (Figure 1(c)). The abundance of Lactobacillus species in group N was >75%. The abundance of Gardnerella species in group B was ~50%. NMDS for analyses of species diversity revealed remarkable differences among the three groups (Figure 1(b)). Group N and group B could be distinguished readily. whereas group M lay intermediate between group N and B. The stress was 0.146. Each point in (Figure 1(b)) represents a sample, and the distance between points represents the degree of difference. Stress <0.2 indicates that NMDS may reflect the degree of difference in different samples accurately.
Figure 1.
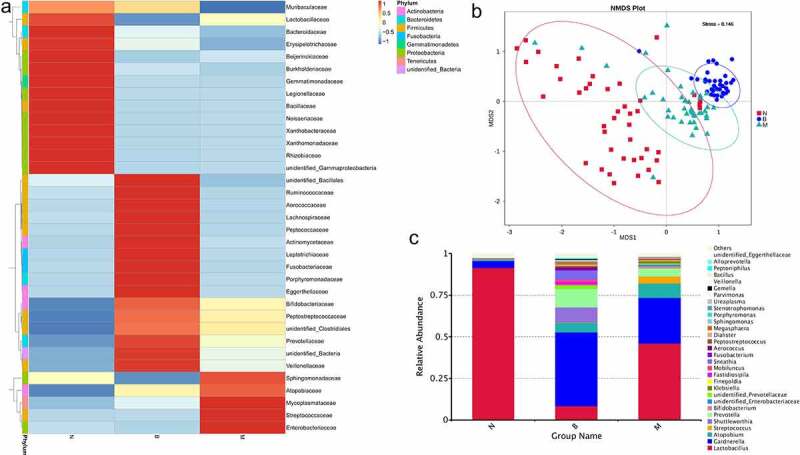
The detection of differential abundance of the healthy, BV and BV intermediate groups by 16S rRNA sequencing.
A: Heatmap of bacteria at the genus level.B: The analysis of NMDS based on OUT levelC: Relative abundance of the top-30 bacteria at the genus level.
PCR results
We combined 16S rRNA-seq results with data for microorganism species that could be detected using commercial kits. Lactobacillus crispatus, Lactobacillus iners, Lactobacillus jensenii, Lactobacillus gasseri, G. vaginalis, Atopobium vaginae, BVAB-2, Megasphaera-1, Megasphaera-2, Prevotella bivia, Mobiluncus curtisii, Mobiluncus mulieris, Mycoplasma hominis, and Ureaplasma urealyticum were the main species detected in 109 VSs by PCR. The test results were logarithm-converted (if a test result was negative, a value of 1 was used), followed by analyses by the Spearman rank test. Results indicated that only L. crispatus was negatively associated with BV grade among the four species of Lactobacillus, with r = −0.651. Other microflora had a significant (P < 0.05) positive relationship with BV grade, with a range of 0.206 to 0.748. G. vaginalis had the highest positive correlation (r = 0.748) (Table 1).
Table 1.
The analysis of the correlation between 14 common vaginal microorganisms and BV
|
Lactobacillus crispatus |
Lactobacillus iners |
Lactobacillus jensenii |
Lactobacillus gasseri |
Gardnerella vaginalis |
Atopobium vaginae |
BVAB-2 | |
| Spearman coefficient | −0.651 | −0.107 | 0.032 | −0.107 | 0.748 | 0.57 | 0.63 |
| P | <0.01 | 0.266 | 0.741 | 0.266 | <0.01 | <0.01 | <0.01 |
| Megasphaera-1 | Megasphaera-2 |
Prevotella bivia |
Mobiluncus curtisii |
Mobiluncus mulieris |
Mycoplasma hominis |
Ureaplasma urealyticum |
|
| Spearman coefficient | 0.578 | 0.536 | 0.209 | 0.422 | 0.266 | 0.513 | −0.098 |
| P | <0.01 | <0.01 | 0.029 | <0.01 | 0.005 | <0.01 | 0.313 |
Test results were log-transformed. Based on the convenience of drawing and because zero cannot be log-transformed, if the test result was lower than the limit of detection, we used a value of 10. We undertook the Kruskal–Wallis test using α = 0.05 as the detection level (Figure 2). L. crispatus, G. vaginalis, A. vaginae, BVAB-2, Megasphaera-1, Megasphaera-2, M. curtisii, M. mulieris, and M. hominis in group B and group N were present in significant amounts. Considering that there are many negative results of Mobiluncus and Mycoplasma hominis, and some positive results were mostly less than 100 copies, and the sensitivity, repeatability as well as accuracy were not ideal in our experiment. Therefore, the top-five most abundant bacterial species were selected for further analyses.
crispatus was negatively related (r = −0.651) with the BV grade, whereas G. vaginalis, A. vaginae, BVAB-2, Megasphaera-1, and Megasphaera-2 had a positive association with BV. The PCR results for these bacteria were log-transformed using L. crispatus as the numerator and other BV-related bacteria as the denominator. Four new indicators were found: log L/G = log (L. crispatus/G. vaginalis), log L/A = log (L. crispatus/A. vaginae), log L/B = log (L. crispatus/BVAB2), and log L/M = log (L. crispatus/Megasphaera 1/2). The denominator cannot be zero, and zero cannot be log-transformed so, if the test result was lower than the limit of detection, we used a value of 1 (Figure 3). Figure 3 shows that group B and group N were well differentiated through use of the four indicators stated above. Using the Nugent Score as the standard classification and the four indicators as the threshold value, ≥0 was diagnosed as ‘BV-negative’, whereas <0 was diagnosed as ‘BV-positive’. The sensitivity, specificity, positive predictive value, and negative predictive value for each indicator for diagnosing BV are shown in (Table 3). After analyses using a receiver operating characteristic (ROC) curve and comparing it with each indicator, log L/G was selected as the best indicator for diagnosing BV (Figure 4).
Figure 2.
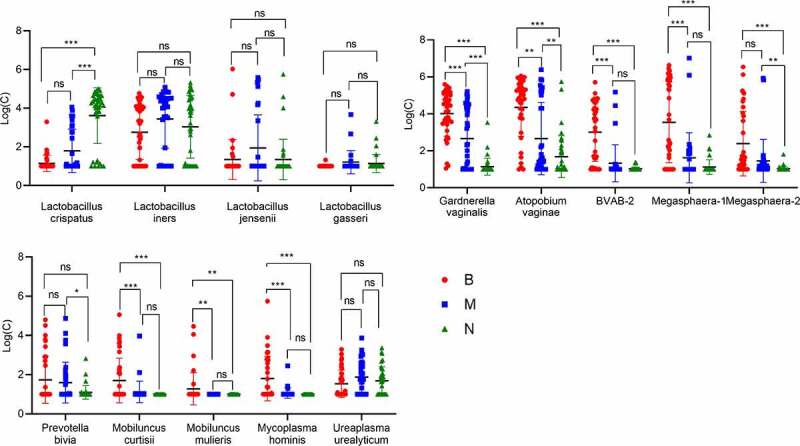
Comparison of RT-qPCR results for bacteria in groups B, M and N. ns:p > 0.05,*:P < 0.05, **:P < 0.01, ***:P < 0.001.
Figure 3.
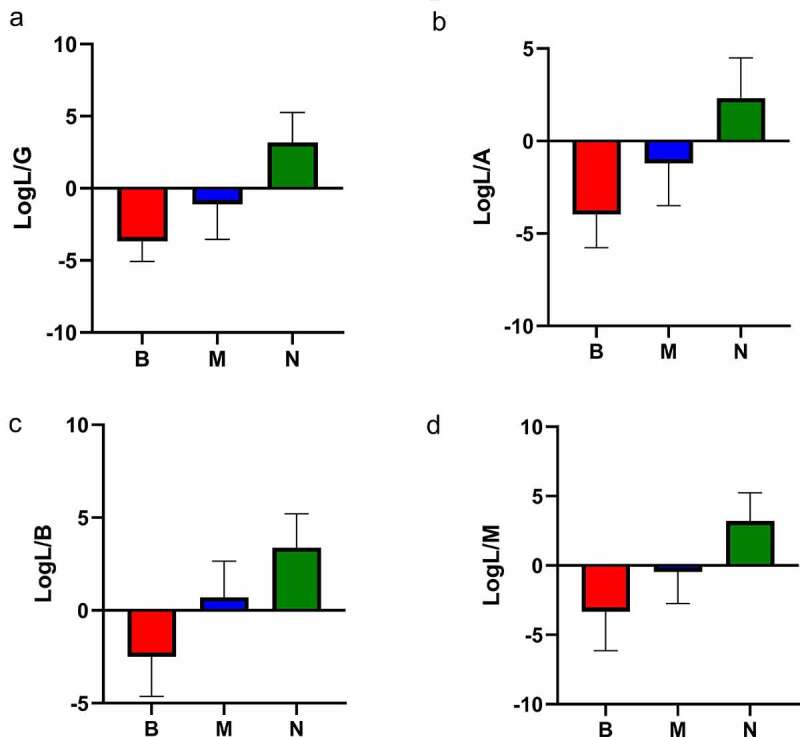
Indicators of log L/G, log L/A, log L/B and log L/M among the three groups.
Table 3.
Use of the four indicators to evaluate a BV diagnosis
| New indicator | BV positive* case | BV negative* case | Sensitivity | False- positive rate | Positive predicted value | Specificity | False-negative rate | Negative predicted value | Consistency | Youden Index | Kappa | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L/G | BV positive | 36 | 3 | 97.3% | 7.5% | 92.3% | 92.5% | 2.7% | 97.4% | 94.81% | 89.80% | 0.896 |
| BV negative | 1 | 37 | ||||||||||
| L/A | BV positive | 35 | 7 | 94.6% | 17.5% | 83.3% | 82.5% | 5.4% | 94.3% | 88.31% | 77.09% | 0.767 |
| BV negative | 2 | 33 | ||||||||||
| L/M | BV positive | 30 | 3 | 81.1% | 7.5% | 90.9% | 92.5% | 18.9% | 84.1% | 87.01% | 73.58% | 0.739 |
| BV negative | 7 | 37 | ||||||||||
| L/B | BV positive | 30 | 2 | 81.1% | 5.0% | 93.8% | 95.0% | 18.9% | 84.4% | 88.31% | 76.08% | 0.765 |
| BV negative | 7 | 38 | ||||||||||
| L/G& | BV positive | 86 | 3 | 93.5% | 2.8% | 96.6% | 97.2% | 6.5% | 94.6% | 95.50% | 0.907 | 0.910 |
| BV negative | 6 | 106 | ||||||||||
*BV diagnosis was made using the Nugent score.
#BV diagnosis was made using logL/G, <0 diagnosed as BV positive, ≥0 indicates BV negative.
&Compare two BV diagnostic indexes logL/G with Nugent score.
Figure 4.
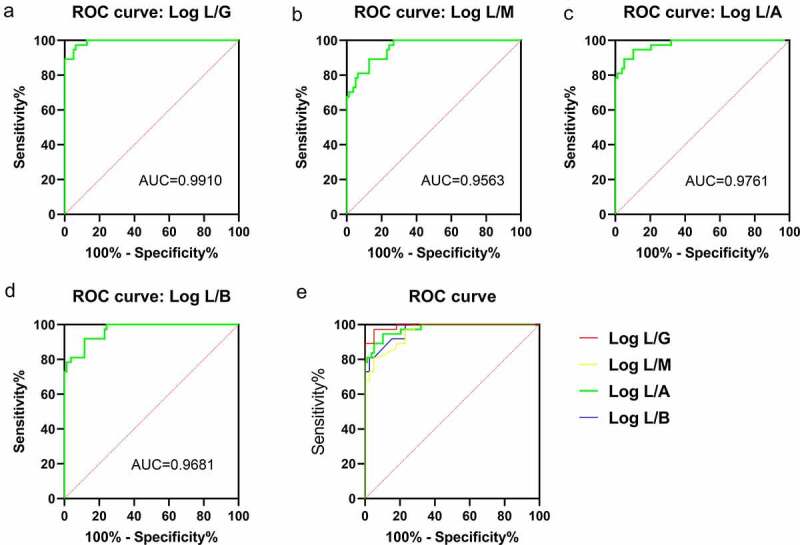
ROC curves for log L/G, log L/A, log L/B and log L/M used for a BV diagnosis.
Clinical verification of the indicator log L/G
From December 2020 to February 2021, case information during indicator verification stage was presented in (Table 2).
Table 2.
The case characteristics analysis of the BV-negative and BV-positive groups
| Item | BV-negative | BV-positive |
|---|---|---|
| Patients (number) | 103 | 92 |
| Candida-positive (number) | 2 | 12 |
| Age (years) | 33.1 ± 8.1 | 34.4 ± 9.9 |
| Cleanliness of secretions | I–II | III–IV |
| pH of secretion | 4.0 ± 0.2 | 4.9 ± 0.3 |
| Nugent Score | 0–3 | 7–10 |
| Cystitis (number) | 31 | 50 |
| Cervicitis (number) | 2 | 1 |
| Pelvic inflammation (number) | 4 | 3 |
| Uterine non-inflammatory disease (number) | 21 | 11 |
| Pregnancy (number) | 32 | 8 |
| Physical examination (number) | 8 | 9 |
| Other (number) | 5 | 10 |
We used log L/G on VSs collected from 103 cases classified as not suffering from BV and 92 cases suffering from BV. When using log L/G < 0 as the criterion, test results demonstrated a sensitivity of 93.5%, specificity of 97.2%, positive predictive value of 96.6, and negative predictive value of 94.6% (Table 3).
Discussion
The stability of the vaginal microbiota is influenced by the menstrual cycle, sex life, and other factors [29,30]. Therefore, different studies may elicit different results when studying the vaginal microbiota. Our 16S rRNA-seq results indicated that the most abundant bacteria were from the genus Lactobacillus in group B, and G. vaginalis in group N, data which are similar to those in the literature [31]. Also, our 16S rRNA-seq results were consistent with our PCR results. The abundance of sequencing results was not further analyzed at the species level in this study because 16S rRNA sequencing technology may have large errors at the species level. Subsequently, RT-qPCR was employed to detect Lactobacillus species.
Several studies have stated that G. vaginalis participates in the morbidity and immune-modification in women suffering from BV. Also, irrespective of whether culture or molecular diagnostics are employed, the sensitivity of detection of G. vaginalis is ≤100%, whereas the specificity is only 50% [3], Because G.vaginalis was present in the vaginal secretions of both BV and non-BV females [32,33], The difference may be in the significantly higher number of Gardenella in BV patients compared with healthy women. For women with BV symptoms, using 5 × 105 as the threshold value, BV could be diagnosed thanks to G. vaginalis detection using the Affirm VP Microbial Identification System (Beckton Dickinson, Franklin Lakes, NJ, USA). The sensitivity of that system is 90% and specificity is 97% [34]. Cartwright and colleagues proposed combining detection of A. vaginae, BVAB-2, and Lactobacillus species with that of Megasphaera-1. Considering the specificity, G. vaginalis was excluded, and they obtained a sensitivity of 96.7% and specificity of 92.2% for patients with clinical symptoms [35]. Hilbert and coworkers suggested combining detection of G. vaginalis with that of Megasphaera-1 and Megasphaera-2 besides L. crispatus for diagnosing BV, and obtained a sensitivity of 92% and specificity of 95% [36].
After RT-qPCR, we found that other common Lactobacillus species have no relationship with BV besides L. crispatus. Data for A. vaginae, BVAB-2, G. vaginalis, and Megasphaera-1 and 2 were combined with the Nugent Score and vaginal-flora characteristics of BV. Then, the data were log-transformed into four new indicators: log L/G, log L/B, log L/M, and log L/A. After comparison of these four indicators, log L/G < 0 was found to be the best indicator for a BV diagnosis with a sensitivity of 97.3% and specificity of 92.5%. During clinical verification of this indicator, the sensitivity and specificity was 93.5% and 97.2%, respectively, which are similar to those for some commercial reagent kits for diagnosing BV through detection of various bacteria types [37]. However, only two bacteria species were examined in this study.
At a similar sensitivity and specificity, compared with single detection of G. vaginalis or combining data for numerous bacteria, log L/G is more appropriate for the flora imbalance seen in BV. Compared with other bacteria associated with BV, L. crispatus and G. vaginalis have a higher relative abundance and copies in VSs, which makes it better linear range and repeatability in the process of detection. The ratio of abundance of L. crispatus to that of G. vaginalis was used, so the amount of VS sampled had little effect on log L/G. Compared with the Nugent Score as the ‘gold standard’, log L/G can be quantified accurately, monitored continuously, and avoids the subjectivity inherent in the morphologic assessment of bacteria.
In some VSs, bacterial recognition based on morphology can be difficult, and the Nugent Score recognizes bacteria only from the genera Lactobacillus, Gardnerella, and Mobiluncus, while other bacteria causing dysbacteriosis are not distinguished [38] Therefore, using this evaluation system, these patients are easily classified into BV intermediate type, which hampers the clinical diagnosis and treatment. Using log L/G for detection of the M group elicits the same problem in this study. In the intermediate group, there were both cases with LogL/G > 0 and cases with LogL/G < 0. Some of the cases in the intermediate group were in the transitional stage of dysbiosis, but a small number of cases could be further differentiated into the healthy or BV group, which needs to be combined with more clinical evidence such as clinical presentation and patient outcome. Thus, this study still needs to be expanded and further studies are needed to verify the clinical value of LogL/G in identifying intermediate groups.
Conclusions
Combination of data for 16S rRNA-seq and RT-qPCR revealed four indicators for BV detection. Of these, log L/G < 0 was the best indicator. Creating a molecular-diagnostic system independent of the Nugent Score for BV could have an important impact on the clinical management of BV.
Acknowledgements
We thank Thermo Fisher Scientific for providing a bacterial PCR detection kit free of charge. We also thank Dr. Xiangquan Song for his valuable suggestions.
Funding Statement
This work was supported by the National Natural Science Foundation of China (81802084) and the Postdoctoral Science Foundation of China (2020M681399).
Highlights
logL/G can better measure present vaginal microecology.
logL/G is quantitative, continuous, and reproducible.
logL/G has a strong linear range, sensitivity and specificity.
log L/G may be utilized to detect BV early.
log L/G can be used as a novel marker for early diagnosis of BV.
Author contributions
Conceptualization: AS. Methodology: TD. Validation: YZ. Formal analyses: YZ, and WW. Investigation: TD, YZ, and WW. Writing-original draft preparation: TD. Writing-review and editing: TD, and AS. Supervision: WW. All authors contributed to the article and approved the submitted version.
Ethics statement
Studies involving human participants were approved by the Human Ethics Committee of Tongde Hospital. Patients/participants provided written informed consent to take part in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Xiao B, Niu X, Han N, et al. Predictive value of the composition of the vaginal microbiota in bacterial vaginosis, a dynamic study to identify recurrence-related flora. Sci Rep. 2016;6:26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chang HH, Larson J, Blencowe H, et al. Preventing preterm births: analysis of trends and potential reductions with interventions in 39 countries with very high human development index. Lancet. 2013;381(9862):223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Onderdonk AB, Delaney ML, Fichorova RN.. The human microbiome during bacterial vaginosis. Clin Microbiol Rev. 2016;29(2):223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Koumans EH, Kendrick JS. CDC bacterial vaginosis working group, preventing adverse sequelae of bacterial vaginosis: a public health program and research agenda. Sex Transm Dis. 2001;28(5):292–297. [DOI] [PubMed] [Google Scholar]
- [5].Wiesenfeld HC, Hillier SL, Krohn MA, et al. Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clin Infect Dis. 2003;36(5):663–668. [DOI] [PubMed] [Google Scholar]
- [6].Klebanoff MA, Hillier SL, Nugent RP, et al. Is bacterial vaginosis a stronger risk factor for preterm birth when it is diagnosed earlier in gestation. Am J Obstet Gynecol. 2005;192(2):470–477. [DOI] [PubMed] [Google Scholar]
- [7].Marrazzo JM. A persistent(ly) enigmatic ecological mystery: bacterial vaginosis. J Infect Dis. 2006;193(11):1475–1477. [DOI] [PubMed] [Google Scholar]
- [8].Cohen CR, Lingappa JR, Baeten JM, et al. Bacterial vaginosis associated with increased risk of female-to-male HIV-1 transmission: a prospective cohort analysis among African couples. PLoS Med. 2012;9(6):e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Larsson PG, Forsum U. Bacterial vaginosis–a disturbed bacterial flora and treatment enigma. APMIS. 2005;113(5):305–316. [DOI] [PubMed] [Google Scholar]
- [10].Marrazzo JM, Thomas KK, Fiedler TL, et al. Relationship of specific vaginal bacteria and bacterial vaginosis treatment failure in women who have sex with women. Ann Intern Med. 2008;149(1):20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Varma R, Gupta JK. Antibiotic treatment of bacterial vaginosis in pregnancy: multiple meta-analyses and dilemmas in interpretation. Eur J Obstet Gynecol Reprod Biol. 2006;124(1):10–14. [DOI] [PubMed] [Google Scholar]
- [12].McDonald HM, Brocklehurst P, Gordon A. Antibiotics for treating bacterial vaginosis in pregnancy. Cochrane Database Syst Rev. 2007;1:CD000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol. 1991;29(2):297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].McMillan A, Rulisa S, Sumarah M, et al. A multi-platform metabolomics approach identifies highly specific biomarkers of bacterial diversity in the vagina of pregnant and non-pregnant women. Sci Rep. 2015;5:14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Watson E, Reid G. Metabolomics as a clinical testing method for the diagnosis of vaginal dysbiosis. Am J Reprod Immunol. 2018;80(2):e12979. [DOI] [PubMed] [Google Scholar]
- [16].Yen S, Shafer MA, Moncada J, et al. Bacterial vaginosis in sexually experienced and non-sexually experienced young women entering the military. Obstet Gynecol. 2003;102(5 Pt 1):927–933. [DOI] [PubMed] [Google Scholar]
- [17].Klebanoff MA, Schwebke JR, Zhang J, et al. Vulvovaginal symptoms in women with bacterial vaginosis. Obstet Gynecol. 2004;104(2):267–272. [DOI] [PubMed] [Google Scholar]
- [18].Schwebke JR, Desmond R. Natural history of asymptomatic bacterial vaginosis in a high-risk group of women. Sex Transm Dis. 2007;34(11):876–877. [DOI] [PubMed] [Google Scholar]
- [19].Coleman JS, Gaydos CA, Kraft CS. Molecular diagnosis of bacterial vaginosis: an update. J Clin Microbiol. 2018;56(9). DOI: 10.1128/JCM.00342-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burton JP, Reid G. Evaluation of the bacterial vaginal flora of 20 postmenopausal women by direct (Nugent score) and molecular (polymerase chain reaction and denaturing gradient gel electrophoresis) techniques. J Infect Dis. 2002;186(12):1770–1780. [DOI] [PubMed] [Google Scholar]
- [21].Ferris MJ, Masztal A, Aldridge KE, et al. Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect Dis. 2004;4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353(18):1899–1911. [DOI] [PubMed] [Google Scholar]
- [23].Zhou X, Brown CJ, Abdo Z, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007;1(2):121–133. [DOI] [PubMed] [Google Scholar]
- [24].Menard JP, Fenollar F, Henry M, et al. Molecular quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clin Infect Dis. 2008;47(1):33–43. [DOI] [PubMed] [Google Scholar]
- [25].Zozaya-Hinchliffe M, Lillis R, Martin DH, et al. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol. 2010;48(5):1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vitali B, Pugliese C, Biagi E, et al. Dynamics of vaginal bacterial communities in women developing bacterial vaginosis, candidiasis, or no infection, analyzed by PCR-denaturing gradient gel electrophoresis and real-time PCR. Appl Environ Microbiol. 2007;73(18):5731–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Srinivasan S, Morgan MT, Fiedler TL, et al. Metabolic signatures of bacterial vaginosis. mBio. 2015;6(2). DOI: 10.1128/mBio.00204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vitali B, Cruciani F, Picone G, et al. Vaginal microbiome and metabolome highlight specific signatures of bacterial vaginosis. Eur J Clin Microbiol Infect Dis. 2015;34(12):2367–2376. [DOI] [PubMed] [Google Scholar]
- [29].Gajer P, Brotman RM, Bai G, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4(132):132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jespers V, van de Wijgert J, Cools P, et al. The significance of Lactobacillus crispatus and L. vaginalis for vaginal health and the negative effect of recent sex: a cross-sectional descriptive study across groups of African women. BMC Infect Dis. 2015;15:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].van der Veer C, van Houdt R, van Dam A, et al. Accuracy of a commercial multiplex PCR for the diagnosis of bacterial vaginosis. J Med Microbiol. 2018;67(9):1265–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Schwebke JR, Flynn MS, Rivers CA. Prevalence of Gardnerella vaginalis among women with lactobacillus-predominant vaginal flora. Sex Transm Infect. 2014;90(1):61–63. [DOI] [PubMed] [Google Scholar]
- [33].van de Wijgert JH, Borgdorff H, Verhelst R, et al. The vaginal microbiota: what have we learned after a decade of molecular characterization. PLoS One. 2014;9(8):e105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Briselden AM, Hillier SL. Evaluation of affirm VP microbial identification test for Gardnerella vaginalis and Trichomonas vaginalis. J Clin Microbiol. 1994;32(1):148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cartwright CP, Lembke BD, Ramachandran K, et al. Development and validation of a semiquantitative, multitarget PCR assay for diagnosis of bacterial vaginosis. J Clin Microbiol. 2012;50(7):2321–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hilbert DW, Smith WL, Chadwick SG, et al. Development and Validation of a Highly Accurate Quantitative Real-Time PCR Assay for Diagnosis of Bacterial Vaginosis. J Clin Microbiol. 2016;54(4):1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Richter SS, Otiso J, Goje OJ, et al. Prospective evaluation of molecular assays for diagnosis of vaginitis. J Clin Microbiol. 2019;58(1). DOI: 10.1128/JCM.01264-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tamrakar R, Yamada T, Furuta I, et al. Association between Lactobacillus species and bacterial vaginosis-related bacteria, and bacterial vaginosis scores in pregnant Japanese women. BMC Infect Dis. 2007;7:128. [DOI] [PMC free article] [PubMed] [Google Scholar]


