Molecular mimicry can induce autoimmunity. By developing and using a bioinformatic tool to analyze molecular mimicry between tumor and viral antigens, the authors show this phenomenon can also play a role in antitumor immune responses.
Abstract
Molecular mimicry is one of the leading mechanisms by which infectious agents can induce autoimmunity. Whether a similar mechanism triggers an antitumor immune response is unexplored, and the role of antiviral T cells infiltrating the tumor has remained anecdotal. To address these questions, we first developed a bioinformatic tool to identify tumor peptides with high similarity to viral epitopes. Using peptides identified by this tool, we demonstrated that, in mice, preexisting immunity toward specific viral epitopes enhanced the efficacy of cancer immunotherapy via molecular mimicry in different settings. To understand whether this mechanism could partly explain immunotherapy responsiveness in humans, we analyzed a cohort of patients with melanoma undergoing anti-PD1 treatment who had a high IgG titer for cytomegalovirus (CMV). In this cohort of patients, we showed that high levels of CMV-specific antibodies were associated with prolonged progression-free survival and found that, in some cases, peripheral blood mononuclear cells (PBMC) could cross-react with both melanoma and CMV homologous peptides. Finally, T-cell receptor sequencing revealed expansion of the same CD8+ T-cell clones when PBMCs were expanded with tumor or homologous viral peptides. In conclusion, we have demonstrated that preexisting immunity and molecular mimicry could influence the response to immunotherapies. In addition, we have developed a free online tool that can identify tumor antigens and neoantigens highly similar to pathogen antigens to exploit molecular mimicry and cross-reactive T cells in cancer vaccine development.
Introduction
CD8+ T cells have a key role in the detection and elimination of cells that present abnormal peptides on their surface as a result of viral infection or malignant transformation. Because of the promiscuity of the T-cell receptor (TCR), T cells recognize a large variety of targets. Thus, a relatively small number of T cells can recognize multiple peptide–MHC (pMHC) molecules that can represent a threat (1, 2). A downside of TCR promiscuity is that the immune response directed against a pathogen might result in recognition of self-antigens. This can cause a deleterious off-target effect mediated by cross-reactive T cells. The result of this process, which is known as molecular mimicry, is well established in the field of autoimmunity (3); however, it has thus far not been fully explored in cancer.
The best prognostic markers for successful outcome of immunotherapy are thought to be high tumor mutational burden and abundant T-cell infiltration, according to the rationale that a tumor that has a high number of mutations will have a higher chance of being recognized and eliminated by infiltrating T cells (4,5,6,7,8). Nevertheless, pivotal studies show that the qualitative properties of tumor neoantigens might be more important than their quantity. It is thought that tumor antigens are more likely to be immunogenic if they resemble infectious disease–associated antigens, because they are more likely to be recognized by a T cell (4, 8). Consistent with this idea, antiviral T cells populate the tumor microenvironment (9, 10), but whether their role is active or not is still unclear.
Herein, we tested our hypothesis that tumors might present peptides that share a high degree of homology with viral peptides and thereby might enable cross-reactive T cells to recognize and kill tumor cells via molecular mimicry. To do this, we developed a bioinformatic tool called homology evaluation of xenopeptides (HEX) to identify tumor-specific peptides highly similar to viral-derived peptides. We then used several sets of murine and human tumor-specific peptides highly homologous to viral peptides identified by HEX to examine the role of viral preexisting immunity in antitumor T-cell responses through molecular mimicry.
Materials and Methods
HEX
HEX is a novel in silico platform that can be used to compare similarity between tumor peptides (reference peptides) and viral peptides (query peptides). It utilizes several metrics to expediate candidate peptide selection. This is done by incorporating both novel methods (peptide scoring and alignment scoring algorithm) and preexisting methods (MHC class I–binding prediction). HEX comes with a number of precompiled databases of known proteins, such as proteins derived from viral pathogens and the human proteome (11).
Peptides are ranked by a score (referred to here as B-score) as described previously (11), which represents the log-likelihood of the viral peptide being recognized by a T cell. The associated scoring matrix is generated ad hoc based on the amino acid composition of the reference peptide, as opposed to experimentally. In the matrix, rows represent the amino acid position in the peptide and columns represent each of the 20 standard amino acids, amino acid positions of the reference peptide are assigned the same high score and other positions are assigned the same low score. The B-score is the agonist log-likelihood score for this special matrix and is given by:
 |
Alignments are computed pairwise between peptides in the query set against the reference set. For a given pair of peptides, their alignment is calculated by summing the distance scores between pairs of amino acids in the same position. Scoring is weighted to prioritize similarity between more central amino acids in the peptide. HEX supports both BLOSUM and PAM substitution matrices across several evolutionary distances.
Predictions of MHC class I–binding affinity are made using NetMHC (12) via the Application programming interface of Immune Epitope Database (IEDB; http://tools.iedb.org/main/tools-api/) and are then parsed and collated within the tool. The user can specify their desired scoring method or return a number of recommended results. Predictions for a number of human and murine MHC class I alleles are supported.
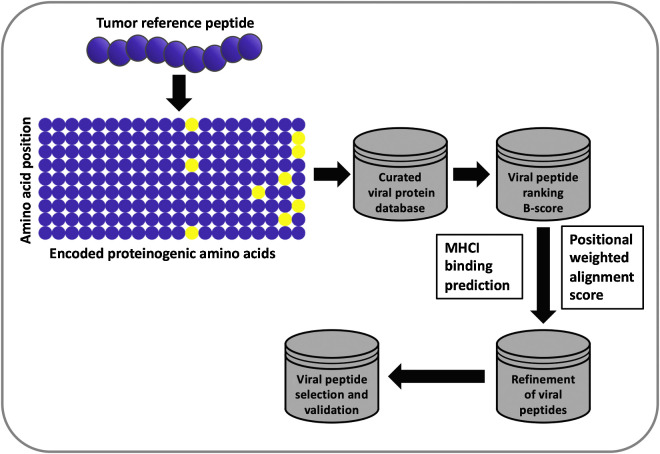
Users are able to select peptides by their own criteria or allow peptides to be selected by a random forest model. The random forest was trained on experimental outcomes of peptides chosen by us. Feature importance was determined by out-of-bag increase in mean squared error and cross-validated on an unseen sample of the peptides. HEX was developed as a web application using the R package Shiny and is accessible at https://picpl.arcca.cf.ac.uk/hex/app/ without user registration. The source code is available at https://github.com/whalleyt/hex. An explanatory schematic of the software pipeline can be found in Fig. 1.
Figure 1.
Flowchart of the HEX algorithm. When using the HEX software, a matrix is generated on the basis of the amino acid composition of a tumor peptide (referred to as the reference peptide). This matrix is then used to scan the viral database, and resulting viral peptides are ranked in order of log-likelihood of recognition (B-score). Each viral peptide is assigned an alignment score and a score for the predicted MHC class I–binding affinity. The candidate viral peptides are ranked on the basis of the following criteria: MHC class I–binding affinity prediction score > alignment score > B-score; the highest scoring peptides are analyzed experimentally.
Patients and samples
A total of 18 patients with stage IV metastatic melanoma were treated with anti-PD1 mAb in the Helsinki University Central Hospital (HUCH; Helsinki, Finland) Comprehensive Cancer Center. Patients were randomly selected to receive either nivolumab (n = 8) infusions every second week or pembrolizumab (n = 10) infusions every third week. The study was approved by the HUCH ethical committee (Dnro 115/13/03/02/15). Written informed consent was received from all patients and the study was conducted in accordance with the Declaration of Helsinki. For detailed patient characteristics, see Supplementary Table S1.
Peripheral blood samples (3 mL EDTA blood, 50 mL Heparin blood) were collected at three time points: before initiation of treatment, and then after 1 and 3 months of treatment. From these samples, the plasma was separated by centrifuging and then stored at −70°C, peripheral blood mononuclear cells (PBMC) were isolated from Heparin samples using Ficoll-Paque density gradient centrifugation (catalog no. 17-1440-03, GE Healthcare) according to the manufacturers' instructions. Levels of CMV- and Epstein–Barr virus (EBV)–specific IgG were measured from thawed EDTA plasma samples using VIDAS CMV IgG (catalog no. 30204, BioMérieux) and Siemens Enzygnost Anti-EBV/IgG kits (catalog no. ZOWIS155, Siemens Healthcare Diagnostics) according to the manufacturers' instructions. The levels of each immunoglobulin class (IgA, IgM, IgG) in thawed Heparin plasma were measured in the central laboratory of the Helsinki University Central Hospital (HUSLAB).
Cell lines and human samples
The murine melanoma cell line B16-F10 was purchased from the ATCC in 2016. Cells were cultured in RPMI (catalog no. 21875091, Gibco, Thermo Fisher Scientific) supplemented with 10% FBS (catalog no. 10500064, Life Technologies), 1% GlutaMAX (catalog no. 35050061, Gibco, Thermo Fisher Scientific), and 1% penicillin and streptomycin (catalog no. 15140122, Gibco, Thermo Fisher Scientific), henceforth referred to as “complete medium,” at 37°C/5% CO2.
The cell line B16-OVA, a mouse melanoma cell line modified to constitutively express chicken ovalbumin (OVA), was kindly provided by Richard Vile (Mayo Clinic) in 2016. These cells were cultured in complete medium supplemented with 1% Geneticin (catalog no. 10-131-027, Gibco, Thermo Fisher Scientific) at 37°C/5% CO2. All cell lines were put in culture approximately 1 week and passed two to three times before being injected in mice for tumor engraftment.
All cells were tested for Mycoplasma contamination with a commercial detection kit (MycoAlert, catalog no. LT07-118, Lonza). Isolated human PBMCs were frozen in FBS supplemented with 10% DMSO (catalog no. D2438-50ML, Sigma-Aldrich) and then maintained in liquid nitrogen until use.
Cryopreserved PBMCs were thawed and rested overnight at 37°C/5% CO2 in complete medium over night before plating them for ELISpot and cross-reactivity assays.
Peptides
All peptides used in this study were purchased from Zhejiang Ontores Biotechnologies Co. or GenScript. A total of 5 mg of each peptide was ordered with a guaranteed purity of >90%. To produce the PeptiCRAds (see below) poly-K tailed versions of the peptides of interest were purchased. The sequences of the peptides used for in vivo experiments are found in Supplementary Table S2. The sequences of the peptides used for in vitro experiments with human samples are found in Supplementary Table S3.
PeptiCRAd preparation
PeptiCRAd is a plug-and-play and cloning-free vaccine platform described previously (13). It consists of a peptide-coated adenovirus. In this approach, the virus represents an immunogenic moiety and was used as an adjuvant (14). The cloning-free setup allowed us to easily substitute the peptides coating the virus when the study required it, rather than cloning a new vaccine.
The adenovirus used in this study was a Human adenovirus serotype 5 with a deletion of 24 bp in the E1 gene and enriched in CpG motifs (Ad5-Δ24-CpG) generated previously (15). The deletion in the E1 gene enables the adenovirus to selectively replicate in human tumors, hence it is oncolytic. Although the virus is infective in mice, it generally does not replicate in murine tumor models (16, 17).
All viruses used in this study were propagated and characterized using previously described procedures (13, 18).
All PeptiCRAd complexes described in this work were prepared as follows. Briefly, 1 × 109 viral particles (vp) were mixed with 20 μg poly-Lysine (poly-K) tailed peptides (resuspended in water); after brief vortexing, the mixture was incubated at room temperature for 15 minutes after which, PBS was added up to the injection volume (50 μL/mouse). For the TRP2-PeptiCRAd, 1 × 109 vp were mixed with 20 μg of 6K-TRP2180–188 peptide. The Viral-PeptiCRAd was prepared using 1 × 109 vp mixed with 5 μg of each viral 6K-peptide homologous for TRP2180–188. gp100-PeptiCRAd was prepared using 1 × 109 vp mixed with 20 μg of 6K-GP10025–33 peptide.
New PeptiCRAds were prepared before each experiment using fresh reagents. All dilutions of virus and peptides required, before incubation for PeptiCRAd preparation, were performed in sterile PBS or water.
Animal experiments and ethical permits
All animal experiments were reviewed and approved by the Experimental Animal Committee of the University of Helsinki (Helsinki, Finland) and the Provincial Government of Southern Finland. All in vivo models were carried using C57BL/6JOlaHsd (C57BL/6J) mice obtained from Scanbur.
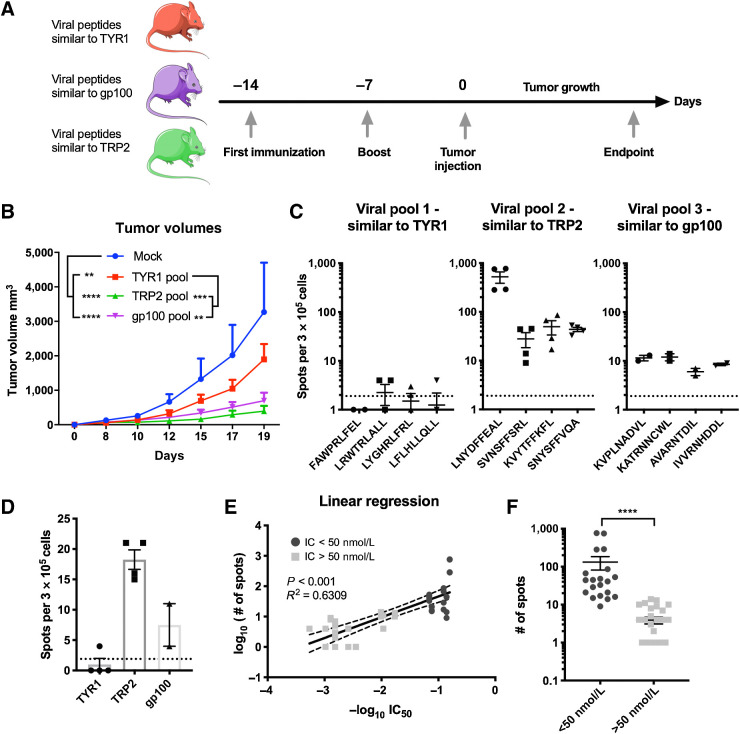
For the first animal experiment (results depicted in Fig. 2), 8 to 9 weeks old immune competent female C57BL/6J mice were divided in four groups. N = 3 mice were used as mock group, n = 7 mice were used in each of the three different treatment groups. Each group was immunized with a different pool of viral-derived peptides homologous to tumor epitopes. Mice were vaccinated twice, and injections were performed at 1-week interval (days 0 and 7) at the base of the tail with 40 μg of peptides (10 μg each peptide) and 40 μg of adjuvant [VacciGrade poly(I:C), catalog no. vac-pic, Invivogen] in a final injectable volume of 100 μL. Mice in the mock group were injected with PBS. At day 14, mice were injected with 3 × 105 B16-OVA cells on the right flank and tumor growth was followed until endpoint was reached.
Figure 2.
Immunization with viral peptides homologous to tumor antigens slows tumor growth. A, Scheme of the animal experiment: To assess whether viral peptides similar to tumor peptides can impact tumor growth, four groups of C57BL/6J mice were formed. A group of naïve mice was used as mock (n = 3); the other three groups (n = 5 mice per group) were each immunized with a different pool of viral peptides. The mice were immunized at two time points, 14 and 7 days, before the engraftment of the tumor. Two weeks after the first immunization, mice were injected subcutaneously with 3 × 105 murine melanoma B16-OVA cells. After the engraftment, tumor growth was measured with a digital caliper every second day for 19 days. B, Average tumor growth shown as mean ± SEM of each treatment group. P value was calculated using two-way ANOVA multiple comparison with Tukey correction. C, Mice were euthanized when the endpoint was reached. Splenocytes from the mice of each group were collected and pooled for an ELISpot assay. Each pool of splenocytes was pulsed with the indicated viral peptides (viral peptides homologous to TYR1208–216, viral peptides homologous to TRP2180–188, or viral peptides homologous to gp10025–33) to assess the response to the treatment. The dotted line shows the background produced by the negative control. D, To assess the response toward the respective original tumor epitope, splenocytes from each group of mice were pooled together and pulsed with the corresponding tumor peptide (dotted line represents background signal). E, Correlation between data from IFNγ response and predicted MHC class I–binding affinity. F, Stratification of peptides based on their affinity and ability to stimulate IFNγ production. High-affinity peptides (IC50 < 50 nmol/L) fostered significantly higher production of IFNγ compared with intermediate/low-affinity peptides (IC50 > 50 nmol/L). The range of P value is labeled with asterisks according to the following criteria: >0.05 (ns), ≤0.05 (*), ≤0.01 (**), ≤0.001 (***), ≤0.0001 (****).
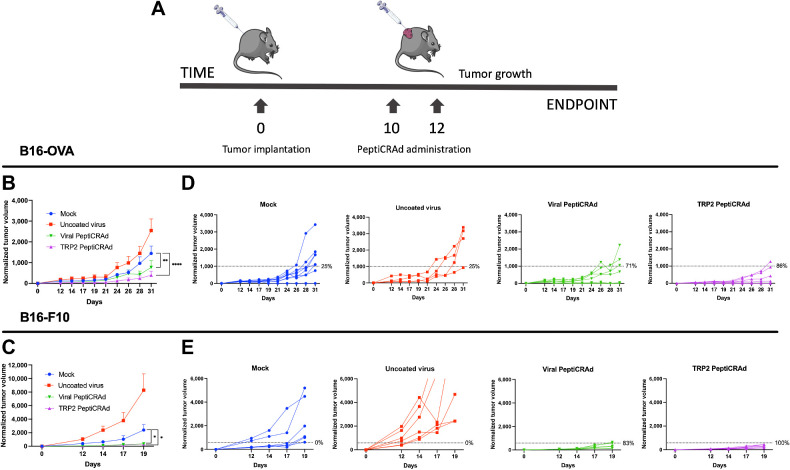
For the treatment of established tumors (results depicted in Fig. 3), we tested two different murine melanoma cell lines: the B16-OVA cell line and the more aggressive B16-F10 cell line. A total of 3 × 105 B16-OVA cells and 1 × 105 B16-F10 were injected subcutaneously on the right flank of 8 to 9 weeks old immune competent female C57BL/6J mice. The mice were randomly divided in to four groups of 7 to 8 mice for each tumor cell line. A mock group was treated with PBS; a second group was treated with uncoated adenovirus (Ad5-Δ24-CpG); a third and a fourth group were treated with the same adenovirus (Ad5-Δ24-CpG) coated with TRP2180–188 (TRP2–PeptiCRAd) or with TRP2-homologous viral peptides (Viral–PeptiCRAd), respectively (peptides shown in Supplementary Table S2). Mice were intratumorally treated twice and injections were performed at 2-day interval (starting from day 10 and 12 from tumor engraftment) with a final volume of 50 μL per injection.
Figure 3.
Viral peptides homologous to tumor antigens can reduce tumor growth in already established tumors. A, C57BL/6J mice were subcutaneously injected with B16-OVA or B16-F10 cells at day 0. As soon the tumors were visible, the mice were treated with either PBS (mock; n = 8), Ad5-Δ24-CpG alone (uncoated virus; n = 8), Ad5-Δ24-CpG coated with viral-derived peptides similar to TRP2180–188 (Viral-PeptiCRAd; n = 8), or Ad5-Δ24-CpG coated with TRP2180–188 peptide (TRP2-PeptiCRAd; n = 8). Normalized B16-OVA (B) and B16-F10 (C) tumor volume is shown as mean ± SEM (statistical analysis by two-way ANOVA with multiple comparison). Normalized B16-OVA (D) and B16-F10 (E) tumor volume curves of individual mice per each group. The dotted line identifies the therapeutic success threshold and represents the median of the normalized volumes measured at the endpoint. P value is represented as follows: >0.05 (ns), ≤0.05 (*), ≤0.01 (**), ≤0.001 (***), ≤0.0001 (****).
Tumor growth in all experiments was measured every 2 days with a digital caliper until endpoint was reached. Tumor volume was calculated according to the following formula:
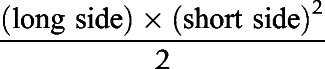 |
The median of the tumor volume measurement of the last day identified the therapeutic success threshold, shown as a dotted line in the graphs. Mice with tumor volume below the threshold at the endpoint were considered responders, while mice above it were considered as nonresponders.
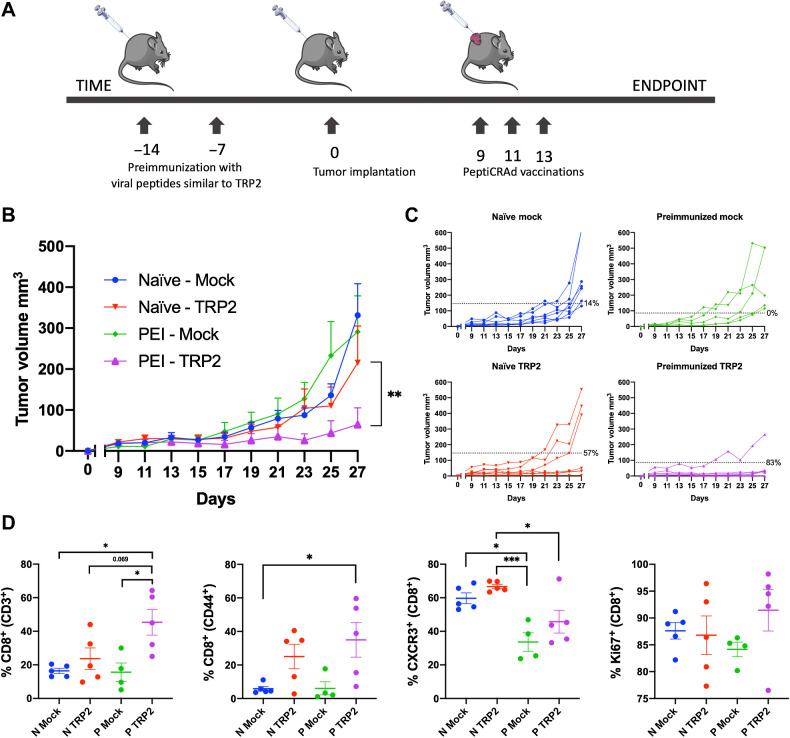
For the animal experiment depicted in Fig. 4, we used a combination of all the above-mentioned methods. Half of the cohort of C57BL/6J female mice (8 to 9 weeks old) were preimmunized with the pool of viral-derived peptides similar to TRP2 (Supplementary Table S2) according to the protocol described above. Two weeks after the first immunizations, mice were subcutaneously injected with 3 × 105 B16-OVA cells on the right flank. When the tumor was visible, mice were divided into six groups (n = 8–10 mice per group) and were intratumorally injected three times (2 days apart) with either PBS (mock group), gp100-coated PeptiCRAd (gp100 group) or TRP2-coated PeptiCRAd (TRP2 group). Tumor growth was followed as described above until the endpoint of the experiment was reached.
Figure 4.
Preexisting immunity toward viral-derived epitopes mimicking tumor antigens increases efficacy of cancer immunotherapy. A, We preimmunized half of a cohort of mice with viral-derived peptides similar to TRP2180–188 identified using HEX. After preimmunization, all the mice were engrafted with syngeneic B16-OVA cells and mice that developed palpable tumors were randomized and treated as follows: PBS (mock; n = 8), PeptiCRAd-gp100 (no preexisting immunity for this peptide; n = 8), and PeptiCRAd-TRP2 (peptide with homologous preexisting immunity; n = 8). B, Average tumor growth shown as mean ± SEM of each treatment group. Statistical analysis by two-way ANOVA. C, Tumor volume curves of individual mice per each treatment group. The dotted line identifies the threshold of the therapeutic success rate and represents the median of the normalized volumes measured at the endpoint for the naïve groups and the preimmunized (PEI) groups, respectively. D, Flow cytometry analysis of tumors collected at the endpoint (N stands for naïve, and P stands for preimmunized). The gating strategy used for the analysis is shown in Supplementary Fig. S1. P value is represented as follows: >0.05 (ns), ≤0.05 (*), ≤0.01 (**), ≤0.001 (***), ≤0.0001 (****).
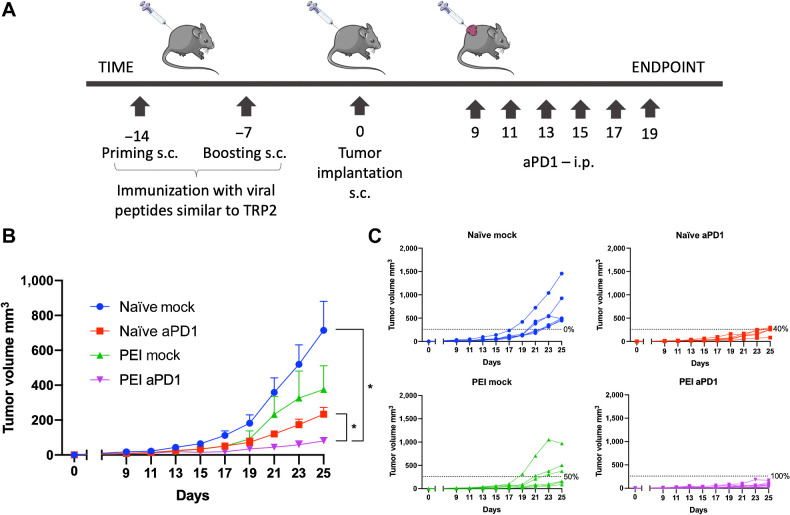
For the last animal experiment (results depicted in Fig. 5), half of the cohort of mice was preimmunized with the pool of viral-derived peptides similar to TRP2 (Supplementary Table S2) using the protocol described above; the other half did not receive any immunization. Two weeks after the first immunization, mice were subcutaneously injected with 3 × 105 B16-OVA cells on their right flank. When the tumors were palpable, nonimmunized and preimmunized mice were each randomly divided into two groups (n = 8 mice per group). Mice in one nonimmunized and one preimmunized group were treated six times by intraperitoneal injection (100 μL) with PBS (referred to as naïve mock and PEI mock, respectively). Mice in the other nonimmunized and preimmunized group were treated six times by intraperitoneal injection (100 μL) with 100 μg/mouse/treatment of PD1 antibody (catalog no. BE0146, BioxCell; referred to as naïve-aPD1 and PEI-aPD1, respectively). Tumor growth was followed as described above until the endpoint of the experiment was reached.
Figure 5.
Synergistic effect of preimmunization status and ICI therapy. A, Schematic of the experiment. Half of a cohort of C57BL/6J mice were preimmunized using virus-derived peptides similar to TRP2180–188 before syngeneic B16-OVA tumor engraftment (naïve groups/PEI groups). Half of the mice in each group were subsequently treated with anti-PD1 (aPD1 groups; n = 8 each) and half with PBS (mock groups; n = 8 each). B, Average tumor volume growth shown as mean ± SEM for each treatment group. Statistical analysis by two-way ANOVA. C, Tumor volume curves of individual mice per each treatment group. The dotted line identifies the threshold of the therapeutic success rate and represents the median of the volumes measured at the endpoint for the naïve anti-PD1 group. P value is represented as follows: >0.05 (ns), ≤0.05 (*), ≤0.01 (**), ≤0.001 (***), ≤0.0001 (****).
ELISpot assay
To assess the extent of the activation of antigen-specific T cells, IFNγ secretion was measured by ELISpot assay using kits from IMMUNOSPOT (catalog no. MIFNGP 1M/5, CTL-Europe GmbH) for murine IFNγ and MABTECH (catalog no. 3420-4HPT-2, Mabtech AB) for human IFNγ.
For murine IFNγ, 3 × 105 fresh splenocytes collected at the endpoint of the animal experiments were plated per well at day 0 in CTL Medium (catalog no. CTLT-010, CTL-Europe GmbH) supplemented with 1% GlutaMAX (catalog no. 35050061, Gibco, Thermo Fisher Scientific). Cells were stimulated with 2 μg/well of peptides. After 3 days of incubation at 37°C/5% CO2, plates were developed according to the kit manufacturer's protocol.
For human IFNγ, human PBMCs were thawed and rested overnight at 37°C/5% CO2 in complete RPMI medium. The following day, 2.5 × 105 PBMCs/well were plated and stimulated with 2 μg/well of peptides. After 48 hours of incubation at 37°C/5% CO2, the plates were developed according to the kit manufacturer's protocol. Plates were sent to CTL-Europe GmbH to be analyzed.
Flow cytometry analysis
After the endpoint of the experiment, mice were euthanized, and tumors were collected in cold PBS. Collected tumors were further reduced into single-cell suspensions by passing them through 70-μmol/L cell strainers using syringe plungers. Intracellular staining was performed using FOXP3 Fixation/Permeabilization Buffer (catalog no. 421403, BioLegend) following the manufacturer's instructions. Briefly, tumor cell suspensions were transferred to a U-shaped 96-well plate. Cells were centrifuged (600 rcf, 4°C for 5 minutes) and washed with cold PBS twice. Subsequently, cells were fixed using Foxp3 Fixation/Permeabilization working solution and incubated in the dark for 45 minutes at room temperature. Next, cells were washed with Permeabilization Buffer once and centrifuged as above. Supernatant was removed and the cell pellet was resuspended in 50 μL antibody cocktail (PBS + cell-surface and intracellular antibodies). Cells were incubated at room temperature for an additional 30 minutes protected from light. Finally, cells were washed twice and ultimately resuspended with PBS and transferred into round-bottom tubes for data acquisition.
The following antibodies were used: TruStain fcX (anti-mouse CD16/32; catalog no. 101320, BioLegend), CD3-BV711 (catalog no. 563123, BD Biosciences), CD4-PECF594 (catalog no. 562285, BD), CXCR3-APC (catalog no. 562266, BD Biosciences), CD44-V450 (catalog no. 560451, BD Biosciences), Ki67-PECy7 (catalog no. 561283, BD Biosciences), CD8a (KT15)-FITC (catalog no. 1705F/33790, Proimmune).
The data were acquired using BD LSR-FORTESSA flow cytometer and subsequently analyzed using FlowJo software v10. The gating strategy for flow cytometry data analysis is depicted in Supplementary Fig. S1.
Docking and three-dimensional alignment
The docking of the two pMHC complexes was performed using the online tool DockTope (19) hosted by IEDB (http://tools.iedb.org/docktope/inicio.php). Visualization and alignment of the generated three-dimensional (3D) structures was performed using the Protein Data Bank Mol* 3D viewer (https://www.rcsb.org/3d-view) using default parameters.
Cross-reactivity assay
The cross-reactivity of T cells against viral and tumor antigens sharing high similarity was studied by first expanding PBMCs from CMV seropositive healthy donor (buffy coat provided by the Histocompatibility Testing Laboratory, Finnish Red Cross Blood Service) in vitro using the matching viral and tumor antigens (V2/T2). The assay was later repeated for the V2/T2 peptides using cryopreserved PBMCs isolated from blood samples collected before the initiation of anti-PD1 therapy from 2 patients with CMV seropositive melanoma (patients no. 17 and 18 in Supplementary Table S1).
Cryopreserved PBMCs were thawed in 37°C RPMI and then rested overnight at 37°C/5% CO2 in complete RPMI medium supplemented with 10% FBS, 1% l-glutamine, 1% penicillin-streptomycin, before stimulation with viral or tumor antigens. After resting overnight, on day 0 the PBMCs were plated at 6 × 106 cells/well in 6-well plates with the selected peptides in final concentration of 4 μmol/L/mL and incubated 48 hours at 37°C/5% CO2 in complete RPMI medium, duplicate samples were used for each peptide stimulation. NLVPMVATV peptide (NLV, hCMV pp65495–504) was used as a positive control; for a negative control, no peptide stimulation was used. On day 2, half of the medium was replaced with fresh complete RMPI with IL2 for a final concentration of 20 IU/mL IL2. On day 8, GolgiStop (catalog no. 554724, BD Biosciences) was added according to the manufacturer's instructions and cells were incubated overnight.
On day 9, the cells were collected and stained with antibodies purchased from BD Biosciences that were specific for surface markers—CD3 (catalog no. 345767), CD8 (catalog no. 563919), CD45 (catalog no. 560178), and CD56 (catalog no. 557747)—and NLV pentamer (catalog no. F008-2B-E, Proimmune—hCMV, pp65495–504) followed by fixing and permeabilization using BD Cytofix/Cytoperm (catalog no. 554714), and staining for intracellular IFNγ (catalog no. 560371). The IFNγ-positive and IFNγ-negative CD8+ cells were sorted with Influx (BD Biosciences) for DNA extraction using NucleoSpin Tissue XS kit (catalog no. 740901.50, Macherey-Nagel) according to the manufacturer's protocol, followed by TCRβ deep sequencing, which was performed at the Institute for Molecular Medicine Finland (Helsinki, Finland). The workflow of cross-reactivity assay is illustrated in Supplementary Fig. S2.
Analysis of TCRβ deep-sequencing data
TCRβ deep-sequencing data (Supplementary Data S1) was analyzed in ImmunoSEQ Analyzer 3.0 program (Adaptive Biotechnologies). The combined rearrangements data display the clones shared between the samples and how abundantly they are found in each sample. From these data, we selected the top three shared clones based on the sum of productive frequency among the IFNγ-positive T-cell pools and compared the productive frequencies of these clones in nonactivated IFNγ-negative and baseline T-cell pools. The productive frequencies of the top three selected clone in each experimental setting were then visualized with GraphPad Prism version 9.1.0 for Mac (GraphPad Software, LLC).
The Venn diagrams were generated in RStudio (Version 1.1.383) using the exported list of amino acid sequences in the rearrangement details view displaying only the productive clones. The healthy control data contained duplicated sample set for each experimental setting, hence, duplicated clones were removed from the pooled data before generating the Venn diagram.
Statistical analysis
All statistical analysis presented in this study were performed using GraphPad Prism version 9.1.0 for Mac (GraphPad Software, LLC). P values below 0.05 were considered significant. Additional information on the statistical tests are found in each figure legend.
Data and materials availability
All data associated with this study are available on request from the corresponding author.
Results
HEX can identify of viral- and tumor-derived peptides with high molecular mimicry
Whether molecular mimicry could drive antiviral T cells to tumors and trigger an antitumor immune response is still unexplored and anecdotal. In fact, a tool for a systematic analysis of molecular mimicry between tumor and viral antigens is lacking.
To address this problem and study whether molecular mimicry between viruses and tumors could impact the antitumor immune response, we developed HEX. HEX is a software that compares input sequences to a custom database of pathogen-derived protein sequences and selects highly homologous candidate pairs of peptides based on three criteria: (i) B-score, which corresponds to the likelihood of the identified peptides being recognized by a given TCR, (ii) the positional weighted alignment score to prioritize the similarity in the area of interaction with the TCR, and (iii) the prediction of MHC class I–binding affinity (Fig. 1).
To validate the efficiency of the software in selecting homologous peptides with real biological mimicry, we designed an experiment where tumor growth was followed in mice preimmunized with viral peptides similar to known tumor antigens. For this experiment, we considered three murine melanoma–associated antigens that have been applied successfully in a number of vaccination studies: TRP2180–188 (amino acids 180–188 of tyrosinase-related protein 2), gp10025–33 [amino acids 25–33 of premelanosome protein (PMEL)] and TYR1208–216 (amino acids 208–216 of tyrosinase 1; refs. 20,21,22,23). The selected TYR1 sequence was not predicted to bind to murine C57BL/6J MHCs, and thus, was considered as an “irrelevant peptide.” Using HEX, we identified viral peptides that shared a high degree of homology with the input tumor epitopes. Pools of four viral-derived peptides per each original tumor epitope (Supplementary Table S2) were chosen for evaluation in vivo.
C57BL/6J mice were preimmunized with selected viral peptide pools followed by the tumor engraftment (Fig. 2A). All preimmunized mice showed reduction in tumor growth compared with nonpreimmunized mice and significant differences between the treatment groups were observed. Tumor growth was most reduced in mice preimmunized with viral peptides homologous to TRP2180–188 or gp10025–33 (Fig. 2B).
To further investigate the contribution of the selected viral peptides to the reduction of tumor growth, we collected splenocytes at the endpoint of the experiment for ELISpot analysis (Fig. 2C). Splenocytes from mice preimmunized with viral peptide pools homologous to TRP2180–188 or gp10025–33 fostered higher IFNγ release compared with splenocytes from mice preimmunized with control viral peptides homologous to TYR1208–216, which was consistent with the tumor growth data.
We further investigated the reactivity of splenocytes from preimmunized mice toward their respective cognate tumor antigen and found that the response toward the original TRP2180–188 epitope was the highest compared with both the original gp10025–33 and TYR1208–216 tumor epitope (Fig. 2D). In addition, we retrospectively assessed the affinities of each tumor antigen and their homologous viral-peptide pools to C57BL/6J MHC class I molecules (Supplementary Fig. S3) and observed significant correlation with their respective ability to stimulate IFNγ secretion (Fig. 2E). Following the guidelines of the IEDB, we divided the peptides into high and low affinity using the threshold of 50 nmol/L (http://tools.iedb.org/mhci/help/) and observed that peptides with high MHC class I–binding affinity fostered the production of more IFNγ compared with peptides with low MHC class I–binding affinity (Fig. 2F). Taken together, these results validate the predictive efficiency of HEX to identify homologous peptides with biologically relevant mimicry.
To further validate the antitumor efficacy of the HEX-selected peptides, we designed an experiment to assess whether the molecular mimicry between viral and tumor antigens could affect the growth of established tumors in naïve mice. To this end, C57BL/6J mice were engrafted with either B16-OVA cells or the more aggressive and immunosuppressive B16-F10 cells and subsequently treated with the PeptiCRAd vaccines consisting of adenoviruses coated with specific MHC class I–restricted peptides (Fig. 3A). For this experiment, vaccines were prepared by coating the Ad5-Δ24-CpG with the original TRP2180–188 epitope (TRP2-PeptiCRAd) or the corresponding pool of viral-derived peptides similar to TRP2180–188 (Viral PeptiCRAd). Intratumoral administration of PeptiCRAd significantly reduced tumor progression compared with treatment with saline buffer or uncoated virus both for B16-OVA (Fig. 3B) and B16-F10 tumors (Fig. 3C). Mice treated with the original tumor antigen (TRP2-PeptiCRAd) or the viral-derived peptides homologous to TRP2180–188 (Viral PeptiCRAd) showed higher therapeutic success rate in both tumor models (Fig. 3D and E). Taken together, this set of experiments suggests that HEX is efficient in selecting peptides that share molecular mimicry and that molecular mimicry has biological relevance in the immune responses that control tumor growth.
Preexisting antiviral immunity enhances peptide-based cancer immunotherapy via molecular mimicry
After having established the efficiency of HEX and the efficacy of HEX-identified peptides at controlling tumor growth, we next investigated whether preexisting immunity to a given virus would be beneficial when mice are treated with a cancer vaccine based on peptides homologous to that same virus.
To mimic preexisting antiviral immunity, we preimmunized half of a cohort of mice with viral-derived peptides similar to TRP2180–188 identified using HEX. After preimmunization, all the mice were engrafted with syngeneic B16-OVA cells and mice that developed palpable tumors were randomized and treated as follows: PBS (mock), PeptiCRAd-gp100 (no preexisting immunity for this peptide), and PeptiCRAd-TRP2 (peptide with homologous preexisting immunity; Fig. 4A). We observed that mice treated with PeptiCRAd-gp100 displayed no statistically significant difference in tumor volume regardless of their preimmunization status (Supplementary Fig. S4A and S4B). In contrast, mice preimmunized with peptides similar to TRP2180–188 had better control of tumor growth (Fig. 4B) and a higher therapeutic success rate (83% vs. 57%) compared with the nonpreimmunized mice when treated with TRP2-coated PeptiCRAd (Fig. 4C). At the experimental endpoint, mice were euthanized, and flow cytometry analysis was performed on the collected tumors. We observed an increase in CD8+ and memory CD8+ T cells in the tumors of preimmunized mice treated with TRP2 PeptiCRAd, together with increased proliferative activity (Ki67+), indicating that the antiviral immunity had conditioned the tumor to become more responsive to the cancer therapy. We also observed a decrease in the intratumoral CD8+ CXCR3+ population in preimmunized mice, suggesting a more memory-like phenotype in the preimmunized mice (Fig. 4D). These results support the hypothesis that preexisting antiviral immunity could boost the effect of immunotherapies in an antigen-specific fashion when the preexisting antigens and the antigens used for the therapeutic vaccine share a high degree of homology.
MHC-I epitope mimicry and cross-reactive T cells contribute to prolonged survival of anti-PD1–treated patients with metastatic melanoma
As demonstrated above, the similarity between tumor antigens and viral-derived antigens can play a crucial role in tumor clearance via cross-reactive T cells in murine model of cancer. Thus, to investigate our hypothesis in a setting that was closer to a clinical scenario, we evaluated the effect of preexisting immunity on immune checkpoint inhibitor (ICI) therapy in a preclinical model and studied whether there was a synergistic effect between these two. To this end, we preimmunized half of a cohort of C57BL/6J mice using virus-derived peptides similar to TRP2180–188 before the syngeneic B16-OVA tumor engraftment. Half the mice in each group were subsequently treated with anti-PD1 (aPD1 group) and half with PBS (mock group) according to the scheme in Fig. 5A. We observed that tumors in preimmunized mice treated with anti-PD1 were significantly smaller than tumors in nonpreimmunized mice treated with anti-PD1 (Fig. 5B) and the therapeutic success rate increased from 40% (for naïve mice treated with anti-PD1) to 100% (for preimmunized mice treated with anti-PD1; Fig. 5C).
To investigate whether molecular mimicry could also play a role in patients with cancer, we studied a cohort of patients with metastatic melanoma undergoing treatment with anti-PD1 monotherapy (Supplementary Table S1). We collected peripheral blood samples from patients before initiation of therapy, and after 1 and 3 months of therapy and tested their serologic status for CMV and EBV at each time point. We used Cox regression analysis to study the association of pretreatment serum CMV- and EBV-specific IgG levels with progression-free survival (PFS). Our results indicated that patients with pretreatment titer of serum anti-CMV IgG higher than the median had significantly longer PFS when compared with patients with anti-CMV IgG levels lower than the median [HR = 0.34, 95% confidence interval (CI): 0.08–1.4, P = 0.04] (Supplementary Fig. S5A). In contrast, anti-EBV IgG levels were not associated with prolonged PFS (HR = 0.75, 95% CI: 0.2–3.1, P = ns; Supplementary Fig. S5B).
To evaluate the more general immunologic status of the patients, quantitative assessment of serum IgA, IgG, and IgM was performed at each time point. The pretreatment levels of these immunoglobulins showed no differences between the responders (R) and nonresponders (NR), indicating that there was no general increased immune-reactive status in patients before initiation of ICI therapy (Supplementary Fig. S5C–S5E). Furthermore, after initiation of anti-PD1 treatment, the responders' IgG levels were significantly higher compared with nonresponders' IgG levels (mean: R1mo 12.8 g/L vs. NR1mo: 9.4 g/L, P = 0.02, R3mo 12.7 g/L vs. NR3mo: 9.9 g/L; Supplementary Fig. S5D). Taken together, our data indicate that anti-CMV, but not anti-EBV immunity, correlated with prolonged PFS in patients with metastatic melanoma undergoing ICI therapy.
These findings presented us with an opportunity to study whether molecular mimicry between CMV and tumor antigens could explain the differences in prognosis. We hypothesized that, T cells cross-reactive between known melanoma antigens and CMV might explain the better PFS in some patients. Therefore, we selected a pool of melanoma-associated proteins (24) and compared them with the CMV proteome using HEX, which generated a list of melanoma-associated peptides highly similar to CMV (Supplementary Table S3). The cross-reactivity of patient PBMCs between tumor and CMV peptides was first assessed by ELISpot assay. We observed that these PBMCs were stimulated by both tumor peptides and their CMV homologous counterparts, suggesting that CMV infection could expand viral-specific T-cell clones that could attack and kill tumor cells, making seropositive patients more prone to react toward melanoma-specific epitopes similar to CMV (Supplementary Fig. S6).
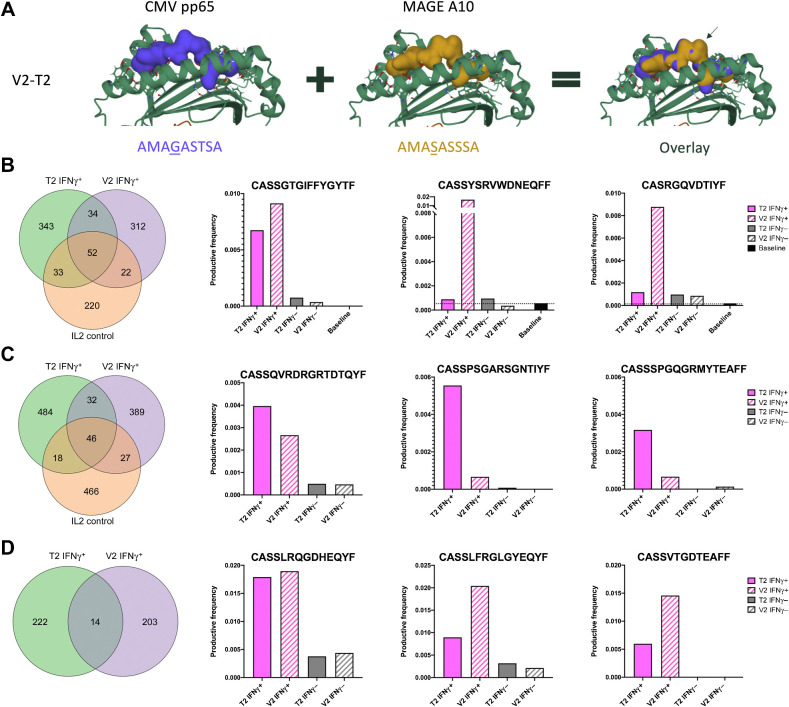
To study cross-reactive T cells and whether homologous peptides can activate and expand the same T-cell clones, we designed an additional cross-reactivity experiment using PBMCs from healthy donors and patients with metastatic melanoma seropositive for CMV. In these individuals, we investigated whether the same T-cell clones could be expanded when PBMCs were pulsed with viral (V) and tumor (T) peptides with high molecular mimicry. We selected the epitope pair V2 and T2 which, in the position exposed to the TCR, differed by only one amino acid between the two sequences (Fig. 6A, mismatch highlighted by an arrow). Next, we activated PBMCs collected from CMV seropositive donors using the V2/T2 viral and tumor antigen pair. After the activation, we sorted the IFNγ-positive and negative CD8+ T cells for further analysis with TCRβ sequencing. From the TCRβ data, we identified clones that were present in both activated T-cell pools, pulsed either with V2 or T2, but not present in T-cell pools stimulated with IL2 only (negative control). We observed that among the healthy donor IFNγ-positive CD8+ T cells, 34 T-cell clones (9.9% of T2 and 10.9% of V2 expanded clones) were expanded by both T2 and V2 and not by IL2 alone. Of these shared clones, the top three (CASSGTGIFFYGYTF, CASSYSRVWDNEQFF, and CASRGQVDTIYF) with highest productive frequency among IFNγ-positive CD8+ T cells did not appear to be as frequently represented in IFNγ-negative and baseline CD8+ T-cell pools (Fig. 6B). Similar observations were made using IFNγ-positive CD8+ T cells from 2 patients with melanoma. In patient 18 (Supplementary Table S1), a total of 32 T-cell clones (6.6% of T2 and 8.2% of V2-expanded clones) were expanded by stimulation with T2 and V2 after filtering out the IL2-expanded clones (Fig. 6C). In patient 17 (Supplementary Table S1), IL2 activation alone led to a low number of IFNγ-positive CD8+ T cells and hence no TCR data were obtained. However, among the IFNγ-positive CD8+ T cells stimulated with T2 or V2, a total of 14 (6.3% of T2 and 6.9% of V2-expanded clones) clones were shared (Fig. 6D). Of these shared clones, the top three candidates based on the sum of productive frequency (patient 18: CASSQVRDRGRTDTQYF, CASSPSGARSGNTIYF, and CASSSPGQGRMYTEAFF, patient 17: CASSLRQGDHEQYF, CASSLFRGLGYEQYF, and CASSVTGDTEAFF) appeared to be enriched in the activated IFNγ-positive CD8+ T-cell pool (Fig. 6C and D) when compared with IFNγ-negative CD8+ T cells. Taken together, these data suggest that homologous peptides predicted by HEX can be recognized by the same TCR and that molecular mimicry could play role in mediating antitumor immune responses. This could potentially be exploited to design more efficient anticancer immunotherapies based on the preexisting immunity of the patient.
Figure 6.
T-cell cross-reactivity between viral and tumor antigens sharing a high degree of homology. A, Docking overlay of the V2 and T2. B, Cross-reactive T-cell clones generated using healthy donor PBMCs. C, Cross-reactive T-cell clones generated using PBMCs isolated before initiation of anti-PD1 therapy from a patient with metastatic melanoma (patient 18). D, Cross-reactive T-cell clones generated using PBMCs isolated before initiation of anti-PD1 therapy from a patient with metastatic melanoma (patient 17). B–D, Venn diagrams represent the number of shared clones in IFNγ-positive CD8+ T cells between the indicated settings. The bar plots represent the three clones with the highest productive frequency among the IFNγ-positive CD8+ T cells that were expanded after T2 and V2 stimulation, but not when stimulated with IL2 alone. Pink bars represent the productive frequency in IFNγ-positive CD8+ T-cell pool, grey bars represent the productive frequency in the IFNγ-negative CD8+ T-cell pool, and black bar represents the productive frequency in baseline CD8+ T cells without any experimental stimulation. Black dotted line represents the baseline productive frequency of each clone.
Discussion
In this study, we present a bioinformatic tool (HEX) to identify tumor-specific MHC class I–restricted peptides with high similarity to viral-specific peptides. Using peptides identified by this tool, we showed that, in murine tumor models, preexisting antiviral immunity enhanced the efficacy of cancer immunotherapy via molecular mimicry. We further report, that in a cohort of human patients with melanoma with high humoral response to CMV, molecular mimicry between CMV and tumor antigens potentially played a role in the response to anti-PD1 therapy via activation of cross-reactive T cells. We also demonstrate that in healthy donors and patients with melanoma with preexisting immunity to CMV, melanoma-specific peptides similar to CMV-specific peptides activated and expanded the same T-cell clones. Our findings support the hypothesis that viral molecular mimicry favorably modifies the tumor immune microenvironment, improving efficacy of ICI therapy.
The TCR is a highly promiscuous receptor and thus allows T cells to recognize a large variety of targets (1, 2). The idea that an immune response toward certain pathogens could lead to cross recognition of self-antigens is well established in the field of autoimmune diseases, a process known as “molecular mimicry” (3). Here, we investigated whether a similar mechanism could also drive the antitumor immune response via cross-reactive T cells.
Some studies have speculated that pathogen-specific cross-reactive T cells could be responsible for the extraordinary antitumor immune responses and prolonged PFS observed in some patients with cancer (4, 8, 10). Although intriguing, this hypothesis has not been systematically studied, nor has it impacted clinical practice, partly due to the lack of a proper tool to identify molecular mimicry between tumor and pathogen antigens. Hence, we developed a unique software to rapidly identify tumor-specific MHC class I–restricted peptides with high homology to pathogen-derived MHC class I–restricted peptides and named it HEX. Rather than simply aligning sequences, this software assesses physicochemical similarity at a molecular level between an input sequence (single peptide or a list of peptides) and a list of pathogen-derived antigens (11) and provides a positionally weighted (25) alignment score to prioritize similarities occurring in the central section of the peptide, which is the region of the peptide primarily involved in interacting with the TCR (26). Moreover, to increase the chances of identifying naturally presented epitopes, it predicts the MHC class I–binding affinity of both the input and the cognate viral peptides using state of the art predictors of MHC class I–binding affinity (NetMHC-API from IEDB servers; ref. 12).
We found that preimmunization with viral peptides similar to well-characterized tumor peptides (20,21,22,23), efficiently slowed down the growth of subcutaneously injected melanoma tumor cells in mice, indicating that preexposure to viral-derived peptides can affect tumor growth. Rosato and colleagues have reported that virus-specific memory T cells can populate tumors, enhancing ICI therapy (9). Herein, we propose the underlying mechanism to be molecular mimicry between viral and tumor antigens. We also observed that the tumor-homologous viral-derived peptides have therapeutic effect on already established tumors, suggesting that a viral infection occurring during tumor progression could influence tumor response to treatment. Mice intratumorally injected with the peptide-uncoated virus showed no antitumor effect in contrast to what has been shown by Newman and colleagues (27). However, viruses coated with highly homologous peptides produced a marked antitumor response, showing that the effect was antigen specific and based on molecular mimicry. The apparent discrepancy between our findings and those of Newman and colleagues could be explained by the use of different viruses, with differing antigen repertoire and different degree of homology between virus and tumor, in the two studies. Tumor regression following viral administration or natural infection has been reported previously. This could be due to a generic mechanism, such as an “adjuvant” effect of the virus with consequent recruitment of T cells to the tumor, or to a more specific mechanism of shared antigens and activation of cross-reactive T cells (28, 29). Many coincidences have to occur to trigger potent mimicry-mediated tumor regression spontaneously, and, as for autoimmune diseases, the mimicry-mediated mechanisms may not be enough to trigger a biologically relevant antitumor T-cell response, without other concomitant circumstances (3). However, we envision that with the right tool to identify optimal homologous antigens, mimicry between tumors and pathogens previously encountered by a patient's immune system could be exploited to specifically boost the efficacy of cancer immunotherapy. Our findings potentially have implications in peptide-selection for cancer vaccines, where peptides able to recruit antiviral memory T cells in the tumor microenvironment might improve the outcome of treatment (4, 8). Recently, several attempts have been made to repurpose existing antiviral T cells or to use general vaccines to boost and/or redirect T cells against tumors. Rosato and colleagues (9) used viral-derived peptides to engage preexisting T cells, Newman and colleagues (27) used direct intratumoral administration of the flu vaccine and Tähtinen and colleagues used tetanus-derived peptides to engage tetanus-specific CD4+ T cells in prevaccinated tumor-bearing mice (30). All these approaches were successful in preclinical models and some of them have proceeded to clinical testing. We suggest that these could be even more efficient if designed on the basis of molecular mimicry with regards to a patient's preexisting immunity.
We also found that molecular mimicry could enhance the effect of ICI therapy in mice, recapitulating and partially explaining the effect of ICIs in human patients with cancer. ICIs such as anti-PD1 have significantly improved survival for patients with solid tumors, especially those who have metastatic melanoma, when compared with other commonly used therapies such as radiation and chemotherapy (31, 32). Despite the enhanced survival and efficient response rates, it is unknown why some patients benefit more than others. The direct administration of viruses to tumor sites enhances the influx of T cells, predisposing ICIs to work more efficiently (28, 29, 33). However, the link between these viral T cells and antitumor responses is still unknown. Molecular mimicry could be the missing link, at least in some cases, explaining why antiviral T cells at the tumor site have a beneficial effect. Therefore, we studied a cohort of patients with metastatic melanoma patients undergoing anti-PD1 therapy and measured their serum anti-CMV and anti-EBV IgG titers, as these are very common viruses and T cells specific for these viruses have been often found in patient tumors (34). In particular, CMV-reactive T cells have previously been reported in patients treated with ICIs (34) and speculated to be associated with a beneficial effect on therapy response (33). Our results indicated that patients with a high titer of CMV-specific IgG had significantly longer PFS compared with patients with a lower titer of CMV-specific IgG. In contrast, EBV-specific IgG levels did not associate with prolonged PFS. No significant differences were observed in the general immune activation between the responder and nonresponder cohorts, supporting the hypothesis that the CMV-specific immune response might contribute to the prolonged survival of patients with melanoma treated with anti-PD1. Strengthening this hypothesis, we observed that PBMCs from a patient with high titer of CMV-specific IgG, reacted with melanoma antigens similar to CMV peptides. In addition to this, a deeper evaluation on the cross-reactivity by TCRβ sequencing indicated that the same TCR clones appeared to be expanded in activated T-cell populations pulsed with mimicking viral- and tumor-derived antigens even when these clones were absent at baseline. These data indicate the possibility that molecular mimicry between viral- and tumor-derived antigens could contribute to antitumor T-cell immunity. Thus, we believe that the molecular mimicry between CMV and melanoma could provide a clinical advantage for patients undergoing ICI therapy.
There are some limitations to our study. First, the HEX tool only predicts binding affinity of MCH class I–restricted epitopes. These were, however, prioritized because CD8+ T cells have a central role in recognizing both virally infected and transformed cells. Second, HEX does not consider whether the peptides are naturally processed and this could affect the number of false positive candidate peptides (35). Another limitation of our study is our exclusive focus on tumor-associated antigens, whereas probably the most interesting application of HEX could have been the analysis of the effect of mutations in cancer neoepitopes in the context of molecular mimicry. The small patient cohort available for this study is an additional limiting factor. Because of limited sample material from patients with melanoma, we had to prioritize the experimental setting in the cross-reactivity assay to include only one viral/tumor pair (T2/V2) together with a negative IL2 control. Hence, in Fig. 6C and D, we do not have the baseline data from patients with melanoma, unlike in healthy donor in Fig. 6B. To further explore molecular mimicry in a clinical setting, studies with larger patient cohorts in controlled clinical trials together with other possible target epitopes are warranted. In addition, in future studies, we will expand our analysis to a subpopulation of tumor-infiltrating viral-specific CD39−CD8+ T cells, because these cells are generally considered to have just a bystander role (36).
The results of this study indicate that viral infections could have an impact on tumor growth and clearance. In addition, we show cross-reactivity of cytotoxic T cells against viral- and homologous tumor-derived antigens selected using our HEX software. Our findings highlight the importance of preexisting antiviral immunity in cancer immunotherapy and suggest the use of HEX to select tumor antigens highly homologous to viral antigens to engage cross-reactive T cells.
Authors' Disclosures
M. Hernberg reports personal fees from Novartis, MSD, and Bristol Myers Squibb outside the submitted work. S. Mäkelä reports personal fees from Bristol Myers Squibb, Merck Sharp & Dohme, Novartis, Merck Group, and Sanofi outside the submitted work. H. Karhapää reports grants from The Finnish Melanoma Group outside the submitted work. S. Mustjoki reports grants from Cancer Foundation Finland and Sigrid Juselius Foundation during the conduct of the study, as well as grants and personal fees from Novartis, Pfizer, and Bristol Myers Squibb outside the submitted work. V. Cerullo is a cofounder of and shareholder at VALO Therapeutics. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors thank all the coauthors for their support and hard work and to the Advanced Research Computing at Cardiff (ARCCA) for hosting and assisting with HEX. The authors also thank the patients, clinicians, and study nurses for their participation.
This work has been supported by European Research Council under the European Union's Horizon 2020 Framework programme (H2020/ERC-CoG-2015 Grant Agreement number 681219). Moreover, this research was supported by the Helsinki Institute of Life Science (HiLIFE), Jane and Aatos Erkko Foundation, and Cancer Society of Finland. In addition, this work has been supported by Finnish Cancer Organizations, Sigrid Juselius Foundation, Relander Foundation, state funding for university-level health research in Finland, Fican South funding, and HiLife Fellow funds from the University of Helsinki.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
#J. Chiaro, H.H.E. Kasanen, A.S. Kreutzman, S. Mustjoki, and B. Szomolay contributed equally to the article.
Cancer Immunol Res 2021;9:981–93
Authors' Contributions
J. Chiaro: Conceptualization, data curation, software, formal analysis, writing–original draft. H.H.E. Kasanen: Conceptualization, formal analysis, validation, writing–original draft. T. Whalley: Software. C. Capasso: Methodology. M. Grönholm: Supervision. S. Feola: Formal analysis, methodology. K. Peltonen: Supervision. F. Hamdan: Formal analysis, methodology. M. Hernberg: Resources. S. Mäkelä: Resources. H. Karhapää: Resources. P.E. Brown: Resources. B. Martins: Data curation, formal analysis. M. Fusciello: Data curation. E.O. Ylösmäki: Supervision, methodology. D. Greco: Conceptualization, writing–review and editing. A.S. Kreutzman: Supervision. S. Mustjoki: Resources, formal analysis, supervision. B. Szomolay: Resources, software, writing–review and editing. V. Cerullo: Conceptualization, resources, formal analysis, supervision, writing–original draft.
References
- 1. Bentzen AK, Such L, Jensen KK, Marquard AM, Jessen LE, Miller NJ, et al. T cell receptor fingerprinting enables in-depth characterization of the interactions governing recognition of peptide–MHC complexes. Nat Biotechnol 2018. Nov 19 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wooldridge L, Ekeruche-Makinde J, Van Den Berg HA, Skowera A, Miles JJ, Tan MP, et al. A single autoimmune T cell receptor recognizes more than a million different peptides. J Biol Chem 2012;287:1168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rojas M, Restrepo-Jiménez P, Monsalve DM, Pacheco Y, Acosta-Ampudia Y, Ramírez-Santana C, et al. Molecular mimicry and autoimmunity. J Autoimmun 2018;95:100–23. [DOI] [PubMed] [Google Scholar]
- 4. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, et al. First-line nivolumab in stage IV or recurrent non-small cell lung cancer. N Engl J Med 2017;376:2415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69–74. [DOI] [PubMed] [Google Scholar]
- 8. Balachandran VP, Luksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017;551:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosato PC, Wijeyesinghe S, Stolley JM, Nelson CE, Davis RL, Manlove LS, et al. Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat Commun 2019;10:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loftus DJ, Castelli C, Clay TM, Squarcina P, Marincola FM, Nishimura MI, et al. Identification of epitope mimics recognized by CTL reactive to the melanoma/melanocyte-derived peptide MART-1(27–35). J Exp Med 1996;184:647–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szomolay B, Liu J, Brown PE, Miles JJ, Clement M, Llewellyn-Lacey S, et al. Identification of human viral protein-derived ligands recognized by individual MHCI-restricted T-cell receptors. Immunol Cell Biol 2016;94:573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016;32:511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Capasso C, Hirvinen M, Garofalo M, Romaniuk D, Kuryk L, Sarvela T, et al. Oncolytic adenoviruses coated with MHC-I tumor epitopes increase the antitumor immunity and efficacy against melanoma. Oncoimmunology 2016;5:e1105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tatsis N, Ertl HC. Adenoviruses as vaccine vectors. Mol Ther 2004;10:616–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cerullo V, Diaconu I, Romano V, Hirvinen M, Ugolini M, Escutenaire S, et al. An oncolytic adenovirus enhanced for toll-like receptor 9 stimulation increases antitumor immune responses and tumor clearance. Mol Ther 2012;20:2076–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ying B, Toth K, Spencer JF, Meyer J, Tollefson AE, Patra D, et al. INGN 007, an oncolytic adenovirus vector, replicates in Syrian hamsters but not mice: comparison of biodistribution studies. Cancer Gene Ther 2009;16:625–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Halldén G, Hill R, Wang Y, Anand A, Liu TC, Lemoine NR, et al. Novel immunocompetent murine tumor models for the assessment of replication-competent oncolytic adenovirus efficacy. Mol Ther 2003;8:412–24. [DOI] [PubMed] [Google Scholar]
- 18. Hamdan F, Martins B, Feodoroff M, Giannoula Y, Feola S, Fusciello M, et al. GAMER-Ad: a novel and rapid method for generating recombinant adenoviruses. Mol Ther Methods Clin Dev 2021;20:625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rigo MM, Antunes DA, Vaz de Freitas M, Fabiano de Almeida Mendes M, Meira L, Sinigaglia M, et al. DockTope: a Web-based tool for automated pMHC-I modelling. Sci Rep 2015;5:18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Colluru VT, Johnson LE, Olson BM, McNeel DG. Preclinical and clinical development of DNA vaccines for prostate cancer. Urol Oncol 2016;34:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bowne WB, Srinivasan R, Wolchok JD, Hawkins WG, Blachere NE, Dyall R, et al. Coupling and uncoupling of tumor immunity and autoimmunity. J Exp Med 1999;190:1717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gold JS, Ferrone CR, Guevara-Patiño JA, Hawkins WG, Dyall R, Engelhorn ME, et al. A single heteroclitic epitope determines cancer immunity after xenogeneic DNA immunization against a tumor differentiation antigen. J Immunol 2003;170:5188–94. [DOI] [PubMed] [Google Scholar]
- 23. Wolchok JD, Yuan J, Houghton AN, Gallardo HF, Rasalan TS, Wang J, et al. Safety and immunogenicity of tyrosinase DNA vaccines in patients with melanoma. Mol Ther 2007;15:2044–50. [DOI] [PubMed] [Google Scholar]
- 24. Weinstein D, Leininger J, Hamby C, Safai B. Diagnostic and prognostic biomarkers in melanoma. J Clin Aesthet Dermatol 2014;7:13–24. [PMC free article] [PubMed] [Google Scholar]
- 25. Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, et al. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res 2008;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharma AK, Kuhns JJ, Yan S, Friedline RH, Long B, Tisch R, et al. Class I major histocompatibility complex anchor substitutions alter the conformation of T cell receptor contacts. J Biol Chem 2001;276:21443–9. [DOI] [PubMed] [Google Scholar]
- 27. Newman JH, Chesson CB, Herzog NL, Bommareddy PK, Aspromonte SM, Pepe R, et al. Intratumoral injection of the seasonal flu shot converts immunologically cold tumors to hot and serves as an immunotherapy for cancer. Proc Natl Acad Sci U S A 2020;117:1119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RHI, Michielin O, et al. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell 2017;170:1109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zamarin D, Holmgaard RB, Subudhi SK, Park JS, Mansour M, Palese P, et al. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Transl Med 2014;6:226ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tähtinen S, Feola S, Capasso C, Laustio N, Groeneveldt C, Ylösmäki EO, et al. Exploiting preexisting immunity to enhance oncolytic cancer immunotherapy. Cancer Res 2020;80:2575–85. [DOI] [PubMed] [Google Scholar]
- 31. Koury J, Lucero M, Cato C, Chang L, Geiger J, Henry D, et al. Immunotherapies: exploiting the immune system for cancer treatment. J Immunol Res 2018;2018:9585614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ugurel S, Röhmel J, Ascierto PA, Flaherty KT, Grob JJ, Hauschild A, et al. Survival of patients with advanced metastatic melanoma: the impact of novel therapies–update 2017. Eur J Cancer 2017;83:247–57. [DOI] [PubMed] [Google Scholar]
- 33. Erkes D, Wilski N, Snyder C. Intratumoral infection by CMV may change the tumor environment by directly interacting with tumor-associated macrophages to promote cancer immunity. Hum Vaccin Immunother 2017;13:1778–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Franklin C, Rooms I, Fiedler M, Reis H, Milsch L, Herz S, et al. Cytomegalovirus reactivation in patients with refractory checkpoint inhibitor-induced colitis. Eur J Cancer 2017;86:248–56. [DOI] [PubMed] [Google Scholar]
- 35. The problem with neoantigen prediction. Nat Biotechnol 2017;35:97. [DOI] [PubMed] [Google Scholar]
- 36. Simoni Y, Becht E, Fehlings M, Loh CY, Koo SL, Teng KWW, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 2018;557:575–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.