Abstract
Purpose:
The consensus molecular subtypes (CMS) represent a significant advance in the understanding of intertumor heterogeneity in colon cancer. Intratumor heterogeneity (ITH) is the new frontier for refining prognostication and understanding treatment resistance. This study aims at deciphering the transcriptomic ITH of colon cancer and understanding its potential prognostic implications.
Experimental Design:
We deconvoluted the transcriptomic profiles of 1,779 tumors from the PETACC8 trial and 155 colon cancer cell lines as weighted sums of the four CMSs, using the Weighted In Silico Pathology (WISP) algorithm. We assigned to each tumor and cell line a combination of up to three CMS subtypes with a threshold above 20%.
Results:
Over 55% of tumors corresponded to mixtures of at least two CMSs, demonstrating pervasive ITH in colon cancer. Of note, ITH was associated with shorter disease-free survival (DFS) and overall survival, [HR, 1.34; 95% confidence interval (CI; 1.12–1.59), 1.40, 95% CI (1.14–1.71), respectively]. Moreover, we uncovered specific combinations of CMS associated with dismal prognosis. In multivariate analysis, ITH represents the third parameter explaining DFS variance, after T and N stages. At a cellular level, combined WISP and single-cell transcriptomic analysis revealed that most colon cancer cell lines are a mixture of cells falling into different CMSs, indicating that ITH may correspond to distinct functional statuses of colon cancer cells.
Conclusions:
This study shows that CMS-based transcriptomic ITH is frequent in colon cancer and impacts its prognosis. CMS-based transcriptomic ITH may correspond to distinct functional statuses of colon cancer cells, suggesting plasticity between CMS-related cell populations. Transcriptomic ITH deserves further assessment in the context of personalized medicine.
Translational Relevance.
The classification into consensus molecular subgroups (CMS) represents the current framework for depicting the transcriptomic heterogeneity of colorectal cancer. This CMS classification has prognostic and predictive value and is viewed as the path toward precision medicine. In this article, we add another layer of complexity to colorectal cancer transcriptional heterogeneity by demonstrating that a large fraction of colorectal cancer samples actually corresponds to CMS mixtures, rather than exhibiting a pure CMS signature. We find that patients with intratumor heterogeneity have a dismal prognosis and that specific combinations of CMS are particularly unfavorable. Our results suggest that intratumor heterogeneity is the result of the co-occurrence of tumor cell populations with distinct transcriptomes, which may synergize to escape immune surveillance. Our study advocates for a refinement of colorectal cancer subtyping taking into account intratumor CMS heterogeneity.
Introduction
Colon cancer is a frequent and severe disease with more than 1.36 million new cases worldwide (1) making it the second most common cancer. Approximately 70% to 80% of patients with colon cancer are diagnosed without metastatic spread and may be cured by surgery. However, 10% to 60% of them will experience disease recurrence depending on the disease stage. To improve long-term outcomes, adjuvant chemotherapy is used in patients with a substantial risk of disease recurrence (high-risk stage II and stage III disease; ref. 2). It is estimated that for patients eligible to adjuvant chemotherapy combining oxaliplatin with a fluoropyrimidine, approximately half of them would have been cured by surgery alone and 25% will relapse despite the use of adjuvant treatment. Altogether, although all patients receiving adjuvant treatment are exposed to short-term and long-lasting toxicities, only 25% of these patients experience therapeutic benefit. For this reason, personalization of cancer treatments is required. Indeed, the heterogeneity in clinical responses points to a biological heterogeneity of the cancers that requires further exploration.
A variety of genetic biomarkers, including mutations and copy-number variations, are being characterized for their impact on patient trajectories (3, 4). Transcriptomic classifications have also been proposed to decipher the intertumor heterogeneity of colon cancer, which has led to the definition of four consensus molecular subtypes (CMS) through an international effort (CMS1..4; ref. 5). These CMSs are robustly associated to several clinical and biological features (e.g., the mesenchymal CMS4 is associated to a poorer prognosis).
Both genetic and transcriptomic studies of colon cancer heterogeneity have highlighted the existence of intratumor heterogeneity (ITH), which could also contribute to the mechanisms of anti-cancer drug resistance (6, 7). While several forms (genetic, epigenetic, transcriptomic, and phenotypic) of ITH have been explored for various types of cancers, genetic ITH has led to more extensive studies (8). In particular genetic ITH was shown to be frequent and associated to poorer prognosis in several cancer types (9, 10), including colon cancer (11, 12).
Concerning transcriptomic ITH, it has been studied in relation to molecular subtypes in different tumor types. In colon cancer, it was notably investigated through intratumor multi-sampling (13) or single-cell RNA sequencing (RNA-seq; refs. 14, 15); however, in small series. In other cancer types, transcriptome deconvolution was recently shown as a powerful method to evaluate ITH and its prognostic value in large series (16).
Here we used transcriptome deconvolution to assess ITH and its possible prognostic impact in a large multicentric colon cancer cohort collected in the frame of a prospective phase III trial (PETACC8, n = 1779, NCT00265811). We considered CMS as representing different tumor cell populations, with the idea to estimate the CMS-based ITH. Similarly, we analyzed the composition of the tumor microenvironment (TME) in terms of different immune and stromal cell populations, features that also characterize ITH. We then assessed whether these tumor and TME-based variables could bring information for the determination of stage III colon cancer prognosis, after adjustment for established prognostic factors.
Materials and Methods
Ethical statements
This study was done in accordance with the Declaration of Helsinki (amended 2000) and the International Conference on Harmonization of Technical Requirements of Pharmaceuticals for Human Use Note for Guidance on Good Clinical Practice and approved by the appropriate Ethics Committees of each participating country. Furthermore, the patients signed a written informed consent for specific translational research, which was obtained for2,043 patients of the 2,559 patients included in PETACC8.
Patients and tumor samples
As depicted in the flow chart (extended data Supplementary Fig. S1), three series of tumors from patients with colorectal cancer were used: (i) the CIT initial series (n = 566 patients), (ii) the CIT extension series (n = 45 patients), and (iii) the PETACC8 series (n = 1,779 patients; refs. 17, 18).
For the CIT initial series, RNA was extracted from frozen tumor samples stored in liquid nitrogen. For the CIT extension series, RNA was extracted from both frozen and formalin-fixed paraffin-embedded (FFPE) tissues. For PETACC08 tumor series, RNA was extracted from FFPE tissues. The following described experiments were performed blinded to the study endpoint.
Transcriptome data
To promote the clinical use of CMS, our aim was to develop a classifier allowing to study ITH in terms of CMS in FFPE tumor samples, and we chose the NanoString technology for this purpose. The full transcriptome profiles and CMS labels of the 566 samples of the CIT initial series (17) were publicly available (GSE39582). For targeted transcriptomic profiling, we selected 196 transcriptomic markers, including 10 housekeeping genes, 81 CMS specific markers identified using the CIT initial series, and 105 other markers, mainly related to microenvironment cell populations (19), immune modulators (20), and oncogenic pathways (5), as recapitulated in extended data Supplementary Table S1. The transcriptomic profile of these 196 genes was acquired using NanoString nCounter Analysis System using 150 ng of total RNA from 1,779 FFPE samples from the PETACC8 series, 144 samples of the CIT initial series [133 fresh frozen (FF) and 11 FFPE], and 90 samples from the CIT extension series (corresponding to 45 patients with paired FF and FFPE tumor samples). Nine batches and four subcodesets were used. The normalization approach was selected to minimize batch/subcodeset variation, while maximizing genes and samples correlation with Affymetrix data and paired samples. Given a sample j, the corresponding vector of raw counts, RAWj, was first normalized to a vector NORM1j, using a scaling factor (SF1j) based on the expression of four housekeeping genes (CLTC, NT5C2, ATP5E, VDAC2): NORM1j = RAWj * SF1j. If we note Fj the geometric mean expression of housekeeping gene for sample j, the scaling factor SF1j corresponds to F*/Fj, where F* denotes the intersample mean of F. To get normalized profiles (NORM2j) relatively to batch/subcodeset bias, we used a scaling factor SF2j based on a calibrator sample whose profile was obtained in the 36 (batch, subcodeset) pairs, and using a reference (batch, subcodeset) pair: NORM2j = NORM1j * SF2j. SF2j is calculated as the ratio of the calibrator sample profile in the reference (batch, subcodeset) pair versus in the (batch, subcodeset) pair corresponding to sample j. Finally, we log2 transformed the NORM2 values, and did not use the NanoString endogeneous positive and negative controls. PETACC8 samples showing either high SF1 (>20) or low raw counts expression (95th percentile < 100) were removed, which left us with 1,779 PETACC8 samples out of 1,809 with available NanoString profiles. Markers with low expression across samples (95th percentile < 6) were removed, yielding 172 retained markers.
Training of a random forest classifier of the CMS
Using normalized counts data of the CIT extension series, we measured, for each of the 172 markers, the correlation across expression profiles derived from the paired FF and FFPE samples. We retained the 23 markers showing both a FF/FFPE correlation above 0.2 and inter-CMS differential expression in the FFPE CIT extension series (ANOVA P < 0.1). These markers were used to train a random forest (RF) classifier using the 133 FF samples from the CIT initial series. To remove the marker specific bias between the FF and FFPE profiles, we performed feature specific quantile normalization (21), using the 90 paired samples from the CIT extension series. We thus derived “pseudo-FFPE” profiles (see Supplementary Materials and Methods) of the 133 FF profiles of the CIT initial series before training the RF classifier. Finally, this RF classifier was applied to the 1,779 normalized profiles of PETACC8 series. In the end, all the samples from the CIT initial and CIT extension series were used in one of the two training steps (variable selection and RF learning) to build the RF classifier, while the PETACC8 series was used solely for validation.
Intratumor CMS heterogeneity measures, based on WISP-supervised deconvolution
The Weighted In Silico Pathology (WISP) deconvolution algorithm (16) sees a tumor as a mixture of the four CMS, each seen as a cell population. WISP provides a vector of four weights (w1, w2, w3, w4), respectively related to CMS1, CMS2, CMS3, and CMS4. Any of these four weights is superior or equal to zero, and the sum of the four weights is 100% (w1+w2+w3+w4 = 1). The WISP algorithm was applied to all 1,779 samples from PETACC8 series, using their 172-marker normalized expression profiles, and the CMS labels predicted with the RF classifier. This yielded a (1,779 × 4) matrix giving the estimated weights (from 0 to 1) of the four CMS for all 1,779 samples. To assess the level of ITH, we counted the (intrasample) number of CMS showing a weight greater than 20%. We may note that after testing for several cutoffs ranging from 10% to 30%, which all yielded similar results, we arbitrarily chose a 20% cutoff.
For any sample, the CMS with the highest weight was designated as major CMS, while the CMS with second highest weight (if > 20%) was designated as minor CMS. The major CMS weight varies from 32% to 100% across the 1779 Petacc8 samples (mean: 74%, median 73%). A sample is defined as pure for its major CMS if all other CMS have weights below 20%.
The application of the WISP algorithm to public datasets is described in Supplementary Materials and Methods.
Intratumor CMS heterogeneity measures in colon cancer cell lines
To probe the existence of intrasample CMS heterogeneity in colorectal cancer cell lines, we used a series (22) of 155 cell lines profiled using HT12 Illumina gene expression platform (GSE59857), of which 55 were also profiled (23) using Affymetrix HGU133Plus2 gene expression platform [Cancer Cell Line Encyclopedia (CCLE) CC series: GSE36133]. On the basis of the 55 common samples between these two series, we calculated gene-level linear models of the deviation between the two platforms (Affymetrix and Illumina), to derive an “Affymetrix-like” version of the GSE59857 Illumina series (22). Then, we selected genes with highest coefficients of variation in the cell line series to calculate CMS centroids on the CIT colon cancer dataset (GSE39582; ref. 17), through the WISP training function. These centroids were finally used to estimate intrasample CMS weights in the 155-cell-line GSE59857 series, using WISP prediction function (as shown in extended data Supplementary Fig. S3A).
Validation of intratumor CMS heterogeneity of Lovo and Mdst8 cell lines, using single-cell RNA-seq
To validate that WISP algorithm brings a reliable measure of intratumor CMS heterogeneity in colon cancer cell lines, we selected two CMS heterogeneous cell lines (Lovo and Mdst8) purchased specifically for this manipulation from ATCC in April 2019 and ECACC in March 2019, respectively. The cell lines were used at P3 and were tested for Mycoplasma by PCR reaction. We analyzed their transcriptome by single-cell RNA-seq. Mixing these two cell lines in three series of relative proportions [Lovo 25%: Mdst8 75% (L25M75); Lovo 50%: Mdst8 50% (L50M50); Lovo 75%: Mdst8 25% (L75M25)], we performed three single-cell RNA-seq analyses using the 10× Genomics standard protocol. The transcriptome of 14,545 cells was obtained (L25M75: 6,921 cells, L50M50: 5,346 cells, L75M25: 2,278 cells). Of these cells, 9,437 (65%) passed the quality check filters: (i) at least 1,000 genes with at least one count each, and (ii) less than 20% of counts related to mitochondrial genes. To normalize the read counts within each cell, we divided them by the total number of reads and multiplied them by 100,000; then we log2 transformed the obtained values. To recover cells, respectively, corresponding to Lovo and Mdst8 cell lines within each mixture, we first selected Lovo-specific and Mdst8-specific markers comparing the bulk transcriptome of these two cell lines (GSE36133) and, retaining markers with a log2 fold change above 7, then we used these two sets of markers to calculate their mean expression within each cell: cells showing both a mean expression of Lovo (respectively Mdst8) markers above 0.5 and a mean expression of Mdts8 (respectively Lovo) markers below 0.5 were classified as Lovo (respectively Mdst8) cells. To predict CMS label of each cell, we used the single sample predictor (SSP) from Guinney and colleagues (5), using only the centroids from the (RNA-seq) The Cancer Genome Atlas series. Of note, 77% of the cells predicted as corresponding to CMS2 did not pass the quality check filters, as compared with 32% for the cells corresponding to the three other CMSs.
Transcriptomic metagenes
To characterize tumor samples in terms of immune and stromal infiltrates, immune functions (e.g., immune checkpoints activity or cytotoxicity), oncogenic pathways [epithelial–mesenchymal transition (EMT), TGFβ, angiogenesis] or an Immunoscore surrogate, we used metagenes scores derived from the MCP-counter method (19) or previously reported gene lists (5, 20). Metagene scores were calculated as the intergene mean of intersample median-centered expression values.
Statistical tests and survival analyses
Analyses were performed using R software (v3.6.1). Association between CMS and qualitative variables were assessed using χ2 tests. Association between CMS and quantitative variables were assessed with ANOVA. The survival R package was used to perform log-rank tests (for difference between Kaplan–Meier survival curves) and Cox univariate and multivariate models. The significant variables in the univariate analyses (P < 0.05) were kept in multivariate analyses. In multivariate analyses, we used recoded variables, based on grouping of modalities. Because major endpoint of PETACC8 was DFS, this grouping was based on DFS univariate analyses and was kept for overall survival (OS) analyses. Because it was an exploratory and descriptive study, no prespecified hypothesis was made.
Results
NanoString RF classifier of the CMS
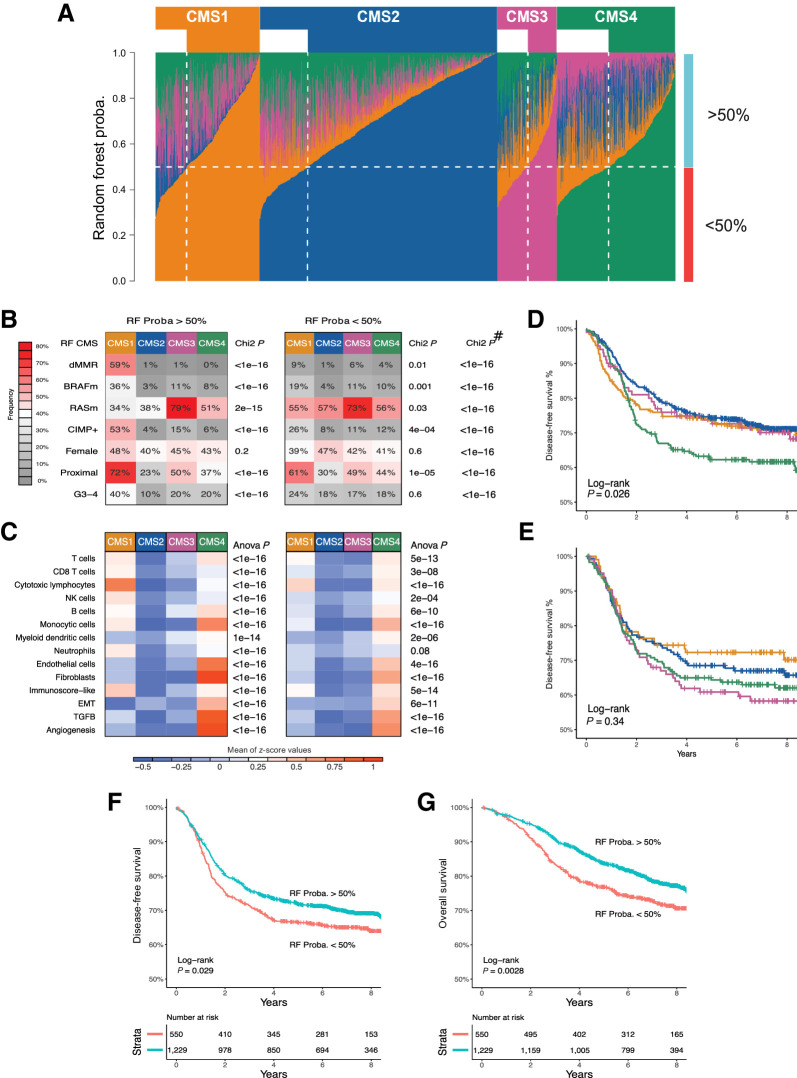
We developed a RF classifier of the four CMS (5) using NanoString measurements of 196 genes (extended data Supplementary Table S1), and classified 1,779 tumors from the PETACC8 trial (18). The CMS classification applied to this new patient series revealed the expected clinical and biological characteristics (extended data Supplementary Fig. S2). Among these 1,779 samples, 1,229 (69%) were attributed a CMS with a RF probability greater than 50%, the 550 (31%) remaining samples showing a RF probability below 50%. Thus, in one hand, roughly one-third of the tumors demonstrated mixed traits rather than the transcriptomic characteristics of one particular CMS (Fig. 1A). On the other hand, less than 15.5% of the tumors showed a probability superior to 90% to belong to a unique attributed CMS. RF probability (>50% vs. <50%) had little impact on the clinicobiological characteristics of colon cancer, although the CMS features were much more pronounced for samples with a RF probability above 50% than below 50% (Fig. 1B and C). Moreover, the RF probability manifestly impacted on the correlation between CMS classification and prognosis. Within tumors with an RF probability greater than 50%, CMS4 had a poorer prognosis than the other three CMS categories (Fig. 1D), as expected (5), but this correlation was lost for tumors with an RF probability below 50% (Fig. 1E). Importantly, for the entire cohort (irrespective of the CMS category), a 10% decrease of RF probability for CMS assignment was linearly related to poorer prognosis [DFS HR = 1.08, 95% confidence interval (CI) = (1.035–1.13), P = 0.0005; OS HR = 1.10, 95% CI = (1.05–1.16), P = 0.0002]. Thus, the prognosis of patients classified in a CMS with an RF probability below 50%, was significantly poorer from those with an RF probability above 50%, both for DFS and OS [DFS HR = 1.21, 95% CI = (1.02–1.44), P < 0.03; OS HR = 1.35, 95% CI = (1.11–1.65), P < 0.003; Fig. 1F and G]. Altogether, these results suggest that the CMS-based classification is not clear-cut and that tumors that do not unambiguously fit into the categories, potentially due to ITH, have a dismal prognosis.
Figure 1.
RF CMS classification, characterization, and survival impact based on related probability. A, Fraction of votes (probability) of the RF classification for each of the four CMSs within each of the 1,779 samples (columns), summing up to one for each sample. Samples are grouped according to RF-predicted CMS. The horizontal dotted white line indicates an RF probability of 50%. The vertical dotted white line delimits, within each CMS, samples with low (<50%) versus high (>50%) probability of CMS assignment according to RF classifier. B, The proportions of samples with dMMR, BRAF mutated (BRAFm), KRAS/NRAS mutated (RASm), CIMP+, female gender, proximal location, and G3/G4 grade are shown according to CMS with either high (left) or low RF probability (right). # indicates the P values of the χ2 tests comparing the distribution of values of each variable between RF > 50% and RF < 50% samples. C, Heatmaps showing the mean scores for eight immune and two stromal cell populations infiltration (based on MCP-counter estimates), a surrogate of the Immunoscore (Immunoscore-like), and three pathways are shown according to CMS with either high (left) or low RF probability (right). The P values of the t test comparing the distribution of values of each variable between RF > 50% and RF < 50% samples are displayed in the extended data Supplementary Table S3. DFS curves according to RF-predicted CMS in PETACC8 samples, based on RF probability of class assignment above 50% (D) or below 50% (E). Kaplan–Meier curves of DFS (F) and OS (G) are shown according to low (<50%) versus high (>50%) RF probability (whatever the predicted CMS). CMS color codes: CMS1 (orange), CMS2 (blue), CMS3 (purple), CMS4 (green).
Assessment of CMS-based intratumor heterogeneity through deconvolution
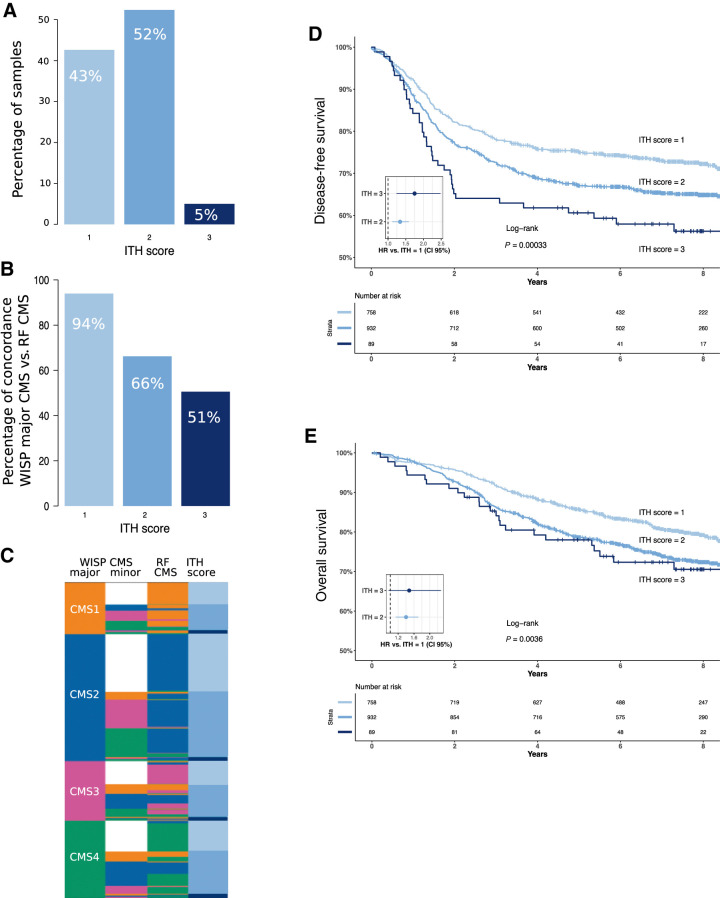
To further assess ITH, we deconvoluted the 1,779 NanoString colon cancer expression profiles, using the WISP supervised deconvolution algorithm (16). We thus decomposed each colon cancer profile as a weighted linear combination of the four CMS profiles. We ended up with a vector of the relative proportions (weights) of the four CMSs within each sample, these proportions summing up to 100%. We then defined an ITH score by recording for each sample the number of CMS reaching weights above 20%. This ITH score ranged from 1 (low ITH) to 3 (high ITH). On the basis of this criterion, only 43% of the PETACC8 tumors exhibited a low ITH score (Fig 2A). The remaining tumors exhibited an ITH score of two (52%) or three (5%). Samples with a low ITH score showed a high level of agreement (94%) between the RF-based and WISP-based (top weight) CMS label, contrary to samples with a high ITH score (51% of agreement; Fig. 2B and C). Of note, intermediate and high ITH scores were found to be associated to worse prognosis (Fig. 2D and E) [DFS HR ITH2 vs. ITH1 = 1.34, 95% CI (1.12–1.59), P = 0.001; DFS HR ITH3 vs. ITH1 = 1.75, 95% CI (1.24–2.47), P < 0.002; OS ITH2 vs. ITH1 HR = 1.40, 95% CI (1.14–1.71), P < 0.002; OS ITH3 vs. ITH1 HR = 1.48, 95% CI (0.96–2.28), P < 0.08]. These results confirm the idea that ITH may compromise the prognosis of patients with colon cancer.
Figure 2.
ITH distribution and impact on survival. A, Barplots showing the proportions of low (i), intermediate (ii), and high (iii) ITH scores in the 1,779 PETACC8 samples. B, Barplots showing the proportion of identical CMS assignment between WISP top weighted CMS (WISP major CMS) and RF-based CMS, according to ITH score. C, Sample level data (lines) representing WISP major CMS and second top weighted CMS (WISP minor CMS), RF CMS and ITH score in the 1,779 PETACC8 samples. Kaplan–Meier curves of DFS (D) and OS (E) are shown according to ITH scores. The P value of the overall log-rank test and the HRs of the univariate Cox model are shown. Color code for ITH score: low (i), light blue; intermediate (ii), blue; high (iii), dark blue.
Validation of ITH measures in cell lines through single-cell analyses
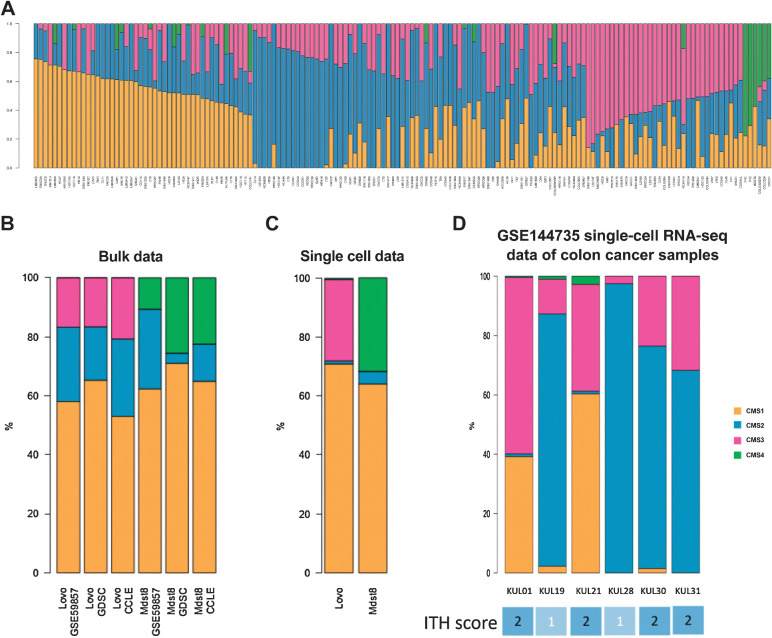
The aforementioned analyses of bulk transcriptomics do not resolve the fundamental doubt as to whether each tumor represents a mixture of CMS types, because each tumor cell, one by one, could represent a mixed CMS type, or whether colon cancers would be composed of cells falling into distinct CMS categories. To solve this problem, we first analyzed the bulk transcriptome of 155 distinct colon cancer cell lines (GSE59857; ref. 22). We discovered that very few of them represented a “pure” CMS type, meaning that the overwhelming majority of them (73%) could not be classified in one single CMS category and thus have an ITH score >1 (Fig. 3A; extended data Supplementary Fig. S3A and B). Again, this result did not completely solve the issue, because each cell line might contain an ensemble of cells that are very similar among each other, each reflecting a mixed CMS phenotype, or—alternatively—a heterogeneous mélange of cells corresponding to various CMS types. For this reason, we selected two CMS-heterogeneous colon cancer cell lines (Lovo, Mdst8), performed their single-cell RNA-seq analysis (extended data Supplementary Fig. S4), and classified each cell into CMS1 to 4 using the CMS SSP (5). For each cell line, the proportions of cells attributed to the different CMS after single-cell transcriptomics was similar to the WISP weights deduced from their bulk transcriptomes (Fig. 3B and C). It must be noted that the SSP CMS classifier (from Guinney and colleagues) used here for cell-level CMS calling was not designed for single-cell data. However, it is remarkable that the deconvolution of bulk Affymetrix profiles using the WISP algorithm brings estimated intrasample proportions of the CMS that are consistent with the cell-level CMS labeling obtained with the SSP classifier on paired single-cell data. This observation confirms the validity of the WISP-based measure of ITH on bulk transcriptomic analyses insofar as it reveals a true intercellular heterogeneity. In addition, reanalysis of a recent colon cancer single-cell RNA-seq series (GSE144735) of freshly obtained colon cancer samples revealed that 4 of 6 tumors had a mixed ITH score, further supporting that ITH is common in colon cancer tumors (Fig. 3D). In conclusion, it appears that the ITH score obtained by bulk transcriptomic analyses that averages mRNA expression across all cells in the tissue reflects an intercellular colon cancer heterogeneity that can be confirmed by single-cell transcriptomics.
Figure 3.
CMS intrasample heterogeneity in colon cancer cell lines. A, CMS ITH was determined in 155 colon cancer cell lines (GSE59857 dataset). Intrasamples WISP weights of CMS1..4 are shown for each cell line (column). Comparison of intrasample CMS1..4 proportions in the Lovo and Mdst8 cell lines, based either on bulk data (B) from three datasets (GSE59857, GDSC, CCLE) or on single-cell data (C). For bulk data, CMS1..4 intrasample proportions are estimated using WISP. For single-cell data, CMS1..4 intrasample proportions are estimated using the SSP from Guinney and colleagues (5), applied to each cell. D, Single-cell RNA-seq data of six colon cancer samples from the GSE144735 dataset. CMS label per cell comes from the online annotation file of this dataset. The intrasample distribution of these CMS labels is shown. ITH scores based on this distribution are shown (bottom).
Clinicobiological characteristics of CMS combinations
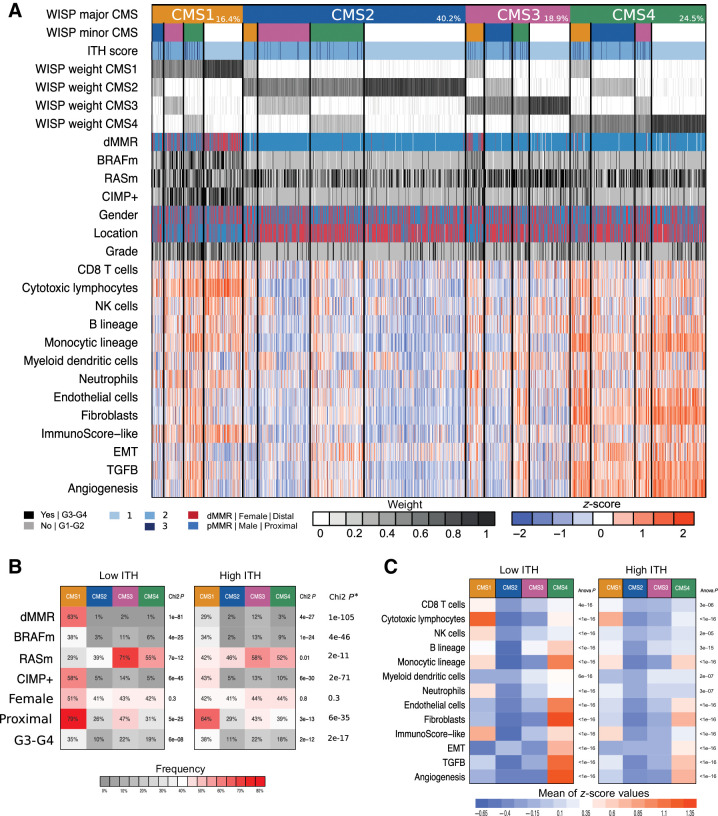
To refine the characteristics of ITH according to intra-sample CMS combinations, we defined 16 different tumor subgroups, including 4 “pure” and 12 “mixed” subcategories (Fig. 4; extended data Supplementary Fig. S5). The four pure subgroups showed the expected (5) molecular features of CMS 1 to 4. However, for the mixed subgroups referring to a pair of CMSs (major and minor) jointly identified within a tumor, the corresponding molecular features are intermediate of those observed in the corresponding pure CMS, as awaited. For example, the CMS3(major)-CMS1(minor) subgroup shows intermediate states of the respective molecular features of pure CMS3 (KRAS mutation) and CMS1 [deficient mismatch repair (dMMR), CpG island methylator phenotype (CIMP+), BRAF mutation] subgroups. Similar results are observed for either immune contexture, or TGFβ, EMT, and angiogenic pathways (Fig. 4; extended data Supplementary Fig. S5). In particular, high expression of genes indicating the presence of fibroblasts and endothelial cells, or related to the TGFβ, EMT, and active angiogenesis pathways, which are expected to be features of CMS4 tumors (5), is observed in all mixed subgroups with a CMS4 component, even if this component is a minor one. High infiltration by cytotoxic lymphocytes, a prominent feature of CMS1 tumors (24), is also clearly more frequent in all mixed subgroups with a CMS1 component.
Figure 4.
A, CMS intrasample heterogeneity, based on WISP deconvolution algorithm, and related characteristics. CMS intrasample heterogeneity, based on WISP deconvolution algorithm, on 1,779 samples from PETACC8 trial. Samples (columns) are grouped according to WISP major CMS (WISP top weighted CMS), and WISP minor CMS (WISP second highest weighted CMS, if weighted above 20%). The CMS color code is as follows: CMS1 (orange), CMS2 (blue), CMS3 (purple), CMS4 (green). “Pure” samples, related to a unique CMS (i.e., with no WISP minor CMS), are shown in white (line 2). The proportions of samples in these categories are also reported (line 1). Further shown are ITH score; intrasample WISP weights for each CMS [from 0 (white) to 1 (black)]; molecular and clinical features, including dMMR, BRAFm, RASm, CIMP+, gender, tumor location, grade, immune and stromal cell populations infiltration (based on MCP-counter), a surrogate of the Immunoscore (Immunoscore-like); and scores related to EMT, TGFβ, and angiogenesis pathways. MCP-counter and pathway scores are trimmed z-scores [low (−2): blue, intermediate (0): white, high (2): red]. Color codes: Black = yes or grade 3/4; gray = no or grade 1/2; blue = pMRR or male or proximal; red = dMMR or female or distal. B, The proportions of samples with dMMR, BRAFm, RASm, CIMP+, female gender, proximal location, and G3/G4 grade are shown according to CMS with either low ITH (ITH score = 1, left) or high ITH (ITH score >1, right). * indicates the P values of the χ2 tests comparing the distribution of values of each variable between low ITH and high ITH samples. C, Heatmaps showing the mean scores for eight immune and two stromal cell populations infiltration (based on MCP-counter estimates), a surrogate of the Immunoscore (Immunoscore-like), and three pathways are shown according to CMS with either low ITH (ITH score = 1, left) or high ITH (ITH score >1, right). The P values of the t test comparing the distribution of values of each variable between low ITH and high ITH samples are displayed in the extended data Supplementary Table S4.
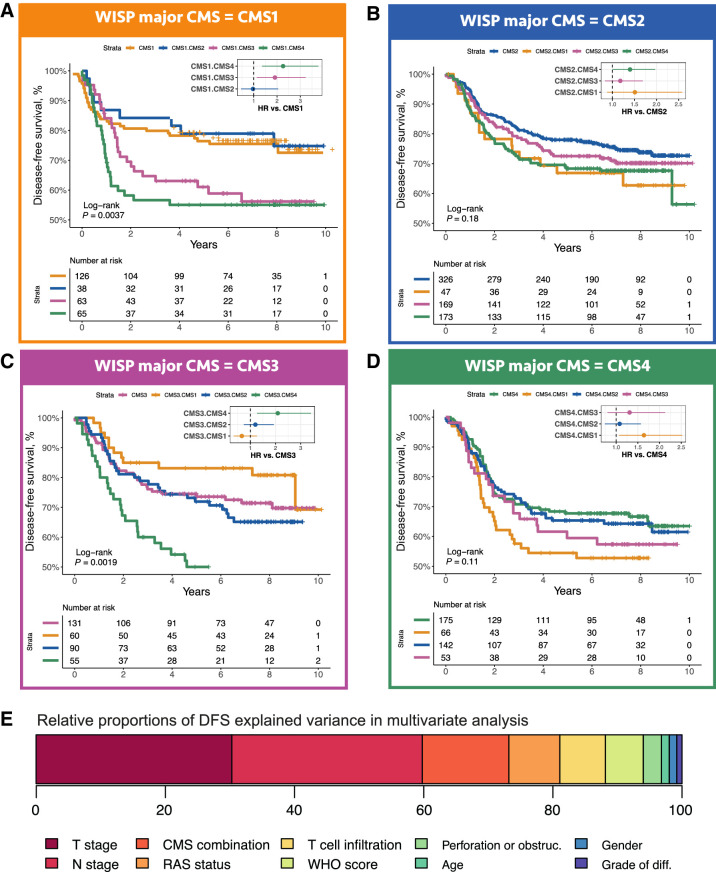
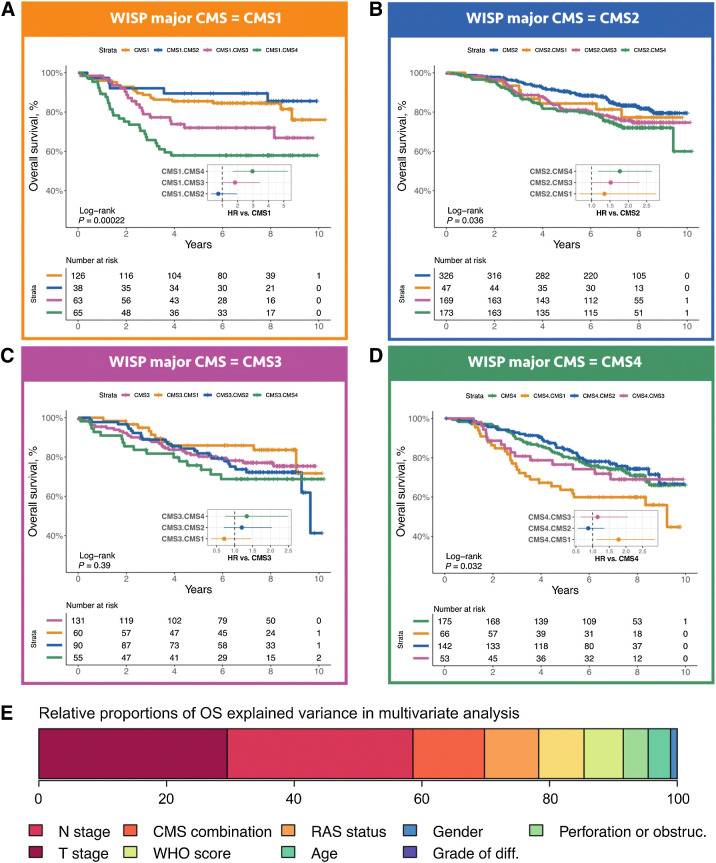
The ITH score being related to prognosis, we then asked whether we could refine this global observation by analyzing survival, considering DFS and OS, according to the 16 subgroups mentioned above (Figs. 5 and 6). Mixed subgroups with a minor CMS2 component never showed a prognosis significantly worse than related pure subgroups. Other mixed subgroups, notably all those with a minor CMS4 component, mostly showed worsen prognosis as compared to the related pure subgroup. Strikingly, the worse prognosis of mixed subgroups as compared to the related pure subgroup is even observed for CMS4-CMS1, that is, when combining a minor (good prognosis) CMS1 component with a major (poor prognosis) CMS4 component.
Figure 5.
DFS according to intrasample CMS combinations. The DFS of 1,779 samples from PETACC8 trial are shown according to WISP top and second top weighted CMS. Kaplan–Meier curves for DFS from the samples classified as CMS1 (respectively, CMS2, CMS3, CMS4) based on WISP top weight, and segregated according to WISP second top weighted CMS (if above 20%) are shown in A (respectively, B, C, and D). E, Relative proportions of DFS explained variance, for all prognostic factors included in the DFS Cox multivariate model, based on the χ2 proportion test. The variable “CMS combination” is derived from the 16 possible combinations of the two top weighted CMS according to WISP deconvolution, and is equal to 1 for the following combinations: CMS1.CMS3, CMS1.CMS4, CMS3.CMS4, CMS4.CMS1, 0 otherwise.
Figure 6.
OS according to intrasample CMS combinations. The OS of 1,779 samples from PETACC8 trial are shown according to WISP top and second top weighted CMS. Kaplan–Meier curves for OS from the samples classified as CMS1 (respectively CMS2, CMS3, CMS4) based on WISP top weight, and segregated according to WISP second top weighted CMS (if above 20%) are shown in A (respectively B, C, D). The P value of the overall log-rank test, and the HRs of the univariate Cox model, are shown within each panel. E, Relative proportions of OS explained variance, for all prognostic factors included in the OS Cox multivariate model, based on the χ2 proportion test. The variable “CMS combination” is derived from the 16 possible combinations of the two top weighted CMS according to WISP deconvolution, and is equal to 1 for the following combinations: CMS1.CMS3, CMS1.CMS4, CMS3.CMS4, CMS4.CMS1, 0 otherwise.
Finally, we assessed the potential added prognostic value of the CMS-based ITH, through multivariable DFS and OS Cox modelling. We first built a DFS multivariable reference model, treatment-arm-stratified, including all prognostic factors shown to be significant in univariate analysis, that is, age, gender, obstruction, and/or perforation, T and N stages, tumor differentiation grade, World Health Organization performance status, KRAS/NRAS mutation, and an mRNA-based surrogate of the Immunoscore (extended data Supplementary Tables S2 and S3). Then, using a DFS Cox univariate model, we selected the CMS combinations related to the poorest survival (CMS1-CMS4, CMS4-CMS1, CMS3-CMS4 and CMS1-CMS3, P ≤ 0.002; extended data Supplementary Table S2) to define a variable set to “high risk” for these subgroups, and to “low risk” for the remaining subgroups. Adding this variable to the DFS reference multivariable model yielded a significantly improved model [DFS HR = 1.74, 95% CI (1.38–2.19), likelihood ratio test P < 0.0001; Table 1]. This new variable explained 13.4% of the variance of the DFS model (Fig. 5E). Similar results were obtained for OS (Fig. 6E; extended data Supplementary Table S2E and S2F).
Table 1.
Multivariate analysis of DFS including clinical, pathologic, mutational, immunologic variables, and CMS combination (model 1).
| Term | HR | 95% CI | P |
|---|---|---|---|
| Age: | |||
| >70 y vs. ≤70 y | 1.22 | 0.93–1.59 | 0.15 |
| Gender: | |||
| Male vs. female | 1.14 | 0.95–1.37 | 0.17 |
| WHO score: | |||
| 1–2 vs. 0 | 1.40 | 1.13–1.73 | 0.002 |
| Bowel obstruction or perforation: | |||
| Yes vs. no | 1.26 | 1.02–1.55 | 0.035 |
| T stage: | |||
| T4 vs. T1–T3 | 2.05 | 1.69–2.49 | <0.0001 |
| N stage: | |||
| N2 vs. N1 | 1.92 | 1.6–2.3 | <0.0001 |
| Grade: | |||
| G1–G2 vs. G3–G4 | 1.14 | 0.91–1.41 | 0.25 |
| RAS statusa | |||
| RAS mutated vs. RAS wildtype | 1.40 | 1.17–1.68 | 0.0003 |
| T-cell tumor infiltration: | |||
| Low and intermediate vs. high infiltration | 1.42 | 1.16–1.74 | 0.0006 |
| CMS combination: | |||
| High-risk vs. low-risk CMS combination | 1.74 | 1.38–2.19 | <0.0001 |
Note: R2 = 0.118; likelihood ratio = 194.7; degree of freedom = 10; Harrell C-index = 0.685. Model 1 versus model 0 likelihood ratio test P < 0.0001 (χ2 test); C-indexes are statistically significantly different (P < 0.03).
Abbreviations: WHO, World Health Organization; y, years.
aKRAS or NRAS.
Discussion
The development of NanoString-based signatures allows the determination of CMS based on RNA extracted from paraffin-embedded tissues (25, 26). Here, this strategy allowed us to characterize the transcriptomic ITH of a large series of colon cancers. Several forms of ITH are reported in the literature: phenotypic, histopathologic, genetic, epigenetic, and transcriptomic (27). ITH is mostly studied at the genetic level, referring then to subclonality, and was in particular documented through multi-regions intratumor DNA sequencing (11, 28). In colon cancer, it was shown that subclonality is related to higher risk of metastasis (11). Beyond genetic forms of ITH, Meir and colleagues (15) recently showed, through single-cell analyses, that a variety of stable epigenetic and transcriptomic states can develop in clonal colon cancer cell populations. Transcriptomic ITH was also detected in colon cancer tumor samples, by means of single-cell (14) or multi-region bulk transcriptomic analyses (13), in small series (n < 30 patients). While these three studies highlighted the existence of transcriptomic ITH in colon cancer, the limited number of patient samples precluded conclusions regarding its prognostic impact.
We recently explored transcriptomic ITH through supervised deconvolution of bulk transcriptome profiles from 450 mesotheliomas, with the WISP software, and described its major impact on prognosis (16). Here, we used the WISP method to analyze transcriptomic ITH in 1,779 samples from the stage III colon cancer PETACC8 trial. Based on the strength of the international consensus classification of colon cancer initially proposed by Guinney and colleagues (5), we explored the hypothesis that the four corresponding consensus molecular subtypes (CMS1..4) might actually represent distinct colon cancer cell populations, and, using WISP, deconvoluted each of the 1,779 transcriptome profiles as linear combinations of the four CMS centroids.
As WISP supervised deconvolution needs classes on which to be trained, we used a RF CMS classifier to get CMS labels for each tumor sample. We observed that one-third of these stage III colon cancer samples were attributed to a CMS with a RF probability less than 50%, supporting the existence of ITH in these tumors. The previously reported clinical and molecular characteristics of the four CMS (5) were much more contrasted across CMS, and statistically significant, in the samples with a RF probability of CMS assignment above 50% (RF.High), than in those with a RF probability below 50% (RF.Low). For example, considering RF.High samples, 59% of CMS1 samples were related to dMMR versus 0% to 1% in any other CMS; while for RF.low samples, the range of dMMR across the four CMSs ranged between 1% and 9%. Regarding the known prognostic differences across the four CMSs, they were clearly observed for RF.High samples but not for RF.Low samples. We further found that the RF probability of CMS assignment, as a continuous variable, was by itself related to prognosis. Altogether these data support the existence of ITH in colon cancer PETACC8 samples and brings up its possible impact on prognosis.
Class prediction approaches such as RF essentially suppose that each sample belongs to a unique class; in addition, some of these approaches, in particular RF, assess the certainty of class assignment through a probabilistic approach. A supervised deconvolution approach, such as WISP, is totally different, as WISP intrinsically considers that each sample is composed of a mixture of classes and estimates the relative proportions of these classes in the sample (29). As a consequence, supervised deconvolution algorithms are perfectly fit to measure ITH from bulk transcriptome data, contrary to class prediction approaches.
Here we choose to use the WISP-supervised deconvolution algorithm, given its unique ability during the training phase to automatically remove poorly informative samples from the training set (https://github.com/cit-bioinfo/WISP; ref. 16). We trained WISP on the CMS labels obtained from the RF classifier, thus obtaining for each of the 1,779 PETACC8 samples the intrasample relative proportions of the four CMS. We then defined a discrete score of ITH, ranging from 1 (low ITH) to 3 (high ITH), representing the number of CMS within a sample reaching a WISP weight above 20%. We showed that this ITH score was related to dismal prognosis. Half of the tumors (52%) showed an intermediate ITH score, and 5% showed a high ITH score. Intermediate or high ITH was found to be even more prevalent in colon cancer cell lines (>70%), suggesting that the cooperation of different cancer cell types may favor their survival in culture (30). Indeed, single-cell transcriptomic analysis revealed comparable degrees of ITH for colon cancer cell lines that had been cultured for months in vitro as for freshly obtained colon cancer tumors ex vivo, confirming the conjecture that colon cancer ITH is characterized by the simultaneous presence of cancer cells falling into distinct CMS categories. This observation fully fits in with the recent description of intracellular transcriptomic heterogeneity across multiple cancer cell lines (31) or in breast cancer cell lines (32).
To refine and further explore ITH in colon cancer tumors, we defined 16 subgroups (4 pure, 12 mixed) according to the two main intrasample CMS components if reaching WISP weights above 20%. Pure subgroups showed the more contrasted molecular characteristics, while mixed subgroups showed intermediate characteristics of their CMS components, as expected. For example, dMMR was observed in 63% of CMS1 pure samples, 1% to 2% in the other pure CMS, in 0% to 4% of the mixed subgroups without a CMS1 component, and in 9% to 39% in the mixed subgroups with a CMS1 component. Similar results were obtained for BRAF mutations, CIMP status, or microenvironment contexture. While CMS2 and CMS3 are usually considered as immune cold tumors (24), this does not apply to mixed subgroups that incorporate either a CMS1 or CMS4 minor component. This observation could have some relevance regarding the development of immune checkpoint immunotherapy in Microsatellite stable tumors (33, 34).
We then explored the prognosis of these 16 subgroups. While having a combination of two or more CMS components in a tumor is globally related to shorter survival, we observed contrasted situations depending on the CMS combinations. On one hand, some CMS combinations were not associated with any prognostic aggravation and showed similar prognosis to that of the pure related CMS. This applies in particular to combinations including a minor CMS2 component (e.g., CMS1.CMS2 shows a prognosis similar to that of pure CMS1). On the other hand, most of the other CMS combinations were associated to much poorer survival than the related pure CMS, suggesting that CMS combinations do affect colon cancer prognosis. This is notably the case for CMS4.CMS1 tumors, which exhibit a poorer prognosis than both pure CMS1 and pure CMS4 tumors. This latter observation is intriguing, because one would not expect that adding a minor (good prognosis) CMS1 component to a major (poor prognosis) CMS4 component would aggravate the prognosis of the later. However, CMS1 tumors are known to show the shortest survival after recurrence, implying that immune control escape of CMS1 tumors leads to strong aggressiveness (5). In the specific case of mixed CMS4.CMS1 tumors, it is tempting to speculate that the CMS4 component releases TGFβ, favoring immune escape of the highly genetically instable CMS1 component (35). This kind of (hypothetical) cooperation among colon cancer cells belonging to different CMS categories might explain the new biological properties acquired by these mixed tumors.
Finally, we assessed the possible value for stage III colon cancer prognostication of ITH, in the frame of a multivariable Cox model including all the established prognostic factors. Having pooled the CMS combinations related to shorter survival, the corresponding variable contributed to 13.4% of the explained DFS variance in the multivariable Cox model. Of note, this model already incorporates a transcriptomic surrogate of the Immunoscore, previously shown to be relevant for colon cancer prognostication (20, 36).
One limitation of our study is the restriction to stage III colon cancer. To address this issue, we leveraged public databases to investigate the ITH score, the enrichment in CMS4 and the enrichment of the CMS1.CMS4 and CMS4.CMS1 combinations in synchronous and metachronous stage IV disease. We found similar distributions of ITH scores in metastatic and non-metastatic disease and an enrichment in CMS4 (53% stage IV vs. 20% for other CMS) in metastatic disease. In addition, the CMS1.CMS4 or CMS4.CMS1 combinations are overrepresented in metastatic disease (39%) as compared with the other CMS combinations (27%; Extended data Supplementary Table S5).
In conclusion, our analysis pipeline reveals that transcriptomic ITH, defined in terms of CMS, is highly prevalent in stage III colon cancer and actually affects its clinical evolution. Few studies analyzed retrospectively the predictive value of CMS for various treatments (37). Our observations support the idea that such studies could be revisited and refined according to CMS ITH status.
Authors' Disclosures
C. Lepage reports personal fees from Bayer, Amgen, Ipsen, Pierre Fabre, and Novartis outside the submitted work. V. Boige reports grants, personal fees, and non-financial support from Merck Serono; personal fees and non-financial support from Bayer, Roche, Ipsen, Merck MSD, and Amgen; non-financial support from Sanofi; and personal fees from BMS, Eisai, and Novartis outside the submitted work. J.-F. Emile reports personal fees from Pierre Fabre, Merck Sharp & Dohme, Merck Serono, Amgen, Novartis, and Bristol Myers Squibb outside the submitted work. P. Laurent-Puig reports grants from Federeation francophones de cancerologie disgestive and Ligue National de luttte contre le cancer during the conduct of the study, as well as personal fees from Amgen, Boehringer Ingelheim, Sanofi, MSD, Imedex, Pierre Fabre, and Institut Servier; grants and personal fees from Biocartis, BMS, and Roche; and grants from Institut Roche, Bio-Rad, and Servier outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
We thank Guido Kroemer for fruitful discussion.
This study was supported by the program “Cartes d'Identité des Tumeurs” (CIT) from the Ligue Nationale Contre le Cancer (LNCC), by the Institut National du Cancer (INCa) with the program Heterogeneity of tumors and ecosystem (HTE program) within the sub program “deciphering the heterogeneous genome-microenvironment interplay in colon and hepatocellular carcinomas (HETCOLI)”, by the ITMO CANCER through the program single cell COCAHEMISCLE, by the integrated site of cancer research “cancer research for personalized medicine” (SIRIC CARPEM, INCa-DGOS-Inserm_12561) and the Fédération Francophone de Cancérologie Digestive (FFCD). We thank the biological resource center EPIGENETEC (BB-0033-00055) for their support.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
#Y. Blum, J. Taieb, A. de Reyniès, and P. Laurent-Puig contributed equally to this article.
Clin Cancer Res 2021;27:4768–80
Authors' Contributions
L. Marisa: Data curation, software, formal analysis, validation, visualization, methodology, writing–original draft. Y. Blum: Data curation, software, formal analysis, validation, methodology. J. Taieb: Conceptualization, formal analysis, funding acquisition, validation, investigation, visualization, methodology, writing–original draft. M. Ayadi: Resources, data curation, supervision, visualization, writing–review and editing. C. Pilati: Data curation, software, formal analysis, validation, visualization, methodology, writing–original draft. K. Le Malicot: Conceptualization, data curation, formal analysis, validation, methodology, writing–review and editing. C. Lepage: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology. R. Salazar: Data curation, supervision, investigation, visualization, writing–review and editing. D. Aust: Data curation, formal analysis, supervision, investigation, visualization. A. Duval: Resources, data curation, formal analysis, supervision, investigation, visualization. H. Blons: Data curation, software, formal analysis, supervision, validation, investigation, methodology, writing–original draft. V. Taly: Conceptualization, data curation, formal analysis, supervision, validation, methodology. D. Gentien: Investigation, methodology. A. Rapinat: Investigation, visualization, methodology. J. Selves: Data curation, formal analysis, validation, investigation, visualization, writing–review and editing. S. Mouillet-Richard: Conceptualization, supervision, validation, investigation, methodology, writing–original draft. V. Boige: Conceptualization, data curation, validation, methodology, writing–review and editing. J.-F. Emile: Resources, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. A. de Reyniès: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft. P. Laurent-Puig: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018;391:1023–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol 2016;27:1386–422. [DOI] [PubMed] [Google Scholar]
- 3. Taieb J, Le Malicot K, Shi Q, Penault-Llorca F, Bouché O, Tabernero J, et al. Prognostic value of BRAF and KRAS mutations in MSI and mss stage III colon cancer. J Natl Cancer Inst 2017;109:djw272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smeby J, Sveen A, Merok MA, Danielsen SA, Eilertsen IA, Guren MG, et al. CMS-dependent prognostic impact of KRAS and BRAFV600E mutations in primary colorectal cancer. Ann Oncol 2018;29:1227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marusyk A, Janiszewska M, Polyak K. Intratumor Heterogeneity: the rosetta stone of therapy resistance. Cancer Cell 2020;37:471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613–28. [DOI] [PubMed] [Google Scholar]
- 8. Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, et al. The evolutionary history of 2,658 cancers. Nature 2020;578:122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2016;22:105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morris LGT, Riaz N, Desrichard A, Şenbabaoğlu Y, Hakimi AA, Makarov V, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 2016;7:10051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joung J-G, Oh BY, Hong HK, Al-Khalidi H, Al-Alem F, Lee H-O, et al. Tumor heterogeneity predicts metastatic potential in colorectal cancer. Clin Cancer Res 2017;23:7209–16. [DOI] [PubMed] [Google Scholar]
- 12. Oh BY, Shin H-T, Yun JW, Kim K-T, Kim J, Bae JS, et al. Intratumor heterogeneity inferred from targeted deep sequencing as a prognostic indicator. Sci Rep 2019;9:4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunne PD, McArt DG, Bradley CA, O'Reilly PG, Barrett HL, Cummins R, et al. Challenging the cancer molecular stratification dogma: intratumoral heterogeneity undermines consensus molecular subtypes and potential diagnostic value in colorectal cancer. Clin Cancer Res 2016;22:4095–104. [DOI] [PubMed] [Google Scholar]
- 14. Lee H-O, Hong Y, Etlioglu HE, Cho YB, Pomella V, Van den Bosch B, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet 2020;52:594–603. [DOI] [PubMed] [Google Scholar]
- 15. Meir Z, Mukamel Z, Chomsky E, Lifshitz A, Tanay A. Single-cell analysis of clonal maintenance of transcriptional and epigenetic states in cancer cells. Nat Genet 2020;52:709–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blum Y, Meiller C, Quetel L, Elarouci N, Ayadi M, Tashtanbaeva D, et al. Dissecting heterogeneity in malignant pleural mesothelioma through histo-molecular gradients for clinical applications. Nat Commun 2019;10:1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013;10:e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taieb J, Tabernero J, Mini E, Subtil F, Folprecht G, Van Laethem J-L, et al. Oxaliplatin, fluorouracil, and leucovorin with or without cetuximab in patients with resected stage III colon cancer (PETACC-8): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:862–73. [DOI] [PubMed] [Google Scholar]
- 19. Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marisa L, Svrcek M, Collura A, Becht E, Cervera P, Wanherdrick K, et al. The balance between cytotoxic T-cell lymphocytes and immune checkpoint expression in the prognosis of colon tumors. J Natl Cancer Inst 2018;110. [DOI] [PubMed] [Google Scholar]
- 21. Franks JM, Cai G, Whitfield ML. Feature specific quantile normalization enables cross-platform classification of molecular subtypes using gene expression data. Bioinformatics 2018;34:1868–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun 2015;6:7002. [DOI] [PubMed] [Google Scholar]
- 23. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Becht E, de Reyniès A, Giraldo NA, Pilati C, Buttard B, Lacroix L, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res 2016;22:4057–66. [DOI] [PubMed] [Google Scholar]
- 25. Piskol R, Huw L, Sergin I, Kljin C, Modrusan Z, Kim D, et al. A clinically applicable gene-expression classifier reveals intrinsic and extrinsic contributions to consensus molecular subtypes in primary and metastatic colon cancer. Clin Cancer Res 2019;25:4431–42. [DOI] [PubMed] [Google Scholar]
- 26. Morris JS, Luthra R, Liu Y, Duose DY, Lee W, Reddy NG, et al. Development and validation of a gene signature classifier for consensus molecular subtyping of colorectal carcinoma in a CLIA-certified setting. Clin Cancer Res 2021;27:120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sharma A, Merritt E, Hu X, Cruz A, Jiang C, Sarkodie H, et al. Non-genetic intra-tumor heterogeneity is a major predictor of phenotypic heterogeneity and ongoing evolutionary dynamics in lung tumors. Cell Rep 2019;29:2164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gaujoux R, Seoighe C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics 2013;29:2211–2. [DOI] [PubMed] [Google Scholar]
- 30. Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 2014;508:113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kinker GS, Greenwald AC, Tal R, Orlova Z, Cuoco MS, McFarland JM, et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat Genet 2020;52:1208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen F, Ding K, Priedigkeit N, Elangovan A, Levine KM, Carleton N, et al. Single-cell transcriptomic heterogeneity in invasive ductal and lobular breast cancer cells. Cancer Res 2021;81:268–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chalabi M, Fanchi LF, Dijkstra KK, Van den Berg JG, Aalbers AG, Sikorska K, et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat Med 2020;26:566–76. [DOI] [PubMed] [Google Scholar]
- 34. Fukuoka S, Hara H, Takahashi N, Kojima T, Kawazoe A, Asayama M, et al. Regorafenib plus nivolumab in patients with advanced gastric or colorectal cancer: an open-label, dose-escalation, and dose-expansion phase Ib trial (REGONIVO, EPOC1603). J Clin Oncol 2020;38:2053–61. [DOI] [PubMed] [Google Scholar]
- 35. Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538–43. [DOI] [PubMed] [Google Scholar]
- 36. Pagès F, Mlecnik B, Marliot F, Bindea G, Ou F-S, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 2018;391:2128–39. [DOI] [PubMed] [Google Scholar]
- 37. Lenz H-J, Ou F-S, Venook AP, Hochster HS, Niedzwiecki D, Goldberg RM, et al. Impact of consensus molecular subtype on survival in patients with metastatic colorectal cancer: results from CALGB/SWOG 80405 (alliance). J Clin Oncol 2019;37:1876–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.