This work demonstrates that circBART2.2 binding to RIG-I is essential for the regulation of PD-L1 and subsequent immune escape in nasopharyngeal carcinoma.
Abstract
Epstein–Barr virus (EBV) infection is an established cause of nasopharyngeal carcinoma (NPC) and is involved in a variety of malignant phenotypes, including tumor immune escape. EBV can encode a variety of circular RNAs (circRNA), however, little is known regarding the biological functions of these circRNAs in NPC. In this study, EBV-encoded circBART2.2 was found to be highly expressed in NPC where it upregulated PD-L1 expression and inhibited T-cell function in vitro and in vivo. circBART2.2 promoted transcription of PD-L1 by binding the helicase domain of RIG-I and activating transcription factors IRF3 and NF-κB, resulting in tumor immune escape. These results elucidate the biological function of circBART2.2, explain a novel mechanism of immune escape caused by EBV infection, and provide a new immunotherapy target for treating NPC.
Significance:
This work demonstrates that circBART2.2 binding to RIG-I is essential for the regulation of PD-L1 and subsequent immune escape in nasopharyngeal carcinoma.
Introduction
The programmed cell death-1/programmed cell death-ligand 1 (PD-1/PD-L1) signaling pathway is an important mechanism mediating tumor immunosuppression (1). PD-L1 is often expressed on the surface of tumor cells and immunosuppressive cells in the tumor microenvironment and interacts with PD-1 on T cells. This immune checkpoint prevents tumor antigen–specific T cells from being effectively activated and killing tumor cells, resulting in tumor immune escape. Based on this mechanism, antibodies targeting PD-1 and PD-L1 protein have been rapidly developed for the treatment of cancer, with some showing remarkable curative effects in immune-related tumors (2). However, the accumulated data from clinical trials for solid tumors revealed that the antitumor response rate of PD-1 inhibitors is not significantly high (3,4,5,6). Therefore, it is urgent to clarify the mechanisms regulating PD-1/PD-L1 expression in cancer.
Nasopharyngeal carcinoma (NPC) is a type of malignant tumor that originates in the nasopharyngeal epithelium, with distinct ethnic and regional differences in its incidence and common malignant tumors in South China and Southeast Asia (7, 8). Genetic susceptibility and environmental factors [including Epstein–Barr virus (EBV) infection] are the leading causes of NPC (9,10,11). EBV infects more than 95% of adults worldwide (12, 13) and is associated many kinds of tumors including NPC and gastric cancer (14). EBV infection is involved in NPC proliferation, migration, invasion, and tumor immune escape, through EBV-encoded proteins or noncoding RNAs such as microRNAs (miRNA) and long noncoding RNAs (15, 16). EBV can also encode various circular RNAs (circRNA), which are noncoding RNAs with a circular structure (17). Toptan and colleagues first reported that three gene loci (BART, LMP2, and BHLF1) encode circRNAs in EBV (17). LMP2 and BHLF1 genes can encode circLMP2 and circBHLF1, whereas the BART gene has abundant variable splicing sites and can encode four different circBARTs, namely, circBART1.1 (711 nt, including exons II, IIIa, IIIb, IV, and intron IIIa), circBART1.2 (509 nt, including exons II, IIIa, IIIb, and IV), circBART2.1 (609 nt, including exons IIIa, IIIb, IV, and intron IIIa), and circBART2.2 (399 nt, including exons IIIa, IIIb, and IV). However, the functions of these circRNAs in NPC remain unclear (18, 19).
circRNAs are a class of ncRNAs characterized by a covalent closed-loop structure without 5- or 3-ends and not easily degraded by exonucleases (20). Several circRNAs are reported to dysregulate in different types of cancers and function as competitive endogenous RNAs (ceRNA), protein-binding or protein-coding RNAs to regulate the gene expression and are involved in tumorigenesis and progression (21, 22). In NPC, multiple circRNAs participate in the regulation of tumor cell biological functions through the ceRNA mechanism, including circARHGAP12 (23), circSETD3 (24), or even EBV-encoded circRPMS1 (also circBART2.2; refs. 25,26,27). However, the mechanism of circRNAs reported in NPC mainly focuses on tumor malignant biological properties. Further insight into the role of circRNAs in tumor immune escape may contribute to understanding the development of NPC.
RIG-I (retinoic acid-inducible gene I) is a cytoplasmic sensor of double-stranded RNA due to the unique structural combination of an N-terminal caspase recruitment domain (CARD) and a C-terminal DExD/H RNA helicase domain. It plays an important role in the host defense response to eliminate invading viruses. It has been reported that EBV-encoded RNA EBERs, LMP1, and EBV-encoded miR-BART6-3p are recognized by RIG-I (28,29,30) and evade immune surveillance.
In this study, we assessed the function of EBV-encoded circBART2.2 in NPC and found that RIG-I through its helicase domain recognized the nucleotides from 114–165 bp of circBART2.2, which promoted PD-L1 expression through activating canonical transcription factors including interferon regulator factor 3 and NF-κB in NPC, resulting in tumor immune escape. The role of circBART2.2–RIG-I interaction in triggering tumor immune escape through the PD-1 immune-checkpoint pathway may shed light on NPC treatment, which will provide new therapeutic targets focusing on circBART2.2-activated tumor immunotherapy.
Materials and Methods
Patient samples
A total of 72 NPC and 20 nontumor nasopharyngeal epithelial tissues from patients with chronic nasopharyngeal inflammation (Supplementary Table S1) were collected for quantitative RT-PCR. And 52 NPC and 36 noncancerous nasopharyngeal epithelial tissue samples from patients with chronic nasopharyngeal inflammation or the adjacency of NPC tissues were collected for in situ hybridization or IHC (Supplementary Table S2). All the clinical samples were approved by the Research Ethics Committee of the Second Xiangya Hospital, Central South University according to the ethical and legal standards Declaration of Helsinki. The patients were provided with the informed written consent before surgery.
Cell lines, reagents, plasmids, and cell transfection
EBV-uninfected (HONE1 and HK1) and -infected (HONE1-EBV and HK1-EBV) NPC cell lines, and EBV-positive NPC cell line C666-1 (RRID: CVCL_7949) were gifted by Professor George Sai Wah Tsao, University of Hong Kong and Professor Xin Li, Southern Medical University (31). EBV-negative Akata and EBV-positive Akata were cultured and amplified in RPMI-1640 medium in our laboratory, which was supplemented with 10% fetal bovine serum (Invitrogen), penicillin (100 U/mL, Sigma-Aldrich), and streptomycin (100 U/mL, Sigma-Aldrich). All cells were confirmed to be Mycoplasma negative before culture (TaKaRa) and were authenticated by the STR Multi-amplification Kit (Goldeneye- DNA ID system 20A, Peoplespot) every six months. The length of time between thawing and use is less than 3 months.
Prediction of the secondary stem loop structure of circBART2.2 was performed using the RNAfold database (http://rna.tbi.univie.ac.at/), and three well-characterized stem loop structures were selected for deletion mutants. Full-length circBART2.2 and deletion mutants were cloned into pcDNA3.1+ circRNA mini vectors containing tandem repeat sequences at both ends of the inserted sequence to aid circRNA looping, followed by Sanger sequencing. Prediction of the RIG-I function domain was performed using the UniProt database (https://www.uniprot.org/, Universal Protein Resource, RRID: SCR_002380). Additionally, flag-tagged full-length RIG-I and its truncations were synthesized and cloned into the pcDNA3.1 vector (Invitrogen, RRID: Addgene_79663).
The nontarget scrambled siRNA controls were provided by GenePharma. Three siRNAs specifically targeting the splice site of circBART2.2 were also synthesized and transfected into HONE1-EBV and HK1-EBV. Two siRNAs of RIG-I were used to knockdown of RIG-I. Lipofectamine RNAiMAX Reagent (Invitrogen) with OptiMEM medium (Invitrogen) was used for transfection.
For RNase R (RNR07250, Epicentre) treatment, total RNA were incubated with RNase R (20 U/μL) at 37°C for 30 minutes and at 70°C for 10 minutes to inactivation and then for quantitative real-time PCR (qRT-PCR) detection.
For actinomycin D (A4262, Sigma) treatment, cells were treated with actinomycin D (1 μg/mL) for 0, 6, 12, and 24 hours and then for qRT-PCR detection.
For treatment with anti–PD-L1 antibody, sufficient anti–PD-L1 antibody (5 mg/mL, atezolizumab, AbMole) was added and cocultured for 3 hours with activated primary T cells.
Quantitative real-time PCR and regular PCR
Total RNA was extracted using TRIzol reagent (15596026, Invitrogen) and reverse transcribed using the HiScript cDNA Synthesis kit (R323-01, Vazyme). qRT-PCR was performed using the Universal SYBR qPCR Master Mix (Q511-02, Vazyme). The 2−ΔΔCt method was used to normalize the data.
Regular PCR experiments were carried out using the Golden Star T6 Super PCR Mix (TSE101, Tsingke) according to the instructions. Primers are listed in Supplementary Table S3. To amplify four different circBART variants, isoform-specific primers that could be discriminated each other were designed (Fig. 1A). The primers for circBART2.2 were designed according to its circular splice site spanning exons IV and IIIA of the BART gene, which were specific to circBART2.2 and the linear forms could not be detected. circBART2.1 and circBART2.2 could also be discriminated.
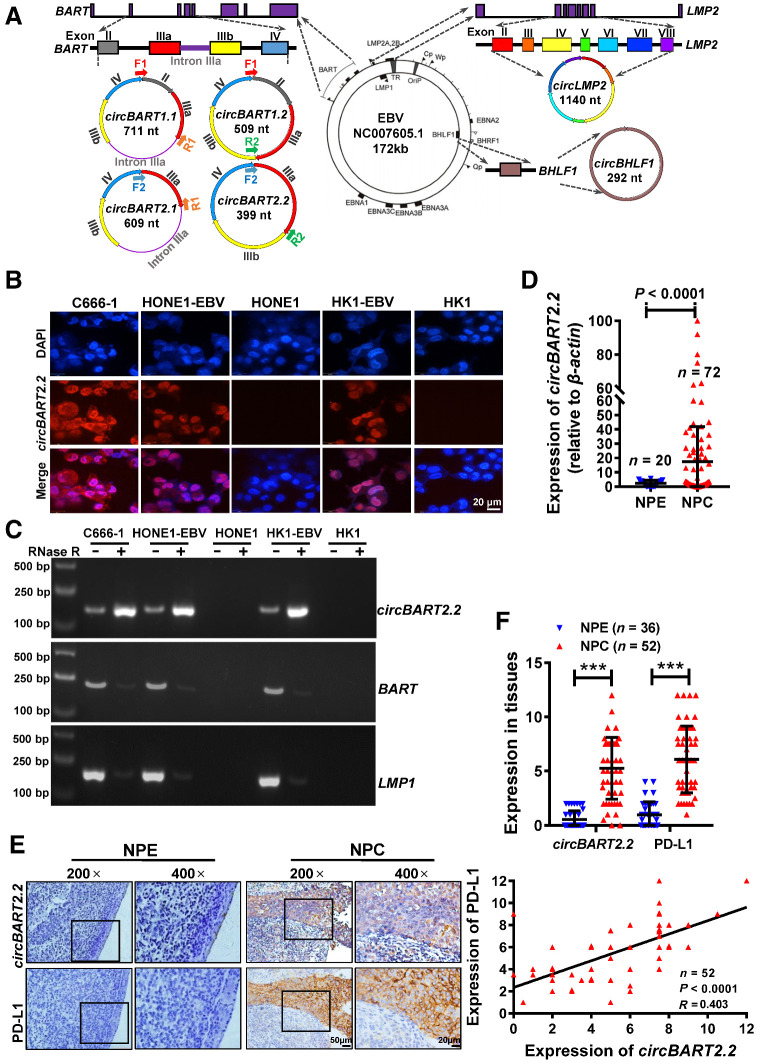
Figure 1.
EBV-encoded circBART2.2 was highly expressed in NPC and positively correlated with PD-L1. A, Schematic diagram of EBV-encoded circRNAs in EBV (NC007605.1, 172 kb). The LMP2 gene encodes circLMP2 (1140 nt), and BHLF1 encodes circBHLF1 (292 nt). The BART gene encodes four different circBARTs: circBART1.1 (711 nt, including exons II, IIIa, IIIb, and IV and intron IIIa), circBART1.2 (509 nt, including exons II, IIIa, IIIb, and IV), circBART2.1 (609 nt, including exons IIIa, IIIb, and IV and intron IIIa), and circBART2.2 (399 nt, including exons IIIa, IIIb, and IV). F1, F2, R1, and R2: primer sites for detecting circBARTs are indicated. B, Intracellular localization of circBART2.2 (red) in the EBV-positive NPC cell lines C666-1, HONE1-EBV, and HK1-EBV, as determined by FISH using a biotin-labeled circBART2.2 probe. EBV-negative HONE1 and HK1 were used as the negative controls. Nuclei were stained with DAPI (blue). Magnification, ×400. Scale bar, 20 μm. C, The stability of circBART2.2 was detected in RNase R–treated C666-1, HONE1-EBV, and HK1-EBV cells for 24 hours by qPCR. EBV-negative HONE1 and HK1 were used as negative controls. The linear BART and LMP2 mRNA were also used as controls. D, circBART2.2 expression was measured in 72 NPC tissues and 20 noncancerous nasopharyngeal epithelial (NPE) tissue samples by RT-PCR. β-Actin was used as an internal reference (P < 0.0001). E, Representative images of circBART2.2 and PD-L1 expression in 52 NPC tissues and 36 noncancerous NPE tissues by in situ hybridization for circBART2.2 and IHC for PD-L1 protein. Magnification, ×200; scale bar, 50 μm; ×400; scale bar, 20 μm. F, Statistical analysis of the expression and correlation of circBART2.2 and PD-L1 according to the expression in 52 NPC tissues and 36 noncancerous NPE tissue samples by in situ hybridization or IHC. ***, P < 0.001.
Immunofluorescence and fluorescence in situ hybridization
Immunofluorescence was performed according to the manufacturer's instructions. Images were captured using the Operetta CLS High-Content Fluorescence Analysis System (PerkinElmer).
The digoxigenin-labeled specific probe for circBART2.2 was designed and synthesized by Tsingke Co. The fluorescence in situ hybridization (FISH) experiment was performed according to the manufacturer's instructions (RiboBio). The nuclei were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI; D1306, Invitrogen). Cells were imaged using a confocal laser scanning microscope (PerkinElmer).
Western blotting
The total protein was extracted and separated by 10% twelve alkyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride membrane (IEVH07850, Millipore). The membrane was incubated antibodies, and the ECL detection reagent (Millipore) was used to detect. GAPDH was used as a protein control. Antibodies are shown in Supplementary Table S4.
IHC and in situ hybridization
IHC was performed using the Elivision plus Polyer HRP (Mouse/Rabbit) IHC Kit (KIT-9902, Maxim), and the primary antibodies used are shown in Supplementary Table S4.
In situ hybridization was performed using the Enhanced Sensitive ISH Detection kit I (POD; MK1030, BOSTER, China) kit using the digoxigenin-labeled circBART2.2 probe (Tsingke) according to the manufacturer's instructions. The staining intensity score was determined as 0 = negative, 1 = weak, 2 = moderate, and 3 = strong, and the positive rate score was determined as 0 = negative, 1 = 1%–25%, 2 = 26%–50%, 3 = 51%–75%, and 4 = 76%–100%. The total score was obtained by multiplying the intensity and positive rate scores of the stained cells and completed by two experienced pathologists. Images were captured using an Olympus BX51 fluorescence microscope (Olympus).
Enzyme linked immunosorbent assay
The concentration of IFNγ in serum of nude mice or CD8-positive T cells was measured by the IFNγ ELISA Kit (KHC4021, eBioscience) according to the manufacturer's guidelines.
Flow cytometry analysis
An Annexin V–FITC Apoptosis Detection Kit (BMS500FI, Invitrogen) was used to detect apoptosis by flow-cytometric analysis. Cells were incubated with Alexa Fluor 488 Annexin V and propidium iodide (PI) for 15 minutes and performed using the DX athenatm flow cytometry. The FlowJo software (FlowJo, RRID: SCR_008520) was used for data analysis. Unstained cells and fluorescence minus controls were used for cytometry and gating set up. Q1: Annexin V−/PI+, Q2: Annexin V+/PI+, Q3: Annexin V−/PI−, Q4: Annexin V+/PI−.
RNA pulldown and liquid chromatography coupled to tandem mass spectrometry
An RNA pulldown assay was performed using a magnetic RNA protein pulldown kit (20164, Thermo Scientific) according to the manufacturer's instructions. In short, biotin-labeled specific circBART2.2 probe or its fragments (367–11 nt for DEL1, 114–165 nt for DEL2, and 62–89 nt for DEL3) that deleted the core structural area of circBART2.2 were designed. The circBART2.2 or its fragment overexpression vectors were transfected into cells. After 24 hours, the biotin-labeled or unbiotin-labeled probes were transfected into cells for another 24 hours. The cell lysates were incubated with biotin-affinity magnetic beads for 2 hours at room temperature to bind the RNA-associated proteins. The purified proteins were separated using SDS-PAGE gels and then stained with Coomassie blue staining and analyzed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) using an UltiMate 3000 RSLCnano system coupled to an LTQ OrbitrapVelos Pro mass spectrometer (Thermo Scientific).
RNA immunoprecipitation analysis
Cells were incubated with anti-RIG-I or rabbit IgG control-coupled magnetic beads. RNA in immunoprecipitates was isolated with TRIzol reagent and analyzed by qRT-PCR.
Luciferase reporter analysis
Vectors for the reporter genes detecting the ISRE and NF-κB activities were purchased from Beyotime Biotechnology (pISRE-TA-luc, D2179, pNF-κB-luc, and D2206). For the PD-L1 promoter analysis, different lengths of the PD-L1 promoter and corresponding mutant fragments were cloned into the PGL3 basic vector (E1751, Promega). The ratio of the luminescence of the firefly luciferase to that of the Renilla luciferase was calculated using the Dual-Glo Luciferase Assay System (E2980, Promega) kit.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) analysis was performed using the Pierce Magnetic ChIP Kit (26157, Thermo Scientific). The NF-κB (p65) and IRF3 antibodies were used to enrich the PD-L1 promoter region fragments, and then qPCR was used to analyze the expression of the enriched fragments.
Preparation of dendritic cells and primary T cells
After testing, HONE1 was classified as HLA-A2+. The peripheral blood of HLA-A2+ healthy donors was used to prepare dendritic cells. First, peripheral blood mononuclear cell was separated using the Ficoll (45-001-749, Cytiva) separation solution and added to 10% FBS-1640 medium containing 50 ng/m LGM-CSF (PHC2013, Invitrogen) and 20 ng/mL IL4 (PHC0044, Invitrogen) for 5 days. To promote the maturation of dendritic cells, 25 ng/mL IFNγ (Invitrogen) was added and cultured for 1 day, and then cocultured with HONE1 lysate for another day. To expand T cells in vitro, T cells were added with CD3/CD28 amplified magnetic beads (Miltenyi Biotec) and 15 ng/mL IL2 (PHC0026, Invitrogen), 5 ng/mL IL7 (PHC0073, Invitrogen), and 10 mL IL15 (PHC9151, Invitrogen) for 8-day culture. To generate tumor-specific cytotoxic T-lymphocytes, the prepared dendritic cells and expanded T cells were cocultured in a medium supplemented with IL2, IL7, and IL15 for 5 days at a ratio of 1:5.
Real-time tracking T-cell activity status using the high-content cell imaging analysis system
HONE1 cells stained with the CellTracker CM-DiI Dye (C7000, Invitrogen) and activated human primary T cells labeled with live-cell fluorescent dye CMFDA (5-chloromethylfluorescein diacetate, C7025, Invitrogen) at the ratio of 1: 10 were cocultured for 3 hours. The Operetta CLS high-content cell imaging analysis system (PerkinElmer) was used to track the fluorescence status of T cells in real time.
Nude mouse model in vivo
HONE1 cells (5 × 106) transfected with the circBART2.2 overexpression vector or the empty vector were injected subcutaneously into the right thigh root of 4 weeks old and female nude mice (22 mice per group). Tumor formation was observed macroscopically 7 days later. Dendritic cells were first cocultured with HONE1 cell lysate for 24 hours, and then cocultured with T cells to present HONE1-specific tumor antigens to T cells to enable them to produce HONE1-specific T cells. For one part, human primary T cells that had received tumor-specific antigen presentation were labeled with Deep Red live-cell fluorescent dye (DiR, Thermo Fisher) and injected into the mice through the tail vein (3 mice per group) to evaluate T cells distribution in nude mice using the small animal in vivo imaging system (Bruker).
For the second part, T cells that had received tumor-specific antigen presentation without Deep Red live-cell fluorescent dye were injected into the mice (5 mice per group) to evaluate T cells survival. The mice peripheral blood was extracted for qRT-PCR, ELISA, or flow-cytometric analysis after 7 days of adoptive T cells treatment.
For the third part, T cells without fluorescent dye were also injected into nude mice (7 mice per group) for 32 days. Tumor volume, size, weight, and body weight of nude mice were measured. In situ hybridization or IHC were performed to examine the expression of circBART2.2, RIG-I, PD-L1, cleaved caspase-3, and cleaved-PARP. All animal studies were approved by the Ethics Committee of the Xiangya Hospital, Central South University.
Data analysis and statistical analysis
The NPC gene-expression data set (GSE12452) was downloaded from the Gene-Expression Omnibus (GEO, RRID: SCR_005012) database. Statistical analysis was performed using the GraphPad Prism 7.0 (GraphPad Prism, Inc.; RRID: SCR_002798). Two-tailed t tests were used to analyze the data. P values < 0.05 were considered statistically significant.
Availability of data and materials
All data that support the findings of this study are available from the corresponding authors upon reasonable request.
Results
EBV-encoded circBART2.2 was highly expressed and positively correlated with PD-L1 expression in NPC
To identify the role of EBV-encoded circRNAs in the development of NPC, qRT-PCR was performed in six EBV-positive NPC clinical samples and EBV-positive cell lines. Isoform-specific primers that could be discriminated from each other were designed for circRNAs. For four circBART variants, the forward primer was overlapped with the back splice sequence (either backsplice of exon IV to II for 1.1 and 1.2 variants or backsplice of exon IV to IIIa for 2.1 and 2.2) variants and the reverse primers overlapped the exon-exon junctions exon IIIb to IIIa (for variants 1.2 and 2.2) or Intron IIIa to exon IIIa (for variants 1.1 and 2.1; Fig. 1A). All four different variants were discriminated and expressed in the EBV-positive Akata cell line, which was consistent with Toptan and colleagues (17). circBART2.2 was detected at high levels in all six NPC clinical tissues and EBV-positive NPC cell lines, including C666-1, HONE1-EBV, and HK1-EBV. circBART2.1 was weakly detected in C666-1, HONE1-EBV, and HK1-EBV, but was not expressed in six NPC tissues. circBART1.1, circBART1.2, circLMP2, and circBHLF1 were only detected in Akata; additionally, there were no specific electrophoretic bands in six NPC clinical samples and other EBV-positive NPC cells (Supplementary Fig. S1A). These data suggest that circBART2.2 may be expressed in EBV-positive NPC with the high levels, thereby affecting the development of NPC.
Using RNA FISH (Fig. 1B), the expression of circBART2.2 was detected in C666-1, HONE1-EBV, and HK1-EBV, but not in HONE1 and HK1. circBART2.2 was resistant to RNase R degradation and more stable than linear BART mRNA in C666-1, HONE1-EBV, and HK1-EBV after RNase R (Fig. 1C) or actinomycin D treatments (Supplementary Fig. S1B). Additionally, high expression of circBART2.2 was confirmed in 72 NPC tissues and 20 nasopharyngeal epithelial tissues by qRT-PCR (Fig. 1D) and in 52 NPC and 36 noncancerous samples by in situ hybridization (Fig. 1E and F).
To examine whether circBART2.2 is involved in the PD-L1–regulated immune escape in NPC, we analyzed the correlation between circBART2.2 and PD-L1. PD-L1 was highly expressed in the same NPC tissues by IHC and significantly positively correlated with the expression of circBART2.2 (Fig. 1E and F).
circBART2.2 inhibited T-cell killing of NPC cells by upregulating PD-L1
To further validate the correlation between circBART2.2 and PD-L1 in NPC, a circBART2.2 overexpression vector was transfected into HONE1 and HK1 (Supplementary Fig. S2A). The expression of circBART2.2 was stable and resistant to RNase R degradation after overexpression of circBART2.2 in HONE1 and HK1, compared with GAPDH mRNA (Supplementary Fig. S2B). Additionally, three siRNAs specifically targeting the splice site of circBART2.2 were transfected into HONE1-EBV and HK1-EBV, and knockdown of circBART2.2 (Supplementary Fig. S2C) reduced circBART2.2 expression, but not linear BART. siRNAs targeting circBART2.2 also had some effect on the circBART2.1 expression because they shared the same back splice site. However, compared with circBART2.2, the expression of circBART2.1 in HONE1-EBV and HK1-EBV was extremely low (Supplementary Fig. S2C). In contrast, PD-L1 expression is not affected by circBART2.1 overexpression in HONE1 and HK1 cells (Supplementary Fig. S2D). We speculate that circBART2.1 may have little influence on the regulation of PD-L1 in NPC.
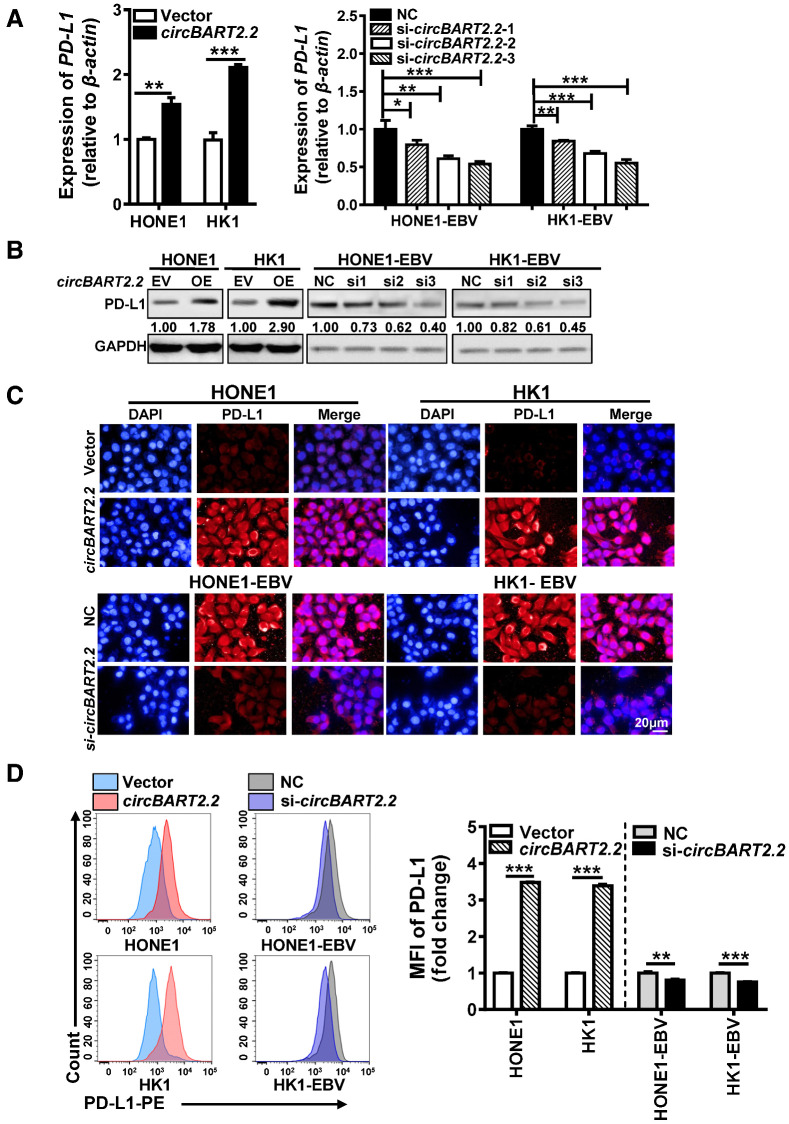
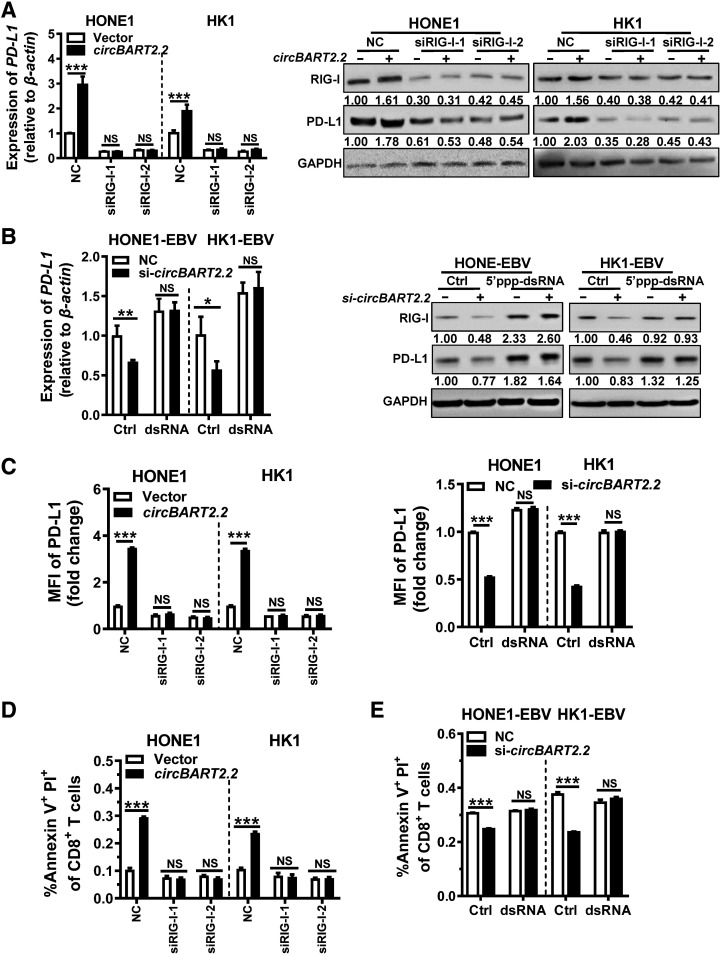
RT-PCR and Western blotting showed that overexpression of circBART2.2 promoted PD-L1 expression at the mRNA (Fig. 2A) and protein levels (Fig. 2B), while PD-L1 expression was diminished after circBART2.2 siRNA treatment. Immunofluorescence (Fig. 2C) and flow cytometry (Fig. 2D) using anti–PD-L1 antibody also showed that circBART2.2 promoted PD-L1 expression. In summary, circBART2.2 is a potent inducer of PD-L1 in EBV-associated NPC.
Figure 2.
circBART2.2 significantly upregulated PD-L1 expression. A, The mRNA expression of PD-L1 was examined in HONE1 and HK1 cells after circBART2.2 overexpression or in HONE1-EBV and HK1-EBV cells transfected with three circBART2.2 siRNAs, respectively, as demonstrated by RT-PCR. *, P < 0.05; **, P < 0.01; ***, P < 0.001. B, The expression of PD-L1 protein was examined in HONE1 and HK1 cells after circBART2.2 overexpression or in HONE1-EBV and HK1-EBV cells transfected with three circBART2.2 siRNAs, respectively, as demonstrated by Western blotting. C, PD-L1 expression (red) was measured in EBV-negative or -positive NPC cells after circBART2.2 overexpression or knockdown by immunofluorescence using anti–PD-L1 antibody. Nuclei were stained with DAPI (blue). Magnification, ×400. Scale bar, 20 μm. D, PD-L1 expression was measured in EBV-negative or -positive NPC cells after circBART2.2 overexpression or knockdown using flow-cytometric analysis using APC-stained anti–PD-L1 antibody. Left, original flow cytometry results; right, statistical results. Three independent replicates were evaluated for each group. MFI: mean fluorescence intensity. **, P < 0.01; ***, P < 0.001.
To determine whether circBART2.2 inhibits T-cell function and mediates NPC immune escape through PD-L1, circBART2.2 was overexpressed or knocked down in NPC cells. Then cells were stained with DeepRed live-cell fluorescent dye and cocultured with activated primary cultured T cells labeled with the live-cell fluorescent dye CMFDA. The T-cell viability status was tracked in real time using a high-content cell imaging analysis system (Supplementary Fig. S3A). The fluorescence intensity of T cells decreased faster than that of control cells in NPC cells when circBART2.2 was overexpressed, whereas knockdown of circBART2.2 in EBV-infected NPC cells inhibited the decrease of T cells' fluorescence intensity, suggesting circBART2.2 effectively promoted T-cell apoptosis in NPC. By contrast, the fluorescence intensity of NPC cells decreased in a slower manner than that of control cells after overexpression of circBART2.2 by flow cytometry (Supplementary Fig. S3B), suggesting that circBART2.2 promoted the immune escape of NPC cells.
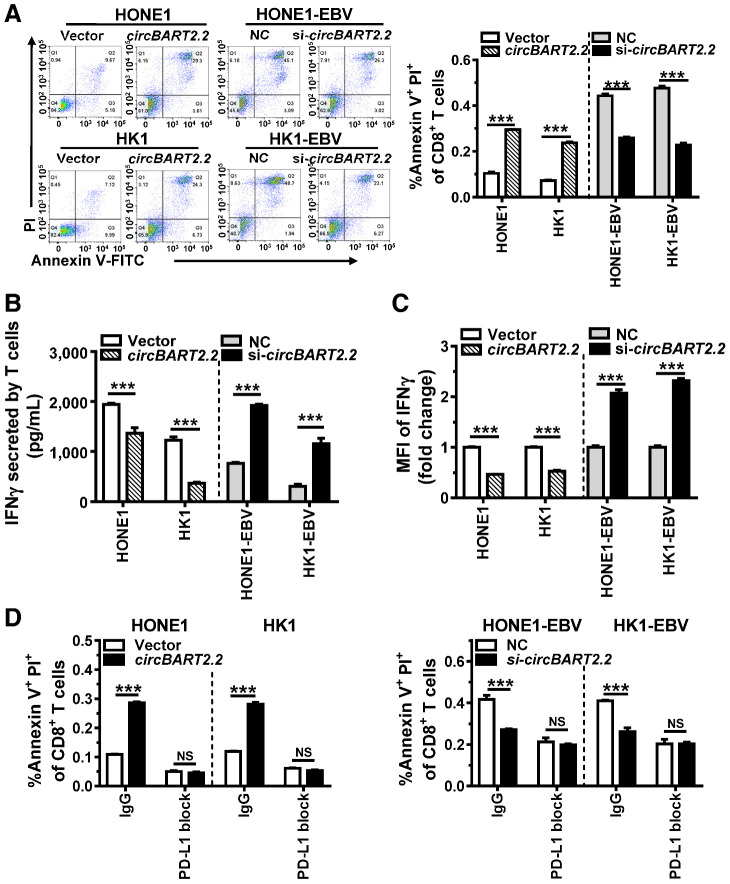
CD8+ T cells can be used to measure the activity of T cells. Flow cytometry using anti-CD8 antibody showed that T-cell apoptosis (Annexin V+PI+) in coculture with NPC cells was significantly increased after overexpression of circBART2.2 and significantly decreased after knockdown of circBART2.2 (Fig. 3A). Cytotoxic T cells inhibit tumor growth through IFNγ secretion. Overexpression of circBART2.2 in NPC cells significantly inhibited IFNγ secretion of T cells, and knockdown of circBART2.2 restored the ability of T cells to secrete IFNγ (Fig. 3B). Flow cytometry using anti-IFNγ antibody showed that overexpression of circBART2.2 significantly inhibited IFNγ accumulation in T cells, whereas knockdown of circBART2.2 partially restored the ability of T cells to produce IFNγ (Fig. 3C; Supplementary Fig. S3C). Notably, a sufficient anti–PD-L1 antibody to block PD-L1 expression inhibited the promoting effects of circBART2.2 on T-cell apoptosis in HONE1 and HK1 after overexpression of circBART2.2 by flow cytometry (Fig. 3D; Supplementary Fig. S3D). Based on these, we concluded circBART2.2 inhibited T-cell function and mediated NPC immune escape through PD-L1.
Figure 3.
circBART2.2 promoted T-cell apoptosis and inhibited IFNγ secretion of T cells through PD-L1. A, The degree of T-cell apoptosis was measured by flow cytometry in EBV-negative or -positive NPC cells after circBART2.2 overexpression or knockdown after incubation with Alexa Fluor 488 Annexin V and PI. Left, original flow cytometry results; right, statistical results. ***, P < 0.001. Q1, Annexin V−/PI+; Q2, Annexin V+PI+; Q3, Annexin V−/PI−; Q4, Annexin V+/PI−. B, IFNγ secretion from T cells was measured by ELISA in EBV-negative or -positive NPC cells after circBART2.2 overexpression or knockdown. ***, P < 0.001. C, The concentration of IFNγ in cell culture medium was examined by flow-cytometric analysis in T cells cocultured with EBV-negative or -positive NPC cells after circBART2.2 overexpression or knockdown using APC-stained anti-IFNγ antibody. Each experiment was independently repeated three times, and the original results are shown in Supplementary Fig. S4A. MFI, mean fluorescence intensity; ***, P < 0.001. D, Annexin V+ PI+ cells of CD8-positive active T cells were measured by flow cytometry in T cells cocultured with EBV-negative or -positive NPC cells after circBART2.2 overexpression or knockdown. NPC cells were treated with a sufficient anti–PD-L1 antibody to block PD-L1 activity. Each experiment was independently repeated three times, and the original results are shown in Supplementary Fig. S4B. NS, not significant; ***, P < 0.001.
circBART2.2 bind to the helicase domain of RIG-I protein around nucleotides 114–165
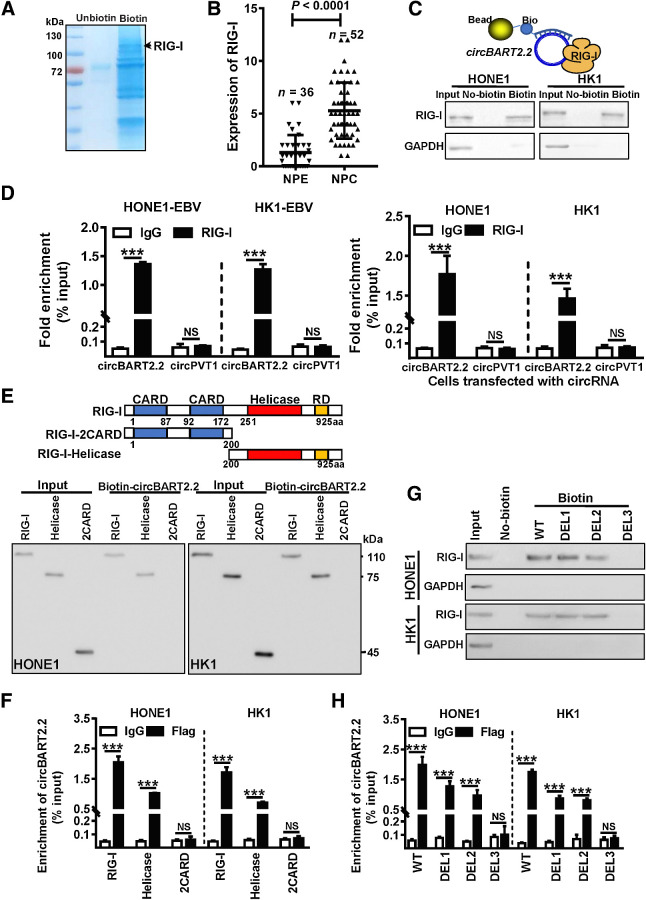
To explore the molecular mechanisms through which circBART2.2 affects PD-L1, RNA pulldown, followed by the mass spectrum identification, was performed in HONE1 using biotin-labeled circBART2.2 (Fig. 4A). A total of 172 potential proteins were precipitated and identified (Supplementary Table S5). Then the GSE12452 GEO data set was used to analyze PD-L1 correlated genes. A total of 326 genes were positively correlated to PD-L1 expression (r > 0.5) while 247 genes were negatively correlated to PD-L1 (r < −0.5; Supplementary Fig. S4A). RIG-I (DDX58) was pulled down by circBART2.2 and also positively correlated to PD-L1 expression. Both of RIG-I and PD-L1 genes were highly expressed and positively correlated with each other in GSE12452 (Supplementary Fig. S4B). IHC confirmed that RIG-I was highly expressed (Fig. 4B) and positively correlated with circBART2.2 and PD-L1 protein expression in 52 NPC tissues, compared with that in 36 noncancerous samples (Supplementary Fig. S4C).
Figure 4.
circBART2.2 binds to RIG-I protein through the helicase domain. A, Biotin-labeled or unbiotin-labeled circBART2.2 probes were transfected into HONE1 cells after the overexpression of circBART2.2. The cell lysates were incubated with biotin-affinity magnetic beads for 2 hours. The precipitated proteins were resolved by SDS-PAGE, followed by Coomassie blue staining. Next, the differential band in the biotin-labeled circBART2.2 probe lane was identified by LC-MS/MS. The unbiotin-labeled circBART2.2 probe served as a control. RIG-I was also identified. B, The expression of RIG-I in 52 NPC tissues and 36 noncancerous NPE tissues by IHC using an anti-RIG-I antibody. P < 0.0001. C, Binding of circBART2.2 to RIG-I protein was detected in HONE1 and HK1 cells after overexpression of circBART2.2 using RNA pulldown assays with a biotin-labeled circBART2.2 probe. The unbiotin-labeled circBART2.2 probe was used for control. D, Direct binding of RIG-I protein to circBART2.2 was evaluated in HONE1 and HK1 cells after overexpression of circBART2.2 or in EBV-positive HONE1-EBV and HK1-EBV cells by RNA immunoprecipitation using anti–RIG-I antibody, followed by qPCR analysis for circBART2.2. CircPVT1 was used as a negative control. ***, P < 0.001. E, The binding between circBART2.2 and the helicase domain of RIG-I protein was detected in HONE1 and HK1 cells after cotransfection of the circBART2.2 overexpression vector and the RIG-I full-length or truncated fragments using RNA pulldown assays with a biotin-labeled circBART2.2 probe. F, The binding between circBART2.2 and the helicase domain of RIG-I protein was examined in HONE1 and HK1 cells after the cotransfection of the circBART2.2 overexpression vector and the Flag-tagged RIG-I full-length or truncated fragments by RNA immunoprecipitation using anti-Flag-RIG-I antibody. IgG was used as a control. G, The 114–165 nt of circBART2.2 was crucial for the binding between circBART2.2 and RIG-I proteins. HONE1 and HK1 cells were transfected with the full-length circBART2.2 (WT) or deletion mutants (367–11 nt for DEL1, 62–89 nt for DEL2, 114–165 nt for DEL3). RNA pulldown assays were performed using a biotin-labeled circBART2.2 probe, followed by Western blotting using anti–RIG-I antibody. Unbiotin-labeled circBART2.2 or mutant probes were used as controls. H, RIG-I protein directly binds to the 114–165 nt of circBART2.2 in HONE1 and HK1 cells. Cells were cotransfected with the FLAG-tagged RIG-I vector and the full-length circBART2.2 (WT) or deletion mutants (DEL1, DEL2, and DEL3). RNA immunoprecipitation was performed using anti-Flag-RIG-I antibody, followed by qPCR analysis for circBART2.2. NS, not significant; ***, P < 0.001.
RNA pulldown experiments further confirmed biotin-labeled circBART2.2 precipitated RIG-I protein in circBART2.2-overexpressing HONE1 and HK1 (Fig. 4C). RNA immunoprecipitation showed that anti-RIG-I–antibody-enriched exogenous circBART2.2 in circBART2.2-overexpressing HONE1 and HK1 or endogenous circBART2.2 in HONE1-EBV and HK1-EBV (Fig. 4D).
To explore the molecular mechanism underlying the promotion of RIG-I signaling by circBART2.2, in vitro RNA pulldown and RNA immunoprecipitation (RIP) assays were performed to determine the domain of RIG-I that binds to circBART2.2. circBART2.2 binds to the helicase domain with RIG-I but not to the CARD domain in HONE1 and HK1 (Fig. 4E and F). Meanwhile, three well-characterized stem loop structures of circBART2.2 (367–11 nt, 62–89 nt, and 114–165 nt) were predicted to bind with RIG-I protein (Supplementary Fig. S5A and S5B). RNA pulldown assay and RIP indicated the 114 to 165 nt region (DEL3) of circBART2.2 was the critical region to bind to RIG-I in HONE1 and HK1 after overexpression of the circBART2.2 WT vector or three truncated circBART2.2 fragments (DEL1, DEL2, or DEL3; Supplementary Fig. S5C; Fig. 4G and H).
circBART2.2 induced PD-L1–associated NPC immune escape by binding to RIG-I
To explore whether circBART2.2 regulates PD-L1 by binding to RIG-I, RIG-I was knocked down in HONE1 and HK1 using two siRNAs or activated using 5′ppp-dsRNA (tlrl-3prna, Invitrogen) in HONE1-EBV and HK1-EBV (Supplementary Fig. S6A and S6B). qRT-PCR and Western blotting showed circBART2.2 significantly upregulated PD-L1, whereas upregulation of PD-L1 by circBART2.2 was inhibited by two siRIG-Is (Fig. 5A). Conversely, activation of RIG-I reversed the inhibition of PD-L1 by si-circBART2.2 in HONE1-EBV and HK1-EBV (Fig. 5B). Flow cytometry (Fig. 5C; Supplementary Fig. S6C) and immunofluorescence (Supplementary Fig. S6D) also validated the above experimental results using an anti–PD-L1 antibody. These results suggested circBART2.2 upregulated PD-L1 expression through binding with RIG-I protein.
Figure 5.
circBART2.2 regulates PD-L1 expression through binding to RIG-I. A, PD-L1 expression was examined in HONE1 and HK1 cells after circBART2.2 overexpression or knockdown of RIG-I simultaneously using RT-PCR or Western blotting. B, PD-L1 expression was examined in HONE1-EBV and HK1-EBV cells after circBART2.2 knockdown and RIG-I activator 5′-ppp-dsRNA treatment using RT-PCR or Western blotting. C, The expression of PD-L1 was measured in HONE1 and HK1 cells after circBART2.2 overexpression and simultaneous RIG-I knockdown or HONE1-EBV and HK1-EBV cells after circBART2.2 knockdown and simultaneous RIG-I activator 5′-ppp-dsRNA treatment by flow-cytometric analysis using anti–PD-L1 antibody. Each experiment was independently repeated three times; the original results are shown in Supplementary Fig. S6C. MFI, mean fluorescence intensity. D, Annexin V+ PI+ cells of CD8-positive active T cells were measured by flow cytometry in T cells cocultured with EBV-negative HONE1 and HK1cells after circBART2.2 overexpression or knockdown of RIG-I simultaneously. Each experiment was independently repeated three times; the original results are shown in Supplementary Fig. S7B. E, Proportions of CD8-positive active T cells were measured by flow cytometry in T cells cocultured with HONE1-EBV and HK1-EBV cells after circBART2.2 knockdown and simultaneous RIG-I activator 5′-ppp-dsRNA treatment. Each experiment was independently repeated three times; the original results are shown in Supplementary Fig. S7C. NS, not significant; ***, P < 0.001.
To identify whether RIG-I affects the function of circBART2.2 in T-cell apoptosis, HONE1 and HK1 were cotransfected with the circBART2.2 overexpression vector and siRIG-I and then cocultured with activated human primary T cells. High-content cell imaging analysis showed knockdown of RIG-I inhibited the decrease in T-cell fluorescence intensity caused by circBART2.2 (Supplementary Fig. S7A). Flow cytometry using anti-CD8 antibody showed the ability of circBART2.2 to induce T-cell apoptosis was inhibited after knockdown of RIG-I. Conversely, the activation of RIG-I reversed the inhibitory effects of circBART2.2 knockdown on T-cell apoptosis in HONE1-EBV and HK1-EBV (Fig. 5D and E; Supplementary Fig. S7B and S7C), suggesting that circBART2.2 regulates T-cell apoptosis and tumor immune escape through the RIG-I signaling pathway.
circBART2.2 upregulated PD-L1 by activating the transcription factors IRF3 and NF-κB
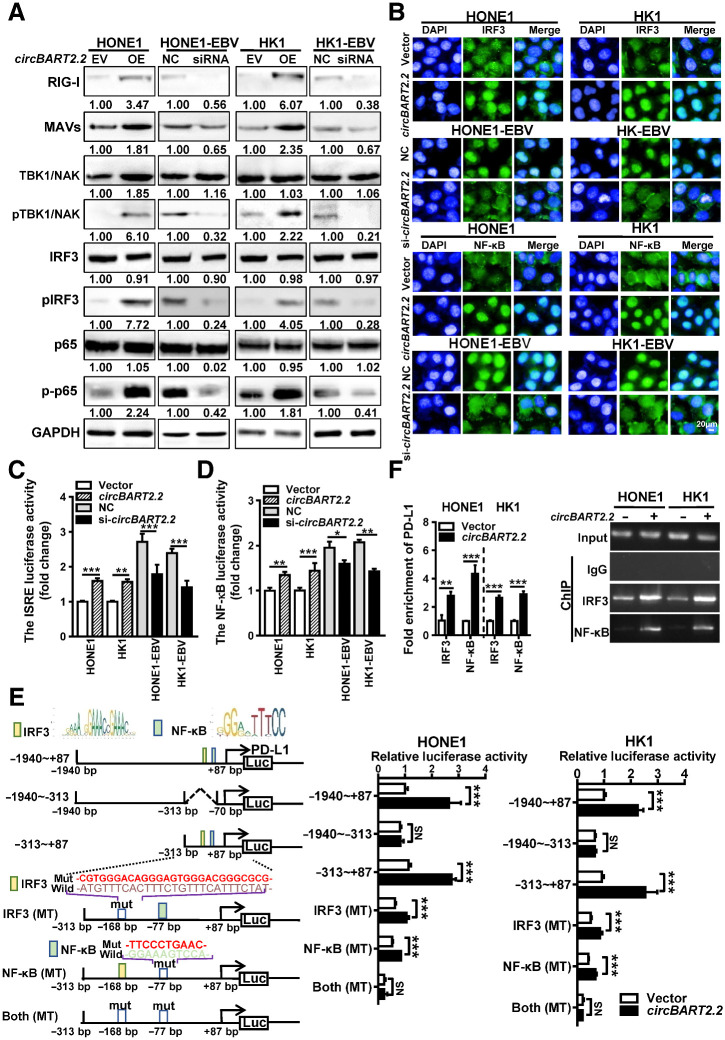
To explore whether circBART2.2 regulates the RIG-I pathway, the expression of related molecules in the RIG-I pathway was detected by Western blotting in HONE1 and HK1 after overexpression of circBART2.2 or in HONE1-EBV and HK1-EBV following knockdown of circBART2.2. Overexpression of circBART2.2 increased the levels of RIG-I, mitochondrial antiviral-signaling protein (MAVS), TANK binding kinase 1 (TBK1)/NF-κB activating kinase (NAK), phospho-TBK1/phospho-NAK, phospho-IRF3, and phospho-NF-κB (p-p65), whereas knockdown of circBART2.2 reduced the levels of these factors (Fig. 6A). circBART2.2 significantly promoted the nuclear entry of transcription factors NF-κB and IRF3, whereas knockdown of circBART2.2 decreased it by immunofluorescence (Fig. 6B). Luciferase reporter activity assays showed that overexpression of circBART2.2 promoted the transcriptional activities of IRF3 and NF-κB in HONE1 and HK1, whereas knockdown of circBART2.2 decreased the activities of them in HONE1-EBV and HK1-EBV (Fig. 6C and D).
Figure 6.
circBART2.2 upregulated PD-L1 expression by activating the transcriptional activity of IRF3 and NF-κB. A, The expression levels of RIG-I, MAVS, TBK1/NAK, IRF3, NF-κB (p65), phospho-TBK1/phospho-NAK, phospho-IRF3, and phospho-NF-KB (p-p65) were examined by Western blotting in HONE1 and HK1 cells after overexpression of circBART2.2 or in HONE1-EBV and HK1-EBV cells after knockdown of circBART2.2. B, Immunofluorescence analysis was used to evaluate the nuclear entry of the transcription factors IRF3 or NF-κB (green) in HONE1 and HK1 cells after overexpression of circBART2.2 or in HONE1-EBV and HK1-EBV cells after knockdown of circBART2.2. Nuclei were stained with DAPI (blue). Magnification, ×400. Scale bar, 50 μm. C, The transcriptional activity of IRF3 was measured using the IRSE luciferase reporter assays in HONE1 and HK1 cells after circBART2.2 overexpression or in HONE1-EBV and HK1-EBV cells after circBART2.2 knockdown. **, P < 0.01; ***, P < 0.001. D, The transcriptional activity of NF-κB was measured using the NF-κB luciferase reporter assays in HONE1 and HK1 cells after circBART2.2 overexpression or in HONE1-EBV and HK1-EBV cells after circBART2.2 knockdown. *, P < 0.05; **, P < 0.01; ***, P < 0.001. E, A series of luciferase reporter vectors was constructed according to the PD-L1 promoter sequence. Luciferase reporter activity was analyzed in HONE1 and HK1 cells after circBART2.2 overexpression. The binding site of IRF3 transcription factor at −168 bp and NF-κB transcription factor at −77 bp on the PD-L1 promoter region were predicted according to the PD-L1 promoter (left); the results of luciferase reporters activity in HONE1 and HK1 cells after circBART2.2 overexpression (right). ***, P < 0.001. F, Binding of the transcription factors IRF3 and NF-κB on the PD-L1 promoter region was detected by ChIP assays according to the predicted IRF3 and NF-κB binding sites. NS, not significant; **, P < 0.001; ***, P < 0.001. Left, the statistical results; right, the gel electrophoresis.
To identify if circBART2.2 regulates RIG-I expression at the transcription level, the RIG-I mRNA expression was also examined by qRT-PCR in HONE1 and HK1 or HONE1-EBV and HK1-EBV after circBART2.2 overexpression or knockdown. circBART2.2 significantly upregulated RIG-I expression at the mRNA level (Supplementary Fig. S8A). The Jaspar software showed there was a binding site of NF-κB near −342 bp of the RIG-I promoter. ChIP experiment confirmed that circBART2.2 promoted the enrichment of NF-κB on the RIG-I promoter in HONE1 and HK1 (Supplementary Fig. S8B), suggesting that circBART2.2 can also promote activation of RIG-I at the transcription level. Taken together, these results suggested that circBART2.2 as an exogenous circRNA molecule encoded by EBV activated the RIG-I signaling pathway in NPC.
To further explore whether circBART2.2 regulates PD-L1 through the RIG-I signaling pathway, transcription factor binding sites on the PD-L1 promoter were analyzed. Binding sites for IRF3 at −168 bp and for NF-κB at −77 bp were shown on the PD-L1 promoter. Luciferase reporter activity revealed that the core promoter region of PD-L1 was located from −313 to + 87 bp. Overexpression of circBART2.2 increased the luciferase reporter activity at positions −313 to + 87 bp on the PD-L1 promoter in HONE1 and HK1; however, this effect was blocked by mutation of the binding sites for IRF3 or NF-κB (Fig. 6E). ChIP experiments also showed that circBART2.2 promoted the enrichment of IRF3 or NF-κB on the PD-L1 promoter (Fig. 6F). Collectively, it is conceivable that the binding of circBART2.2 to RIG-I results in PD-L1 upregulation through an IRF3- and NF-κB–initiated transcriptional program.
circBART2.2 promoted immune escape of NPC cells in vivo
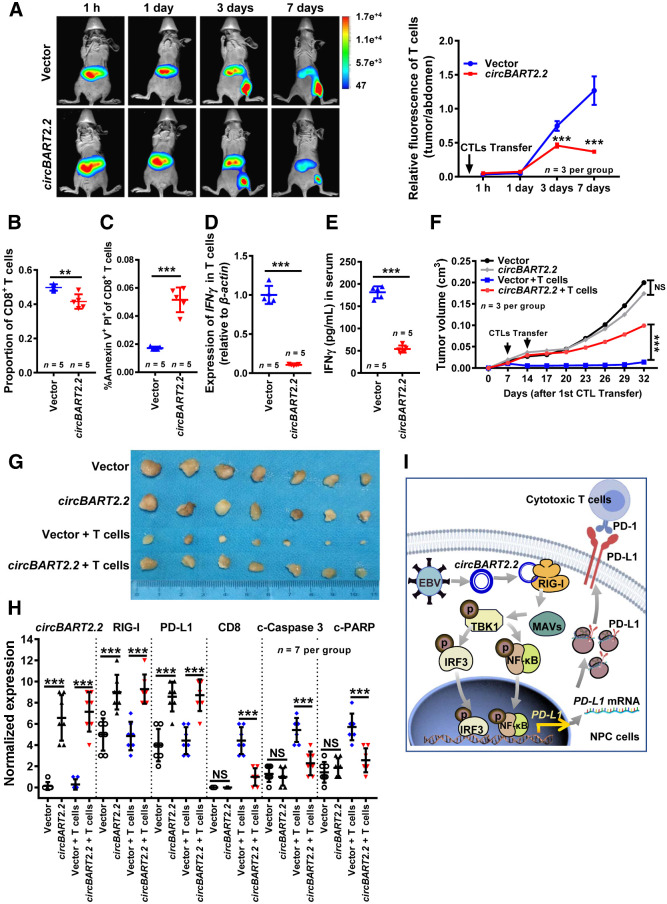
To test whether circBART2.2 promotes the immune escape of NPC in vivo, HONE1 cells after overexpression of circBART2.2 were inoculated into nude mice to establish a xenograft model. As shown in Supplementary Fig. S9A, T cells were significantly aggregated toward the site of migrating tumors on day 3, and circBART2.2 significantly decreased the fluorescence intensity of T cells at the site of migration tumors on day 7, suggesting that circBART2.2 could promote T-cell apoptosis in vivo, leading to immune escape (Fig. 7A). Additionally, circBART2.2 decreased the proportion of CD8-positive T cells among CD3-positive T cells and increased the proportion of apoptotic T cells by flow cytometry (Fig. 7B and C; Supplementary Fig. S9B and S9C). qRT-PCR (Fig. 7D) and ELISA (Fig. 7E) showed that circBART2.2 inhibited the ability of T cells to secrete IFNγ in T cells and mice serum.
Figure 7.
circBART2.2 induced NPC immune escape in nude mice. A, Living animal imaging analysis was used to detect the survival and distribution of human primary T cells in xenograft tumor mice (n = 3 per group) after injection of T cells. T cells were labeled with DeepRed. Left, representative images. Right, statistical results according to the DiR fluorescence signal ratio in mice collected at 1 day, 3 days, 5 days, and 7 days post injection (the DiR fluorescence signal ratio, T-cell fluorescence intensity in tumors of the root of the right thigh: T-cell fluorescence intensity in the abdomen of mice). The color scales indicate the DiR fluorescence intensity of T cells in mice. B, The proportion of CD8+ T cells to all CD3+ T cells was measured by flow cytometry using PE-Cy7–stained anti-CD8 antibody in human primary T cells of mice serum in HONE1 cell-derived xenograft mice models. A density of 5 × 106 HONE1 cells was transfected with the circBART2.2 overexpression vector or the negative control (NC). Activated T cells were injected into nude mice, and after 14 days the peripheral blood was extracted for flow cytometry; n = 5 per group. C, Annexin V+ PI+ cells of CD8-positive T cells were measured by flow cytometry in HONE1 cell–derived xenograft mice models. Mice peripheral blood was extracted for flow cytometry after incubation with PE-Cy7–stained anti-CD8 antibody, n = 5 per group. D, qRT-PCR analysis of IFNγ mRNA levels in peripheral blood after injection of activated T cells. β-Actin was used as an internal control; n = 5 per group. E, The IFNγ secretion in peripheral serum was detected by ELISA after injection of activated T cells; n = 5 per group. F, The tumor volumes for each group were measured after 32 days of tumor cells injection; n = 7 per group. ***, P < 0.001. G, The tumor images for each group were captured after 32 days of tumor cells injection; n = 7 per group. H, The statistical results for the expression of circBART2.2, RIG-I, PD-L1, CD8, cleaved caspase-3, and cleaved-PARP were analyzed using in situ hybridization or IHC in each mice tissue section. **, P < 0.01; ***, P < 0.001. I, Working model of circBART2.2 promotes the RIG-I signaling pathway. In EBV-infected NPC cells, circBART2.2 binds to the helicase domain of RIG-I protein through the binding sequence around 114 to 165 nt of circBART2.2, forming a complex to activate RIG-I. Activated RIG-I promotes PD-L1 transcription through its downstream transcription factors IRF3 and NF-κB, resulting in tumor immune escape in EBV-infected NPC cells. NS, not significant.
There were no significant differences in tumor size or body weight in nude mice between the circBART2.2 group and the control group without T-cell injection; however, significant differences were observed in tumor volume, tumor size, tumor weight, and body weight between the circBART2.2 group and the control group with T-cell injection (Fig. 7F and G; Supplementary Fig. S9D and S9E), suggesting that circBART2.2 induced tumor immune escape in nude mice. IHC showed that RIG-I and PD-L1 were highly expressed in mice tumors in the circBART2.2 group, regardless of whether T cells were injected. The numbers of CD8-positive T cells and apoptotic tumor cells [cleaved-caspase-3, cleaved-poly (ADP ribose) polymerase (PARP)] in the circBART2.2 group injected with T cells were significantly lower than those in the control group injected with T cells (Fig. 7H; Supplementary Fig. S9F). These results underscore significance of RIG-I in the process of the circBART2.2 -triggered tumor immune escape in promoting PD-L1 expression in vivo.
In this study, EBV-encoded circBART2.2 was found to be highly expressed in NPC tissues and induced the upregulation of PD-L1 in NPC by binding with RIG-I protein to activate its signaling pathway, leading to immune escape in NPC. The binding sequence around 114–165 bp was crucial for the function of circBART2.2. circBART2.2 interacts with the helicase domain of RIG-I, consequently strengthening RIG-I–mediated PD-L1 transcription and downstream signaling to initiate tumor immune escape in NPC (Fig. 7I).
Discussion
EBV was the first virus shown to be associated with human tumors and closely related to the pathogenesis of NPC, gastric cancer, Burkitt lymphoma, and other tumors (32). Several proteins and miRNAs encoded by EBV are involved in tumor progression and malignant phenotypes (33, 34). EBV is also found to be one of the first viruses to encode circRNAs (17, 35). Among them, circLMP2 was reported to induce stemness in EBV-associated gastric cancer cells (36). circBART2.2 (EBV-circRPMS1) showed that it was expressed in EBV-positive cell lines and tissues representing latency types I, II, and III, including EBV-positive post-transplant lymphoproliferative disorder (PTLD), Burkitt lymphoma, EBV-associated gastric carcinoma, NPC, and AIDS-associated lymphoma (17, 36). Liu and colleagues found that circBART2.2 was increased in metastatic NPC and was associated with a short survival time. However, the molecular mechanisms of circBART2.2 still needs to be further clarified (25). In this study, we detected the high expression of circBART2.2 in NPC clinical samples and cell lines. Although other EBV-encoded circRNAs were not detected or lowly expressed in EBV-positive NPC cells, they may be expressed in other EBV-associated cancer cells. The high expression of circBART2.2 suggested that circBART2.2 may play important biological roles in NPC.
EBV infection is closely related to tumor immune escape through upregulation of PD-L1 expression in various tumors (3, 37, 38). EBV-encoded LMP1 (39), EBNA2 (40), and miR-BART5–5p (41) have been reported to cooperate with PD-L1, affect T-cell immune recognition and clearance, and ultimately promote tumor immune escape (42). In this study, we made some preliminary exploration regarding the possible function of circBART2.2, which could bind with RIG-I to regulate PD-L1 and promote tumor immune escape, highlighting this new role of EBV infection in NPC.
RIG-I has generally been used as a key sensor of viral infection (43). Activation of RIG-I results in activation of PD-L1 through IRF3 and NF-kB transcription factors (28,44,45,46). Exogenous circRNAs generally lead to activation of RIG-I (47, 48). In NPC, EBV infection is tightly associated with RIG-I–mediated inflammation (28, 49). EBV-encoded EBER1 (50), miR-BART6-3p (30), and LMP1 (29) regulate the activity of RIG-I in NPC. In this study, circBART2.2, as an exogenous circRNA of host cells, could bind with RIG-I protein to activate PD-L1 and promote tumor immune escape. Further studies are needed to determine whether circBART2.2 can affect other biological phenotypes of NPC through RIG-I.
The siRNAs for circBART2.2 were designed to span the circular splice sites of exons IV and IIIA of the BART gene, which is specific only to circular forms, and BART mRNA cannot be amplified. Theoretically, both circBART2.1 and circBART2.2 could be knocked down because they share the same back splice site. However, our data showed that only circBART2.2 was highly expressed, and circBART2.1 was not or very lowly expressed in NPC cells. Overexpression of circBART2.1 in NPC did not regulate the expression of PD-L1. RNA pulldown and RIP assays showed that the 114 to 165 nt region (DEL3) of circBART2.2 was the critical region to bind to RIG-I. This 114 to 165 nt region is unique to the circBART2.2 sequence. Therefore, we speculate that circBART2.1 may have little influence on the regulation of PD-L1 through the RIG-I pathway in NPC.
In conclusion, we identified the functions of circBART2.2 in NPC immune escape for the first time. As an exogenous circRNA, circBART2.2 promoted PD-L1 expression by binding to RIG-I protein and activating the RIG-I pathway, leading to the immune escape of NPC cells. The interplay between circBART2.2 and RIG-I has important implications for EBV-associated NPC therapy. Targeting circBART2.2 therapy generates a superior antitumor immune response that is critically dependent on PD-L1 expression, highlighting the role of circBART2.2 in the antitumor immunity of EBV-infected NPC, which will help establish a novel approach to efficiently reprogram or restore protective antitumor immunity for NPC immunotherapy.
Authors' Disclosures
No disclosures were reported.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Natural Science Foundation of China (81772928, 81803025, 81972776, and U20A20367) and the Natural Science Foundation of Hunan Province (2019JJ50872). The authors thank Professors George Sai Wah Tsao, Xin Li, and Lunquan Sun for providing EBV-positive cell lines.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Cancer Res 2021;81:5074–88
Authors' Contributions
J. Ge: Conceptualization, resources, data curation, software, formal analysis, visualization, methodology, writing–original draft. J. Wang: Resources, data curation, formal analysis. F. Xiong: Resources, data curation. X. Jiang: Resources, data curation. K. Zhu: Resources, data curation. Y. Wang: Software, formal analysis. Y. Mo: Resources, data curation. Z. Gong: Data curation. S. Zhang: Formal analysis. Y. He: Software, formal analysis. X. Li: Formal analysis. L. Shi: Formal analysis. C. Guo: Data curation. F. Wang: Data curation. M. Zhou: Formal analysis. B. Xiang: Data curation. Y. Li: Formal analysis, writing–original draft. G. Li: Formal analysis, writing–original draft. W. Xiong: Writing–original draft, writing–review and editing. Z. Zeng: Conceptualization, supervision, funding acquisition, validation, project administration, writing–review and editing.
References
- 1. Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer 2019;18:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ren D, Hua Y, Yu B, Ye X, He Z, Li C, et al. Predictive biomarkers and mechanisms underlying resistance to PD1/PD-L1 blockade cancer immunotherapy. Mol Cancer 2020;19:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ma BBY, Lim WT, Goh BC, Hui EP, Lo KW, Pettinger A, et al. Antitumor activity of nivolumab in recurrent and metastatic nasopharyngeal carcinoma: an international, multicenter study of the mayo clinic phase 2 consortium (NCI-9742). J Clin Oncol 2018;36:1412–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity 2018;48:434–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fan C, Zhang S, Gong Z, Li X, Xiang B, Deng H, et al. Emerging role of metabolic reprogramming in tumor immune evasion and immunotherapy. Sci China Life Sci 2021;64:534–47. [DOI] [PubMed] [Google Scholar]
- 6. Wei F, Wang D, Wei J, Tang N, Tang L, Xiong F, et al. Metabolic crosstalk in the tumor microenvironment regulates antitumor immunosuppression and immunotherapy resisitance. Cell Mol Life Sci 2021;78:173–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu C, Li M, Meng H, Liu Y, Niu W, Zhou Y, et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci 2019;62:640–7. [DOI] [PubMed] [Google Scholar]
- 8. Tang T, Yang L, Cao Y, Wang M, Zhang S, Gong Z, et al. LncRNA AATBC regulates Pinin to promote metastasis in nasopharyngeal carcinoma. Mol Oncol 2020;14:2251–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tu C, Zeng Z, Qi P, Li X, Guo C, Xiong F, et al. Identification of genomic alterations in nasopharyngeal carcinoma and nasopharyngeal carcinoma-derived Epstein-Barr virus by whole-genome sequencing. Carcinogenesis 2018;39:1517–28. [DOI] [PubMed] [Google Scholar]
- 10. Tu C, Zeng Z, Qi P, Li X, Yu Z, Guo C, et al. Genome-wide analysis of 18 Epstein-Barr viruses isolated from primary nasopharyngeal carcinoma biopsy specimens. J Virol 2017;91:e00301–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiong F, Deng S, Huang HB, Li XY, Zhang WL, Liao QJ, et al. Effects and mechanisms of innate immune molecules on inhibiting nasopharyngeal carcinoma. Chin Med J 2019;132:749–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu Y, Wei F, Tang L, Liao Q, Wang H, Shi L, et al. Herpesvirus acts with the cytoskeleton and promotes cancer progression. J Cancer 2019;10:2185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao J, Guo C, Xiong F, Yu J, Ge J, Wang H, et al. Single cell RNA-seq reveals the landscape of tumor and infiltrating immune cells in nasopharyngeal carcinoma. Cancer Lett 2020;477:131–43. [DOI] [PubMed] [Google Scholar]
- 14. Wu Y, Wang D, Wei F, Xiong F, Zhang S, Gong Z, et al. EBV-miR-BART12 accelerates migration and invasion in EBV-associated cancer cells by targeting tubulin polymerization-promoting protein 1. FASEB J 2020;34:16205–23. [DOI] [PubMed] [Google Scholar]
- 15. Wang D, Zeng Z, Zhang S, Xiong F, He B, Wu Y, et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell proliferation through the LOC553103-STMN1 axis. FASEB J 2020;34:8012–27. [DOI] [PubMed] [Google Scholar]
- 16. He B, Li W, Wu Y, Wei F, Gong Z, Bo H, et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis 2016;7:e2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, Lee N, et al. Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci U S A 2018;115:E8737–E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li P, Zhu K, Mo Y, Deng X, Jiang X, Shi L, et al. Research progress of circRNAs in head and neck cancers. Front Oncol 2021;11:616202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y, Mo Y, Peng M, Zhang S, Gong Z, Yan Q, et al. The influence of circular RNAs on autophagy and disease progression. Autophagy 2021:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan CM, Wang JP, Tang YY, Zhao J, He SY, Xiong F, et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci 2019;110:2180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer 2020;19:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018;17:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan C, Qu H, Xiong F, Tang Y, Tang T, Zhang L, et al. CircARHGAP12 promotes nasopharyngeal carcinoma migration and invasion via ezrin-mediated cytoskeletal remodeling. Cancer Lett 2021;496:41–56. [DOI] [PubMed] [Google Scholar]
- 24. Tang L, Xiong W, Zhang L, Wang D, Wang Y, Wu Y, et al. circSETD3 regulates MAPRE1 through miR-615-5p and miR-1538 sponges to promote migration and invasion in nasopharyngeal carcinoma. Oncogene 2021;40:307–21. [DOI] [PubMed] [Google Scholar]
- 25. Liu Q, Shuai M, Xia Y. Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manag Res 2019;11:8023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang JT, Chen JN, Gong LP, Bi YH, Liang J, Zhou L, et al. Identification of virus-encoded circular RNA. Virology 2019;529:144–51. [DOI] [PubMed] [Google Scholar]
- 27. Nahand JS, Jamshidi S, Hamblin MR, Mahjoubin-Tehran M, Vosough M, Jamali M, et al. Circular RNAs: new epigenetic signatures in viral infections. Front Microbiol 2020;11:1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duan Y, Li Z, Cheng S, Chen Y, Zhang L, He J, et al. Nasopharyngeal carcinoma progression is mediated by EBER-triggered inflammation via the RIG-I pathway. Cancer Lett 2015;361:67–74. [DOI] [PubMed] [Google Scholar]
- 29. Xu C, Sun L, Liu W, Duan Z. Latent membrane protein 1 of Epstein-Barr virus promotes RIG-I degradation mediated by proteasome pathway. Front Immunol 2018;9:1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu Y, Qin Z, Wang J, Zheng X, Lu J, Zhang X, et al. Epstein-Barr virus miR-BART6-3p inhibits the RIG-I pathway. J Innate Immun 2017;9:574–86. [DOI] [PubMed] [Google Scholar]
- 31. Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J, et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun 2015;6:7353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y, et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J Cancer 2018;9:2852–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, et al. Identification of microRNAs of the herpesvirus family. Nat Methods 2005;2:269–76. [DOI] [PubMed] [Google Scholar]
- 34. Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science 2004;304:734–6. [DOI] [PubMed] [Google Scholar]
- 35. Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, Cao S, et al. The Epstein Barr virus circRNAome. PLoS Pathog 2018;14:e1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gong LP, Chen JN, Dong M, Xiao ZD, Feng ZY, Pan YH, et al. Epstein-Barr virus-derived circular RNA LMP2A induces stemness in EBV-associated gastric cancer. EMBO Rep 2020;21:e49689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thompson ED, Zahurak M, Murphy A, Cornish T, Cuka N, Abdelfatah E, et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut 2017;66:794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carbone A, Gloghini A, Carlo-Stella C. Are EBV-related and EBV-unrelated Hodgkin lymphomas different with regard to susceptibility to checkpoint blockade? Blood 2018;132:17–22. [DOI] [PubMed] [Google Scholar]
- 39. Bi XW, Wang H, Zhang WW, Wang JH, Liu WJ, Xia ZJ, et al. PD-L1 is upregulated by EBV-driven LMP1 through NF-kappaB pathway and correlates with poor prognosis in natural killer/T-cell lymphoma. J Hematol Oncol 2016;9:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anastasiadou E, Stroopinsky D, Alimperti S, Jiao AL, Pyzer AR, Cippitelli C, et al. Epstein-Barr virus-encoded EBNA2 alters immune checkpoint PD-L1 expression by downregulating miR-34a in B-cell lymphomas. Leukemia 2019;33:132–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yoon CJ, Chang MS, Kim DH, Kim W, Koo BK, Yun SC, et al. Epstein-Barr virus-encoded miR-BART5-5p upregulates PD-L1 through PIAS3/pSTAT3 modulation, worsening clinical outcomes of PD-L1-positive gastric carcinomas. Gastric Cancer 2020;23:780–95. [DOI] [PubMed] [Google Scholar]
- 42. Goodman A, Patel SP, Kurzrock R. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat Rev Clin Oncol 2017;14:203–20. [DOI] [PubMed] [Google Scholar]
- 43. Jiang M, Zhang S, Yang Z, Lin H, Zhu J, Liu L, et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 2018;173:906–19. [DOI] [PubMed] [Google Scholar]
- 44. Antonangeli F, Natalini A, Garassino MC, Sica A, Santoni A, Di Rosa F. Regulation of PD-L1 expression by NF-kappaB in cancer. Front Immunol 2020;11:584626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ritprajak P, Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol 2015;51:221–8. [DOI] [PubMed] [Google Scholar]
- 46. Wang W, Chapman NM, Zhang B, Li M, Fan M, Laribee RN, et al. Upregulation of PD-L1 via HMGB1-activated IRF3 and NF-kappaB contributes to UV radiation-induced immune suppression. Cancer Res 2019;79:2909–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell 2019;76:96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, et al. Sensing self and foreign circular RNAs by intron identity. Mol Cell 2017;67:228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jangra S, Yuen KS, Botelho MG, Jin DY. Epstein-Barr virus and innate immunity: friends or foes? Microorganisms 2019;7:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheng S, Li Z, He J, Fu S, Duan Y, Zhou Q, et al. Epstein-Barr virus noncoding RNAs from the extracellular vesicles of nasopharyngeal carcinoma (NPC) cells promote angiogenesis via TLR3/RIG-I-mediated VCAM-1 expression. Biochim Biophys Acta Mol Basis Dis 2019;1865:1201–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available from the corresponding authors upon reasonable request.