Abstract
Histone deacetylase inhibitors (HDACi) induce hyperacetylation of histones by blocking HDAC catalytic sites. Despite regulatory approvals in hematological malignancies, limited solid tumor clinical activity has constrained their potential, arguing for better understanding of mechanisms of action (MOA). Multiple activities of HDACis have been demonstrated, dependent on cell context, beyond the canonical induction of gene expression. Here, using a clinically relevant exposure duration, we established DNA damage as the dominant signature using the NCI-60 cell line database and then focused on the mechanism by which hyperacetylation induces DNA damage. We identified accumulation of DNA–RNA hybrids (R-loops) following romidepsin-induced histone hyperacetylation, with single-stranded DNA (ssDNA) breaks detected by single-cell electrophoresis. Our data suggest that transcription-coupled base excision repair (BER) is involved in resolving ssDNA breaks that, when overwhelmed, evolve to lethal dsDNA breaks. We show that inhibition of BER proteins such as PARP will increase dsDNA breaks in this context. These studies establish accumulation of R-loops as a consequence of romidepsin-mediated histone hyperacetylation. We believe that the insights provided will inform design of more effective combination therapy with HDACis for treatment of solid tumors.
Implications:
Key HDAC inhibitor mechanisms of action remain unknown; we identify accumulation of DNA–RNA hybrids (R-loops) due to chromatin hyperacetylation that provokes single-stranded DNA damage as a first step toward cell death.
Introduction
Histone deacetylase inhibitors (HDACi) were introduced to the laboratory as differentiating agents in 1975, with the observation that the addition of sodium butyrate could increase the production of fetal hemoglobin in erythroleukemia cells (1). Differentiation was recognized in multiple different cell types, including colon cancer and poorly differentiated thyroid adenocarcinoma (2,3,4), and linked to increased histone acetylation (5) and inhibition of HDACs. Once introduced to the clinic, the agents were found to have little activity in solid tumors, but a dramatic and sometimes long-lasting effect in T-cell lymphomas. This led to FDA approvals in cutaneous T-cell lymphomas (CTCL) and peripheral T-cell lymphomas (PTCL) (6,7,8), and multiple myeloma (MM) (9). Despite success in the clinic, the mechanism by which HDACis induce differentiation and cell death is not fully understood.
Gene transcription is regulated in part by chromatin accessibility. Acetyl groups added to positively charged lysine residues via the activity of histone acetyl transferase enzymes interfere with binding of negatively charged DNA, resulting in a more accessible chromatin structure that favors transcription (10). This process is then reversed by the activity of HDAC enzymes that encourage chromatin compaction (11). The classical description of the mechanism of action for HDACis is that the increased histone acetylation opens chromatin, leading to differentiation. However, many other mechanisms of action of HDACis have been described, with diverse mechanisms, including protein acetylation, direct induction of apoptosis, and DNA damage (11, 12).
DNA damage has long been observed following the administration of HDACis, but the mechanism by which that damage occurs is not known. Explanations include transcriptional downregulation of several repair proteins such as RAD50, RAD51, RAD52, BRCA1, CHK1, KU70, KU80, MRE11, as well as impaired recruitment or acetylation of repair proteins such as P53 and KU70 (11, 13). The administration of HDACis has been shown to result in double-strand break (DSB), and this has been ascribed to the generation of reactive oxygen species or alterations in chromatin structure (11, 14, 15).
In the data that follow, we evaluated the dominant mechanism of action, among the multiple proposed. We begin with results from our investigation on the mechanism of action of romidepsin using CellMiner (http://discover.nci.nih.gov/cellminer) to probe the NCI-60 cell line database where we observed a striking DNA-damage signature.
We then investigated the mechanism by which DNA damage is caused by HDACis, using romidepsin (also called depsipeptide, NSC 630176 or FK228) as our model HDACi. We hypothesized that a unique form of DNA damage occurs preferentially at sites that are actively transcribed, and genes are “trapped” by histone modifications due to the activity of HDACis and prolonged chromatin accessibility. We further hypothesized that DNA damage at sites of active transcription would be associated with R-loops, three strand nucleic acid structures that form during active gene transcription, where the nascent RNA is hybridized to its complementary DNA sequence leaving displaced and vulnerable single-stranded DNA (ssDNA). R-loops are known to be sources of genome instability and sites of DNA damage. They have also been reported to form at other cellular events such as replication and immunoglobulin class-switch recombination (16,17,18). Here, we show that the hyperacetylation of histone lysine residues that follows romidepsin exposure provokes the formation of R-loops, a molecular structure giving rise to the initiation of damage to DNA in the form of single-stranded breaks (SSB). When R-loops are not resolved and SSB are not repaired, DNA damage evolves to DSBs and ultimately cell death.
Materials and Methods
Reagents and cell culture
Romidepsin (depsipeptide, NSC 630176) was obtained from the National Cancer Institute Anticancer Drug Screen (Bethesda, MD) and from (Sigma-Aldrich Cat # SML1175) and Olaparib from (Selleckchem Cat # S1060).
Cell lines from the NCI-60 were obtained from the NCI Anticancer Drug Screen, these cell lines are regularly authenticated by the NCI; HUT-78 and LOXIMVI were purchased from the ATCC. Cell line Authentication and Mycoplasma testing was performed yearly at the Johns Hopkins University Genetic Resources Core Facility (GRCF). Routine Mycoplasma testing was also performed using MycoAlert Mycoplasma detection kit (Lonza, Cat#LT07–118). Cells were cultured in appropriate cell culture medium (Gibco Laboratories) supplemented with 10% FBS (Gibco Laboratories), and 1% glutamine (Gibco Laboratories) and cell lines were passaged at a maximum of 30 passages between thawing and using in experiments.
RNaseH1-expressing LOXIMVI cells were generated by transfecting cells with 2 μg of plasmid DNA using FuGENE HD Transfection Reagent (Lonza, Cat # E2311) according to the manufacturers' instructions, using pFRT-TODestGFP_RNAseH1 plasmid, purchased from Addgene (Addgene, Cat # 65784). Following transfection GFP+ cells were selected by cell sorting using BD Influx cell sorter (BD Biosciences) and stable selection with 0.5 to 1 μg mL−1 blasticidine and clonal selection.
Annexin V assay
Apoptosis was measured as previously described (19) using the Annexin V-fluorescein isothiocyanate (Annexin V-FITC) Apoptosis Detection Kit (BD Biosciences) according to the manufacturer's instructions. Following treatment, Annexin-positive cells were quantitated by FACS analysis (Becton Dickinson) using FlowJo Software (Tree Star, Inc.) and FCS Express 7 Software (De Novo Software Inc.). The percentage of Annexin positive was calculated and the delta between treated and untreated control was determined.
Pattern comparison and statistical analysis
To perform pattern comparison, a “pattern comparison input template file” was downloaded from the CellMiner website: http://discover.nci.nih.gov/cellminer/. Compared with the untreated, the percentage of increase in cell death following romidepsin treatment was used as the input pattern of interest. “Pattern in 60 element array” was selected as the type of input. We performed a two-sided Fisher's exact test (alpha 0.05) for each mechanism, comparing the proportion of the experimental data having a correlation greater than 0.35 with the proportion of the given mechanism out of the total with known mechanisms in the CellMiner database.
Microarray gene expression analysis
HUT-78, LOXIMVI, M14, A549, MDA231, ACHN, and PC3 cell lines were treated with 25 ng/mL (46 nmol/L) of romidepsin for 6 hours. Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. RNA quality was assessed on Agilant Bioanalyzer (Agilent Technologies). Global gene expression analysis was performed at the NCI-Frederick LMT Microarray Center (Frederick, MD) according to protocol using the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array covering +47.000 transcripts. Microarray gene expression data were summarized by applying the RMA (robust multi-array average) and quantile normalization workflow as implemented in the Expression Console 1.4 software (Affymetrix). Differentially expressed genes (DEG) were identified by ANOVA, and significance was adjusted for multiple testing by estimating FDRs (20). Data were visualized in Qlucore Omics Explorer v. 3.4 (Qlucore AB), including principal component analysis (PCA), heat maps, and unsupervised hierarchical clustering. DNA repair gene lists were pulled from the KEGG Pathway database (https://www.genome.jp/kegg/pathway.html) and from the Molecular Signatures Database v6.2 (MSigDB) at BROAD Institute (http://software.broadinstitute.org/gsea/msigdb/index.jsp). The expression data are deposited in Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE133120.
Western blotting
Western blotting was carried out as previously described (19). Membranes were incubated with primary antibodies: Anti–phospho-histone H2A.X (Ser139) antibody, clone JBW301(1:1,000, Millipore Sigma Cat # 05–636), FANCD2, FI17 (1:500, Santa Cruz Biotechnology Cat # sc-20022), GAPDH (1:10,000, Abcam Cat # ab8245); and secondary antibodies IRDye 680RD Goat anti-Mouse (LI-COR Biosciences Cat #P/N 925–68070)- IRDye 800CW Goat anti-Rabbit (LI-COR Biosciences #P/N 925–32211). Signal quantitated using the Odyssey CLx imaging system (LI-COR Biosciences), and various exposures within the linear range captured using ImageStudio software V3.1 (LI-COR Biosciences).
Comet assay
DNA SBs were analyzed using a commercial comet assay (Trevigen, Inc.) following the manufacturer's protocol. Slides from both alkaline and neutral conditions were stained with Vista Green DNA Dye (CELL BIOLABS, INC.) according to the manufacturer's recommendations. Alkaline and neutral assay control cells (Trevigen Cat # 4256–010-CC and # 4257–010-NC, respectively) were used as DNA-damage controls. Images of cells in different fields of view were collected using EVOS FL Auto microscope (Life Technologies). At least 50 cells analyzed per sample. Tail moments (= tail length × DNA in the tail/total DNA) were analyzed using the Tritek Comet Score software.
Immunocytochemistry of R-loops
Immunofluorescence using S9.6 antibody (Kerafast Inc.) was performed as previously described (21). Cells were incubated with primary antibodies, anti-DNA–RNA hybrid (S9.6) antibody (1:500, Kerafast Cat # ENH001), anti-nucleolin antibody (1:1,000, Abcam Cat # ab22758), anti-PARP1 antibody (1:1,000, Abcam Cat # ab32138 and 1:500 Cat # ab191217), anti-XRCC1 antibody (1:1,000, Cell Signaling Technology Cat #27350), anti-DNA Polymerase beta antibody (1:1,000, Abcam Cat # ab26343), anti-CYCLIN A antibody (1:200, Sigma Cat #C4710), and anti-RPA70/RPA1 antibody (1:50, Cell Signaling Technology Cat #2267S) followed by incubation in secondary antibodies Alexa Fluor 488 goat anti-mouse (1:1,000, Invitrogen Cat #A11034) or Alexa Fluor 594 goat anti-rabbit (1:1,000, Invitrogen Cat #A11005). DNA was stained with DAPI. Images were captured at ×40 magnification with a Zeiss Confocal Microscope (Zeiss LSM 700). Fiji software (version: 2.0.0-rc-69/1.52i, open source image processing software) used for analysis of images. As a control, cells were treated with 10 U of RNase H enzyme (New England Biolabs Cat # M0297L), before staining with S9.6 antibody.
siRNA transfection assay
On-TargetPlus siRNAs and non-targeting control siRNA were purchased from GE Healthcare Dharmacon Inc. For transfection, 200,000 cells were seeded per well in a 6-well plate and transfected using DharmaFECT 1 Transfection Reagent (GE Healthcare Dharmacon Inc., Cat #T-2001–01) according to the manufacturer's instructions, using 25 nmol/L siRNA in a final volume of 2 mL of culture medium without antibiotics. A pool of four different siRNAs was used. A non-targeting siRNA pool and GAPD siRNA pool (GE Healthcare Dharmacon) were used as controls. The target sequences were:
FANCD2:
5′UGGAUAAGUUGUCGUCUAU3′
5′CAACAUACCUCGACUCAUU3′
5′GGAUUUACCUGUGAUAAUA3′
5′GGAGAUUGAUGGUCUACUA3′
Nontargeting control pool:
5′UGGUUUACAUGUCGACUAA3′
5′UGGUUUACAUGUUGUGUGA3′
5′UGGUUUACAUGUUUUCUGA3′
5′UGGUUUACAUGUUUUCCUA3′
GAPD Control Pool:
5′UGGUUUACAUGUCGACUAA3′
5′UGGUUUACAUGUUGUGUGA3′
5′UGGUUUACAUGUUUUCUGA3′
5′UGGUUUACAUGUUUUCCUA3′
DNA–RNA immunoprecipitation
DNA–RNA immunoprecipitation (DRIP) assay was performed in LOXIMVI and siFANCD2-transfected LOXIMVI cells according to the method described earlier (22). Briefly, following gentle extraction of the genomic DNA, cells were subjected to digestion using a cocktail of restriction enzymes, treated with or without RNaseH (New England Biolabs Cat # M0297L). DNA–RNA hybrids were then immunoprecipitated from the digested genomic DNA using S9.6 antibody (Kerafast Inc.). Quantitative PCR was performed at the R-loop–positive loci RPL13A, CALM3, and TFPT genes and R-loop–negative locus SNRPN gene with the primers listed in Supplementary Table S1. Means and SEM from three independent experiments were calculated.
ChIP-qPCR
The chromatin immunoprecipitation (ChIP) assay was performed on LOXIMVI cells using the Zymo-Spin ChIP kit (Zymo Research) according to the manufacturer's instructions. Following treatment for 6 hours with romidepsin, cells were harvested, and chromatin was cross-linked using formaldehyde and sheared mechanically by sonication on ice for four 30 second cycles, at 40% amplitude using a sonic dismembrator model 500 (Thermo Fisher Scientific). Protein–DNA complexes were precipitated using control IgG; normal Rabbit IgG (Cell Signaling Technology #2729), anti–acetyl-histone H3 (Lys9; C5B11; 1:50, Cell Signaling Technology #9649S), and anti-RPA70/RPA1 (1:25, Cell Signaling Technology #2267S) antibodies, followed by elution, reverse cross-linking and purification of the ChIP-DNA. Quantitative PCR was performed in technical triplicates using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Cat # 1725272) and primers for the RPL13A, CALM3, TFPT, and SNRPN genes, with the primers listed in Supplementary Table S1. ChIP-qPCR signals were calculated as the percentage of input.
Results
Different sensitivity spectrum of the NCI drug screen collection in 6- versus 48-hour assays
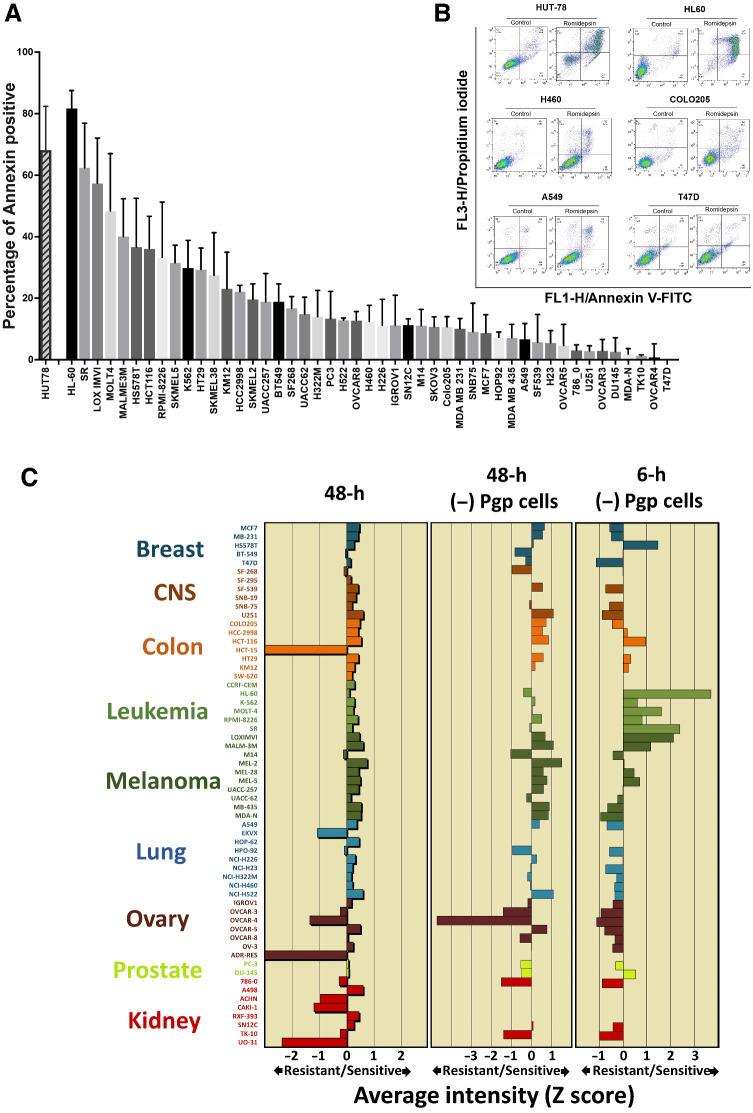
The CellMiner analytic tools in the NCI drug screen allow the inference of a dominant mechanism of action of anticancer agents, based on signatures of drug activity. To generate data for CellMiner, we studied the sensitivity of the NCI-60 collection of cell lines exposing cells to 25 ng/mL romidepsin for 6 hours followed by a 42-hour incubation in romidepsin-free medium. A 6-hour exposure duration was chosen to better emulate the clinical exposure achieved with the 4-hour infusion of a drug that has a 3.5-hour half-life (19, 23, 24). We performed the short-term exposure assay on cell lines of the NCI-60, avoiding those known to express P-glycoprotein, a mechanism of resistance to romidepsin (25,26,27). We included HUT-78 cells as a reference, as our previous studies showed this cell line to be markedly sensitive to HDACis (19). Figure 1A shows a range of sensitivity across the remaining NCI-60 cell lines as well as HUT-78 using a 6-hour treatment platform with HL-60, SR and LOXIMVI cell lines most sensitive and TK10, OVCAR4, and T47D least sensitive to romidepsin among NCI-60 cells determined by Annexin staining. Representative histograms of quantitating Annexin staining are shown in Fig. 1B.
Figure 1.
Different sensitivity spectrum of the NCI drug screen collection in 6- versus 48-hour assays. A, Analysis of apoptotic cell death in NCI-60 cell lines and HUT-78 following romidepsin treatment. Annexin V–positive cells were determined by flow cytometry. Results are baseline mean ± SD from three independent experiments. Each bar represents percentage of increase in cell death compared with untreated control. B, Representative histograms quantitating Annexin staining. C, A graphical representation of z-score of sensitivity of tumor cells to romidepsin following treatment. Drug sensitivity profiles in the NCI-60 cell lines plotted as mean graphs. In these profiles, the mean IC50 value for all cell lines is plotted as the center line. Then the difference in IC50 value for each cell line from the mean is plotted, as Z-scores, with the bars to the left indicating resistance and the bars to the right indicating sensitivity. Left, mean graph showing results following treatment with romidepsin in the conventional NCI-60 48-hour assay. Middle, eliminating cell lines previously shown to express Pgp (SF-295, HCT-15, SW-620, CCRF-CEM, EKVX, HOP-62, NCI-ADR-RES, A498, ACHN, CAKI-1, RXF-393, UO-31) in the 48-hour assay. Right, mean graph showing results of a 6-hour drug exposure after removing the Pgp-expressing cells.
The NCI-60 database includes data from over 50,000 compounds tested on a 48-hour exposure platform in which drugs with similar dominant mechanisms of action (MOA) generate highly correlated sensitivity profiles. Using pattern comparison analyses, the CellMiner platform identifies highly correlating profiles, generating insights into potential mechanisms of sensitivity and resistance (28, 29). The sensitivity profile obtained from the 48-hour assay in the complete NCI-60 cell line panel is shown in the left of Fig. 1C. Pgp-expressing cell lines show marked resistance to romidepsin and drive the overall mean graph; the mean graph without Pgp-expressing lines is shown in the middle. We used the 6-hour exposure to achieve a greater dynamic range among the cell lines, finding it more relevant to the romidepsin clinical profile, with sensitivity in the leukemia and melanoma cell lines (Fig. 1C, right).
Romidepsin 6-hour drug sensitivity profile in the NCI-60 database is highly correlated with that of DNA-damaging agents
Using the greater dynamic range derived from the more clinically relevant 6-hour treatment, we determined correlations in CellMiner with agents with known or proposed MOA (28, 29). Among the 349 compounds with assigned MOA in the NCI-60 database, 80 correlated with the cell sensitivity profile of the 6-hour treatment at a level considered statistically meaningful—Pearson correlation coefficient values r > 0.3. Supplementary Fig. S1A shows the top 27 correlating compounds including methyl-CCNU, carmustine, and multiple other alkylating agents and topoisomerase inhibitors. This profile differed markedly from that found with romidepsin in the 48-hour database (30). Because some MOA are overrepresented in the NCI-60 database, we asked whether the high fraction of DNA-damaging agents among the correlating compounds could simply reflect their high representation in the database. We found the A7 alkylating agents significantly overrepresented in our dataset. HDACis, including vorinostat, were also identified, in the set of compounds with r>0.3. The MOA identified for these hits are plotted in Supplementary Fig. S1B. This analysis suggests the dominant molecular intermediate responsible for romidepsin sensitivity is DNA damage.
Gene expression profiling shows impact on DNA repair genes
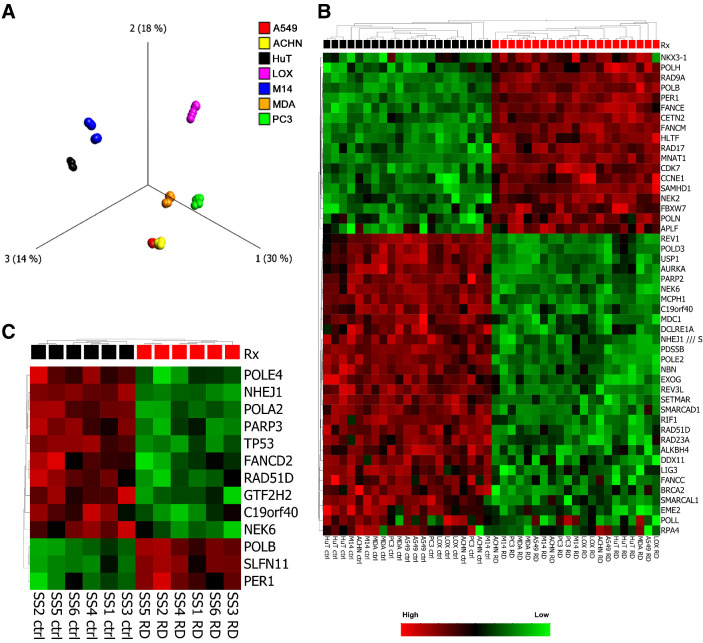
Downregulation of DNA repair gene expression has been suggested as having a role in DNA damage observed with romidepsin (11, 13). We performed a series of Affymetrix cDNA microarray experiments to evaluate gene expression changes in 7 cell lines following treatment with romidepsin for 6 hours. These cell lines were chosen for their spectrum of response to romidepsin, and included HUT-78, LOXIMVI, M14, A549, MDA231, ACHN, and PC3. Initial results demonstrated that, as previously seen in patient samples (7), cell context was the dominant factor in determining the gene expression profile.
A PCA revealed cell lines, not treatment effect as the dominant difference (variance) in gene expression (Fig. 2A). This is also illustrated in a 2-way unsupervised hierarchical clustering of the 48 samples and the 1,441 most variably expressed genes showing samples from the same cell lines clustering together (Supplementary Fig. S2). These two results indicate the dominant difference driving gene expression is cell (and tissue) type. Treatment effect emerged as a driver only when cell type was eliminated as a factor in the analysis (comparable with a paired t test, data not shown). Since our earlier analysis as well as previously published data suggested that DNA damage occurs following romidepsin treatment, we analyzed the microarray data for DEG involved in DNA damage repair (DDR) pathways. Figure 2B; Supplementary Fig. S3 show the impact of romidepsin treatment on DDR genes. Supplementary Fig. S3A shows a heat map of 123 DEGs involved in DDR with P values <0.05 and >1.2-fold-change in expression. After removing the cell line as a factor, DEGs were found in every DNA-repair pathway: Homologous recombination repair (HRR), non-homologous end-joining (NHEJ), nucleotide excision repair (NER), base excision repair (BER), and mismatch repair (MMR). Figure 2B shows the DEGs after applying a more stringent cutoff value of >1.5-fold change in expression. We found downregulation of genes involved in the HRR and NHEJ pathways, including BRCA2, EME2, RAD51D, NBN, NHEJ1, as well as in BER and NER pathways, including POLD2, POLE2, LIG3, and RPA4.
Figure 2.
Gene expression profiling shows impact on DNA repair genes. A, PCA plot showing that samples separate primarily into cell-specific clusters, rather than by treatment. The PCA plot is based on the 1851 probe sets that were found to be most variable across samples [applying a Variance filter of 0.4 sigma/sigma max (s/smax) >0.4 to maximize the projection score]. Samples are colored according to the cell line. B, Heat map and unsupervised 2-way hierarchical clustering of DEGs involved in DNA damage repair (DDR) pathways. Clustering is based on 47 DNA repair genes (out of 252) with P < 0.05, and expression fold change of >1.5-fold. Cell line removed as factor. Primary clustering according to romidepsin 6 hours Rx. C, Heat map and unsupervised 2-way hierarchical clustering of DEGs involved in DDR pathways from patients with Sezary syndrome treated with romidepsin for 24 hours. Clustering is based on 13 DNA repair genes (out of 252) with P < 0.05, and expression fold change of >2-fold.
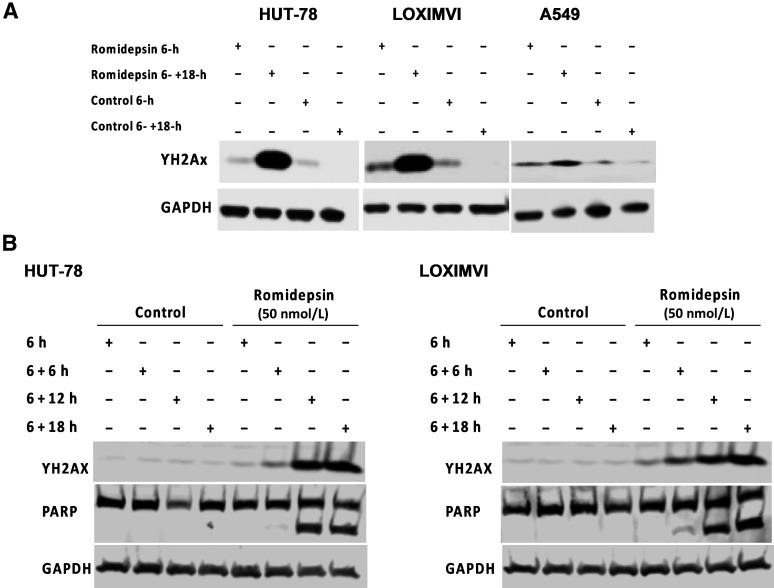
Figure 3.
γ-H2AX signal increases following romidepsin treatment. A, The expression of γ-H2AX following treatment with 100 nmol/L romidepsin in HUT-78, LOXIMVI, and A549 cell lines was detected by western blot assay. B, HUT-78 and LOXIMVI cells treated with 50 nmol/L romidepsin for 6 hours followed by incubation in the drug-free medium for additional time periods of (+6/+12/+18 hours). Presence of γ-H2AX and PARP signals were detected by western blot assay.
In addition, we analyzed the data generated from a recent RNA-sequencing analysis of CD4-positive T cells derived from 6-primary tumors in patients with Sezary syndrome treated ex vivo with romidepsin for 24 hours, available on the NCBI Gene Expression Omnibus (GEO) under accession number GSE110248 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE110248). Figure 2C shows DEGs after applying a cutoff value of >2-fold-change in expression confirming the effect of romidepsin treatment on impairing DNA-repair gene expression in samples from patients with Sezary syndrome similar to that observed in our microarray data.
γ-H2AX signal increases following romidepsin treatment
We looked for the presence of phosphorylated H2AX (γ-H2AX) as a marker of DSBs following a short-term (6 hours) treatment of HUT-78, LOXIMVI, and A549 cells with 100 nmol/L romidepsin followed by incubation in drug-free medium for an additional 18 hours (Fig. 3A). In the HUT-78 and LOXIMVI cells, both highly sensitive to romidepsin, we observed a clear increase in the γ-H2AX signal only after additional 18 hours incubation when compared with the vehicle-treated controls. A similar observation was made in the more resistant A549 cells but to a lesser degree, suggesting that DSBs are not the initial event following short-term romidepsin treatment. To monitor the emergence of DSBs (i.e., γ-H2AX signal), we treated the HUT-78 and LOXIMVI cells with 50 nmol/L romidepsin for 6 hours followed by incubation in drug-free medium over a 24-hour period with samples collected every 6-hour (Fig. 3B). Our data showed that until the 12-hour timepoint (6-hour treatment + 6-hour incubation in drug-free medium), there is minimal increase in the γ-H2AX signal in both cell lines tested. Furthermore, we observed that PARP is not cleaved until the 12- or 18-hour timepoint following treatment, suggesting that DSB is not occurring at early hours following treatment with romidepsin, but rather develops gradually over time, finally leading to apoptosis.
R-loop structures form following romidepsin treatment
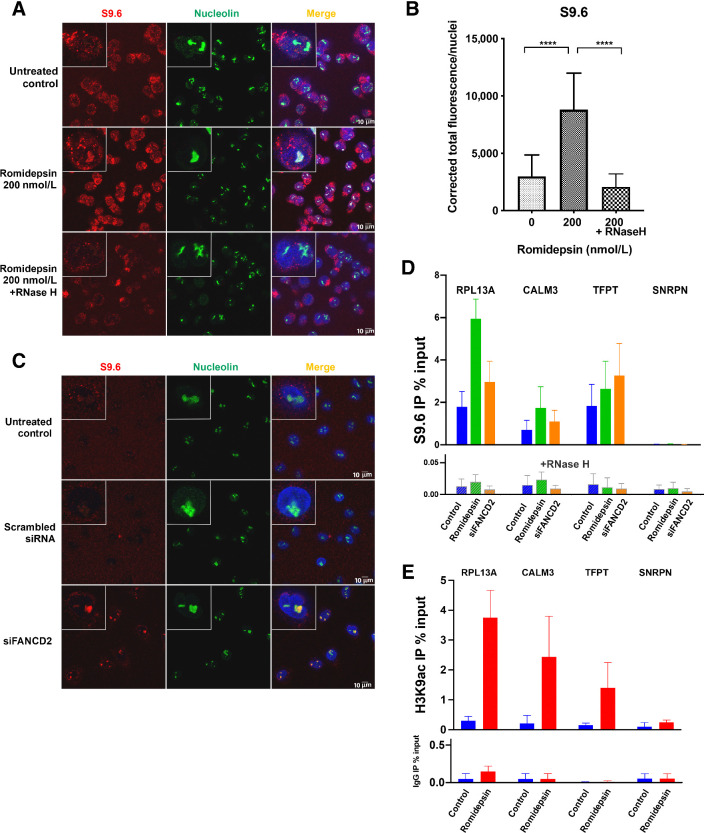
Having observed evidence of DNA damage following romidepsin treatment in the earlier assays, we next asked what mechanism could initiate damage after hyperacetylation of chromatin by romidepsin shortly after the treatment? Found in some actively transcribed regions of DNA, R-loops are three-stranded nucleic acid structures comprising a template DNA hybridized to nascent RNA leaving the non-template ssDNA. Previous studies have shown R-loop accumulation behind stalled elongating RNA polymerases and also during failure in termination of transcription (16). We reasoned that because hyperacetylation of chromatin occurs mostly at transcription start sites where the chromatin structure is open (31, 32), lysine hyperacetylation following treatment with an HDACi could force chromatin to remain open for longer periods resulting in defect in the termination of transcription. This could lead to the accumulation of R-loop structures at stalled transcription bubbles, which if unresolved would result in DNA damage (17). To test these hypotheses, we performed confocal microscopy using an antibody against DNA–RNA hybrids (S9.6) to look at R-loop formation following treatment with romidepsin for 6 hours in the LOXIMVI cell line (Fig. 4). We observed accumulation of R-loops in the nucleus and their co-localization with nucleolin, a ubiquitous nuclear and nucleolar protein that binds DNA–RNA hybrids and, among other functions, is involved in the synthesis of ribosomal RNA and induction of chromatin decondensation (33,34,35,36). We confirmed that R-loops are also identified outside of nucleolar condensates by subtracting the observed nucleolar signal, which does not change after romidepsin treatment (Supplementary Fig. S4). To show that the observed structures were in fact R-loops, we demonstrated their sensitivity to RNase H treatment (Fig. 4A and B). Incubation of romidepsin-treated cells with RNase H before staining with the S9.6 monoclonal antibody resulted in loss of the R-loop structures. As a positive control, we used A549 cells with siRNA knockdown of the FANCD2 gene, an intervention known to induce the formation of R-loop structures (Fig. 4C; Supplementary Fig. S5; ref. 37). To further confirm the accumulation of R-loops following romidepsin treatment, we performed DRIP using the S9.6 antibody for the immunoprecipitation with or without RNase H treatment, followed by qPCR. Gene loci where R-loops have been previously shown to occur during normal transcription were selected for analysis, including the RPL13A, CALM3, and TFPT genes (22, 38). As expected, R-loops were detected in untreated controls in the DRIP assay; increased R-loop accumulation after romidepsin and FANCD2 siRNA was confirmed (Fig. 4D). Romidepsin had the greatest impact at the RPL13A locus; SNRPN negative control locus was similar to the background resulting from RNase H treatment.
Figure 4.
R-loop structures form following romidepsin treatment. A, DNA–RNA hybrid accumulation in romidepsin-treated cells. Staining was carried out using S9.6 and nucleolin antibodies. Nuclei were stained with DAPI. Bottom, treatment with RNase H in the romidepsin-treated cells eliminated the DNA–RNA hybrids. B, Quantification of nuclear S9.6 immunofluorescence signal in LOXIMVI cells treated with or without romidepsin and with RNase H following romidepsin treatment. The S9.6 signal was quantified only in the nuclear regions, DAPI staining. The median of the nuclear S9.6 signal intensity per nucleus is shown. ***, P < 0.001; ****, P < 0.000 (t test, two‐tailed). C, Detection of DNA–RNA hybrids in siFANCD2-transfected A549 cells. Top two demonstrating the untreated control and negative control of siRNA experiment and the bottom is showing positive R-loop staining in siFANCD2 cells. See also Supplementary Fig. S4A. D, DRIP–qPCR signal values at RPL13A, CALM3, TFPT, and SNRPN genes in LOXIMVI cells treated with or without romidepsin for 6 hours. LOXIMVI cells transfected with the FANCD2 siRNAs were included as positive control for R-loops. Cells treated in vitro with (bottom) or without (top) RNase H before immunoprecipitation. The mean ± SEM from three independent experiments is shown. E, ChIP-qPCR was performed with antibody against H3K9ac in the LOXIMVI cells after treatment with romidepsin for 6 hours. Immunoprecipitated chromatin samples were analyzed by qPCR using specific primer pairs as shown on Supplementary Table S1. The mean ± SEM from three independent experiments is shown.
We also performed ChIP-qPCR using H3K9ac antibody to show that R-loop–positive loci from the DRIP experiment, namely RPL13A, CALM3, and TFPT genes were hyperacetylated following 6 hours treatment with romidepsin. As shown in Fig. 4E, all three loci demonstrated increased acetylation in romidepsin-treated samples compared with the control following immunoprecipitation with H3K9ac antibody. In contrast with the positive loci, there was no significant change in acetylation of the R-loop–negative SNRPN locus following romidepsin treatment. Taken together, these data support the thesis that romidepsin-induced hyperacetylation contributes to R-loop formation.
ssDNA breaks following romidepsin treatment
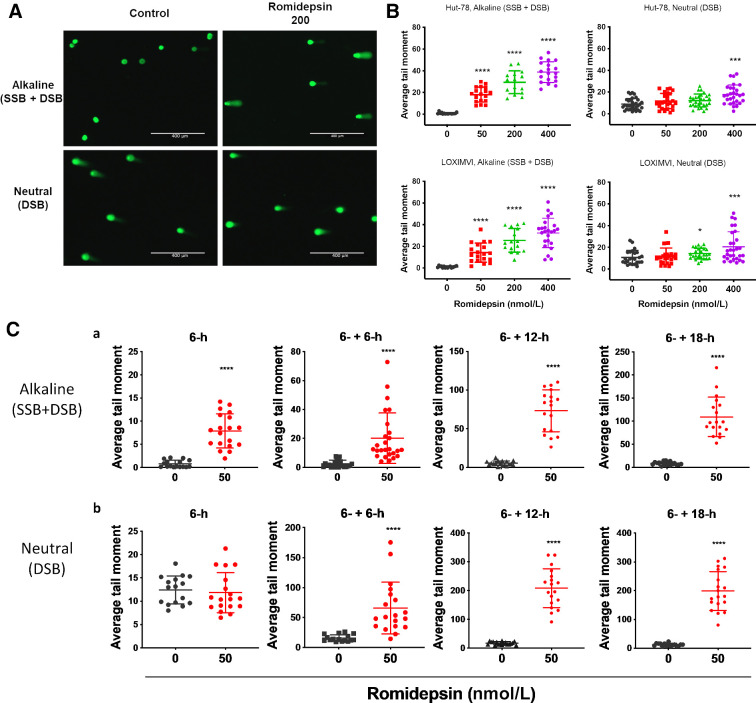
R-loop accumulation, particularly if it remains unresolved, can lead to ssDNA damage. R-loop formation results in a DNA–RNA hybrid that leaves the complimentary ssDNA very sensitive to damage (17). Having observed the formation of R-loops following a 6-hour romidepsin treatment, we hypothesized that we would find damage to ssDNA as the initial lesion following treatment. To test this, we performed a comet assay following a 6-hour treatment with different concentrations of romidepsin. We quantified single and DSBs in the HUT-78 and LOXIMVI cell lines using tandem neutral (pH 8) and alkaline (pH >13) comet assays to detect only DSBs or the sum of SSBs + DSBs, respectively. The results were analyzed by calculating the tail moments, which reflect the extent of DNA breaks in each comet-positive cell. Representative images of the comet assay are shown in Fig. 5A. Following a 6-hour treatment, the tail moments at pH 8 (neutral, DSBs) increased in romidepsin-treated compared with untreated control cells only at a concentration of 400 nmol/L romidepsin, whereas at pH >13 (alkaline, SSBs + DSBs) comet tails increased with increasing romidepsin concentrations beginning at 50 nmol/L (Fig. 5B). We also evaluated the time course, treating cells with 50 nmol/L of romidepsin for 6 hours followed by incubation in drug-free medium over a 24-hour period with sample collection every 6 hours. Similarly, we observed that under alkaline conditions, the increase in the tail moment began at the 6-hour timepoint compared with the vehicle-treated control and expanded over time (Fig. 5C-a). However, under neutral conditions, an increase in the tail moment was observed only after a further 6- to 18-hour incubation in drug-free medium (total 12 to 24 hours; Fig. 5C-b), indicating that romidepsin treatment leads to SSBs initially, with DSBs occurring only at higher concentrations and longer incubation times.
Figure 5.
Single-stranded DNA breaks following romidepsin treatment. DNA damage in HUT-78 and LOXIMVI cells treated with various concentrations of romidepsin for 6 hours was measured by single-cell gel electrophoresis (comet assay). DNA damage examined by fluorescence microscopy. A, Representative images of comet assay. B, Quantitative analysis of DNA damage. Average comet tail moment was calculated for at least 50 cells per sample. Tail moments = tail length × DNA in the tail/total DNA. Significance was calculated based on an unpaired student t test comparing each condition with the control. C, Quantitative analysis of DNA damage in LOXIMVI cells treated with 50 nmol/L romidepsin for 6 hours followed by additional incubation times in drug-free medium. Samples were run in tandem alkaline (A) and neutral (B) assay conditions to determine the type of DNA damage following treatment. *, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (t test, two‐tailed).
R-loop structures co-localize with ssDNA-damage response proteins following romidepsin treatment
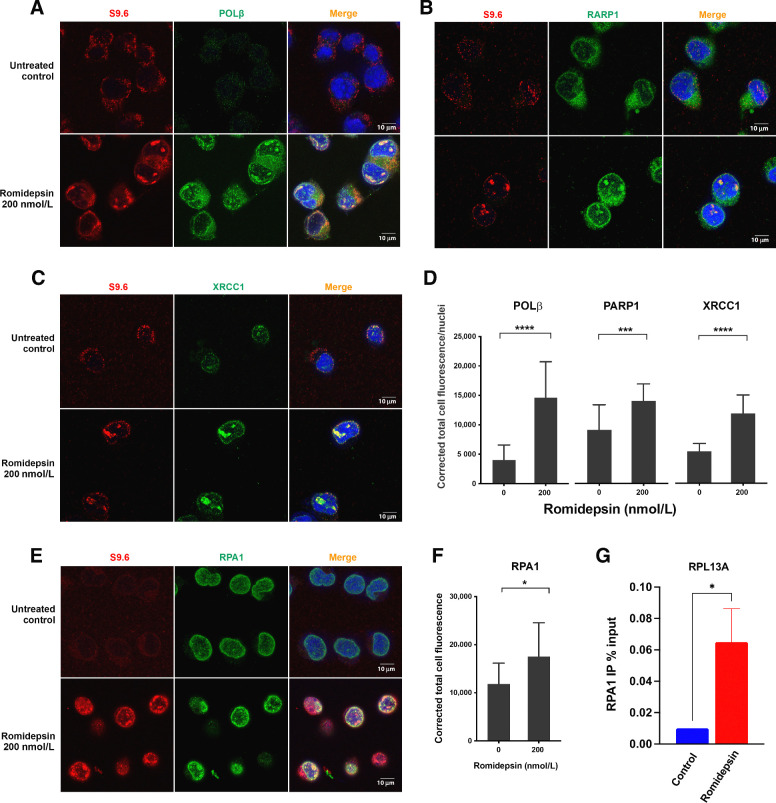
The above was consistent with a hypothesis that envisions hyperacetylation of histone lysin residues leading to stalled transcription complexes and in turn an increase in unscheduled R-loop formation rendering the non-transcribed DNA strand vulnerable to DNA damage. This would initiate the SSBs we observed by comet assay. Hence, we hypothesized that there would be evidence of a cellular response to the SSBs, such as the transcriptional-coupled repair machinery, at the site of R-loop formation. Three pathways that repair SSB are NER, BER, and MMR. Non-transcribed ssDNA in R-loops is likely subject to endogenous DNA damaging agents causing small lesions and AP sites, and BER machinery will be among the initial damage response systems to these lesions. Moreover, we observed upregulation of RNA polymerase beta (Polβ), RAD9A, and CHEK2 genes in our earlier microarray data (Fig. 2; Supplementary Fig. S3A), suggesting the availability of BER pathway proteins. We therefore looked for the presence of several key proteins involved in the BER pathway: PARP1, which binds damaged ssDNA and recruits X-ray repair cross-complementing protein (XRCC1; ref. 39); XRCC1, which recruits downstream repair enzymes to repair SSBs; and polymerase β (POLβ), a DNA polymerase involved in BER (40). We performed confocal microscopy to determine whether any of these proteins accumulated at sites of R-loop formation following romidepsin exposure and observed an increase in the number of foci of POLβ, PARP1, and XRCC1 (Fig. 6A–D). Importantly, as is evident from the images, these foci co-localized with the R-loops. We also found co-localization of R-loops with replication protein A (RPA1), part of a heterotrimeric complex that binds and stabilizes ssDNA (Fig. 6E–F). To test for accumulation of RPA1 and by inference, DNA damage, at the R-loop–positive locus RPL13A following romidepsin treatment, we performed ChIP-qPCR using an antibody against RPA1. As shown in Fig. 6G, an increase in RPA1 binding at the RPL13A locus was detected following treatment with romidepsin. Taken together, these studies support a role for DNA damage and SSB repair in the R-loops induced by romidepsin.
Figure 6.
R-loop structures co-localize with single-stranded DNA damage response proteins following romidepsin treatment. Co-localization of accumulated R-loops in the romidepsin treated cells with SSD repair proteins involved in BER. Nuclei were stained with DAPI. DNA–RNA hybrids accumulated in the nuclei and co-localized with POLβ (A), PARP1 (B), XRCC1 (C), and RPA1 (E) following romidepsin treatment in LOXIMVI cells. D, Quantification of nuclear POLβ, PARP1, and XRCC1 immunofluorescence signal in LOXIMVI cells treated with or without romidepsin for 6 hours. F, Quantification of nuclear RPA1. For each protein, signal was quantified only in the nuclear regions, defined by DAPI staining. The median of the signal intensity per nucleus is shown. ***, P < 0.001; ****, P < 0.000 (t test, two‐tailed). G, ChIP-qPCR was performed with antibody against RPA1 in the LOXIMVI cells after treatment with romidepsin for 6 hours. Immunoprecipitated chromatin samples were analyzed by qPCR using RPL13A primer sets. *, P < 0.03.
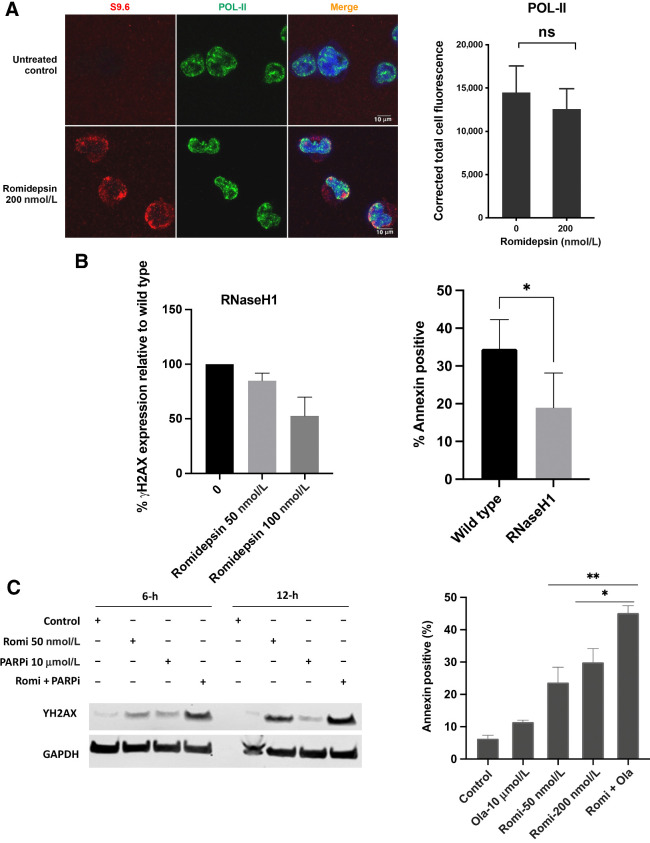
Several additional lines of evidence support a role for R-loop accumulation in the ssDNA damage that follows romidepsin exposure. First, we observed that Pol-II did not accumulate following treatment nor did it co-localize with the R-loops, consistent with the idea that R-loops are accumulating behind the polymerase-II complex during RNA transcription (Fig. 7A; ref. 17). We also reasoned that, following R-loop–induced single-strand breaks, incapacity of BER to deal with excess ssDNA damage could drive damage accumulation in S-phase, as others have shown (41, 42). Accordingly, we co-stained the cells with CYCLIN A, a marker of S phase, and PARP1, and observed that cells with accumulated PARP1 foci also accumulated the CYCLIN A signal indicating accumulation of damage during the S phase in LOXIMVI cells (Supplementary Fig. S6).
Figure 7.
A, Co-localization of accumulated R-loops in the romidepsin treated cells with POL-II. Quantification of nuclear POL-II. Immunofluorescence signal in LOXIMVI cells treated with or without romidepsin for 6 hours. For each protein signal was quantified only in the nuclear regions, defined by DAPI staining. The median of the signal intensity per nucleus is shown; *, P < 0.01; ns, not significant (t test, two‐tailed). B, left, Analysis of γ-H2AX in the RNaseH1-transfected LOXIMVI cell line following romidepsin treatment. γ-H2AX signal was detected by western blot assay and quantified using Image Studio Software, the percentage of decrease in the expression of γ-H2AX signal relative to the wild-type parental cells is shown here. Treatment was for 6 hours followed by incubation in drug-free medium an additional 6 hours. B, right, Analysis of apoptotic cell death in both RNaseH1 transfected and wild-type (parental) LOXIMVI cell lines following romidepsin treatment. Annexin V–positive cells were determined by flow cytometry. Results are baseline mean ± SD from three independent experiments. Each bar represents percentage of increase in cell death compared with untreated control. *, P < 0.04 (t test, two‐tailed). C, LOXIMVI cells treated with romidepsin 50 nmol/L, PARP-inhibitor olaparib (10 μmol/L) and combination of romidepsin and olaparib for 6 hours and harvested immediately (6 hours) or incubated an additional 6 hours before harvest (12-hour time point). The presence of γ-H2AX signal was detected by western blot assay (left). Analysis of apoptotic cell death in the LOXIMVI cell line following treatment with romidepsin (50 and 200 nmol/L), olaparib alone (10 μmol/L), and 50 nmol/L romidepsin in combination with 10 μmol/L olaparib. Annexin V–positive cells were determined by flow cytometry (right).
To test whether disruption of the R-loop with RNaseH would protect cells from DNA damage and cytotoxicity, we transfected LOXIMVI cells with an RNaseH1 plasmid. Using γ-H2AX protein as an indicator of DSBs following exposure to romidepsin for 6 hours followed by incubation for an additional 6 hours (total 12 hours) at 50 and 100 nmol/L concentrations, we observed a 15% and 47% reduction in γ-H2AX protein detected at 50 and 100 nmol/L concentrations, respectively, suggesting that increased expression of RNaseH1 correlated with reduced R-loop accumulation and consequently DNA damage (Fig. 7B, left). Cytotoxicity was also assessed by treating cells with 50 nmol/L romidepsin for 6 hours followed by incubation in drug-free medium for an additional 18 hours (total 24 hours). There was a 45% reduction in Annexin-positive cells in the RNaseH1-transfected population compared with the non-transfected population following treatment (Fig. 7B, right).
Having observed the above DNA-repair response, we postulated that inhibiting proteins involved in the repair of ssDNA would augment the R-loop–initiated DNA lesions, leading to DSBs in the initial 6 hours of treatment. To test this, we combined romidepsin (50 nmol/L) with the PARP-inhibitor (PARPi) olaparib (10 μmol/L) and treated LOXIMVI cells with a single dose and the combination of both drugs for 6 hours, and looked at the emergence of γ-H2AX signal by immunoblot. As seen in Fig. 7C left, γ-H2AX signal increased after a 6-hour treatment in the drug combination samples compared with that of romidepsin or PARPi alone, reaching the level observed after 12 hours. This indicates that targeting SSB repair proteins could augment the initial DNA damage caused by romidepsin and increase its efficacy. We confirmed the cytotoxicity of the combination romidepsin for 6-hours followed by an additional 12-hour incubation with or without olaparib. Figure 7C, right, indicates that the augmented DNA damage resulting from the combination of romidepsin with olaparib also sensitized the cells to cell death from the drug combination.
Discussion
HDACis are antineoplastic agents that cause epigenetic alterations in chromatin. However, their regulatory approval is currently limited to hematological malignancies. Attempts to broaden their reach to the more common solid tumors have been largely unsuccessful. For progress to ensue, a better understanding of their mechanism of action, including the DNA damage that follows their administration, is needed. Here, we studied the DNA damage caused by romidepsin following a more clinically relevant 6-hour treatment schedule. Analysis of the drug sensitivity profile from our experiment in CellMiner revealed a strong correlation with known DNA damaging and alkylating agents. This encouraged us to investigate the mechanism of action and type of DNA damage caused by romidepsin with the 6-hour exposure duration. Although DNA damage after HDACis has long been known, the underlying mechanism has not been explored. We pursued the hypothesis that HDACis such as romidepsin lead to a surge of histone hyperacetylation that disrupts transcription at open chromatin sites by interfering with the reannealing and repackaging of DNA strands that normally occurs behind RNA polymerase. Furthermore, the romidepsin-mediated hyperacetylation creates a favorable environment for a nascent RNA transcript to tread back and hybridize with its DNA template resulting in increased formation of DNA–RNA hybrids, or R-loops, which we document increase following treatment of cells with romidepsin for 6 hours. Accumulation of R-loops in turn renders DNA susceptible to SSBs. Evidence that SSBs occur initially and are followed by the emergence of DSBs was provided by single-cell gel electrophoresis, under both alkaline and neutral conditions to discriminate between SSBs and DSBs. This is consistent with only sporadic co-localization of γH2AX DSB signal with R-loops after 6-hour treatment (Supplementary Fig. S7). A lack of sufficient response to the damaged DNA leads to the accumulation of SSBs over the course of treatment and eventually to DSBs; confirmed by the increase in both the γH2AX signal as well as the tail moment in both alkaline and neutral comet assay conditions (Figs. 3B and 5C). The occurrence of DSBs following treatment with romidepsin or other HDACis has also been previously reported (43,44,45).
We propose that the accumulation of R-loops occurs as a result of the hyperacetylation-induced increase in open chromatin and that this in turn is augmented by the repression of genes involved in DNA repair and in prevention and resolution of R-loops. Normal cells prevent R-loop formation via RNA-binding proteins such as the THO complex or the SRSF1 factor proteins; or resolve R-loop structures via enzymes such as RNase H, senataxin (SETX), and other DNA–RNA helicases (16,17,46,47,48); and indirectly by the action of DNA repair factors, including Fanconi Anemia proteins or BRCA2 and BRCA1 (37, 47, 49, 50). Our microarray data confirm prior observations by showing repressed expression of DDR genes (14, 51, 52), including genes involved in preventing or resolving R-loops such as SRSF1, BRCA2, and TREX1 (Supplementary Fig. S3B).
We next turned to the question of what pathway would be used to repair the SSBs that arise from romidepsin-induced R-loops and reasoned the DNA repair pathways involved in the early response would be a form of transcription-coupled SSB repair such as NER or BER (21). NER is normally responsible for dealing with larger DNA lesions caused by exogenous mutagens such as UV, which are less likely to form during the initial hours of exposure to romidepsin, whereas BER removes base oxidation induced by endogenous ROS, deamination caused by deaminases, and DNA lesions induced by endogenous alkylating agents. Furthermore, it has been suggested that a subset of oxidative lesions occurring during transcriptional arrest is detected by BER and although transcription coupled NER proteins are recruited, these oxidative lesions are eventually repaired by BER rather than NER (53). We therefore envision that by interfering with the resolution of R-loops, romidepsin treatment increases the length of time the non-transcribed DNA persists as a single strand in R-loops and this in turn leads to endogenous DNA damage with the BER machinery likely contributing to the initial damage response. Indeed, by confocal microscopy, we observed an increase in the number of PARP1, XRCC1, RPA1, and POLβ foci; in all cases co-localizing with R-loops following a 6-hour romidepsin treatment consistent with involvement of the BER machinery in the early response to romidepsin-induced R-loop arrest (Fig. 6A–E). Interestingly, in the microarray studies, the RNA expression of PARP1, XRCC1, and RPA1 is preserved after romidepsin, whereas POLβ expression is induced.
Our data are supported by a recent study demonstrating that Sin3A, a core component of the Sin3–HDAC complex, prevents R-loop accumulation and R-loop–mediated instability (54). This study showed that the RNA-binding THO complex, which prevents co-transcriptional R-loop accumulation, physically interacts with the Sin3A–deacetylase complex. The HDACi trichostatin A prevented this interaction (54). Depletion of Sin3A caused replication fork stalling, which was rescued by overexpression of RNase H1 (54) arguing that DNA breaks may occur as a consequence of replication traversing the R loops, and thus might also explain the high DSB occurrence following long-term romidepsin treatment as determined by γH2AX (Fig. 3A and B). Whether replication fork stalling would be in part a consequence of a previously occurring ssDNA break caused by the incapacity of BER to deal with excess ssDNA damage is an open question.
The accumulation of R-loops in romidepsin-treated cells has several implications. Although R-loops arise during normal gene transcription, in romidepsin-treated cells the delay in R-loop resolution leads to DNA damage. Interestingly, accumulation of R-loops has also been linked to the silencing of genes involved in neurogenerative disease. They are thought to reduce gene expression by impeding the activity of RNA polymerase II during transcription and activation of the repressive chromatin mark H3K9me2 via direct recruitment of methyltransferase enzymes (21,55,56,57,58). It is plausible that some of the observed effects of romidepsin on silencing genes are due to the accumulation of R-loops. Investigations in this area could provide more evidence for this hypothesis, and further clarify the mechanism of action of HDACis.
We believe that the mechanistic insight provided by this work provides opportunities for exploiting HDACis in solid tumors. Combinations of romidepsin with drugs targeting the proteins involved in the formation and resolution of R-loops could enhance the antitumor activity of these compounds, as could identifying tumors with defects in DDR, that render cells more sensitive to HDAC inhibition. For example, topoisomerase I suppresses R-loop formation by preventing the annealing of RNA with DNA through resolving the negative torsional stress behind RNA polymerase II and relaxing the DNA supercoils (59). Inhibition of topoisomerase I has been shown to stabilize and increase R-loop formation; camptothecin, for instance, stabilizes R-loops (17, 60, 61). HDAC inhibition could augment R-loop accumulation, and potentially explain the reported activity of an HDAC and topoisomerase I inhibitor combination (62, 63). Identifying and targeting the proteins involved in the repair of ssDNA damage could also be another strategy to enhance the efficacy of HDACis; inhibitors of PARP1 have been shown to generate DNA lesions in the absence of BRCA1 and BRCA2 (64). We observed both an increase in γH2AX signal and sensitivity of the cells with a combination of a PARPi and romidepsin compared with that of the individual agents alone (Fig. 7C). The romidepsin effect on R-loops could explain multiple studies that have reported increased efficacy combining PARP1 inhibitors with romidepsin (65, 66).
In conclusion, this study provides evidence for a novel mechanism of DNA damage occurring following romidepsin treatment. We show for the first time, accumulation of DNA–RNA hybrids following romidepsin treatment, leading to the initiation of single-strand DNA damage. Furthermore, we provide evidence that BER is likely the first line of response to the DNA damage that occurs in romidepsin induced R-loops. By having an impact on both the generation and repair of SSBs, romidepsin leads to an increase in lethal DSBs and cell death.
Authors' Disclosures
T. Litman reports personal fees from LEO Pharma A/S outside the submitted work. A. Basseville reports grants from the European Commission during the conduct of the study. O.A. O'Connor reports grants from Celgene outside the submitted work. S.E. Bates reports grants from Celgene during the conduct of the study; as well as a patent for Depsipeptide for therapy of kidney cancer issued and Deacetylase inhibitor therapy issued. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors acknowledge the important and vital contributions of Victoria Luchenko, who died before this work was completed; her efforts were critical in establishing the dominant effect of DNA damage and importance of cell context in the response to HDAC inhibitors. The authors would like to thank Sudhir Varma for his help in analyzing the CellMiner data. The authors would like to acknowledge the support of the research and imaging facility at the James J. Peters VA Medical Center. Part of this work was supported by the Center for Cancer Research, Intramural Research Program of the National Cancer Institute; (1ZIABC010621–12; to S.E. Bates) and (BC-006161 and BC-006150; to W.C. Reinhold and Y. Pommier), and the European Research Council (ERC2014 AdG669898 TARLOOP; to A. Aguilera).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Mol Cancer Res 2021;19:1361–74
Authors' Contributions
M. Safari: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. T. Litman: Data curation, software, formal analysis, investigation, visualization, methodology, writing–review and editing. R.W. Robey: Data curation, formal analysis, investigation, writing–review and editing. A. Aguilera: Funding acquisition, validation, visualization, methodology, writing–review and editing. A.R. Chakraborty: Data curation, writing–review and editing. W.C. Reinhold: Resources, formal analysis, writing–review and editing. A. Basseville: Data curation, writing–review and editing. L. Petrukhin: Data curation, investigation, methodology. L. Scotto: Resources, writing–review and editing. O.A. O'Connor: Resources, writing–review and editing. Y. Pommier: Funding acquisition, writing–review and editing. A.T. Fojo: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, writing–review and editing. S.E. Bates: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Leder A, Leder P. Butyric acid, a potent inducer of erythroid differentiation in cultured erythroleukemic cells. Cell 1975;5:319–22. [DOI] [PubMed] [Google Scholar]
- 2. Mickley LA, Bates SE, Richert ND, Currier S, Tanaka S, Foss F, et al. Modulation of the expression of a multidrug resistance gene (mdr-1/P-glycoprotein) by differentiating agents. J Biol Chem 1989;264:18031–40. [PubMed] [Google Scholar]
- 3. Bates SE, Mickley LA, Chen YN, Richert N, Rudick J, Biedler JL, et al. Expression of a drug resistance gene in human neuroblastoma cell lines: modulation by retinoic acid-induced differentiation. Mol Cell Biol 1989;9:4337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kitazono M, Robey R, Zhan Z, Sarlis NJ, Skarulis MC, Aikou T, et al. Low concentrations of the histone deacetylase inhibitor, depsipeptide (FR901228), increase expression of the Na+/I− symporter and iodine accumulation in poorly differentiated thyroid carcinoma cells. J Clin Endocrinol Metabol 2001;86:3430–5. [DOI] [PubMed] [Google Scholar]
- 5. Ohlsson-Wilhelm BM, Farley BA, Kosciolek B, La Bella S, Rowley PT. K562 human erythroleukemia cell variants resistant to growth inhibition by butyrate have deficient histone acetylation. Am J Hum Genet 1984;36:1225–38. [PMC free article] [PubMed] [Google Scholar]
- 6. Lopez AT, Bates S, Geskin L. Current status of HDAC inhibitors in cutaneous T-cell lymphoma. Am J Clin Dermatol 2018;19:805–819. [DOI] [PubMed] [Google Scholar]
- 7. Bates SE, Eisch R, Ling A, Rosing D, Turner M, Pittaluga S, et al. Romidepsin in peripheral and cutaneous T-cell lymphoma: mechanistic implications from clinical and correlative data. Br J Haematol 2015;170:96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shustov A, Coiffier B, Horwitz S, Sokol L, Pro B, Wolfson J, et al. Romidepsin is effective and well tolerated in older patients with peripheral T-cell lymphoma: analysis of two phase II trials. Leuk Lymphoma 2017;58:2335–41. [DOI] [PubMed] [Google Scholar]
- 9. Richardson PG, Schlossman RL, Alsina M, Weber DM, Coutre SE, Gasparetto C, et al. PANORAMA 2: panobinostat in combination with bortezomib and dexamethasone in patients with relapsed and bortezomib-refractory myeloma. Blood 2013;122:2331–7. [DOI] [PubMed] [Google Scholar]
- 10. Bates SE. Epigenetic therapies for cancer. N Engl J Med 2020;383:650–63. [DOI] [PubMed] [Google Scholar]
- 11. Bose P, Dai Y, Grant S. Histone deacetylase inhibitor (HDACI) mechanisms of action: emerging insights. Pharmacol Ther 2014;143:323–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Piekarz RL, Bates SE. Epigenetic modifiers: basic understanding and clinical development. Clin Cancer Res 2009;15:3918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen CC, Huang JS, Wang TH, Kuo CH, Wang CJ, Wang SH, et al. Dihydrocoumarin, an HDAC inhibitor, increases DNA damage sensitivity by inhibiting Rad52. Int J Mol Sci 2017;18:2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robert C, Rassool FV. Chapter Three—HDAC inhibitors: roles of DNA damage and repair. In: Grant S., editor. Advances in Cancer Research: Academic Press; 2012. p. 87–129. [DOI] [PubMed] [Google Scholar]
- 15. Petruccelli LA, Dupéré-Richer D, Pettersson F, Retrouvey H, Skoulikas S, Miller WH, Jr. Vorinostat induces reactive oxygen species and DNA damage in acute myeloid leukemia cells. PLoS One 2011;6:e20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell 2012;46:115–24. [DOI] [PubMed] [Google Scholar]
- 17. Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 2014;28:1384–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 2011;25:2041–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luchenko VL, Litman T, Chakraborty AR, Heffner A, Devor C, Wilkerson J, et al. Histone deacetylase inhibitor-mediated cell death is distinct from its global effect on chromatin. Mol Oncol 2014;8:1379–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 2003;19:368–75. [DOI] [PubMed] [Google Scholar]
- 21. Sollier J, Stork Caroline T, García-Rubio María L, Paulsen Renee D, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 2014;56:777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanz LA, Chédin F. High-resolution, strand-specific R-loop mapping via S9. 6-based DNA–RNA immunoprecipitation and high-throughput sequencing. Nat Protoc 2019;14:1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Byrd JC, Marcucci G, Parthun MR, Xiao JJ, Klisovic RB, Moran M, et al. A phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood 2005;105:959–67. [DOI] [PubMed] [Google Scholar]
- 24. Jain S, Zain J. Romidepsin in the treatment of cutaneous T-cell lymphoma. J Blood Med 2011;2:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee JS, Paull K, Alvarez M, Hose C, Monks A, Grever M, et al. Rhodamine efflux patterns predict P-glycoprotein substrates in the National Cancer Institute drug screen. Mol Pharmacol 1994;46:627–38. [PubMed] [Google Scholar]
- 26. Robey RW, Zhan Z, Piekarz RL, Kayastha GL, Fojo T, Bates SE. Increased MDR1 expression in normal and malignant peripheral blood mononuclear cells obtained from patients receiving depsipeptide (FR901228, FK228, NSC630176). Clin Cancer Res 2006;12:1547. [DOI] [PubMed] [Google Scholar]
- 27. Xiao JJ, Huang Y, Dai Z, Sadée W, Chen J, Liu S, et al. Chemoresistance to depsipeptide FK228 [(E)-(1S,4S,10S,21R)-7-[(Z)-Ethylidene]-4,21-diisopropyl-2-oxa-12,13-dithia-5,8,20,23-tetraazabicyclo[8,7,6]-tricos-16-ene-3,6,9,22-pentanone] is mediated by reversible MDR1 induction in human cancer cell lines. J Pharmacol Exp Ther 2005;314:467–75. [DOI] [PubMed] [Google Scholar]
- 28. Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, et al. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res 2012;72:3499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reinhold WC, Sunshine M, Varma S, Doroshow JH, Pommier Y. Using CellMiner 1.6 for systems pharmacology and genomic analysis of the NCI-60. Clin Cancer Res 2015;21:3841–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sandor V, Robbins AR, Robey R, Myers T, Sausville E, Bates SE, et al. FR901228 causes mitotic arrest but does not alter microtubule polymerization. Anticancer Drugs 2000;11:445–54. [DOI] [PubMed] [Google Scholar]
- 31. Balakrishnan L, Milavetz B. Histone hyperacetylation in the coding region of chromatin undergoing transcription in SV40 minichromosomes is a dynamic process regulated directly by the presence of RNA polymerase II. J Mol Biol 2007;365:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grunstein M. Histone acetylation in chromatin structure and transcription. Nature 1997;389:349. [DOI] [PubMed] [Google Scholar]
- 33. Erard MS, Belenguer P, Caizergues-Ferrer M, Pantaloni A, Amalric F. A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone Hl. Eur J Biochem 1988;175:525–30. [DOI] [PubMed] [Google Scholar]
- 34. Sirri V, Roussel P, Hernandez-Verdun D. The AgNOR proteins: qualitative and quantitative changes during the cell cycle. Micron 2000;31:121–6. [DOI] [PubMed] [Google Scholar]
- 35. Tajrishi MM, Tuteja R, Tuteja N. Nucleolin: the most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol 2011;4:267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang IX, Grunseich C, Fox J, Burdick J, Zhu Z, Ravazian N, et al. Human proteins that interact with RNA/DNA hybrids. Genome Res 2018;28:1405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. García-Rubio ML, Pérez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, et al. The fanconi anemia pathway protects genome integrity from r-loops. PLos Genet 2015;11:e1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barroso S, Herrera-Moyano E, Muñoz S, García-Rubio M, Gómez-González B, Aguilera A. The DNA damage response acts as a safeguard against harmful DNA–RNA hybrids of different origins. EMBO Rep 2019;20:e47250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nassour J, Martien S, Martin N, Deruy E, Tomellini E, Malaquin N, et al. Defective DNA single-strand break repair is responsible for senescence and neoplastic escape of epithelial cells. Nat Commun 2016;7:10399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Idriss HT, Al-Assar O, Wilson SH. DNA polymerase β. Int J Biochem Cell Biol 2002;34:321–4. [DOI] [PubMed] [Google Scholar]
- 41. Gómez-González B, Felipe-Abrio I, Aguilera A. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol Cell Biol 2009;29:5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sviković S, Crisp A, Tan-Wong SM, Guilliam TA, Doherty AJ, Proudfoot NJ, et al. R-loop formation during S phase is restricted by PrimPol-mediated repriming. EMBO J 2019;38:e99793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Valdez BC, Brammer JE, Li Y, Murray D, Liu Y, Hosing C, et al. Romidepsin targets multiple survival signaling pathways in malignant T cells. Blood Cancer J 2015;5:e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Conti C, Leo E, Eichler GS, Sordet O, Martin MM, Fan A, et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins and induces DNA damage. Cancer Res 2010;70:4470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gaymes TJ, Padua RA, Pla M, Orr S, Omidvar N, Chomienne C, et al. Histone deacetylase inhibitors (HDI) cause DNA damage in leukemia cells: a mechanism for leukemia-specific HDI-dependent apoptosis? Mol Cancer Res 2006;4:563–73. [DOI] [PubMed] [Google Scholar]
- 46. Domínguez-Sánchez MS, Barroso S, Gómez-González B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLos Genet 2011;7:e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 2014;511:362. [DOI] [PubMed] [Google Scholar]
- 48. Wahba L, Amon Jeremy D, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell 2011;44:978–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 2015;57:636–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwab RA, Nieminuszczy J, Shah F, Langton J, Martinez DL, Liang C-C, et al. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol Cell 2015;60:351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sousa FG, Matuo R, Tang SW, Rajapakse VN, Luna A, Sander C, et al. Alterations of DNA repair genes in the NCI-60 cell lines and their predictive value for anticancer drug activity. DNA Repair 2015;28:107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci U S A 2004;101:540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JHJ. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 2014;15:465. [DOI] [PubMed] [Google Scholar]
- 54. Salas-Armenteros I, Pérez-Calero C, Bayona-Feliu A, Tumini E, Luna R, Aguilera A. Human THO–Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J 2017;36:3532–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Groh M, Lufino MM, Wade-Martins R, Gromak N. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLos Genet 2014;10:e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang W-Y, Gerhardt J, et al. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 2014;343:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Loomis EW, Sanz LA, Chédin F, Hagerman PJ. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLos Genet 2014;10:e1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Castellano-Pozo M, Santos-Pereira JM, Rondón AG, Barroso S, Andújar E, Pérez-Alegre M, et al. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell 2013;52:583–90. [DOI] [PubMed] [Google Scholar]
- 59. Pommier Y, Sun Y, Huang SN, Nitiss JL. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 2016;17:703–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sordet O, Redon CE, Guirouilh-Barbat J, Smith S, Solier S, Douarre C, et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep 2009;10:887–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marinello J, Chillemi G, Bueno S, Manzo SG, Capranico G. Antisense transcripts enhanced by camptothecin at divergent CpG-island promoters associated with bursts of topoisomerase I-DNA cleavage complex and R-loop formation. Nucleic Acids Res 2013;41:10110–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gray J, Cubitt CL, Zhang S, Chiappori A. Combination of HDAC and topoisomerase inhibitors in small cell lung cancer. Cancer Biol Ther 2012;13:614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guerrant W, Patil V, Canzoneri JC, Yao L-P, Hood R, Oyelere AK. Dual-acting histone deacetylase-topoisomerase I inhibitors. Bioorg Med Chem Lett 2013;23:3283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917. [DOI] [PubMed] [Google Scholar]
- 65. Li X, Li C, Jin J, Wang J, Huang J, Ma Z, et al. High PARP-1 expression predicts poor survival in acute myeloid leukemia and PARP-1 inhibitor and SAHA-bendamustine hybrid inhibitor combination treatment synergistically enhances antitumor effects. EBioMedicine 2018;38:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yalon M, Tuval-Kochen L, Castel D, Moshe I, Mazal I, Cohen O, et al. Overcoming resistance of cancer cells to PARP-1 inhibitors with three different drug combinations. PLoS One 2016;11:e0155711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.