Abstract
Three fluorescent nucleic acid binding dyes—propidium iodide, TO-PRO-1, and SYTOX green—were evaluated, and their abilities to distinguish between bacterial cells with and without an intact cytoplasmic membrane were compared. Each dye was readily able to discriminate between healthy and permeabilized cells of Escherichia coli, although SYTOX green showed a greater enhancement in fluorescence intensity on staining-compromised, as opposed to healthy, cells in log-phase growth, than either PI or TO-PRO-1. Flow cytometric analysis of E. coli stained with these dyes after exposing them to several antimicrobial agents showed that all three dyes were able to detect antimicrobial action. Notably, however, the intensity of the cell-associated fluorescence was related to the mechanism of action of the antimicrobial agent. Large changes in fluorescence intensity were observed for all the dyes subsequent to β-lactam antibiotic action, but smaller changes (or no change) were seen subsequent to exposure to antimicrobials acting directly or indirectly on nucleic acid synthesis. Furthermore, cell-associated fluorescence did not relate to loss of viability as determined by plate counts. Despite offering much insight into antimicrobial mechanisms of action, these fundamental problems become relevant to the development of rapid antimicrobial susceptibility tests if colony formation is used as the standard.
Numerous fluorescent dyes have been suggested to act as viability probes because they can detect changes in the physiology or metabolism of both eukaryotic and prokaryotic cells as discussed in the Molecular Probes handbook (Molecular Probes Inc., Eugene, Oreg.). The behavior of such dyes has been increasingly studied in combination with flow cytometric techniques (1), and some have been shown to be useful tools for evaluating the effects of antimicrobial agents when combined with flow cytometry (4, 8, 10, 16, 17, 20, 21). These techniques may detect changes in individual bacterial cells in less than 60 min of exposure to antimicrobial agents (15, 16). The combination of flow cytometry with such probes would thus appear to be ideal for the development of rapid bacterial susceptibility testing.
The bacterial cytoplasmic membrane is important in determining which molecules enter or leave the cytoplasm. A change in the ability of the membrane to control such molecular traffic may compromise the cell and its ability to survive. Many probes used in the determination of cytoplasmic membrane integrity rely on the premise that they are fluorescent only when bound to nucleic acids. Healthy cells exclude such probes and will not be rendered fluorescent, unlike dead or damaged cells, which can no longer exclude dye and are therefore fluorescent. While propidium iodide (PI) has frequently been used for this purpose, alternative dyes have more recently been proposed for this role (7). To our knowledge, little or no data exist comparing such alternative probes for their ability to discriminate between cells in a healthy state and those damaged by antimicrobial agents. This study was accordingly designed to investigate the behavior of these probes in the presence of several antimicrobial agents with different mechanisms of action in clinically relevant concentrations.
We studied three nucleic acid binding probes—PI, TO-PRO-1, and SYTOX green. These dyes share the properties of being incapable of permeating (eukaryotic) cells and of fluorescing subsequent to binding nucleic acids. PI is a classical phenanthridinium intercalating dye, and TO-PRO-1 is a monomeric cyanine dye with a single cationic side chain with little base selectivity. Although generally used as a DNA electrophoresis stain, it possesses the chemical characteristics necessary for a viability probe (Molecular Probes handbook). SYTOX green is also a cyanine dye and shows little base selectivity; its advantage lies in its ability to fluoresce 1,000 times more brightly when bound to nucleic acid. It has recently been reported as a promising and perhaps superior alternative to PI (11, 21). There are also some reports of TO-PRO-1 and other cyanine dye analogues being used in flow cytometric studies (12, 13, 22). Antimicrobial agents were selected to provide examples of various mechanisms of action from inhibitors of cell wall synthesis to inhibitors of protein synthesis.
MATERIALS AND METHODS
Bacterial strains, media, and antibiotics.
Escherichia coli NCTC 10418 was maintained on blood agar slopes and grown as a suspension in Iso-Sensitest broth (filtered through a 0.2-μm-pore-size filter). Ceftazidime, chloramphenicol, ciprofloxacin, and rifampin were purchased as powders, and gentamicin was purchased as a stock solution, 10 mg/ml in deionized water (Sigma). Stock solutions (1 mg/ml) of the powdered antibiotics were prepared in sterile distilled water with added ethanol, NaOH, or HCl where required for solubility. All further dilutions were done with sterile distilled water.
Permeabilization of the cells.
Aliquots (1 ml) of cells (107 CFU/ml) were pelleted and resuspended in 70% ethanol (1 ml) for 1 h. The cells were then washed twice in phosphate-buffered saline and finally resuspended in fresh Iso-Sensitest broth. A further 1 ml of cells suspended in Iso-Sensitest broth was immersed in boiling water for 10 min, washed, and resuspended in broth. Permeabilized cells were analyzed by flow cytometry with each dye as described below.
MIC determination.
Doubling dilutions in Iso-Sensitest broth of each antimicrobial agent (100 μl) was made in a 96-well round-bottom plate. Each dilution was inoculated with 11 μl of a 106 CFU/ml suspension (determined using McFarland standards) to give a starting bacterial concentration of 105 CFU/ml in each well and incubated overnight at 37°C. The lowest concentration of antimicrobial showing no visible accumulation of cells in the bottom of the well was considered the MIC. The MICs (in micrograms per milliliter) of the following drugs were determined to be as indicated: ceftazidime, 0.125; chloramphenicol, 2; ciprofloxacin, 0.0075; gentamicin, 0.5; and rifampin, 8.
Susceptibility to antimicrobial agents.
A 1 in 100 dilution (vol/vol) of an overnight culture grown to stationary phase at 37°C was made in Iso-Sensitest broth and grown to logarithmic phase (1.5 h) on a shaker (200 rpm) at 37°C. The culture was divided into three, and antimicrobials at the MIC and 10 times the MIC were added to two of the portions, while the final portion was used as a control. Incubation of the cultures was continued for a further 2 h with samples (0.5 ml) being removed at 30-min intervals. The samples were pelleted and washed with fresh broth to remove the antimicrobial, and finally resuspended in fresh broth. Three aliquots (108 μl) were stained with each one of the three probes (see below), and the final aliquot was used for colony counting.
The experiments were performed on three or more separate occasions: typical results are shown.
Viable counts.
The number of CFU/ml was estimated using the technique of Miles et al. (18). Briefly, aliquots of the serially diluted sample were placed on nutrient agar, and colonies were counted after 24 h of incubation at 37°C.
Staining of cells.
A stock solution of PI (100 μg/ml) (Sigma Chemical, Poole, United Kingdom) was prepared in deionized water. TO-PRO-1 (1 mM) and SYTOX green (5 mM) (both from Molecular Probes) in dimethyl sulfoxide were diluted in Tris-HCl (10 mM) buffer to give working stock solutions of 50 μM. Portions (12 μl) of the probe stock solutions were added to aliquots of cells (108 μl) to give a final concentration of 10 μg of PI per ml or 5 μM for both TO-PRO-1 and SYTOX. These were incubated at room temperature for 3 to 5 min.
Flow cytometric analysis.
The cell populations were analyzed on a dual-parameter Bryte HS (Bio-Rad, Hemel Hempstead, United Kingdom) with a xenon arc lamp as the light source. The instrument is equipped with two light scatter detectors (<15° and >15°) and two fluorescence detectors for green and red fluorescence (beam split at 520 nm). A fluorescein isothiocyanate filter block with the following characteristic wavelengths was used to excite each of the dyes: excitation, 470 to 490 nm; band stop, 510 nm; and emission, >520 nm. SYTOX green and TO-PRO-1 were both detected by detector FL1 which detects emitted light with wavelengths between 515 to 565 nm (green fluorescence) and PI was detected by FL2 (red fluorescence). The detection of light by FL2 is limited by a band-pass filter to wavelengths in excess of 565 nm. All detectors were set on logarithmic amplification. Sample flow and sheath fluid pressure were at 1.5 μl/min and 0.7 kPa/cm2, respectively. Optical and electronic noise were eliminated by setting appropriate electronic gating thresholds to both light scatter detectors.
RESULTS
Fluorescence intensity of intact and permeabilized cells after staining.
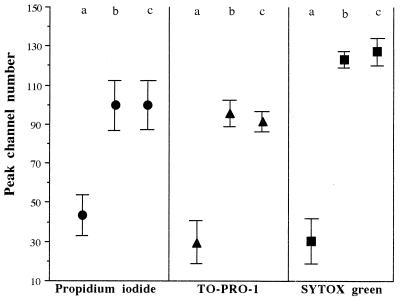
Permeabilized cells showed greater fluorescence intensity than log-phase cultures upon staining with all three dyes, but the enhancement in fluorescence intensity varied between the three probes as indicated by the peak channel number of the populations stained (Fig. 1). The fluorescence scale is divided into 4 log orders with peak channel numbers between 1 and 64 in the first log order, those between 65 and 128 in the second log order, and so forth. SYTOX green showed the greatest difference in fluorescence intensity between permeabilized and log-phase cells.
FIG. 1.
Peak channel numbers of untreated (a), heated (b), and ethanol-fixed (c) cultures of E. coli stained with PI, TO-PRO-1, or SYTOX green. The symbols represent the mean peak channel numbers calculated from six separate cell samples from different days, and the bars indicate the coefficients of variation for these samples.
Cells which had undergone alcohol fixation or heat treatment were used as positive controls to enable a region of interest to be set which equated to fluorescent positive cells.
Antibiotic susceptibility tests. (i) Viable counts.
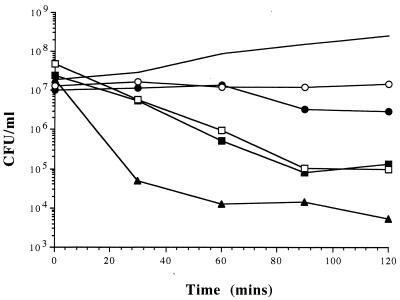
The log-phase culture showed an increase in colony counts of 2 log orders over a 2-h period while the suspensions treated with gentamicin, rifampin, and ciprofloxacin at 10 times the MIC showed between a 2 to 4 log order reduction in CFU/ml (Fig. 2). Ceftazidime caused only a small decline in CFU/ml at both 10 times the MIC and MIC (results not shown), while chloramphenicol was bacteriostatic.
FIG. 2.
Number of CFU for cultures of E. coli over time. Results are shown for an untreated culture (solid line) and cultures treated at 37°C for 120 min at 10 times the MIC of ceftazidime (●), chloramphenicol (○), ciprofloxacin (□), gentamicin (▴), and rifampin (■).
(ii) Changes in bacterial fluorescence.
With each probe, less than 5% of the population of cells in log-phase growth allowed penetration of the dye, and this remained constant over 2 h. Similarly, there was no change to the proportion of cells in the population showing fluorescence with any dye during exposure to chloramphenicol at both its MIC and 10 times the MIC for 2 h (results not shown).
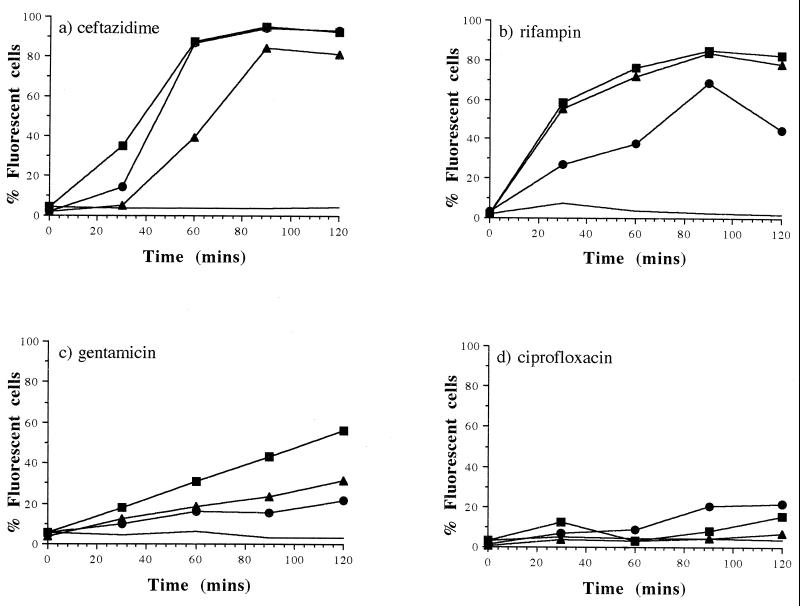
After only 1 h of exposure to 10 times the MIC of ceftazidime, PI and SYTOX green rendered nearly 90% of the population fluorescent; TO-PRO-1, however, rendered only 40% of the population fluorescent. This percentage doubled after 90 min of exposure (Fig. 3a). At the MIC of ceftazidime, TO-PRO-1 also showed less cell-associated fluorescence than the other probes at each time interval (results not shown). Rifampin-treated cells (10 times the MIC) showed a similar uptake of SYTOX and TO-PRO-1 over the 2-h period. With this antibiotic, however, PI caused only approximately half the percentage of fluorescence observed with the other two probes (Fig. 3b). A gradual increase in the number of fluorescent cells was observed over 2 h with all three probes in a population treated with gentamicin (10 times the MIC), although SYTOX tended to render a slightly higher percentage of cells fluorescent than TO-PRO-1 or PI (Fig. 3c). There was little change in the cell-associated fluorescence for all the probes after ciprofloxacin exposure at 10 times the MIC (Fig. 3d). In some instances, fluorescence was slightly greater with each probe when the population was treated with ciprofloxacin at its MIC, but the response was variable and typically the percentage uptake remained low (results not shown).
FIG. 3.
Percentage of fluorescent E. coli cells in the presence of PI (●), TO-PRO-1 (▴), and SYTOX green (■) after treatment with ceftazidime (a), rifampin (b), gentamicin (c), and ciprofloxacin (d) all at 10 times the MIC. The solid line represents the maximum percentage of fluorescent cells stained by any of the dyes in an untreated culture.
It was notable also that the quantity of fluorescent cells within a population appeared to be unrelated to the changes observed in CFUs.
(iii) Distribution of fluorescence intensity in the bacterial population.
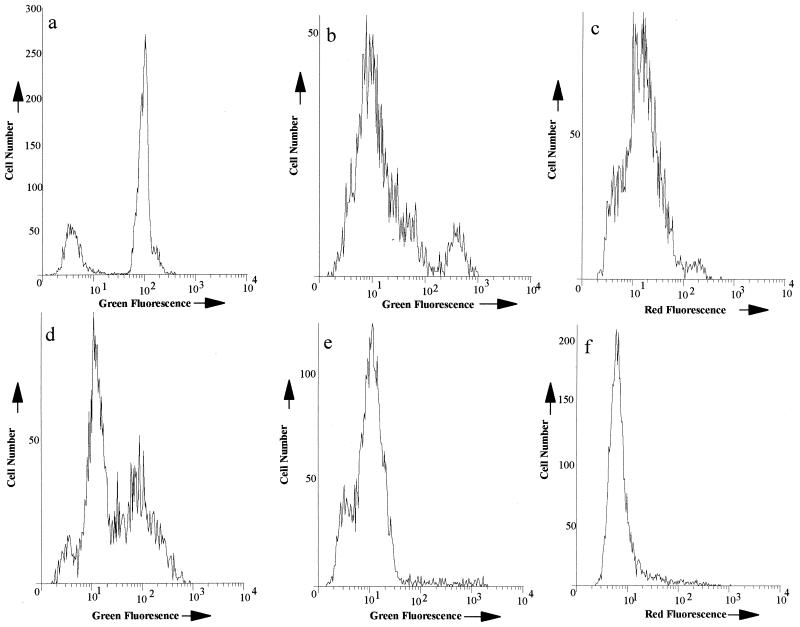
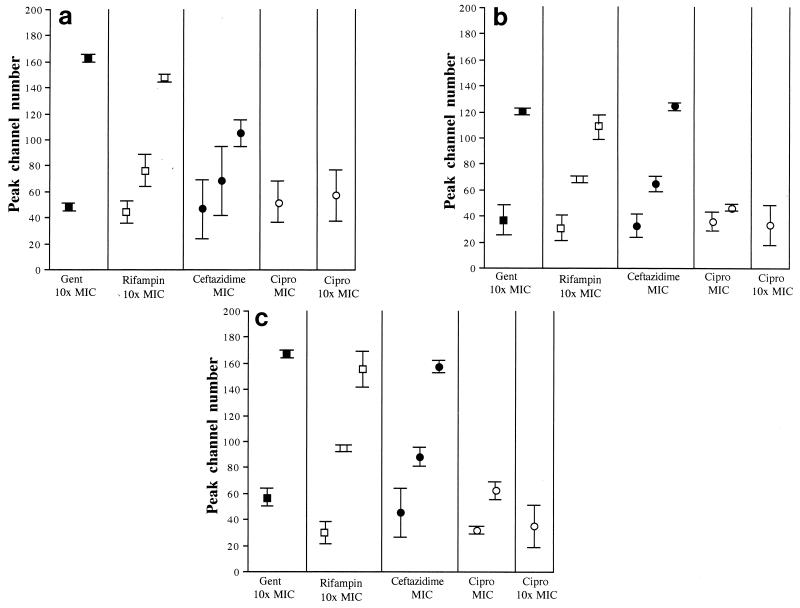
Staining a suspension containing a mixture of permeabilized (fixed or heated) and log-phase cells with any of the probes showed fluorescence within the population distributed around two distinct narrow peaks (Fig. 4a). During treatment with the antimicrobial agents, however, the distribution of fluorescence was less well defined, with some antimicrobials causing the population to distribute around two peaks while for others the fluorescence intensity of the bacteria was distributed around three distinct peaks (Fig. 4 and 5). Gentamicin-treated suspensions showed fluorescence distributed around two peaks, although the peaks were broader than those observed in the artificially mixed population (Fig. 4b). In the case of PI and TO-PRO-1, the peak channel number of the lower peak was similar to that of a log-phase culture (i.e., cells in an unperturbed state), but with SYTOX green, this peak was shifted slightly to the right from that of an untreated population, suggesting that some dye was able to associate with these cells (Fig. 5). Interestingly, for all three dyes, the cells distributed around the higher fluorescent peak were more fluorescent than those permeabilized by either ethanol or heat. Cells exposed to rifampin (10 times the MIC) and ceftazidime (MIC) usually showed three fluorescent peaks within 90 min of treatment (Fig. 4c and e and 5) for each dye. These three peaks suggested the population was divided between cells showing no dye uptake, those showing uptake less than that of a permeabilized population, and some showing fluorescence comparable to, or in excess of, a permeabilized population. As indicated previously, uptake of the dyes in a ciprofloxacin-treated population was minimal. After 120 min of exposure to MICs of this antibiotic, two peaks were nevertheless observed when the population was stained with either SYTOX or TO-PRO-1 (Fig. 4e), although the peak channel number of the most fluorescent peak indicated that the cells in this peak were less fluorescent than those in a fully permeabilized population (Fig. 5). We observed a single peak only for a PI-stained population in the presence of ciprofloxacin at the MIC and for all dyes at 10 times the MIC. The channel number of this peak was related to that of an unpermeabilized population (Fig. 4f and 5).
FIG. 4.
Representative examples of fluorescence distributions (dyes shown in brackets below) from E. coli treated differently. (a) An artificially mixed population of untreated and ethanol-fixed E. coli cells [SYTOX green] and (b to f) E. coli cells treated with 10 times the MIC of gentamicin [SYTOX] (b), 10 times the MIC of rifampin [PI] (c), the MIC of ceftazidime [TO-PRO-1] (d), the MIC of ciprofloxacin [SYTOX] (e) and 10 times the MIC of ciprofloxacin [PI] (f) are shown.
FIG. 5.
Peak channel numbers of E. coli cultures stained with PI (a), TO-PRO-1 (b), and SYTOX green (c) after treatment for 120 min with gentamicin (Gent) (10 times the MIC), rifampin (10 times the MIC), ceftazidime (MIC), ciprofloxacin (Cipro) (MIC), and ciprofloxacin (10 times the MIC). The symbols represent the mean peak channel numbers calculated from six separate cell samples from different days, and the bars indicate the coefficients of variation for these samples. If there were two or three distinct peaks, the mean channel number for each of the peaks is shown.
DISCUSSION
One of the most important criteria for a rapid antimicrobial susceptibility test is that it should be applicable to a wide variety of clinically important organisms and relevant antimicrobial agents. All three probes were able to distinguish between untreated and permeabilized cell suspensions for this gram-negative bacterial species. SYTOX green is reported to have a >500-fold fluorescence enhancement on binding with nucleic acids (21) and accordingly showed a larger difference between the peak channel numbers for the untreated and permeabilized populations than PI, which has only a 20- to 30-fold fluorescence enhancement on binding to its target (discussed in the Molecular Probes handbook).
The probes are of similar molecular weights (PI, 668; SYTOX, 600; TO-PRO-1, 645), and they bind readily to nucleic acids with enhancement in fluorescence normally being greater upon binding with double-stranded DNA than with single-stranded DNA or RNA (Molecular Probes handbook). In addition, they are all believed to show little base selectivity (7; Molecular Probes handbook). Hence, not surprisingly after exposure to an antimicrobial agent, the proportion of cells in the population showing dye-associated fluorescence was similar for each probe. However, there were notable exceptions to this, in particular the smaller percentage of cells rendered fluorescent by PI than those of TO-PRO-1 and SYTOX in a rifampin-treated population and the lower percentage of ceftazidime-exposed cells fluorescing with TO-PRO-1 compared to the other two dyes. Bacterial efflux pumps have been associated with the removal of ethidium bromide, a molecule structurally similar to PI, and this may explain the poorer staining by PI (9). Ethidium bromide extrusion from Pseudomonas aeruginosa has recently been used to investigate efflux pump activity (5). Furthermore, in the case of TO-PRO-1, even when the cytoplasmic membrane is damaged, the steric properties of the fluorochrome may in some instances still prevent the molecule penetrating to its binding site (Molecular Probes handbook).
Antimicrobial agent-related differences in both the percentage of stained cells and the intensity of fluorescence has already been observed by Gant et al. (4). In this study we demonstrate that the percentage of fluorescent cells in an antibiotic-exposed population bears no relation to the ability to form colonies. It is possible that the extent and intensity of fluorescence within a bacterial population after exposure to antimicrobials may not relate entirely to the ability of a dye to penetrate and remain within the cell. Factors other than active efflux, such as the quantity and accessibility of intracellular binding sites, could also influence fluorescence intensity. Alternatively, there may be more nucleic acid per cell due to continued replication in the absence of division. This suggestion would be consistent with our findings of higher fluorescence intensity in some antibiotic-treated populations than in populations containing fixed cells. Finally, such mechanisms of fluorescence might be influenced by the conformational state of nucleic acids, highly supercoiled as opposed to more relaxed states. In this respect, the quantum yield and wavelength emission of several other asymetrical cyanine DNA binding dyes varies with nucleic acid secondary structure; the use of such dyes may identify the factors responsible for our results (Molecular Probes handbook). Finally, nucleic acids may leak out of the cell, causing a reduction in the number of available dye binding sites. Some clues as to the relative contribution of these possibilities are provided by our results. Although the appearance of cells exposed to ceftazidime and ciprofloxacin when viewed under the microscope are similar in that they induce the formation of filaments (2), the staining properties of cells exposed to these two antimicrobial agents were very different. The interference to cell wall synthesis by ceftazidime primarily by binding to the transpeptidase enzyme PBP3 appears to have allowed the penetration of the probes into both filamented and microscopically normal cells. In contrast, neither filaments nor normal-sized particles became highly fluorescent after exposure to ciprofloxacin, even though quinolones are reported to induce permeability changes (3). In this case a reduction in the number of binding sites could explain our results, since this antibiotic inhibits DNA gyrase, an enzyme responsible for conformational changes in DNA. Furthermore, the bactericidal activity of ciprofloxacin may result from the leakage of cellular contents, which might include dye binding sites (3). Gentamicin was notable in that it produced a rapid loss of viability in terms of CFU, but there was no rapid increase in the population fluorescence intensity. Hancock (6) indicated that major disruption to the cytoplasmic membrane by aminoglycosides occurs only after the lethal event that prevents cell recovery. This is consistent with our data where dye uptake is apparent only at the later sampling times in contrast to the immediate decline in CFUs. Finally, the contribution of cell volume to our results remains to be determined as Novo and colleagues suggest that correct fluorescence values for individual cells can be obtained only by using a ratiometric carbocyanine dye method (19).
In conclusion, this study has highlighted a number of issues relevant to the use of non-cell-permeating nucleic acid binding dyes for determining antimicrobial susceptibility. The major obstacle is that the mechanism of action of the antibiotic appears to influence the degree of staining observed with damaged cells, and consequently there is no easily interpretable relationship between dye uptake and the bacteriostatic or bactericidal activity of the antimicrobial agents. Furthermore, it is not possible to predict the behavior of these dyes with other bacterial strains until the nature and importance of the mechanisms discussed above have been formally elucidated. Lebaron et al. (11) recently voiced a word of caution for using SYTOX green, as well as other molecules with similar targets, in assessing the viability of starved population due to the degradation or modification of binding sites during the starvation period. They suggested that similar problems may occur with antimicrobials targeted at nucleic acid. These results support their suspicions. We suggest that the ability of flow cytometry to detect heterogeneous responses in apparently homogeneous log-growth-phase cultures mitigates against the establishment of simple decisions concerning susceptibility of antimicrobials.
ACKNOWLEDGMENT
We thank the C. W. Maplethorpe Trust Fund for providing the financial support for this project.
REFERENCES
- 1.Davey H M, Kell D B. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analysis. Microbiol Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diver J M, Wise R. Morphological and biochemical changes in Escherichia coli after exposure to ciprofloxacin. J Antimicrob Chemother. 1986;18(Suppl. D):31–41. doi: 10.1093/jac/18.supplement_d.31. [DOI] [PubMed] [Google Scholar]
- 3.Dougherty T J, Saukkonen J J. Membrane permeability changes associated with DNA gyrase inhibitors in Escherichia coli. Antimicrob Agents Chemother. 1985;28:200–206. doi: 10.1128/aac.28.2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gant V A, Warnes G, Phillips I, Savidge G F. The application of flow cytometry to the study of bacterial responses to antibiotics. J Med Microbiol. 1993;15:147–154. doi: 10.1099/00222615-39-2-147. [DOI] [PubMed] [Google Scholar]
- 5.Germ M, Yoshihara E, Yoneyama H, Nakae T. Interplay between the efflux pump and the outer membrane permeability barrier in fluorescent dye accumulation in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1999;261:452–455. doi: 10.1006/bbrc.1999.1045. [DOI] [PubMed] [Google Scholar]
- 6.Hancock R E W. Aminoglycoside uptake and mode of action—with special reference to streptomycin and gentamicin. J Antimicrob Chemother. 1981;8:429–445. doi: 10.1093/jac/8.6.429. [DOI] [PubMed] [Google Scholar]
- 7.Hirons G T, Fawcett J J, Crissman H A. TOTO and YOYO: new very bright fluorochromes for DNA content analyses by flow cytometry. Cytometry. 1994;15:129–140. doi: 10.1002/cyto.990150206. [DOI] [PubMed] [Google Scholar]
- 8.Humphreys M J, Allman R, Lloyd D. Determination of the viability of Trichomonas vaginalis using flow cytometry. Cytometry. 1994;15:343–348. doi: 10.1002/cyto.990150410. [DOI] [PubMed] [Google Scholar]
- 9.Jernaes M W, Steen H B. Staining of Escherichia coli for flow cytometry—influx and efflux of ethidium bromide. Cytometry. 1994;17:302–309. doi: 10.1002/cyto.990170405. [DOI] [PubMed] [Google Scholar]
- 10.Langsrud S, Sundheim G. Flow cytometry for rapid assessment of viability after exposure to a quaternary ammonium compound. J Appl Bacteriol. 1996;81:411–418. doi: 10.1111/j.1365-2672.1996.tb03527.x. [DOI] [PubMed] [Google Scholar]
- 11.Lebaron P, Catala P, Parthuisot N. Effectiveness of SYTOX green stain for bacterial viability assessment. Appl Environ Microbiol. 1998;64:2697–2700. doi: 10.1128/aem.64.7.2697-2700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W K W, Jellet J F, Dickie P M. DNA distribution in planktonic bacteria stained with TOTO and TO-PRO. Limnol Oceanogr. 1995;40:1485–1495. [Google Scholar]
- 13.Marie D, Vaulot D, Partensky F. Application of the novel nucleic acid dyes YOYO-1, YO-PRO-1, and PicoGreen for flow cytometric analysis of marine prokaryotes. Appl Environ Microbiol. 1996;62:1649–1655. doi: 10.1128/aem.62.5.1649-1655.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez O V, Gratzner H G, Malinin T I, Ingram M. The effect of some β-lactam antibiotics on Escherichia coli studied by flow cytometry. Cytometry. 1982;3:129–133. doi: 10.1002/cyto.990030211. [DOI] [PubMed] [Google Scholar]
- 15.Mason D J, Allman R, Stark J M, Lloyd D. Rapid estimation of bacterial antibiotic susceptibility. J Microsc. 1994;176:8–16. doi: 10.1111/j.1365-2818.1994.tb03494.x. [DOI] [PubMed] [Google Scholar]
- 16.Mason D J, Gant V A. The application of flow cytometry to the estimation of antibiotic susceptibility. J Antimicrob Chemother. 1995;36:441–443. doi: 10.1093/jac/36.2.441. [DOI] [PubMed] [Google Scholar]
- 17.Mason D J, Power E G M, Talsania H, Phillips I, Gant V A. Antibacterial action of ciprofloxacin. Antimicrob Agents Chemother. 1995;39:2752–2758. doi: 10.1128/aac.39.12.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miles A A, Misra S S, Irwin J O. The estimation of the bactericidal power of the blood. J Hyg. 1938;38:732–749. doi: 10.1017/s002217240001158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novo D, Perlmutter N G, Hunt R H, Shapiro H M. Accurate flow cytometric membrane potential measurement in bacteria using diethyloxacarbocyanine and a ratiometric technique. Cytometry. 1999;35:55–63. doi: 10.1002/(sici)1097-0320(19990101)35:1<55::aid-cyto8>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Ordoñez J V, Wehman N M. Rapid flow cytometric antibiotic susceptibility assay for Staphylococcus aureus. Cytometry. 1993;14:811–818. doi: 10.1002/cyto.990140714. [DOI] [PubMed] [Google Scholar]
- 21.Roth B L, Poot M, Yue S T, Millard P J. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl Environ Microbiol. 1997;63:2421–2431. doi: 10.1128/aem.63.6.2421-2431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Votyakova T V, Kaprelyants A S, Kell D B. Influence of viable cells on the resuscitation of dormant cells in Micrococcus luteus cultures held in an extended stationary phase: the population effect. Appl Environ Microbiol. 1994;60:3284–3291. doi: 10.1128/aem.60.9.3284-3291.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]