Abstract
PURPOSE
Circulating tumor DNA (ctDNA) genotyping may guide targeted therapy for patients with advanced GI cancers. However, no studies have validated ctDNA genotyping for microsatellite instability (MSI) assessment in comparison with a tissue-based standard.
PATIENTS AND METHODS
The performance of plasma-based MSI assessment using Guardant360, a next-generation sequencing–based ctDNA assay, was compared with that of tissue-based MSI assessment using a validated polymerase chain reaction–based method in patients with advanced GI cancers enrolled in GOZILA study, a nationwide ctDNA profiling study. The primary end points were overall percent agreement, positive percent agreement (PPA), and negative percent agreement. The efficacy of immune checkpoint inhibitor therapy was also evaluated.
RESULTS
In 658 patients with advanced GI cancers who underwent both plasma and tissue testing for MSI, the overall percent agreement, PPA, and negative percent agreement were 98.2% (95% CI, 96.8 to 99.1), 71.4% (95% CI, 47.8 to 88.7), and 99.1% (95% CI, 98.0 to 99.7), respectively. In patients whose plasma samples had a ctDNA fraction ≥ 1.0%, the PPA was 100.0% (15/15; 95% CI, 78.2 to 100.0). Three patients with MSI-high (MSI-H) tumors detected only by ctDNA genotyping achieved clinical benefits after receiving anti–programmed cell death 1 therapy with the progression-free survival ranging from 4.3 to 16.7 months. One patient with an aggressive cancer of an unknown primary site benefited from pembrolizumab after rapid detection of MSI-H by ctDNA genotyping.
CONCLUSION
ctDNA genotyping was able to detect MSI with high concordance to validated tissue-based MSI testing, especially in patients with tumors that have sufficient ctDNA shedding. Furthermore, ctDNA genotyping enabled identification of patients with MSI-H tumors who benefited from immune checkpoint inhibitor treatment.
INTRODUCTION
Traditionally, microsatellite instability-high (MSI-H) or deficient mismatch repair can be evaluated by polymerase chain reaction (PCR), immunohistochemical testing (IHC), or next-generation sequencing (NGS) of tissue.1-3 However, tissue-based MSI assessment approaches have long turnaround times, including the time for obtaining samples. This hinders the ability to make rapid decisions with regard to immune checkpoint inhibitor (ICI) therapy, especially when choosing first-line treatment or for patients who require urgent treatment for aggressive disease.
CONTEXT
Key Objective
The convenience and rapid turnaround time of circulating tumor DNA (ctDNA) genotyping can potentially be useful as a microsatellite instability (MSI) testing option, especially when deciding on immunotherapy. Our aim was to compare the performance of plasma-based MSI assessment using a next-generation sequencing–based ctDNA assay with that of a validated tissue-based MSI test in patients with GI cancers.
Knowledge Generated
Our findings demonstrated the overall, positive, and negative percent agreement of 98.2%, 71.4%, and 99.1%, respectively. In patients whose plasma samples had a ctDNA fraction ≥ 1.0%, the positive percent agreement rose to 100.0%. Patients with MSI-high (MSI-H) tumors detected only by ctDNA genotyping achieved clinical benefits after receiving anti–programmed cell death 1 therapy. One patient with an aggressive cancer of an unknown primary site benefited from pembrolizumab after rapid detection of MSI-H by ctDNA genotyping.
Relevance
Our study supports the use of ctDNA genotyping in assessment of MSI in patients with GI cancers.
Circulating tumor DNA (ctDNA) has demonstrated its capability in the detection of genomic alterations when making therapeutic decisions in patients with advanced solid tumors.4,5 Plasma ctDNA genotyping has demonstrated markedly faster turnaround time and an accelerated enrollment in targeted trials while maintaining equivalent efficacy compared with tissue-based genotyping.6,7 NGS-based ctDNA genotyping can detect MSI by assessing the variability in microsatellite loci as used in the tissue NGS methods.8-10 The convenience and rapid turnaround time of ctDNA genotyping can potentially be useful as an MSI testing option, especially when deciding on ICI immunotherapy. However, no validation studies have compared ctDNA genotyping for MSI assessment with a tissue-based standard, and the efficacy of ICI treatment for patients with MSI-H tumors detected by ctDNA genotyping remains unknown.
Herein, we conducted a validation study comparing the ctDNA genotyping panel for MSI assessment with a validated PCR-based MSI test, which has been approved as a companion diagnostic for pembrolizumab in advanced solid tumors and for nivolumab with or without ipilimumab for metastatic colorectal cancer (CRC) in Japan. To evaluate the validity, we adopted the terms positive and negative percent agreements rather than sensitivity and specificity of ctDNA genotyping because the comparator is a nonreference standard method.11
PATIENTS AND METHODS
Study Design and Patients
This validation study aimed to compare the performance of plasma-based MSI assessment using Guardant360 (Guardant Health, Inc, Redwood City, CA), an NGS-based ctDNA assay, in the SCRUM-Japan GOZILA study with that of tissue-based MSI assessment. The MSI test kit (FALCO biosystems, Kyoto, Japan) approved in Japan, the MSI Analysis System Version 1.2 (Promega, Madison, WI), or an investigational use only (IUO) version of the MSI test kit (FALCO) was used. This study included patients with advanced GI cancers enrolled in GOZILA between November 1, 2018, and February 29, 2020, who had available plasma-based MSI results on Guardant360 and tissue-based MSI results.
GOZILA is a nationwide plasma genomic profiling study involving 31 core cancer institutions in Japan. Patients with metastatic GI cancers were eligible. Eligible patients provided written informed consent, including publication of any materials, and ctDNA genotyping was conducted using Guardant360. To avoid the suppression of ctDNA shedding because of chemotherapy, the patients were required to have disease progression during systemic chemotherapy and have not started subsequent therapy at the time of blood sampling. The SCRUM-Japan GI-SCREEN-MSI is a parallel nationwide study involving 26 institutions that assessed the tissue MSI status of patients with metastatic GI cancers. All institutions also participated in GOZILA. Patients with histopathologically confirmed metastatic GI cancer were eligible. This study was started in October 2015, and enrollment was completed in March 2019. MSI assessment was performed using the MSI Analysis System Version 1.2 from October 2015 to March 2018 and an IUO version of the MSI test kit (FALCO), which is identical to the MSI test kit (FALCO) approved in Japan, from April 2018 to March 2019. Eligible patients provided written informed consent, including publication of any materials. In clinical practice, the MSI test kit (FALCO) was used for tissue-based MSI assessment.
These studies were conducted in accordance with the Declaration of Helsinki and the Japanese Ethical Guidelines for Medical and Health Research Involving Human Subjects. All study protocols were approved by the institutional review board of each participating institution and registered at the University Hospital Medical Information Network Clinical Trials Registry (UMIN000042612).
ctDNA Genotyping and MSI Assessment in GOZILA
The NGS analysis of ctDNA was conducted using Guardant360 at Guardant Health, Inc, a Clinical Laboratory Improvement Amendments–certified, College of American Pathologists–accredited, New York State Department of Health–approved laboratory, as previously described.12 Guardant360 detects single-nucleotide variants (SNVs), indels, fusions, and copy number alterations in 74 genes.
Plasma-based MSI assessment was performed by sequencing microsatellite loci, as previously described.10 Briefly, the biologic instability was assessed for 90 microsatellite loci, and the sample was called positive if it had > 6 unstable loci (the MSI score). Plasma-based MSI assessment was included in GOZILA from November 2018.
Tissue MSI Testing
The tissue MSI status was tested using PCR-based methods. The MSI Analysis System Version 1.2 assessed the MSI status by comparing the allelic profiles of microsatellite markers generated by amplifying DNA from matched tumor and normal tissue samples. Alleles that were present in the tumor sample but not in their corresponding normal samples were indicative of the MSI. The system used five mononucleotide markers (NR-21, BAT-25, MONO-27, NR-24, and BAT-26), and tumors were considered as MSI-H if two or more mononucleotide loci varied in length compared with germline DNA. The MSI test kit (FALCO) detected MSI by assessing the unstable alleles of the five mononucleotide markers used in the MSI Analysis System Version 1.2, outside the quasimonomorphic variation range generated by amplifying DNA extracted from only tumor tissue samples. The complete concordance of the MSI status in metastatic CRC was determined between the MSI Analysis System Version 1.2 and an IUO assay of the MSI test kit (FALCO).13
End Points and Statistical Analysis
The primary end points were overall percent agreement (OPA), positive percent agreement (PPA), and negative percent agreement (NPA) between plasma MSI testing using Guardant360 for MSI detection and tissue MSI testing. For patients with tissue MSI results from both the MSI test kit (FALCO) and one of the assays included in the GI-SCREEN-MSI, the result of the MSI test kit (FALCO) was used. At the planning stage, we expected that plasma MSI testing would be considered effective if the OPA, PPA, and NPA were ≥ 90%, ≥ 70%, and ≥ 90%, respectively. These threshold values were determined on the basis of previous studies comparing ctDNA with tissue genotyping for the detection of genomic alterations, including MSI-H, in which a PPA of 60%–80% and a NPA of ≥ 90% were reported.10,14-17
Because MSI cannot be evaluated in plasma samples with a low ctDNA fraction (as measured by the maximum variant allelic fraction),10 the OPA, PPA, and NPA were assessed in a patient subset defined by various cutoff values of ctDNA fraction (≥ 0.1%, ≥ 0.2%, and ≥ 1.0%) as supplemental analyses. The ctDNA agreements were also evaluated compared with the MSI test kit (FALCO) and MSI testing performed in the GI-SCREEN-MSI. The best overall response and progression-free survival were evaluated in patients with MSI-H tumors detected by tissue or ctDNA testing who received anti–programmed cell death 1 (PD-1) therapy. Tumor response was assessed in patients with measurable lesions using the Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST v1.1). All analyses were conducted for all enrolled patients and for the subset of patients with CRC. The 95% CI was calculated using the Clopper-Pearson method. Statistical analyses were conducted using SAS (version 9.4).
RESULTS
Patients
Of 2,385 patients enrolled in GOZILA between November 1, 2018, and February 29, 2020, 671 patients underwent both tissue MSI testing and the Guardant360 test (Data Supplement). After excluding six patients with a failed tissue MSI test, two with a failed Guardant360 test, one with a canceled Guardant360 test, and four with non-GI cancers, 658 patients were finally included in the analysis. The patient characteristics are presented in the Data Supplement. The approved MSI test kit (FALCO) was used to obtain the tissue result for 537 patients, including 158 who also had results from an assay included in the GI-SCREEN-MSI study. The remaining 121 patients were included on the basis of the tissue results from a GI-SCREEN-MSI study assay. The median time from shipping samples to reporting results was seven days for the tissue MSI testing and eight days for the ctDNA genotyping.
Clinical Validation
The results of the MSI testing by tissue and ctDNA testing are presented in the Data Supplement. MSI-H tumors were detected in tumors of 21 (3.2%) of 658 patients using tissue-based tests. On plasma ctDNA genotyping, MSI-H tumors were detected in 71.4% (15 of 21; 95% CI, 47.8 to 88.7) of patients with a tissue MSI-H result but not detected in 99.1% (631 of 637; 95% CI, 98.0 to 99.7) of patients with tissue microsatellite-stable tumors (Table 1A). As a result, there was an OPA of 98.2% (646 of 658; 95% CI, 96.8 to 99.1), a positive predictive value (PPV) of 71.4% (15 of 21; 95% CI, 47.8 to 88.7), and a negative predictive value (NPV) of 99.1% (631 of 637; 95% CI, 98.0 to 99.7; Table 1A). Those with MSI-H tumors detected on ctDNA had significantly more SNVs and indels than those without MSI-H ctDNA (Fig 1). No MLH1 mutations were detected by ctDNA genotyping in patients with MSI-H tumors.
TABLE 1.
Comparison of Tissue and Plasma ctDNA Results for MSI Detection in Patients With GI Cancers
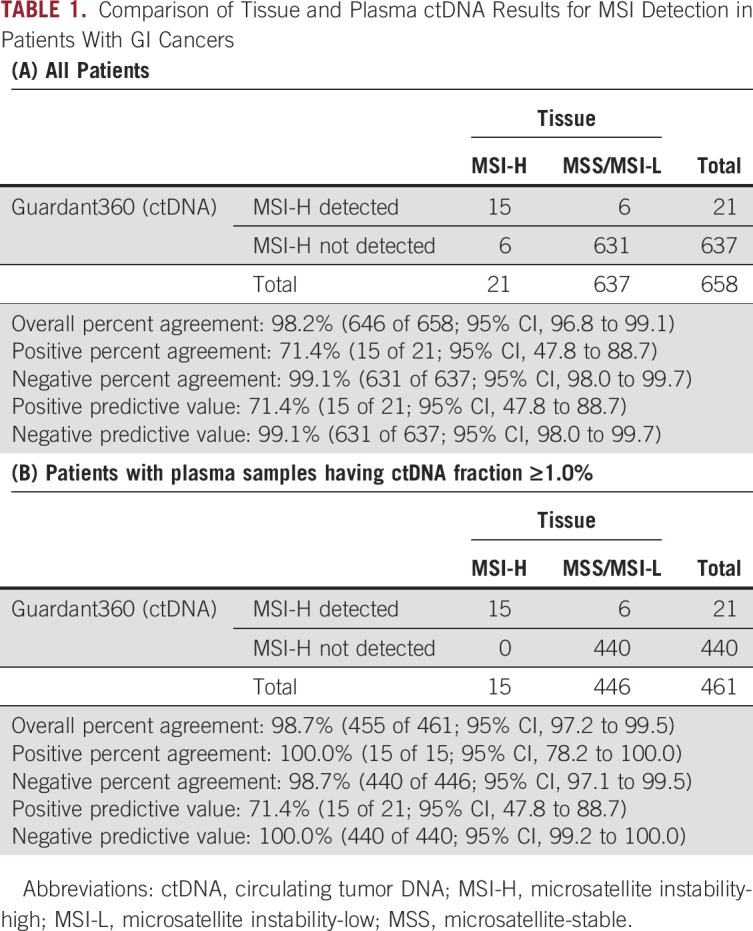
FIG 1.
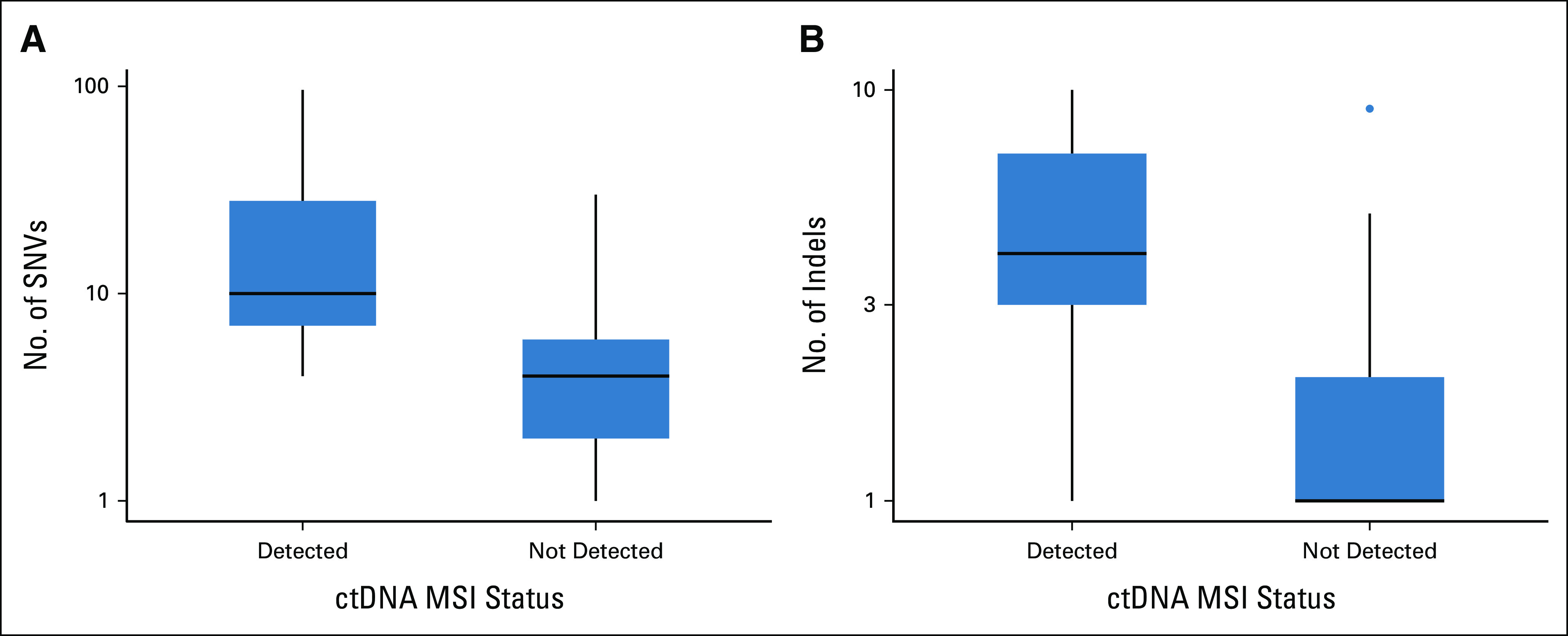
Number of (A) SNVs and (B) indels according to ctDNA MSI status. ctDNA, circulating tumor DNA; indels, insertions and deletions; MSI, microsatellite instability; SNV, single-nucleotide variant.
For the 347 patients with CRC, the OPA, PPA, NPA, PPV, and NPV were 99.4% (345 of 347; 95% CI, 97.9 to 99.9), 80.0% (8 of 10; 95% CI, 44.4 to 97.5), 100.0% (337 of 337; 95% CI, 98.9 to 100.0), 100.0% (8 of 8; 95% CI, 63.1 to 100.0), and 99.4% (337 of 339; 95% CI, 97.9 to 99.9), respectively (Data Supplement). The performance of MSI detection by ctDNA genotyping was better for samples with a higher ctDNA fraction (Table 1B and Data Supplement). In patients whose plasma samples had a ctDNA fraction of ≥ 1.0%, the PPA was 100.0% (15 of 15; 95% CI, 78.2 to 100.0) in the overall population and 100.0% (8 of 8; 95% CI, 63.1% to 100.0) in the CRC subset, but the NPA was similar to the overall data. The agreements were also similar in patients with tissue MSI results confirmed by the MSI test kit (FALCO) and in those tested by one of the assays used in the GI-SCREEN-MSI (Data Supplement).
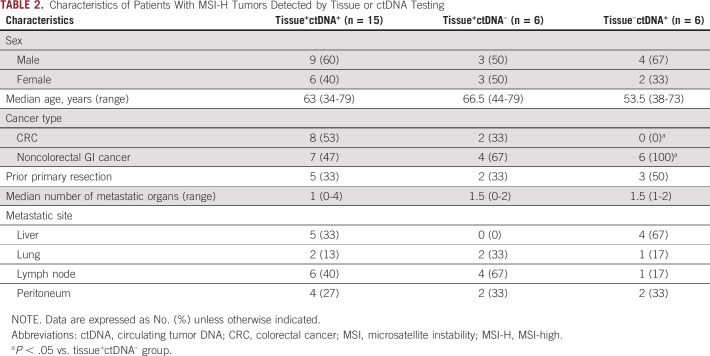
Next, we evaluated the characteristics associated with the discordance of MSI status between tissue and ctDNA testing (Table 2). All six patients with MSI-H tumors detected by ctDNA testing but not tissue testing (tissue–ctDNA+) had noncolorectal GI cancer. No patients with MSI-H in tissue alone (tissue+ctDNA–) had liver metastasis, whereas tissue+ctDNA+ or tissue–ctDNA+ groups included patients with liver metastasis. Consistent with the better performance of MSI detection in samples with a higher ctDNA fraction, the tissue+ctDNA– group had a significantly lower ctDNA fraction than other groups (Fig 2A). Prior treatments in tissue+ctDNA– groups are shown in the Data Supplement. The numbers of SNVs and indels were higher in the order of tissue+ctDNA+, tissue–ctDNA+, and tissue+ctDNA– groups (Figs 2B and 2C).
TABLE 2.
Characteristics of Patients With MSI-H Tumors Detected by Tissue or ctDNA Testing
FIG 2.
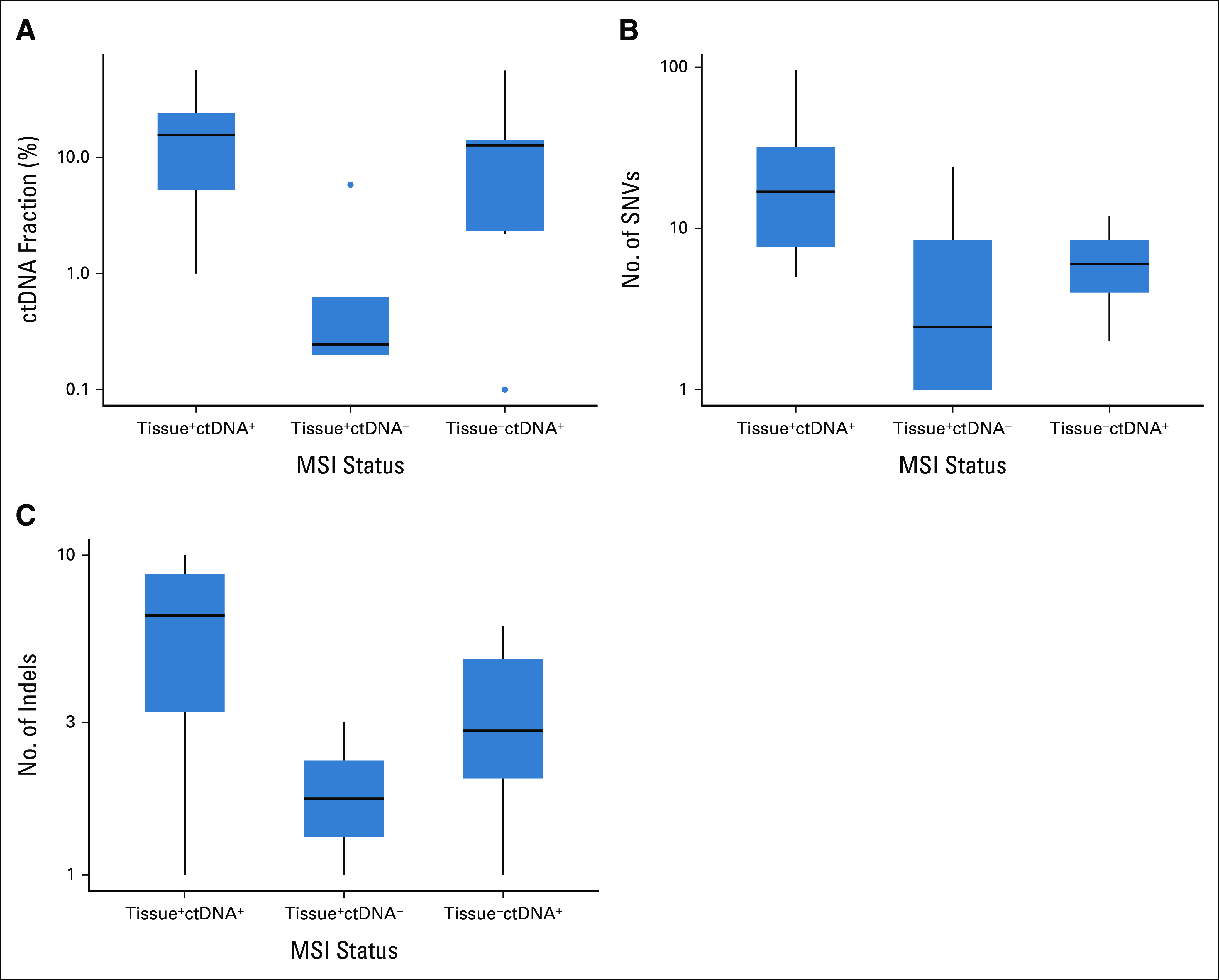
(A) ctDNA fraction and number of (B) SNVs and (C) indels according to tissue and ctDNA MSI status. ctDNA, circulating tumor DNA; indels, insertions and deletions; SNV, single-nucleotide variant.
The Efficacy of Anti–PD-1 Therapy in Patients With MSI-H Tumors Confirmed by ctDNA Genotyping
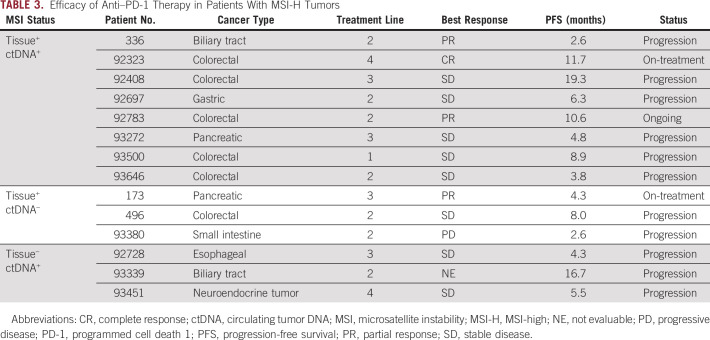
To evaluate the clinical utility of plasma-based MSI assessment, we studied the efficacy of anti–PD-1 therapy in patients with MSI-H tumors detected by tissue or ctDNA testing. In this study cohort, immunotherapy with an anti–PD-1 antibody was given to eight patients with MSI-H tumors confirmed by both tests, three confirmed by tissue MSI testing alone, and three confirmed by ctDNA genotyping alone. Table 3 presents the treatment efficacy in each subgroup. Only one patient had progressive disease as best response assessed per RECIST v1.1 with an obvious clinical disease progression, whereas 11 (78.6%) of 14 patients achieved a progression-free survival (PFS) of more than 4 months. Three patients in whom MSI-H was detected only by ctDNA achieved clinical benefits from anti–PD-1 therapy with the PFS ranging from 4.3 to 16.7 months.
TABLE 3.
Efficacy of Anti–PD-1 Therapy in Patients With MSI-H Tumors
We also present the case of a patient with cancer of an unknown primary site (CUP) with an aggressive clinical course who benefited from anti–PD-1 therapy on the basis of rapid detection of MSI-H by ctDNA genotyping. This patient was not included in the validation study cohort as she was enrolled in the GOZILA outside the study period. A 51-year-old woman presented to a local hospital with a 2-month history of lower back pain because of multiple enlarged lymph nodes. Upper GI endoscopy and colonoscopy did not reveal any abnormalities, but left inguinal lymph node biopsy demonstrated poorly differentiated adenocarcinoma positive for CDX2 and CK7 IHC staining but negative for CK7, TTF1, ER, and S100. She was urgently treated with modified leucovorin, fluorouracil, and oxaliplatin 6 (mFOLFOX6) as first-line treatment for presumptive CRC because her pain worsened with a decline in the Eastern Cooperative Oncology Group Performance Status to 2 (Fig 3). On the day of the start of chemotherapy, a blood sample was immediately collected for the Guardant360 test. Five days later, the ctDNA genotyping results revealed BRAF V600E and MSI-H. Seven days after the initiation of mFOLFOX6, we received a tumor tissue sample from the previous hospital and ordered RAS/BRAF and MSI tests. Tissue BRAF V600E was reported eight days later. Because the patient's status further deteriorated with a larger pleural effusion and lymph node metastases, the treatment was switched to pembrolizumab on the basis of MSI-H results by ctDNA genotyping (Fig 3). On the following day, a nasogastric tube was placed because of duodenal stenosis caused by abdominal lymph node metastases. A negative tissue MSI-H result was reported 19 days after ordering, but the patient's general status gradually improved along with a decreased carbohydrate antigen 19-9 level. The nasogastric tube was removed because of the improvement of the duodenal stenosis 19 days after pembrolizumab initiation. Computed tomography scan obtained 26 days after pembrolizumab initiation revealed improvements in the pleural effusion and decreased size of the lymph node metastases.
FIG 3.
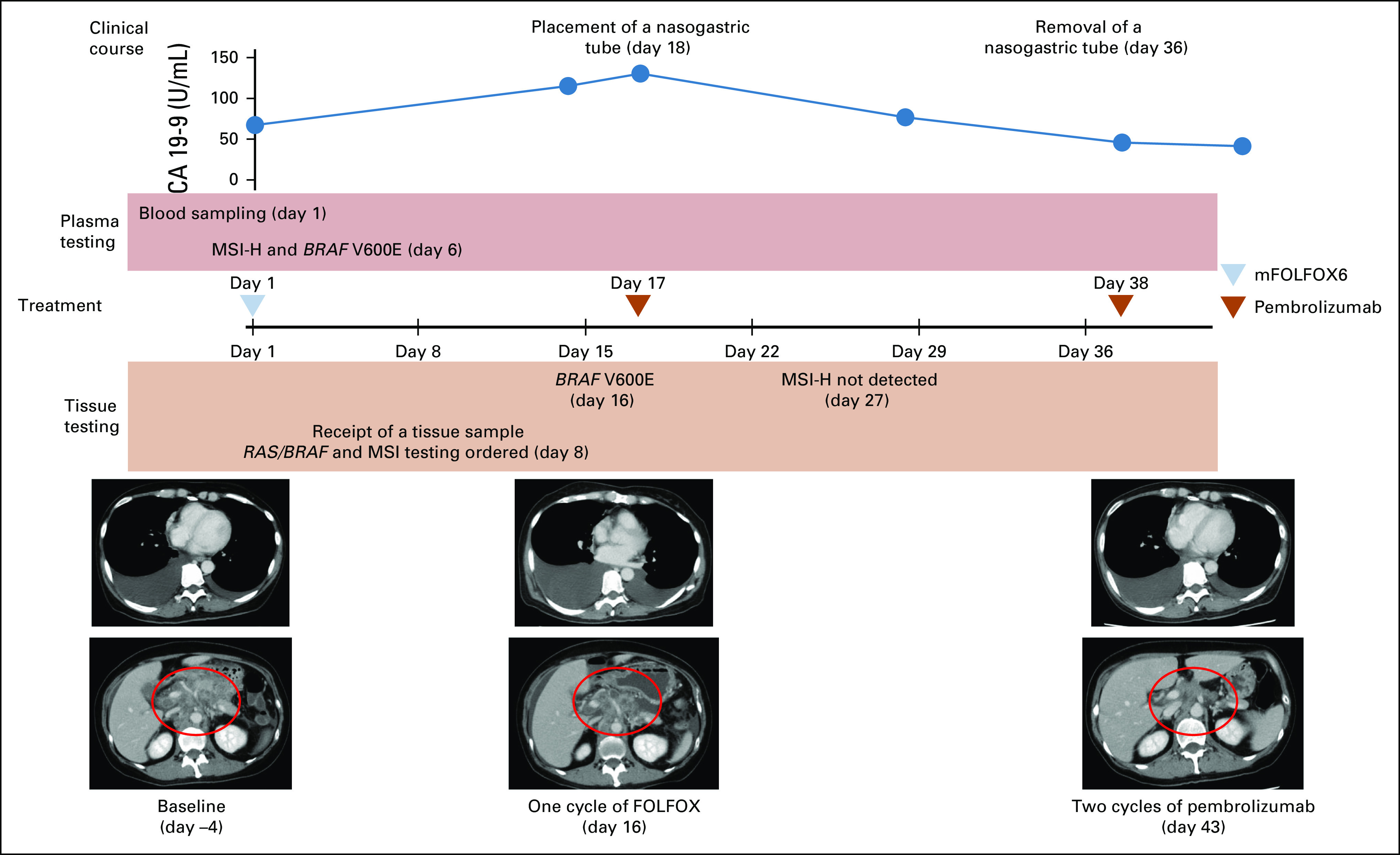
Clinical presentation of patients with MSI-H cancer with an unknown primary site confirmed by ctDNA genotyping. CA 19-9, carbohydrate antigen 19-9; ctDNA, circulating tumor DNA; FOLFOX, infusional fluorouracil, leucovorin, and oxaliplatin; mFOLFOX6, modified FOLFOX6; MSI, microsatellite instability; MSI-H, MSI-high.
DISCUSSION
This validation study of ctDNA genotyping for MSI assessment compared with tissue MSI testing met its primary end point by demonstrating an OPA of 98.2%, a PPA of 71.4%, and a NPA of 99.1% in patients with GI cancers and an OPA of 99.4%, a PPA of 80.0%, and a NPA of 100.0% in those with CRC. The performance of ctDNA genotyping was similar regardless of the specific tissue test comparator, whether it was via the MSI test kit (FALCO) used in standard clinical practice or the MSI testing performed in the GI-SCREEN-MSI. Despite the limited number of patients, 78.6% of those with MSI-H tumors confirmed by either tissue or ctDNA testing clinically benefited from anti–PD-1 therapy, with a PFS longer than 4.0 months. This is similar to the median PFS after pembrolizumab for patients with advanced-stage MSI-H tumors.18,19
MSI-H was not detected using ctDNA genotyping in six of 21 patients with tissue-confirmed MSI-H tumors. Previous validation studies of ctDNA genotyping for detection of genomic alterations (eg, EGFR mutations in non–small-cell lung cancer, RAS mutations in CRC, and MSI status in CRC) versus tissue analysis have indicated PPA values of 60%-80%.10,14-17 These high but imperfect levels of sensitivity are possibly related to variations in the amounts of ctDNA shed into the plasma, with a decline in sensitivity in smaller concentrations of ctDNA. Supporting this hypothesis, the PPA was perfect in patients with a ctDNA fraction of at least 1.0%. Tissue+ctDNA– population in our study had fewer liver metastasis, which is known to be associated with ctDNA shedding,16,20 and a significantly lower ctDNA fraction. These findings indicate that tissue testing may be required in cases wherein ctDNA genotyping is negative for MSI, but there is a low ctDNA fraction (eg, < 1%). Thus, plasma-based MSI assessment may be less likely to detect MSI-H in patients with tumor characteristics, which likely results in a low ctDNA fraction, such as low tumor burden, no liver metastasis, isolated lung metastases, and specific cancer types (eg, primary brain tumors and renal cell carcinoma).20-25
Conversely, some patients had tissue-negative but ctDNA-positive MSI-H tumors. The NGS-based MSI assessment supposedly has higher performance in the evaluation of MSI status in non-CRCs compared with PCR-based testing developed primarily for CRC.1,26 Indeed, all six patients with tissue-negative and ctDNA-positive MSI tumors were non-CRC cases, whereas the NPA in patients with CRC was 100%. Furthermore, the ctDNA analytical results were informed by the molecular characteristics of all tumor cells, which may be heterogeneous within and between metastatic sites. One study demonstrated discrete tumor populations of microsatellite-stable and MSI-H cells assessed by both IHC and PCR in a single tumor mass and discordant MSI status between paired tissue biopsies in the same patient.27 Indeed, numbers of SNVs and indels in the tissue–ctDNA+ group were lower than those in the tissue+ctDNA+ group in our study, suggesting heterogeneity with mixed MSI-H and non–MSI-H cells.
The efficacy of anti–PD-1 therapy as observed in the validation study cohort and in our case report of the patient with CUP suggested its benefit for patients with MSI-H detected by ctDNA genotyping alone. The patient with CUP required urgent and precise treatment because of the aggressive and advanced tumor. Given the time taken for the sample acquisition, the turnaround time for MSI assessment was significantly shorter in ctDNA genotyping than in tissue testing. In this patient, the negative tissue MSI result was reported 21 days after the report of ctDNA genotyping. Even if the tissue MSI result had turned positive, the patient might not have benefited because of the delayed initiation of pembrolizumab owing to the delayed results, considering the rapidly progressive and symptomatic nature of the patient's disease. The speed of plasma-based MSI assessment and its association with the efficacy of anti–PD-1 therapy strongly support the accuracy and the value of the test, especially for the determination of first-line treatment or for patients who require urgent treatment.
Non-GI cancer cases were excluded in this study although around 70% of patients undergoing MSI testing in clinical practice have GI cancer.4 However, in a previous study of MSI assessment using Guardant360, the distribution of MSI score was similar across cancer types and the prevalence of ctDNA-detected MSI-H among various tumor types was consistent with that reported for tissue-based MSI-H testing.10 These findings suggest that this plasma-based MSI assessment can be applied across cancer types.
In conclusion, ctDNA genotyping was able to detect MSI with a high concordance with a validated tissue-based MSI test, especially in patients with tumors that have sufficient ctDNA shedding. Furthermore, NGS-based ctDNA genotyping enabled the identification of patients with MSI-H tumors who can benefit from early ICI treatment, which would have been missed by PCR-based tissue testing. Therefore, plasma-based MSI assessment can be a useful alternative method for detecting MSI-H tumors, especially in patients who require quick therapeutic decisions (eg, first-line treatment) or who have aggressive disease or when an adequate tissue sample is unavailable.
Yoshiaki Nakamura
Research Funding: Taiho Pharmaceutical (Inst), Guardant Health (Inst), Genomedia (Inst), Chugai Pharma (Inst), Guardant Health (Inst), Seattle Genetics (Inst), Roche Diagnostics Japan (Inst)
Wataru Okamoto
Honoraria: Chugai Pharma, Ono Pharmaceutical, Bristol Myers Squibb Japan, Yakult Honsha, Lilly Japan, Thermo Fisher Scientific, Takeda, Novartis, Taiho Pharmaceutical
Research Funding: Janssen Oncology (Inst)
Tadamichi Denda
Honoraria: Sawai Pharmaceutical Co
Speakers' Bureau: Sysmex, Ono Pharmaceutical
Research Funding: MSD (Inst), Ono Pharmaceutical (Inst), Daiichi Sankyo Company, Limited (Inst)
Tomohiro Nishina
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Lilly, Merck Serono, Takeda, Bristol Myers Squibb, Ono Pharmaceutical
Research Funding: Taiho Pharmaceutical (Inst), Chugai Pharma (Inst), Dainippon Sumitomo Pharma (Inst), Lilly Japan (Inst), Merck Serono (Inst), MSD (Inst), Daiichi Sankyo (Inst), Ono Pharmaceutical (Inst), Eisai (Inst)
Yoshito Komatsu
Honoraria: Lilly Japan, Taiho Pharmaceutical, Chugai Pharma, Takeda, Bayer Yakuhin, Bristol Myers Squibb Co, Sanofi/Aventis, Merck, Yakult Honsha, Ono Pharmaceutical, Nipro Corporation, Moroo Co, Asahi Kasei, Mitsubishi Tanabe Pharma, Otsuka, Medical Review Co., Ltd, Daiichi Sankyo
Research Funding: MSD K.K, Taiho Pharmaceutical, Yakult Honsha, Bayer Yakuhin, Daiichi Sankyo Co, Ltd, Ono Pharmaceutical, NanoCarrier, Eisai, Sanofi/Aventis, Sysmex, Shionogi, IQVIA, Parexel International Corporation, Astellas Pharma, Mediscience Planning, Sumitomo Dainippon Pharma Co, Ltd, A2 Healthcare, Incyte, Lilly (Inst), Nipro Corporation (Inst), BeiGene (Inst)
Satoshi Yuki
Honoraria: Chugai Pharma, Takeda, Lilly Japan, Bayer Yakuhin, Taiho Pharmaceutical, Sanofi, Yakult Honsha, Bristol Myers Squibb Japan, Ono Pharmaceutical, Merck, MSD K.K
Hisateru Yasui
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Bristol Myers Squibb Japan, Daiichi Sankyo, Lilly Japan, Yakult Honsha, Bayer Yakuhin, Takeda, Ono Pharmaceutical
Consulting or Advisory Role: Ono Pharmaceutical
Research Funding: MSD (Inst), Daiichi Sankyo (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst)
Taito Esaki
Honoraria: Lilly, Taiho Pharmaceutical, Daiichi Sankyo, Chugai Pharma, Ono Pharmaceutical, Sanofi, MSD
Research Funding: Daiichi Sankyo (Inst), MSD (Inst), Novartis (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst), Lilly (Inst), Bayer (Inst), Nihon Kayaku (Inst), Amgen Astellas BioPharma (Inst), Parexel (Inst), IQVIA (Inst), Quintiles (Inst), Eisai (Inst), Sumitomo Group (Inst), Taiho Pharmaceutical (Inst)
Yu Sunakawa
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Takeda, Yakult Honsha, Bayer Yakuhin, Bristol Myers Squibb Japan, Lilly Japan, Merck, Sysmex, MSD K.K, Ono Pharmaceutical, Daiichi Sankyo
Consulting or Advisory Role: Bristol Myers Squibb Japan, MSD K.K, Merck, Daiichi Sankyo
Research Funding: Taiho Pharmaceutical, Takeda, Chugai Pharma, Lilly Japan, Sanofi, Nihon Kayaku, Otsuka
Makoto Ueno
Honoraria: Taiho Pharmaceutical, Yakult Honsha, AstraZeneca, Merck Serono, MSD, Ono Pharmaceutical, Servier, Chugai Pharma (I)
Research Funding: Taiho Pharmaceutical (Inst), Eisai (Inst), AstraZeneca (Inst), Ono Pharmaceutical (Inst), MSD (Inst), Merck Serono (Inst), Incyte (Inst), Astellas Pharma, Chugai Pharma, Delta-Fly Pharma, JPH Clinical Development
Eiji Shinozaki
Honoraria: Yakult Honsha, Merck Serono, Chugai Pharma, Takeda, Sanofi, Lilly, Taiho Pharmaceutical, Daiichi Sankyo/UCB Japan
Takashi Ohta
Honoraria: Bristol Myers Squibb Japan, Chugai Pharma, Teijin Pharma, Takeda, Taiho Pharmaceutical, Eisai
Ken Kato
Honoraria: Lilly, BMS, Ono Pharmaceutical
Consulting or Advisory Role: Ono Pharmaceutical, BeiGene, MSD, Oncolys BioPharma, Bayer
Speakers' Bureau: Ono Pharmaceutical, Bristol Myers Squibb Japan, MSD
Research Funding: Ono Pharmaceutical (Inst), Shionogi (Inst), MSD Oncology (Inst), BeiGene (Inst), Chugai Pharma (Inst), Bayer (Inst), AstraZeneca (Inst), Taiho Pharmaceutical (Inst)
Hideaki Bando
Honoraria: Taiho Pharmaceutical, Takeda, Chugai Pharma, Sanofi, Yakult Honsha, Merck Serono, Lilly Japan, Ono Pharmaceutical, Bristol Myers Squibb Japan
Hiroki Hara
Honoraria: Chugai Pharma, Taiho Pharmaceutical, Merck Serono, Yakult Honsha, Lilly, Ono Pharmaceutical, Takeda, Bristol Myers Squibb, Sanofi, MSD, Daiichi Sankyo, Kyowa Hakko Kirin, Bayer
Consulting or Advisory Role: Ono Pharmaceutical, MSD, Lilly, Boehringer Ingelheim, Dainippon Sumitomo
Research Funding: AstraZeneca (Inst), Chugai Pharma (Inst), Merck Serono (Inst), MSD (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Boehringer Ingelheim (Inst), Dainippon Sumitomo Pharma (Inst), Daiichi Sankyo (Inst), Pfizer (Inst), Eisai (Inst), Incyte (Inst), BeiGene (Inst), Astellas Pharma (Inst), Bayer (Inst), Elevar Therapeutics (Inst)
Taroh Satoh
Honoraria: Chugai Pharma, Merck Serono, Bristol Myers Squibb, Takeda, Yakult Honsha, Lilly, Bayer Yakuhin, Ono Pharmaceutical, Merck, Astellas Pharma, Taiho Pharmaceutical, Nihon Kayaku, Daiichi-Sankyo
Consulting or Advisory Role: Bayer Yakuhin, Lilly, Ono Pharmaceutical, Takara Bio, Merck Serono, Nihon Kayaku
Research Funding: Yakult Honsha (Inst), Chugai Pharma (Inst), Ono Pharmaceutical (Inst), Sanofi (Inst), Lilly (Inst), Daiichi Sankyo (Inst), Merck (Inst), Merck Serono (Inst), Gilead Sciences (Inst), Dainippon Sumitomo Pharma (Inst), IQVIA (Inst)
Kentaro Yamazaki
Honoraria: Chugai Pharma, Daiichi Sankyo, Yakult Honsha, Takeda, Bayer, Merck Serono, Taiho Pharmaceutical, Lilly, Sanofi, Ono Pharmaceutical, MSD, Bristol Myers Squibb
Research Funding: Taiho Pharmaceutical (Inst)
Yoshiyuki Yamamoto
Honoraria: Taiho Pharmaceutical, Ono Pharmaceutical, Nihon Kayaku, Merck Serono, Yakult Pharmaceutical, Eisai, Takeda, Daiichi Sankyo, Lilly, SERVIER, Bayer
Naohiro Okano
Honoraria: Taiho Pharmaceutical, Bayer Yakuhin, Lilly Japan, Chugai Pharma, Ono Pharmaceutical, Takeda, Kyowa Hakko Kirin, Eisai
Consulting or Advisory Role: GlaxoSmithKline
Tetsuji Terazawa
Employment: Shionogi
Honoraria: Chugai Pharma, Lilly Co, Ltd, Taiho Pharmaceutical, Sanofi Co, Ltd, Chugai Pharma
Takeshi Kato
Honoraria: Chugai Pharma, Yakult Honsha, Ono Pharmaceutical, Takeda, Lilly Japan, Taiho Pharmaceutical, Asahi Kasei
Research Funding: Chugai Pharma
Eiji Oki
Speakers' Bureau: Chugai Pharma, Lilly Japan, Takeda, Ono Pharmaceutical, Bayer Yakuhin, Bristol Myers Squibb Japan
Akihito Tsuji
Speakers' Bureau: Chugai Pharma, Taiho Pharmaceutical, Merck Serono, Takeda Pharmaceutical Company Limited, Lilly Japan, Bristol Myers Squibb Corporation, Sanofi
Research Funding: Taiho Pharmaceutical Co, Ltd (Inst), Sanofi (Inst), Ono Pharmaceutical (Inst)
Yasuo Hamamoto
Consulting or Advisory Role: Dainippon Sumitomo Pharma, Chugai Pharma
Research Funding: Ono Pharmaceutical, Bristol Myers Squibb Japan, MSD Oncology, Lilly Japan
Akihito Kawazoe
Honoraria: Ono Pharmaceutical, Taiho Pharmaceutical, Bristol Myers Squibb
Speakers' Bureau: Taiho Pharmaceutical, Ono Pharmaceutical, Bristol Myers Squibb
Research Funding: Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst)
Hiromichi Nakajima
Employment: Otsuka (I)
Honoraria: Chugai Pharma, Takeda
Speakers' Bureau: Chugai Pharma, Takeda
Shogo Nomura
Employment: Asahi Kasei (I)
Honoraria: AstraZeneca, Chugai Pharma, Kyowa Hakko Bio
Research Funding: Amgen Astellas BioPharma (Inst), AstraZeneca (Inst)
Takayuki Yoshino
Honoraria: Chugai Pharma, Merck, Bayer Yakuhin, Ono Pharmaceutical, Lilly, Taiho Pharmaceutical
Research Funding: Chugai Pharma (Inst), MSD (Inst), Daiichi Sankyo Company, Limited (Inst), Parexel International Inc (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Amgen (Inst), Sanofi (Inst)
No other potential conflicts of interest were reported.
DATA SHARING STATEMENT
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/PO.21.00383.
AUTHOR CONTRIBUTIONS
Conception and design: Yoshiaki Nakamura, Shogo Nomura, Takayuki Yoshino
Administrative support: Hideaki Bando
Provision of study materials or patients: Tomohiro Nishina, Yoshito Komatsu, Satoshi Yuki, Yu Sunakawa, Eiji Shinozaki, Nobuhisa Matsuhashi, Ken Kato, Koushiro Ohtsubo, Hiroki Hara, Taroh Satoh, Kentaro Yamazaki, Naohiro Okano, Takeshi Kato, Eiji Oki, Akihito Tsuji, Yasuo Hamamoto, Akihito Kawazoe
Collection and assembly of data: Yoshiaki Nakamura, Wataru Okamoto, Tadamichi Denda, Tomohiro Nishina, Yoshito Komatsu, Satoshi Yuki, Hisateru Yasui, Taito Esaki, Yu Sunakawa, Makoto Ueno, Eiji Shinozaki, Nobuhisa Matsuhashi, Takashi Ohta, Ken Kato, Koushiro Ohtsubo, Hideaki Bando, Hiroki Hara, Taroh Satoh, Kentaro Yamazaki, Yoshiyuki Yamamoto, Naohiro Okano, Tetsuji Terazawa, Takeshi Kato, Eiji Oki, Akihito Tsuji, Yosuke Horita, Yasuo Hamamoto, Akihito Kawazoe, Hiromichi Nakajima, Ryuta Mitani, Mihoko Yuasa, Takayuki Yoshino
Data analysis and interpretation: Yoshiaki Nakamura, Shogo Nomura
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Yoshiaki Nakamura
Research Funding: Taiho Pharmaceutical (Inst), Guardant Health (Inst), Genomedia (Inst), Chugai Pharma (Inst), Guardant Health (Inst), Seattle Genetics (Inst), Roche Diagnostics Japan (Inst)
Wataru Okamoto
Honoraria: Chugai Pharma, Ono Pharmaceutical, Bristol Myers Squibb Japan, Yakult Honsha, Lilly Japan, Thermo Fisher Scientific, Takeda, Novartis, Taiho Pharmaceutical
Research Funding: Janssen Oncology (Inst)
Tadamichi Denda
Honoraria: Sawai Pharmaceutical Co
Speakers' Bureau: Sysmex, Ono Pharmaceutical
Research Funding: MSD (Inst), Ono Pharmaceutical (Inst), Daiichi Sankyo Company, Limited (Inst)
Tomohiro Nishina
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Lilly, Merck Serono, Takeda, Bristol Myers Squibb, Ono Pharmaceutical
Research Funding: Taiho Pharmaceutical (Inst), Chugai Pharma (Inst), Dainippon Sumitomo Pharma (Inst), Lilly Japan (Inst), Merck Serono (Inst), MSD (Inst), Daiichi Sankyo (Inst), Ono Pharmaceutical (Inst), Eisai (Inst)
Yoshito Komatsu
Honoraria: Lilly Japan, Taiho Pharmaceutical, Chugai Pharma, Takeda, Bayer Yakuhin, Bristol Myers Squibb Co, Sanofi/Aventis, Merck, Yakult Honsha, Ono Pharmaceutical, Nipro Corporation, Moroo Co, Asahi Kasei, Mitsubishi Tanabe Pharma, Otsuka, Medical Review Co., Ltd, Daiichi Sankyo
Research Funding: MSD K.K, Taiho Pharmaceutical, Yakult Honsha, Bayer Yakuhin, Daiichi Sankyo Co, Ltd, Ono Pharmaceutical, NanoCarrier, Eisai, Sanofi/Aventis, Sysmex, Shionogi, IQVIA, Parexel International Corporation, Astellas Pharma, Mediscience Planning, Sumitomo Dainippon Pharma Co, Ltd, A2 Healthcare, Incyte, Lilly (Inst), Nipro Corporation (Inst), BeiGene (Inst)
Satoshi Yuki
Honoraria: Chugai Pharma, Takeda, Lilly Japan, Bayer Yakuhin, Taiho Pharmaceutical, Sanofi, Yakult Honsha, Bristol Myers Squibb Japan, Ono Pharmaceutical, Merck, MSD K.K
Hisateru Yasui
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Bristol Myers Squibb Japan, Daiichi Sankyo, Lilly Japan, Yakult Honsha, Bayer Yakuhin, Takeda, Ono Pharmaceutical
Consulting or Advisory Role: Ono Pharmaceutical
Research Funding: MSD (Inst), Daiichi Sankyo (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst)
Taito Esaki
Honoraria: Lilly, Taiho Pharmaceutical, Daiichi Sankyo, Chugai Pharma, Ono Pharmaceutical, Sanofi, MSD
Research Funding: Daiichi Sankyo (Inst), MSD (Inst), Novartis (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst), Lilly (Inst), Bayer (Inst), Nihon Kayaku (Inst), Amgen Astellas BioPharma (Inst), Parexel (Inst), IQVIA (Inst), Quintiles (Inst), Eisai (Inst), Sumitomo Group (Inst), Taiho Pharmaceutical (Inst)
Yu Sunakawa
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Takeda, Yakult Honsha, Bayer Yakuhin, Bristol Myers Squibb Japan, Lilly Japan, Merck, Sysmex, MSD K.K, Ono Pharmaceutical, Daiichi Sankyo
Consulting or Advisory Role: Bristol Myers Squibb Japan, MSD K.K, Merck, Daiichi Sankyo
Research Funding: Taiho Pharmaceutical, Takeda, Chugai Pharma, Lilly Japan, Sanofi, Nihon Kayaku, Otsuka
Makoto Ueno
Honoraria: Taiho Pharmaceutical, Yakult Honsha, AstraZeneca, Merck Serono, MSD, Ono Pharmaceutical, Servier, Chugai Pharma (I)
Research Funding: Taiho Pharmaceutical (Inst), Eisai (Inst), AstraZeneca (Inst), Ono Pharmaceutical (Inst), MSD (Inst), Merck Serono (Inst), Incyte (Inst), Astellas Pharma, Chugai Pharma, Delta-Fly Pharma, JPH Clinical Development
Eiji Shinozaki
Honoraria: Yakult Honsha, Merck Serono, Chugai Pharma, Takeda, Sanofi, Lilly, Taiho Pharmaceutical, Daiichi Sankyo/UCB Japan
Takashi Ohta
Honoraria: Bristol Myers Squibb Japan, Chugai Pharma, Teijin Pharma, Takeda, Taiho Pharmaceutical, Eisai
Ken Kato
Honoraria: Lilly, BMS, Ono Pharmaceutical
Consulting or Advisory Role: Ono Pharmaceutical, BeiGene, MSD, Oncolys BioPharma, Bayer
Speakers' Bureau: Ono Pharmaceutical, Bristol Myers Squibb Japan, MSD
Research Funding: Ono Pharmaceutical (Inst), Shionogi (Inst), MSD Oncology (Inst), BeiGene (Inst), Chugai Pharma (Inst), Bayer (Inst), AstraZeneca (Inst), Taiho Pharmaceutical (Inst)
Hideaki Bando
Honoraria: Taiho Pharmaceutical, Takeda, Chugai Pharma, Sanofi, Yakult Honsha, Merck Serono, Lilly Japan, Ono Pharmaceutical, Bristol Myers Squibb Japan
Hiroki Hara
Honoraria: Chugai Pharma, Taiho Pharmaceutical, Merck Serono, Yakult Honsha, Lilly, Ono Pharmaceutical, Takeda, Bristol Myers Squibb, Sanofi, MSD, Daiichi Sankyo, Kyowa Hakko Kirin, Bayer
Consulting or Advisory Role: Ono Pharmaceutical, MSD, Lilly, Boehringer Ingelheim, Dainippon Sumitomo
Research Funding: AstraZeneca (Inst), Chugai Pharma (Inst), Merck Serono (Inst), MSD (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Boehringer Ingelheim (Inst), Dainippon Sumitomo Pharma (Inst), Daiichi Sankyo (Inst), Pfizer (Inst), Eisai (Inst), Incyte (Inst), BeiGene (Inst), Astellas Pharma (Inst), Bayer (Inst), Elevar Therapeutics (Inst)
Taroh Satoh
Honoraria: Chugai Pharma, Merck Serono, Bristol Myers Squibb, Takeda, Yakult Honsha, Lilly, Bayer Yakuhin, Ono Pharmaceutical, Merck, Astellas Pharma, Taiho Pharmaceutical, Nihon Kayaku, Daiichi-Sankyo
Consulting or Advisory Role: Bayer Yakuhin, Lilly, Ono Pharmaceutical, Takara Bio, Merck Serono, Nihon Kayaku
Research Funding: Yakult Honsha (Inst), Chugai Pharma (Inst), Ono Pharmaceutical (Inst), Sanofi (Inst), Lilly (Inst), Daiichi Sankyo (Inst), Merck (Inst), Merck Serono (Inst), Gilead Sciences (Inst), Dainippon Sumitomo Pharma (Inst), IQVIA (Inst)
Kentaro Yamazaki
Honoraria: Chugai Pharma, Daiichi Sankyo, Yakult Honsha, Takeda, Bayer, Merck Serono, Taiho Pharmaceutical, Lilly, Sanofi, Ono Pharmaceutical, MSD, Bristol Myers Squibb
Research Funding: Taiho Pharmaceutical (Inst)
Yoshiyuki Yamamoto
Honoraria: Taiho Pharmaceutical, Ono Pharmaceutical, Nihon Kayaku, Merck Serono, Yakult Pharmaceutical, Eisai, Takeda, Daiichi Sankyo, Lilly, SERVIER, Bayer
Naohiro Okano
Honoraria: Taiho Pharmaceutical, Bayer Yakuhin, Lilly Japan, Chugai Pharma, Ono Pharmaceutical, Takeda, Kyowa Hakko Kirin, Eisai
Consulting or Advisory Role: GlaxoSmithKline
Tetsuji Terazawa
Employment: Shionogi
Honoraria: Chugai Pharma, Lilly Co, Ltd, Taiho Pharmaceutical, Sanofi Co, Ltd, Chugai Pharma
Takeshi Kato
Honoraria: Chugai Pharma, Yakult Honsha, Ono Pharmaceutical, Takeda, Lilly Japan, Taiho Pharmaceutical, Asahi Kasei
Research Funding: Chugai Pharma
Eiji Oki
Speakers' Bureau: Chugai Pharma, Lilly Japan, Takeda, Ono Pharmaceutical, Bayer Yakuhin, Bristol Myers Squibb Japan
Akihito Tsuji
Speakers' Bureau: Chugai Pharma, Taiho Pharmaceutical, Merck Serono, Takeda Pharmaceutical Company Limited, Lilly Japan, Bristol Myers Squibb Corporation, Sanofi
Research Funding: Taiho Pharmaceutical Co, Ltd (Inst), Sanofi (Inst), Ono Pharmaceutical (Inst)
Yasuo Hamamoto
Consulting or Advisory Role: Dainippon Sumitomo Pharma, Chugai Pharma
Research Funding: Ono Pharmaceutical, Bristol Myers Squibb Japan, MSD Oncology, Lilly Japan
Akihito Kawazoe
Honoraria: Ono Pharmaceutical, Taiho Pharmaceutical, Bristol Myers Squibb
Speakers' Bureau: Taiho Pharmaceutical, Ono Pharmaceutical, Bristol Myers Squibb
Research Funding: Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst)
Hiromichi Nakajima
Employment: Otsuka (I)
Honoraria: Chugai Pharma, Takeda
Speakers' Bureau: Chugai Pharma, Takeda
Shogo Nomura
Employment: Asahi Kasei (I)
Honoraria: AstraZeneca, Chugai Pharma, Kyowa Hakko Bio
Research Funding: Amgen Astellas BioPharma (Inst), AstraZeneca (Inst)
Takayuki Yoshino
Honoraria: Chugai Pharma, Merck, Bayer Yakuhin, Ono Pharmaceutical, Lilly, Taiho Pharmaceutical
Research Funding: Chugai Pharma (Inst), MSD (Inst), Daiichi Sankyo Company, Limited (Inst), Parexel International Inc (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Amgen (Inst), Sanofi (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Hause RJ, Pritchard CC, Shendure J, et al. Classification and characterization of microsatellite instability across 18 cancer types Nat Med 221342–13502016 [DOI] [PubMed] [Google Scholar]
- 2.Bonneville R, Krook MA, Kautto EA, et al. Landscape of microsatellite instability across 39 cancer types JCO Precis Oncol 20171–152017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Middha S, Zhang L, Nafa K, et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data JCO Precis Oncol 20171–172017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura Y, Yoshino T.Clinical utility of analyzing circulating tumor DNA in patients with metastatic colorectal cancer Oncologist 231310–13182018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakamura Y, Shitara K. Development of circulating tumour DNA analysis for gastrointestinal cancers. ESMO Open. 2020;5:e000600. doi: 10.1136/esmoopen-2019-000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leighl NB, Page RD, Raymond VM, et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer Clin Cancer Res 254691–47002019 [DOI] [PubMed] [Google Scholar]
- 7.Nakamura Y, Taniguchi H, Ikeda M, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies Nat Med 261859–18642020 [DOI] [PubMed] [Google Scholar]
- 8. Ladas I, Yu F, Leong KW, et al. Enhanced detection of microsatellite instability using pre-PCR elimination of wild-type DNA homo-polymers in tissue and liquid biopsies. Nucleic Acids Res. 2018;46:e74. doi: 10.1093/nar/gky251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mayrhofer M, De Laere B, Whitington T, et al. Cell-free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome Med. 2018;10:85. doi: 10.1186/s13073-018-0595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willis J, Lefterova MI, Artyomenko A, et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel Clin Cancer Res 257035–70452019 [DOI] [PubMed] [Google Scholar]
- 11.US Food and Drug Administration . Guidance for Industry and FDA Staff: Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests. Silver Spring, MD: US Food and Drug Administration; 2007. [Google Scholar]
- 12.Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies Clin Cancer Res 243539–35492018 [DOI] [PubMed] [Google Scholar]
- 13.Bando H, Okamoto W, Fukui T, et al. Utility of the quasi-monomorphic variation range in unresectable metastatic colorectal cancer patients Cancer Sci 1093411–34152018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer JAMA Oncol 21014–10222016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray JE, Okamoto I, Sriuranpong V, et al. Tissue and plasma EGFR mutation analysis in the FLAURA trial: Osimertinib versus comparator EGFR tyrosine kinase inhibitor as first-line treatment in patients with EGFR-mutated advanced non-small cell lung cancer Clin Cancer Res 256644–66522019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bando H, Kagawa Y, Kato T, et al. A multicentre, prospective study of plasma circulating tumour DNA test for detecting RAS mutation in patients with metastatic colorectal cancer Br J Cancer 120982–9862019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Woodhouse R, Li M, Hughes J, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One. 2020;15:e0237802. doi: 10.1371/journal.pone.0237802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le DT, Kim TW, Van Cutsem E, et al. Phase II open-label study of pembrolizumab in treatment-refractory, microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: KEYNOTE-164 J Clin Oncol 3811–192020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marabelle A, Le DT, Ascierto PA, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: Results from the phase II KEYNOTE-158 study J Clin Oncol 381–102020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidal J, Muinelo L, Dalmases A, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients Ann Oncol 281325–13322017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grasselli J, Elez E, Caratù G, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer Ann Oncol 281294–13012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thierry AR, El Messaoudi S, Mollevi C, et al. Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment Ann Oncol 282149–21592017 [DOI] [PubMed] [Google Scholar]
- 24. Bachet J-B, Bouche O, Taïeb J, et al. RAS mutations concordance in circulating tumor DNA (ctDNA) and tissue in metastatic colorectal cancer (mCRC): RASANC, an AGEO prospective multicenter study. J Clin Oncol. 2017;35 suppl; abstr 11509. [Google Scholar]
- 25.Kagawa Y, Elez E, Garcia-Foncillas J, et al. Combined analysis of concordance between liquid and tumor tissue biopsies for RAS mutations in colorectal cancer with a single metastasis site: The METABEAM study Clin Cancer Res 272515–25222021 [DOI] [PubMed] [Google Scholar]
- 26. Hempelmann JA, Lockwood CM, Konnick EQ, et al. Microsatellite instability in prostate cancer by PCR or next-generation sequencing. J Immunother Cancer. 2018;6:29. doi: 10.1186/s40425-018-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer Nat Med 241449–14582018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/PO.21.00383.