Abstract
Human immunodeficiency virus (HIV) persists in cells despite antiretroviral therapy; however, the influence of cellular mechanisms such as activation, differentiation, and proliferation upon the distribution of proviruses over time is unclear. To address this, we used full-length sequencing to examine proviruses within memory CD4+ T-cell subsets longitudinally in 8 participants. Over time, the odds of identifying a provirus increased in effector and decreased in transitional memory cells. In all subsets, more activated (HLA-DR–expressing) cells contained a higher frequency of intact provirus, as did more differentiated cells such as transitional and effector memory subsets. The proportion of genetically identical proviruses increased over time, indicating that cellular proliferation was maintaining the persistent reservoir; however, the number of genetically identical proviral clusters in each subset was stable. As such, key biological processes of activation, differentiation, and proliferation influence the dynamics of the HIV reservoir and must be considered during the development of any immune intervention.
Keywords: HIV persistence, HIV reservoir, memory T cells, cellular proliferation, cellular differentiation, HLA-DR, full-length sequencing
The longitudinal analysis of full-length proviruses within memory CD4+ T-cell subsets demonstrates that the biological processes of activation, differentiation, and proliferation influence the dynamics of the HIV reservoir, and must be considered during the development of any immune intervention.
For people living with human immunodeficiency virus (HIV), antiretroviral therapy (ART) must be maintained for a lifetime as replication-competent virus can persist within memory CD4+ T cells [1–4]. Approximately 1%–10% of infected cells contain a provirus with all genetic information necessary to reinitiate infection if therapy ceases [5–9]. These genetically intact, likely replication-competent proviruses represent the barrier to an HIV cure.
The number of cells infected with replication-competent HIV decays slowly during therapy [10–19]. However, the reservoir is also maintained by cellular proliferation [20]. Antigen-stimulated proliferation can occur when a cell encounters its cognate antigen [21, 22]. By contrast, homeostatic proliferation occurs in response to cytokines, such as interleukin 7 (IL-7) and IL-15 [23]. Finally, the integrated HIV provirus has also been postulated to induce proliferation [24, 25].
It has been shown that genetically intact and/or replication-competent HIV is enriched in effector memory (EM) cells [7], TH1 cells [8] and cells displaying activation and exhaustion markers [9, 26–29]. Some of these subsets may be more likely to undergo proliferation, as well as other key processes such as activation or differentiation, which may impact the maintenance of the reservoir during ART [9, 30, 31]. How these cellular processes impact the dynamics of the reservoir within cell subsets over time has not been well described.
As such, to define the stability of genetically intact proviruses within naive and memory T-cell subsets we conducted a longitudinal analysis of full-length proviruses from participants on long-term ART. At the second time point, we co-sorted memory subsets with the HLA-DR receptor to understand the contribution of activated cells to the reservoir. We also examined the role of cellular proliferation in maintaining persistent HIV over time.
METHODS
Ethics Statement
This study was approved by the institutional review board at the Western Sydney Health Department for the Westmead Institute for Medical Research, and the ethics review committees at the University of California San Francisco and Vaccine and Gene Therapy Institute-Florida. Written informed consent was obtained from all participants.
Participants
Leukapheresis samples were obtained from 8 participants at 2 different time points during ART (Table 1). Time point 1 data from 4 participants were included in previous publications [7, 9]; however, all data were reanalyzed as additional participants were included in this study.
Table 1.
Participant Characteristics
| Participant | Sex | Age, y, Time Point 2 | Time Before Initiation of Therapy, mo | CD4 Nadir | Years Between Time Points | Leukapheresis 1, Time Point 1 | Leukapheresis 2, Time Point 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length of Viral Suppression, y | Treatment Regimen | CD4+ T-Cell Count | Viral Load, Copies/mL | Cells Available | Length of Viral Suppression, y | Treatment Regimen | CD4+ T-Cell Count | Viral Load, Copies/mL | Cells Available | ||||||
| 2026 | M | 63 | >12 | 132 | 3.7 | 13.2 | TDF, ABC/3TC, RTV, DRV | 476 | <40 | Naive, CM, TM, EM, HLA-DR+, HLA-DR− | 16.9 | ABC/TCV/3TC | 414 | <40 | Naive, CM/HLA-DR±, TM/ HLA-DR±, EM/HLA-DR± |
| 2046 | M | 53 | >12 | 10 | 2.8 | 15.8 | ECV, EFV/TDF/FTC | 1099 | <40 | CM, TM, EM, HLA-DR+, HLA-DR− | 18.5 | EFV/TDF/FTC, ECV | 528 | <40 | Naive, CM/HLA-DR±, TM/ HLA-DR±, EM/HLA-DR± |
| 1292 | M | 49 | >12 | 342 | 4.6 | 3.3a | EFV/FTC/TDF | 746 | <40 | Naive, CM, TM, EM | 7.9 | EFV/TDF/FTC | 677 | <40 | Naive, CM/HLA-DR±, TM/ HLA-DR±, EM/HLA-DR± |
| 2518 | F | 59 | >12 | 70 | 3.5 | 14.8 | TDF, AZT/3TC, NVP | 432 | <40 | Naive, CM, TM, EM, HLA-DR+, HLA-DR− | 18.3 | TDF, AZT/3TC, NVP | 179 | <40 | Naive, CM/HLA-DR±, TM/ HLA-DR±, EM/HLA-DR± |
| 2013 | M | 70 | >12 | 13 | 4.0 | 17.1 | ABC/3TC, RGV | 819 | <40 | HLA-DR−, HLA-DR+ | 21.2 | ABC/TCV/3TC | 530 | <40 | Naive, CM/HLA-DR±, TM/ HLA-DR±, EM/HLA-DR± |
| 2303 | M | 44 | <6 | 478 | 4.2 | 4.7a | EFV/FTC/TDF | 606 | <40 | Naive, CM, TM, EM | 8.9 | ABC/TCV/3TC | 880 | <40 | Naive, CM/HLA-DR±, TM/ HLA-DR±, EM/HLA-DR± |
| 2302 | M | 32 | <6 | 391 | 4.9 | 4.6a | FPV, RTV, TDF/FTC | 696 | <40 | Naive, CM, TM, EM | 9.5 | EGV/TAF/FTC/COBI | 654 | <40 | Naive, CM/HLA-DR±, TM/ HLA-DR±, EM/HLA-DR± |
| 2286 | M | 52 | <6 | 165 | 5.4 | 9.1a | EFV/FTC/TDF | 381 | <40 | Naive, CM, TM, EM | 14.5 | RPV/TDF/FTC | 400 | <40 | Naive, CM/HLA-DR±, TM/ HLA-DR±, EM/HLA-DR± |
Abbreviations: 3TC, lamivudine; ABC, abacavir; AZT, zidovudine; CM, central memory; COBI, cobicistat; DRV, darunavir; ECV, entecavir; EFV, efavirenz; EGV, elvitegravir; EM, effector memory; ETR, etravirine; FPV, fosamprenavir; FTC, emtricitabine; MVC, maraviroc; NVP, nevirapine; RPV, rilpivirine; RTG, raltegravir; RTV, ritonavir; TAF, tenofovir alafenamide; TCV, dolutegravir; TDF, tenofovir disoproxil fumarate; TM, transitional memory.
aFor 4 participants the time to viral suppression (<40 copies/mL plasma) after ART administration was unknown at time point 1, so for time point 1 the time listed is years on ART.
Time Point 1 Cell Sort
Fluorescence-activated cell sorting (FACSAria; BD Biosciences) was used to sort naive, central (CM), transitional (TM), and effector memory (EM) CD4+ T-cell subsets as previously described [7, 12, 30] (Supplementary Figure 1A). In 4 participants, CD4+CD45RA−HLA-DR+ and CD4+CD45RA−HLA-DR− memory T cells were sorted as previously reported [9] (Supplementary Figure 1B).
Time Point 2 Cell Sort
To obtain naive, CM/HLA-DR±, TM/ HLA-DR±, EM/HLA-DR± cells, peripheral blood mononuclear cells were obtained by Lymphoprep (STEMCELL Technologies) density gradient centrifugation. Total CD4+ T cells were isolated by magnetic negative selection (STEMCELL Technologies). Cells were then sorted using a FACSAria (BD Biosciences) using the following antibodies: CD3-FITC (clone UCHT1, BioLegend), CD4-BV650 (clone RPA-T4, BioLegend), CD45RA-PE (clone HI100, BioLegend), CCR7-PECy7 (clone G043H7, BioLegend), CD27-APC (clone M-T271, BioLegend), HLA-DR-BV421 (clone L243, BioLegend), and Live/Dead Aqua Marker (Fisher Bioscience) (Supplementary Figure 1C).
Full-Length Individual Proviral Sequencing
The full-length individual proviral sequencing (FLIPS) assay was performed and proviral sequences analyzed for defects using a process of elimination [7]. Proviruses not containing a large (>100 bp) deletion, inversion, deleterious stop codon, frameshift mutation, or mutation in the cis-acting region were classified as genetically intact.
Identical Sequence Analysis
Identical sequences were identified using ElimDupes (Los Alamos HIV Database). A cluster of identical sequences was defined as ≥2 100% genetically identical sequences from the same cell subset.
Statistical Methodology
Logistic regression was used to analyze the relationship between cell subset and HIV infection frequency. The contribution of each subset to the reservoir was described using a Wilcoxon signed rank test. The proportion of genetically identical proviruses within cell subsets was estimated using a mixed logistic model. Detailed statistical methods can be found in the Supplementary Material.
RESULTS
The Proviral Landscape Differs Between Cell Subsets
To observe the dynamics of the proviral landscape over time we obtained leukapheresis samples from 8 participants at 2 time points during ART (Table 1). At time point 1, naive, CM, TM, and EM cells were sorted from 7 participants, and HLA-DR ± memory CD4+ T cells sorted from 4 participants. At time point 2, a median of 4.1 years later (interquartile range [IQR], 3.6–4.8), naive, CM/HLA-DR±, TM/ HLA-DR±, EM/HLA-DR± CD4+ T cells were sorted from all 8 participants. We used the FLIPS assay [7] to obtain near full-length (approximately 92%) HIV genomes at the single genome level.
At the first time point, 1124 genomes were isolated and 48 (4.3%) were genetically intact (Supplementary Table 1 and Supplementary Figure 2A). At the second time point, 1654 genomes were isolated and 105 (6.3%) were genetically intact (Supplementary Table 2 and Supplementary Figure 2B). The mutational profile of proviruses differed by subset, with cells considered more differentiated (TM and EM) and cells expressing HLA-DR more likely to contain full-length proviruses, including intact proviruses (Supplementary Figure 2).
HIV Proviruses Increase in EM Cells and Decrease in TM Cells With Time
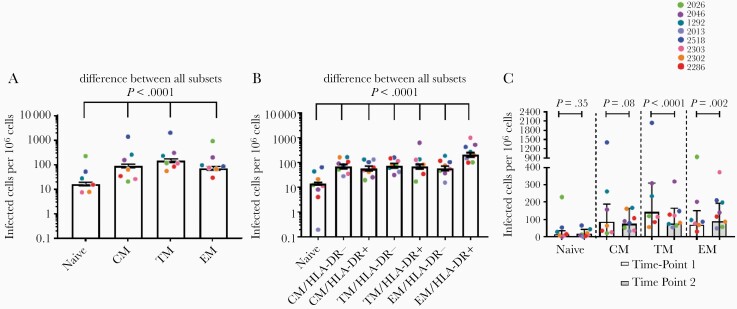
To examine the HIV reservoir in different cell subsets we first estimated the overall HIV infection frequency within each subset during ART. At time point 1, there was a significant difference in the infection frequency between subsets, with strong evidence that the nature of this difference was participant dependent (logistic regression model, P < .0001 for effect modification; Figure 1A). TM cells had the highest estimated infection frequency, 142 cells per million (95% confidence interval [CI], 119–168; Figure 1A and Supplementary Table 3). All other subsets had an estimated infection frequency of <100 cells per million, with a progression TM > CM > EM > naive (Figure 1A and Supplementary Table 3).
Figure 1.
Infection frequencies of cell subsets with any HIV provirus, intact or defective, in the peripheral blood. A, Infection frequency of cell subsets at time point 1. Logistic regression model, estimated total infection frequency, and 95% CI. B, Infection frequency of cell subsets at time point 2. Logistic regression model, estimated total infection frequency and 95% CI. C, Change in the infection frequency of cell subsets between time points 1 and 2. Memory cell subsets for time point 2 recapitulated from the biological proportions of HLA-DR± cells in each subset. Pairwise comparisons, estimated mean, and 95% CI. Abbreviations: CI, confidence interval; CM, central memory; EM, effector memory; HIV, human immunodeficiency virus; TM, transitional memory.
At the second time point, the total infection frequency was again significantly different between subsets, with strong evidence for participant effect modification (logistic regression model, P < .0001; Figure 1B). EM/HLA-DR+ cells had the highest estimated infection frequency, 203 cells per million (95% CI, 165–251), and naive cells had the lowest, 14 infected cells per million (95% CI, 13–16) (Figure 1B and Supplementary Table 3). All other subsets had an estimated infection frequency between 58 and 75 cells per million (Figure 1B and Supplementary Table 3).
To assess if the infection frequencies within each subset changed over 4 years, the CM, TM, and EM subsets at time point 2 were recapitulated using the biological proportions of HLA-DR+ and HLA-DR− cells in each subset (Supplementary Table 4). A 2-fold decrease in the infection frequency of TM cells was observed (odds ratio = 0.53; P < .0001; Figure 1C and Supplementary Table 5). However, HIV-infected EM cells increased by 28% (odds ratio = 1.28; P = .002; Figure 1C and Supplementary Table 5). There was no significant change in the frequency of infected naive or CM cells with time (Figure 1C and Supplementary Table 5).
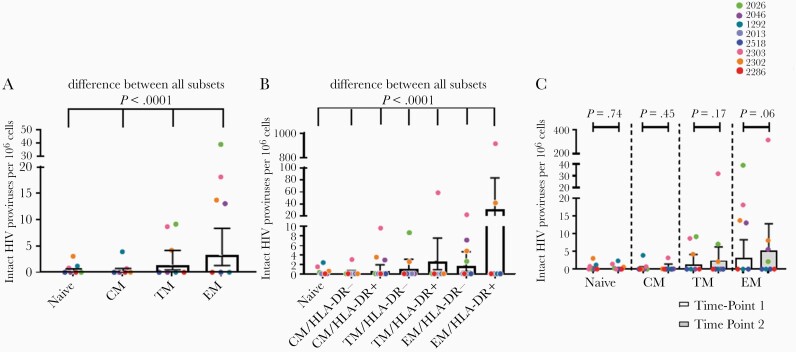
Genetically Intact HIV Proviruses Are Concentrated in Differentiated Cells and/or Cells Expressing HLA-DR
To understand the dynamics of intact proviruses, we then assessed the change in the infection frequency of intact provirus within cell subsets over time. At time point 1 there was a significant difference in the infection frequency with intact provirus between subsets (P < .0001; Figure 2A). The estimated frequency of intact proviruses ranged from 0.2 to 3 per million cells, and increased with differentiation status (CM < TM < EM) (Figure 2A and Supplementary Table 6). At the second time point there was also a significant difference (P < .0001) in the infection frequency between the subsets (Figure 2B); in all subsets an estimated 0–3 cells per million contained an intact provirus, except for EM/HLA-DR+ cells, which had an estimated infection frequency of 31 cells per million (95% CI, 12–84; Figure 2B and Supplementary Table 6). However, only 2 participants had high levels of intact provirus in their EM/HLA-DR+ cells. A stepwise progression was observed in terms of the frequency of intact provirus within the subsets; for both HLA-DR− and HLA-DR+ cells this infection frequency progressed CM < TM < EM (Figure 2B). Moreover, in each subset HLA-DR− cells had a lower estimated frequency of intact proviruses than HLA-DR+ cells (Figure 2B and Supplementary Table 6).
Figure 2.
Infection frequencies of cell subsets with an intact HIV provirus in the peripheral blood. A, Infection frequency with an intact provirus within differentiated cell subsets at time point 1. Logistic regression model, estimated intact infection frequency, and 95% CI. B, Infection frequency with an intact provirus in differentiated cell subsets at time point 2. Logistic regression model, estimated intact infection frequency and 95% CI. C, Change in the infection frequency with an intact provirus within differentiated cell subsets between time points 1 and 2. Cell subsets for time point 2 recapitulated from biological proportions. Pairwise comparisons, estimated mean, and 95% CI. Abbreviations: CI, confidence interval; CM, central memory; EM, effector memory; HIV, human immunodeficiency virus; TM, transitional memory.
We then compared the genetically intact infection frequencies for each subset between time points. After recapitulating the time point 2 memory subsets from the HLA-DR± components (Supplementary Table 4), we found that the intact infection frequency did not change significantly in any subset over time (Figure 2C and Supplementary Table 7).
Memory CD4+ T-Cell Subsets Do Not Contribute Equally to the Peripheral Blood HIV Reservoir
As the proportion of each cell subset within the peripheral blood is different, we assessed the contribution of each subset to the pool of infected cells during therapy taking these proportions into account. That is, we used the biological proportions of each subset within the pool of peripheral blood CD4+ T cells, and the infection frequency within that subset, to calculate their contribution to the reservoir. We assessed this at time point 2 only as each participant had the same subsets sorted, and the contribution of time point 1 cell subsets was determined in previous studies [7, 9].
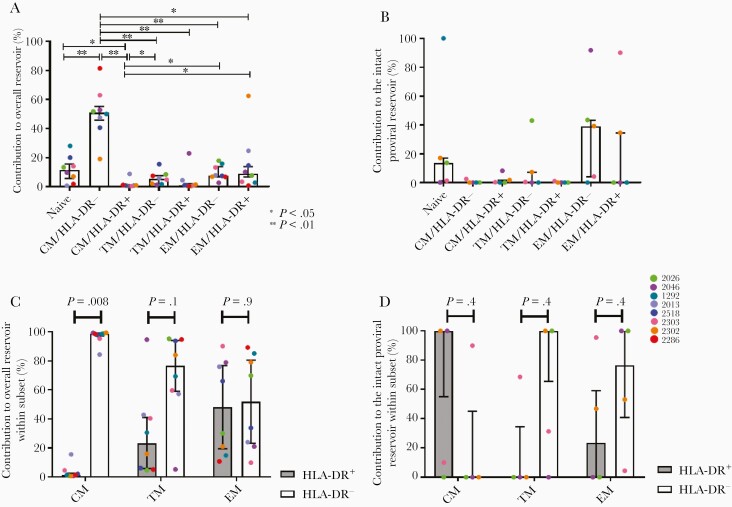
Over half (median, 59.9%; IQR, 45.8%–55.3%) of all HIV proviruses, defective or intact, were found in the CM/HLA-DR− cell subset (Figure 3A), significantly higher than any other subset (Supplementary Table 8). Naive cells contributed a further 11.5% (IQR, 5.9%–15.6%; Figure 3A); however, this contribution was only significantly higher than CM/HLA-DR+ cells (P = .04; Supplementary Table 8). Naive and CM/HLA-DR− cells were the 2 most prevalent peripheral blood CD4+ T-cell subsets (Supplementary Table 9), accounting for their high contribution. All other subsets contributed <10% of the total HIV burden at this time. However, we observed a large amount of variability in this contribution across participants (Figure 3A).
Figure 3.
Contribution of memory cell subsets to the burden of HIV in the peripheral blood. A, Contribution of cell subsets to the population of CD4+ T cells containing any HIV provirus in the peripheral blood at time point 2. B, Contribution of cell subsets to the intact HIV reservoir in the peripheral blood at time point 2. C, Contribution of cells that are HLA-DR+ or HLA-DR− to the total population of each memory cell phenotype that is infected with any HIV provirus at time point 2. Wilcoxon signed rank test. D, Contribution of cells that are HLA-DR+ or HLA-DR− to the population of each memory cell phenotype that contains a genetically intact HIV provirus at time point 2. Wilcoxon signed rank test. All data are median and interquartile range. Abbreviations: CM, central memory; EM, effector memory; HIV, human immunodeficiency virus; TM, transitional memory.
We then compared the relative contribution of the cell subsets to the genetically intact proviral reservoir in the peripheral blood at time point 2. The highest contributor was found to be EM/HLA-DR− cells, contributing a median of 39.2% (IQR, 4.3%–43.3%). Despite their low infection frequency, naive cells were also found to be a high contributor, as has been observed previously [19], contributing a median of 13.7% (IQR, 1.4%–17.2%) (Figure 3B). All other subsets contributed ≤10% to the genetically intact reservoir. However, no subset was found to contribute significantly more than any other (Supplementary Table 10), likely due to the small numbers of intact proviruses found.
Finally, as we had previously observed that HLA-DR+ memory CD4+ T cells contributed over half of all intact proviruses to the peripheral blood reservoir [9], we wanted to establish how these cells contributed to the reservoir found within specific memory cell subsets. We observed that the contribution of HLA-DR+ cells to the total pool of infected cells increased as cells differentiated (Figure 3C). Significantly more CM/HLA-DR− cells contributed to the overall reservoir compared to CM/HLA-DR+ cells (P = .008, Wilcoxon signed rank test); however, in the EM subset HLA-DR+ and HLA-DR− cells contributed approximately equally (Figure 3C). When we assessed the contribution of these cells to the genetically intact reservoir within each subset, we found that HLA-DR+ cells were the highest contributor to the CM subset (Figure 3D), although not significantly. Interestingly, these genetically intact proviruses were not unique to this subset, as 5/7 of the intact proviruses from CM/HLA-DR+ cells were also found within EM cells in the same participant (Figure 3D). By contrast, the contribution of HLA-DR+ cells was lower than HLA-DR− cells in the TM and EM subsets, although not significantly (Figure 3D).
Cellular Proliferation Maintains HIV Proviruses Within the Peripheral Blood Reservoir
We then sought to understand how HIV proviruses were maintained over 4 years of therapy. As intact proviruses may be seeded by the infection of previously uninfected cells, we looked for evidence of this by calculating the change in genetic diversity of intact proviruses over time. In 3 participants where >5 genetically intact proviruses were isolated at both time points, there was no change in genetic diversity with time (P = .20); in fact, genetic diversity decreased in 2 participants, indicating new infections were unlikely to be occurring (Supplementary Figure 3).
To determine if proviruses were being maintained by cellular proliferation, we screened for 100% genetically identical proviruses. We note that while homogenous proviral populations may be formed if ART is initiated during acute/early infection, there was no difference in the proportion of genetically identical proviruses between participants treated during acute/early or chronic infection (time point 1, P = .11; time point 2, P = .15; Supplementary Figure 4). As such, the presence of genetically identical proviruses within this study were unlikely to be influenced by the timing of ART initiation.
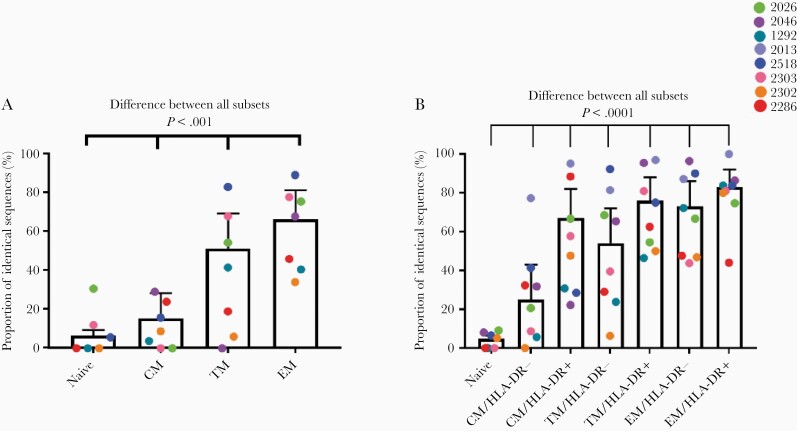
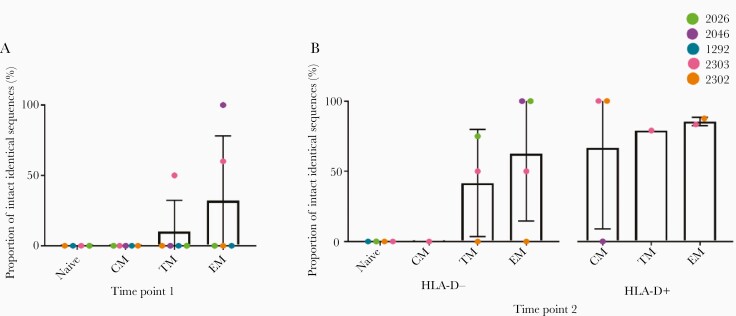
Examples of identical genomes were found in all subsets, and all participants had at least 1 example of a cluster of genetically identical proviruses. At time point 1, the proportion of identical sequences was different between subsets, with evidence that the nature of this difference was participant dependent (logistic regression, P < .0001 for effect modification). Overall, more differentiated subsets were observed to have a higher proportion of genetically identical proviruses (Figure 4A). This pattern was maintained at time point 2, as we again found that the proportion of identical sequences was significantly different between cell subsets, with evidence that the nature of this difference was participant dependent (logistic regression, P < .0001 for effect modification). A stepwise progression was observed as cells differentiated, CM < TM < EM. Moreover, a higher proportion of identical genomes was observed in HLA-DR+ cells compared to HLA-DR− cells of the same subset (Figure 4B). After taking into account the biological proportions of HLA-DR+ and HLA-DR− cells at time point 2 (Supplementary Table 4), we found that the overall proportion of identical sequences increased over time in all subsets, although not significantly (Supplementary Table 11).
Figure 4.
Proportion of identical sequences isolated from different cell subsets at time point 1 (A) and time point 2 (B). Data are mean and 95% confidence interval. Abbreviations: CM, central memory; EM, effector memory; TM, transitional memory.
We then assessed how proliferation may be impacting the landscape of intact proviruses over time. Overall, 23% and 58% of genetically intact sequences at time points 1 and 2 were identical. All 3 participants with >5 intact HIV proviruses isolated at both time points showed an increase in the proportion of genetically identical intact proviruses over time (P = .037; Supplementary Figure 5). At time point 1, genetically identical intact proviruses were identified from 2 participants only; however, the proportion of these increased as cells differentiated (Figure 5A). Similarly, at time point 2, a stepwise progression in terms of the proportion of genetically identical intact proviruses was observed in both the HLA-DR− and HLA-DR+ subsets, EM > TM > CM (Figure 5B). However, we note that interpretations of these data are limited due to the rarity of these sequences.
Figure 5.
Proportion of genetically intact, genetically identical sequences isolated from different cell subsets at time point 1 (A) and time point 2 (B). Data are mean and SD. Abbreviations: CM, central memory; EM, effector memory; TM, transitional memory.
Interestingly, clusters of identical sequences may have impacted our ability to identify intact proviruses in some participants. No intact provirus was isolated from 3 participants at either time point; however, all 3 participants had 1 dominant expansion of defective sequences within their reservoir that was observed at both time points and in all memory cell subsets (Supplementary Figure 6).
The Identity of Cells Undergoing Cellular Proliferation Changes With Time
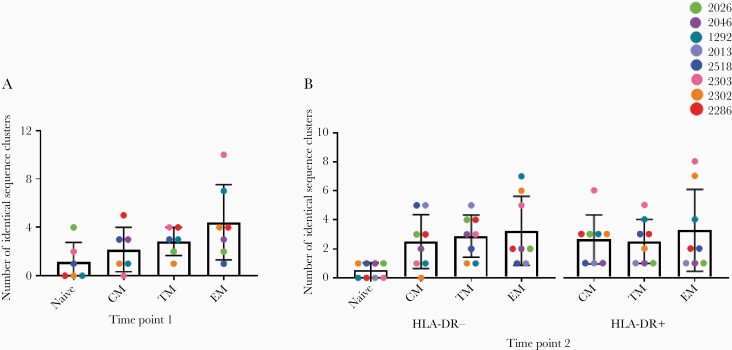
We then investigated the number of unique clusters of identical sequences in each subset—likely representing different proliferative events—to understand the dynamics of identical sequences with time. At both time points we found that the number of clusters of identical sequences, both overall (Figure 6) and intact (Supplementary Figure 7), was higher in more-differentiated cells, although variability in the number of clusters was observed between participants.
Figure 6.
The number of clusters of genetically identical sequences (intact or defective) isolated from different cell subsets at time point 1 (A) and time point 2 (B). Data are mean and SD. Abbreviations: CM, central memory; EM, effector memory; TM, transitional memory.
We then compared the number of clusters of identical sequences within each subset over time to determine if an increase of these clusters may have contributed to the increase in the proportion of identical sequences with time. The number of clusters observed in TM cells was different between time points (P = .0006), with all participants having an increase of clusters at the second time point (Supplementary Figure 8). There was no change in the number of clusters of identical sequences in any other subset (Supplementary Figure 8). Given this stability, we wanted to understand if this was reflected in the proviral populations that were identical at each time point. However, we found that the populations isolated at time points 1 and 2 were different, reflecting underlying fluctuations in the makeup of the reservoir (Supplementary Tables 12–18).
Discussion
In this study, we used full-length sequencing of the HIV genome to analyze the dynamics of the HIV reservoir within CD4+ T-cell subsets over time during therapy. At time point 1, TM and EM cells contained the highest frequencies of genetically intact provirus. This was not surprising as the importance of differentiated cells in harboring genetically intact and/or translation-competent virus has been demonstrated previously [7, 26, 32]. However, this pattern was also observed at a second time point, suggesting that intact HIV genomes may be stably maintained within differentiated cells with time. As EM cells are known to have a shorter lifespan compared to CM [33, 34], there may be a compensatory process that aids in the maintenance of the reservoir. One such mechanism may be the replenishment of the reservoir through the differentiation of infected naive, CM, or TM cells into EM with time. In particular, the finding of intact proviruses within long-lived naive cells at both time points suggests that naive cell differentiation into TM and EM cells after stimulation may be a source of persistent HIV during long-term therapy [19]. While we observed total proviruses to decrease in TM cells and increase in EM cells between time points—suggesting that infected cells move between subsets—the low frequency of cells infected with intact provirus meant we were unable to directly observe the differentiation of cells infected with genetically intact HIV in this study.
Previously, we observed that HLA-DR+ memory cells were enriched for genetically intact provirus [9]. Other studies have shown that markers such as PD-1 [31, 35], TIGIT [28], and OX40 [29] are both expressed on cells that contain a high frequency of provirus and correlate with HLA-DR expression. As such, similar concentrations of HIV within HLA-DR+ memory cells may be likely. However, given the overlap between differentiated cells and HLA-DR+ cells, we were previously unable to determine whether a high proportion of intact genomes in HLA-DR+ cells was due to cellular activation or differentiation [9]. However, within this study there was a higher frequency of genetically intact provirus in HLA-DR+ cells compared to HLA-DR− in all memory cell subsets, indicating both activation and differentiation play a role in maintaining the HIV reservoir. In particular, cellular activation may be important for maintaining the reservoir in less-differentiated cell subsets that do not otherwise contain high frequencies of intact provirus, as demonstrated by the high contribution of HLA-DR+ cells to the intact reservoir in the CM subset.
Cellular proliferation may be a third intrinsic cellular process maintaining the viral reservoir [8, 20, 22–25, 30, 36]. Notably, there may be a strong interplay between proliferation, differentiation, and activation, as examples of identical proviruses were increased in differentiated and/or activated cell subsets at both time points. These identical proviruses were found to increase in all subsets between time points, although not significantly, in contrast to previous studies [37]. Notably, we also found that intact, genetically identical proviruses increased over time, further indicating that cellular proliferation is maintaining the latent reservoir. Recent work has highlighted that intact proviruses undergoing proliferation may not be targeted by the immune system for depletion [16] and instead that cellular proliferation maintains both intact and defective proviruses. Moreover, additional studies have shown that the administration of IL-7 also increases the absolute number of infected cells in the reservoir through an increase in cell proliferation [23]. As such, cell proliferation may have the potential to not only maintain but also expand the reservoir with time.
Finally, similar to what has been observed previously [14, 30, 38], while the number of clusters of identical sequences was stable over time, the identity of these populations changed. As cytokine-driven proliferation is not selective for specific cell populations [38], shifts in the dominant population may be predominately driven by antigenic proliferation. Recent work has highlighted that cell populations reactive to common antigens harbor proviruses during prolonged therapy [21, 22, 36]. During the course of any immune response, these cells (and therefore integrated proviruses) may expand and contract [22], similar to the fluctuations observed both here and previously [38]. As such, changes in the clusters of proviruses that are observed over time may be a reflection of the larger dynamics of the immune system.
However, while inferences can be drawn regarding the roles of cellular activation, differentiation, and proliferation in shaping the reservoir over time, it is important to note that the small number of participants (n = 8) limits the extent to which we can observe patterns within the study, with participant effect modification observed in many of our statistical analysis. Moreover, the number of intact proviruses able to be isolated was limited—including 3 participants with no intact provirus isolated—meaning patterns involving intact proviruses must be interpreted with caution. Furthermore, while there may be a biological basis for the patterns observed herein, evidence of participant effect modification may indicate that there is an additional stochastic nature to this distribution.
Overall, genetically intact HIV proviral infection frequencies are increased in more differentiated and activated cell subsets during long-term ART. Increased proportions of identical sequences are also observed within the same subsets over time. However, underlying shifts in the proviral landscape are evident through an increase in the proportion of identical sequences with time and the different proviruses contributing to these identical sequence clusters between time points. Given that the proliferation of each cell subset—likely driven by the immune response—is influencing the dynamics of the HIV reservoir, clearly taking cellular biology into account—including the cell subset of origin of the provirus and therefore the likelihood of that cell to proliferate with time—is crucial for a complete understanding of the persistent HIV reservoir, and thereby the development of an effective and efficient HIV cure.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the Sydney Informatics Hub (University of Sydney, Sydney, Australia) for support and access to analytical software, and the Ramaciotti Centre for Genomics (University of New South Wales, Sydney, Australia) and the Australian Genome Research Facility (AGRF Sydney Site, the Westmead Institute for Medical Research, Sydney, Australia) for operating the MiSeq. We acknowledge with gratitude the participants who donated samples for this study.
Author contributions. B. A. H., B. H., and K. F. conducted FLIPS on participant samples and analyzed data. R. H., S. D., F. H., J. M. M., S. v. S., L. O., N. C., and R. F. enrolled the participants, collected, and/or sorted cell subsets from the participant samples. B. A. H., B. H., K. F., E. L., S. v. S., and L. O. prepared participant samples. J.-S. E. designed analysis and data visualization workflows. H. M. and T. E. S. conducted the statistical analyses. B. A. H. wrote the original manuscript. S. P. designed the study, supervised the work performed, and edited the manuscript.
Financial support. This work was supported by the Delaney AIDS Research Enterprise DARE to Find a Cure (grant numbers 1U19AI096109 and 1UM1AI126611-01); the Foundation for AIDS Research amfAR Research Consortium on HIV Eradication Collaborative Research Grant (grant number 108074-50-RGRL); University of California San Francisco/Gladstone Institute of Virology and Immunology Center for AIDS Research (grant number P30 AI027763); Australian Centre for HIV and Hepatitis Virology Research (grant number 2015–43); and the Australian National Health and Medical Research Council (grant numbers APP1061681 and APP1149990).
Potential conflicts of interest. N. C. has received consulting fees from Gilead Sciences, and the laboratory of N. C. has received a grant from EMD Serono. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Data and materials availability. Genbank accession numbers are MH843739-MH843916, KY778264–KY778681, KY766150–KY766212, and MZ080627–MZ081008 (time point 1); MW753232–MW754712 (time point 2).
Presented in part: Conference on Retroviruses and Opportunistic Infections 2020, Boston, March 8-11 2020; AIDS2020: Virtual - the 23rd International AIDS Conference, 6-10 July 1010.
References
- 1. Wong JK, Hezareh M, Gunthard HF, et al. . Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278:1291–5. [DOI] [PubMed] [Google Scholar]
- 2. Finzi D, Blankson J, Siliciano JD, et al. . Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999; 5:512–7. [DOI] [PubMed] [Google Scholar]
- 3. Finzi D, Hermankova M, Pierson T, et al. . Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278:1295–300. [DOI] [PubMed] [Google Scholar]
- 4. Chun T- W, Stuyver L, Mizell SB, et al. . Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 1997; 94:13193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ho YC, Shan L, Hosmane Nina N, et al. . Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bruner KM, Murray AJ, Pollack RA, et al. . Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016; 22:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hiener B, Horsburgh BA, Eden J- S, et al. . Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep 2017; 21:813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee GQ, Orlova-Fink N, Einkauf K, et al. . Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Investig 2017; 127:2689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horsburgh BA, Lee E, Hiener B, et al. . High levels of genetically intact HIV in HLA-DR+ memory T cells indicates their value for reservoir studies. AIDS 2020; 34:659–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siliciano JD, Kajdas J, Finzi D, et al. . Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003; 9:727–8. [DOI] [PubMed] [Google Scholar]
- 11. Palmer S, Maldarelli F, Wiegand A, et al. . Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 2008; 105:3879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Josefsson L, von Stockenstrom S, Faria NR, et al. . The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A 2013; 110:E4987–E96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Besson GJ, Lalama CM, Bosch RJ, et al. . HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis 2014; 59:1312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pinzone MR, VanBelzen DJ, Weissman S, et al. . Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nat Commun 2019; 10:728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bruner KM, Wang Z, Simonetti FR, et al. . A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019; 566:120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Antar AA, Jenike KM, Jang S, et al. . Longitudinal study reveals HIV-1-infected CD4+ T cell dynamics during long-term antiretroviral therapy. J Clin Invest 2020; 130:3543–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gandhi RT, Cyktor JC, Bosch RJ, et al. . Selective decay of intact HIV-1 proviral DNA on antiretroviral therapy. J Infect Dis 2021; 223:225– 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peluso MJ, Bacchetti P, Ritter KD, et al. . Differential decay of intact and defective proviral DNA in HIV-1–infected individuals on suppressive antiretroviral therapy. JCI Insight 2020; 5:e132997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Venanzi Rullo E, Pinzone MR, Cannon L, et al. . Persistence of an intact HIV reservoir in phenotypically naive T cells. JCI Insight 2020; 5:e133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chomont N, Far ME, Ancuta P, et al. . HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009; 15:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mendoza P, Jackson JR, Oliveira TY, et al. . Antigen-responsive CD4+ T cell clones contribute to the HIV-1 latent reservoir. J Exp Med 2020; 217:e20200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gantner P, Pagliuzza A, Pardons M, et al. . Single-cell TCR sequencing reveals phenotypically diverse clonally expanded cells harboring inducible HIV proviruses during ART. Nat Commun 2020; 11:4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vandergeeten C, Fromentin R, DaFonseca S, et al. . Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood 2013; 121:4321–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wagner TA, McLaughlin S, Garg K, et al. . Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 2014; 345:570–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maldarelli F, Wu X, Su L, et al. . Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014; 345:179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pardons M, Baxter AE, Massanella M, et al. . Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS Pathog 2019; 15:e1007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fromentin R, Bakeman W, Lawani MB, et al. . CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog 2016; 12:e1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chew GM, Fujita T, Webb GM, et al. . TIGIT marks exhausted T cells, correlates with disease progression, and serves as a target for immune restoration in HIV and SIV infection. PLoS Pathog 2016; 12:e1005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuo H- H, Ahmad R, Lee GQ, et al. . Anti-apoptotic protein BIRC5 maintains survival of HIV-1-infected CD4+ T cells. Immunity 2018; 48:1183–94.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. von Stockenstrom S, Odevall L, Lee E, et al. . Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J Infect Dis 2015; 212:596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee E, Bacchetti P, Milush J, et al. . Memory CD4+ T-cells expressing HLA-DR contribute to HIV persistence during prolonged antiretroviral therapy. Front Microbiol 2019; 10:2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neidleman J, Luo X, Frouard J, et al. . Phenotypic analysis of the unstimulated in vivo HIV CD4 T cell reservoir. eLife 2020; 9:e60933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Macallan DC, Wallace D, Zhang Y, et al. . Rapid turnover of effector-memory CD4+ T cells in healthy humans. J Exp Med 2004; 200:255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bacchus-Souffan C, Fitch M, Symons J, et al. . Relationship between CD4 T cell turnover, cellular differentiation and HIV persistence during ART. PLoS Pathog 2021; 17:e1009214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Psomas C, Younas M, Reynes C, et al. . One of the immune activation profiles observed in HIV-1-infected adults with suppressed viremia is linked to metabolic syndrome: the ACTIVIH study. EBioMedicine 2016; 8:265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simonetti FR, Zhang H, Soroosh GP, et al. . Antigen-driven clonal selection shapes the persistence of HIV-1–infected CD4+ T cells in vivo. J Clin Investig 2021; 131:e145254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cohn LB, Silva IT, Oliveira TY, et al. . HIV-1 integration landscape during latent and active infection. Cell 2015; 160:420–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Z, Gurule EE, Brennan TP, et al. . Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc Natl Acad Sci U S A 2018; 115:E2575–E84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.