Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was the third zoonotic coronavirus to have an outbreak in the first two decades of the 21st century. Human-to-human transmission of this virus has threatened thousands of lives around the world. SARS-CoV-2 shares 79% and 50% sequence homology with severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), respectively. Like SARS-CoV and MERS-CoV infection, evidence has shown that SARS-CoV-2 infection also causes acute tissue damage due to a pathological immune response, particularly in severe cases. T cells play an important role in virus clearance and prevention, and in this paper, we summarize dynamic changes in the T cell count, subsets, phenotype, and function in Coronavirus Disease 2019 (COVID-19) patients based on current clinical reports. This review may help to better understand the pathological immune response of T cells and facilitate making better therapeutic strategies for patients with SARS-CoV-2 infection.
Keywords: COVID-19, SARS-CoV-2, T lymphocyte
1. INTRODUCTION
At the end of 2019, a new respiratory coronavirus, named SARS-CoV-2 by the World Health Organization (WHO), was found in Wuhan, China, and its resulting disease was named COVID-19. As a new virus, SARS-CoV-2 threatens the lives of thousands of people around the world. According to clinical reports and pathological results, it has been suggested that lung immunopathological damage and cytokine storms play an important role in the disease progression of patients with COVID-19.1–9 In addition, T cells play an important role in adaptive immunity for pathogen elimination. As a β-coronavirus, the SARS-CoV-2 genome is similar to that of SRAS-CoV and MERS-CoV, which have previously brought considerable threat. In addition, there have been reports that lymphopenia and cytokine storms occur in SRAS-CoV and MERS-CoV patients.10 However, there is no systematic summary of the characteristics of the T cell immune response in patients with COVID-19. This review might help to provide a better understanding of this disease and assist us in developing better immunotherapeutic strategies for COVID-19 treatment.
1.1. SARS-CoV-2
Coronavirus infection has attracted our attention over the past 20 years, including that of SARS-CoV, MERS-CoV, and SARS-CoV-2, which belong to the β-coronavirus subfamily. The structure of coronaviruses includes non-segmented, positive, single-stranded RNA, and nucleocapsid protein that forms the nucleocapsid, which is surrounded by a phospholipid bilayer and membrane protein. The SARS-CoV-2 RNA virus contains 29,891 nucleotides encoding 9860 amino acids.11 SARS-CoV-2 is a novel family member that is similar to the SARS-like coronaviruses bat-SL-CoVZC45 and bat-SL-CoVZXC21 (with 87.99% and 87.23% identity), and its genome similarity to SARS-CoV and MERS-CoV is ∼79% and 50%, respectively.12 New mutations may contribute to higher infectivity.13 RNA viruses cannot replicate independently; thus, they synthesize virus proteins in infected cells. Neutralizing antibody induction may prevent viruses from entering cells. Specific T cells are needed once a virus enters cells; thus, T cells play an important role in the immune clearance of coronavirus. It has been reported that T cells not only target structural proteins of coronaviruses but are also involved in lung immunopathological damage in SARS-CoV and MERS-CoV.14,15
1.2. T cell response changes in COVID-19 patients
Thus far, there have been many clinical reports of the changes in T cell number and function detected in COVID-19 patients.
1.2.1. T cell counts are reduced in the peripheral blood of COVID-19 patients
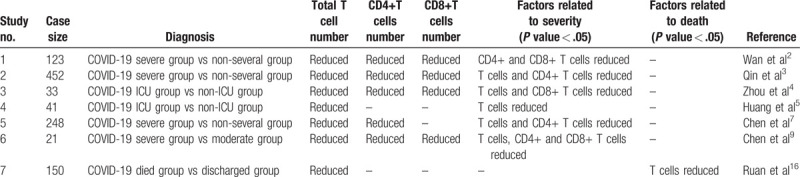
In general, pathogens (including viruses) are recognized, processed, and presented by antigen presenting cells (APCs). After T cell activation, CD4+ T cells mainly differentiate into effector T cells and produce cytokines and chemokines, while CD8+ T cells mainly differentiate into cytotoxic T cells (CTLs) to specifically kill virus-infected target cells. The T cell response to SARS-CoV-2 has great impact on the clinical outcome of patients. Clinical reports have shown that lymphopenia occurs in COVID-19 patients.6–8 Mean T cell count was lower in COVID-19 patients than normal range (541.5/μL vs 955.0–2860.0/μL) and T cell decreased more compared to B cell and NK cell.3 Similarly, 92.9% severe and moderate COVID-19 cases had lower T cell number than normal range.9 There was a significant difference in the absolute number of lymphocytes between patients who died and those who were discharged, which reminds us that lymphopenia may be a predictor of fatal outcome for COVID-19 patients.16 Xu et al found that CD4+ and CD8+ T cells were reduced in the peripheral blood before a patient died of COVID-19.1 More severe patients tend to have lower lymphocyte counts compared to those who are non-severe. CD4+ and CD8+ T cells in COVID-19 patients are reduced together. CD4+ T cells are lower in severe compared with non-severe patients, but there was no significant difference in CD8+ T cells between these two groups.3 In 123 patients diagnosed with COVID-19, the proportion of patients with lower CD4+ T cells between those who were mild and severe was statistically different. Lower CD4+ T cells were found in 95.24% of severe patients compared with 52.90% in mild patients.2 Analysis of 248 patients by multivariate logistical regression demonstrated that the decrease in CD4+ T cell counts was an important factor associated with ICU admission besides age.7 However, Zhou et al found that CD8+ T cells decreased in ICU patients compared with that in non-ICU patients.4 These reports may indicate that COVID-19 patients with a decrease in T cells, including CD4+ and CD8+ T cells, tend to be more severe, and T cells play an important role in coronavirus clearance and preventing reinfection. The relationship of T cell number reduce and outcome of COVID-19 patients from different studies are shown in Table 1.
Table 1.
T cells counts related to severe or death in COVID-19 patients
Regulatory T (Treg) cells are a subgroup of CD4+ T cells that can inhibit the immune response. Researchers have found that naïve regulatory T cells (CD45RA+CD3+CD4+CD25+CD127low+, nTreg) and induced regulatory T cells (CD45RO+CD3+CD4+CD25+CD127low+, iTreg) are reduced in severe patients, but there was no significant difference between severe and non-severe patient groups.3 Chen et al also reported that iTregs and nTregs are reduced in severe and moderate cases. Moreover, nTregs are significantly decreased in severe compared with moderate cases.9 This finding may be a reason for T cell overactivation, which could aggravate the pathological process of immune damage and make a patient's condition more severe.
1.2.2. Dysregulation of T cell in COVID-19 patients
It is known that T cells are composed of heterogeneous groups having different functions. Thus, it is also important to analyze the immunophenotype of T cells in COVID-19 patients in addition to T cell count. The multiple organ damage in patients with SARS-CoV-2 infection, including the lungs in particular, may be caused by the pathological immune response.
Some membrane molecules such as CD69, HLA-DR, and CD38 are expressed after the activation of T lymphocytes. Pathological analysis shows that HLA-DR (CD4 3.47%) and CD38 (CD8 39.4%) double-positive T cells increase in patients with SARS-CoV-2 infection. Xu et al found that CD8+ T cells contain a high concentration of cytotoxic granules, of which 31.6% are perforin positive, 64.2% are granulysin positive, and 30.5% are perforin and granulysin double positive.1 CD28, OX40 (CD134), and 4–1BB (CD137) are costimulatory receptors for T cell activation.17 Another study reported that CD4+ T cells in COVID-19 patients express more CD69, CD38, CD44, and OX40 than CD4+ T cells in healthy controls, and there was a significant increase in OX40 expression in severe ICU patients. CD8+ T cells also express a higher level of CD69, CD38, and CD44. Moreover, 4–1BB expression was increased in ICU patients compared to healthy controls.4 Some researchers have also reported that proinflammatory CCR6+ Th17 cells are increased in CD4+ T cell subset.1 These findings indicate T cell activation in COVID-19 patients.
The higher level of inflammatory cytokines, with the exception of that of IFN-γ, in COVID-19 indicates that T cell overactivation occurs, and the inflammatory cytokine level is related to disease severity. Qin et al found that the level of IL-2R, IL-6, IL-8, IL-10, and TNF-α was significantly higher in severe compared with non-severe patients.3 Similarly, serum levels of IL-2R, IL-6, IL-10, and TNF-α were remarkably higher in severe compared with moderate cases for 21 hospital-admitted patients. The same study also reported a decreased level of IFN-γ produced by CD4+, CD8+, and NK T cells in both moderate and severe cases, and the level of IFN-γ produced by CD4+ T cells was lower in severe compared with moderate cases.9 Overactivated T cells and increased inflammatory cytokines may serve to enhance immunopathological damage.
T cell exhaustion, which often occurs during chronic infection and tumors, is also found in COVID-19 patients. Some studies have reported detection of a subset T cell co-expressing T cell immunoglobulin and mucin domain-3 (Tim-3) and programmed death-1 (PD-1).4 Exhausted CD8+ T cells mainly express PD-1, cytotoxic T lymphocyte-associated antigen-4 (CTLA-4), T cell immunoglobulin and ITIM domain (TIGIT), and Tim-3. TIGIT expression in CD8+ T cells is a factor related to disease progression according to correlation analysis.18 We hypothesize that overactivation of CD8+ T cells might result in excessive loss of these cells in SARS-CoV-2 infection.
Naïve T cells, which express CD45RA and CD62L, are a group of mature T cells that remain in a relatively static state without antigen contact. In contrast, memory T cells are cells that survive for a long time after activation; they can be rapidly activated by the same antigens and mainly express CD45RO. Qin et al discovered that the percentage of naïve CD4+ T cells (CD3+CD4+CD45RA+) increase and memory CD4+ T cells (CD3+CD4+CD45RO+) significantly decrease in severe versus non-severe patients.3 Cytometric analysis of four elderly patients in the symptomatic phase of SARS-CoV-2 infection also demonstrated a high percentage of naïve CD4+ and CD8+ T cells.19 These results implied that the ratio of the T cell subsets could be involved in the process of developing SARS-CoV-2 infection.
1.3. Summary and future direction
The exact mechanisms underlying the immune response after SARS-CoV-2 infection remain unclear. A research claimed that MERS-CoV but not SARS-CoV could infect T cells in peripheral blood causing T cell decrease through apoptosis pathway.20 The accurate reasons of SARS-CoV-2 infection leading to T cell reduce are not clear until now. Besides, we can infer from the above clues that T cells are overactivated in COVID-19 patients, leading to cytokine storm, and decreased Treg cells is a reason for severe disease. A period of excessive T cell activation may cause T cell exhaustion, which would make it more difficult to eradicate a virus. In non-severe COVID-19 patients without chronic complications, T cell activation is appropriately effective against SARS-CoV-2. However, based on differences in severe and non-severe patients, particularly older patients with comorbidities, T cell overactivation followed by T cell exhaustion, lymphocytopenia, increased viral replication, and secondary infection may be reasons leading to death in severe patients.
T cell count reduction in peripheral blood (PB) appears to be a common characteristic of SARS-CoV-2 infection, and this is quite obvious in severe patients with the risk of ICU admission and death. Thus, decreased PB T cell counts, which is independent of age and other complications, might be able to serve as an important warning signal for clinicians. In addition to the decrease in T cell count in COVID-19 patients, there is also disruption of the T cell differentiation balance, including an increased percentage of CCR6+Th17 cells and a decreased percentage of Treg cells, indicating a systematic proinflammatory T cell response was activated, which may aggravate pathological immune damage to the patient. Correlations between the disrupted Th17/Treg differentiation balance and cytokine storm should be elucidated in the future to better understand the pathological role of CD4+ T cells in COVID-19 infection and help to develop appropriate immune treatment. Furthermore, CD8+ T cells play an important role in virus clearance and prevention of recurrence. Excessively activated and exhausted CD8+ T cells are found in COVID-19 patients, particularly in the severe cases, which suggests that the anti-virus adaptive immune response may be triggered at first, but virus replication may gradually lose control due to T cell exhaustion. Future studies focused on discovering anti-virus-specific T cell clones and elucidating immune evasion mechanisms intermediated by SARS-CoV-2 will help in developing new immunotherapeutic “weapons” to fight this threatening disease.
REFERENCES
- [1].Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020;8 (4):420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wan S, Yi Q, Fan S, et al. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP). medRxiv 2020;doi: 10.1101/2020.02.10.20021832. [Google Scholar]
- [3].Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis 2020;doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhou Y, Fu B, Zheng X, et al. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. bioRxiv 2020;doi:10.1101/2020.02.12.945576. [Google Scholar]
- [5].Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395 (10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395 (10223):507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen J, Qi T, Liu L, et al. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect 2020;80 (5):e1–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 2020;8 (5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 2020;doi:10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol 2020;38 (1):1–9. [DOI] [PubMed] [Google Scholar]
- [11].Chan JF, Kok K, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infec 2020;9 (1):221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395 (10224):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Y, Holmes EC. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell 2020;181 (2):223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ng O, Chia A, Tan AT, et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine 2016;34 (17):2008–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol 2017;39 (5):529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intens Care Med 2020;46 (5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Watts TH. TNF/TNFR family members in costimulation of T cell responses. Annu Rev Immunol 2005;23 (1):23–68. [DOI] [PubMed] [Google Scholar]
- [18].Zheng H, Zhang M, Yang C, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol 2020;doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cossarizza A, De Biasi S, Guaraldi G, Girardis M, Mussini C. SARS-CoV-2, the virus that causes COVID-19: cytometry and the new challenge for global health. Cytom Part A 2020;97 (4):340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chu H, Zhou J, Wong BH, et al. Middle east respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis 2016;213 (6):904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]


