Abstract
As one of the best characterized adult stem cells, hematopoietic stem cell (HSC) homeostasis is of great importance to hematopoiesis and immunity due to HSC's abilities of self-renewal and multi-lineage differentiation into functional blood cells. However, excessive self-renewal of HSCs can lead to severe hematopoietic malignancies like leukemia, whereas deficient self-renewal of HSCs may result in HSC exhaustion and eventually apoptosis of specialized cells, giving rise to abnormalities such as immunodeficiency or anemia. How HSC homeostasis is maintained has been studied for decades and regulatory factors can be generally categorized into two classes: genetic factors and epigenetic factors. Although genetic factors such as signaling pathways or transcription factors have been well explored, recent studies have emerged the indispensable roles of epigenetic factors. In this review, we have summarized regulatory mechanisms of HSC homeostasis by epigenetic factors, including DNA methylation, histone modification, chromatin remodeling, non-coding RNAs, and RNA modification, which will facilitate applications such as HSC ex vivo expansion and exploration of novel therapeutic approaches for many hematological diseases.
Keywords: Epigenetics, Hematopoietic stem cells, Homeostasis
1. INTRODUCTION
A type of cells that possess the capabilities of forming colonies in spleens and differentiating into hematopoietic precursors were identified by Ernest McCulloch and James Till in the early 1960s, which are now known as hematopoietic stem cells (HSCs).1 After identifying colony-forming HSCs, Ernest McCulloch and James Till profiled the properties of stem cells such as self-renewal and multi-lineage differentiation, and demonstrated the basis of bone marrow transplantation, which prolongs the lives of patients with leukemia and other hematopoietic malignancies.2 Their pioneering work fostered HSC homeostasis theory, that self-renewal and multi-lineage differentiation of HSCs require a precise balance (Fig. 1). HSC homeostasis is directly linked to the maintenance and functionality of HSCs, and disrupted homeostasis may lead to severe hematopoietic diseases. Excessive self-renewal or differentiation blockage of HSCs can lead to various types of leukemia, while excessive differentiation or exhaustion of HSCs will exert serious negative effects like immunodeficiency and anemia.2 For decades, researchers have been trying to find out the factors that regulate HSC homeostasis. There is an assumption called “hemopoietic inductive microenvironment,”3 which further brings in the “stem cell niche” concept where stimuli are generated and transmitted to regulate HSCs’ activities including self-renewal and differentiation.4 In recent years, progress in epigenetics, focusing on heritable changes of phenotypes with no genotype alterations, including DNA methylation, histone modification, chromatin remodeling, non-coding RNAs (ncRNAs), and RNA modification has revealed that epigenetic regulatory mechanisms greatly contribute to HSC homeostasis (Fig. 1). In this review, we will mainly focus on HSC homeostasis-related epigenetic factors and systematically summarize epigenetic regulation mechanisms of HSC homeostasis (Fig. 2).
Figure 1.
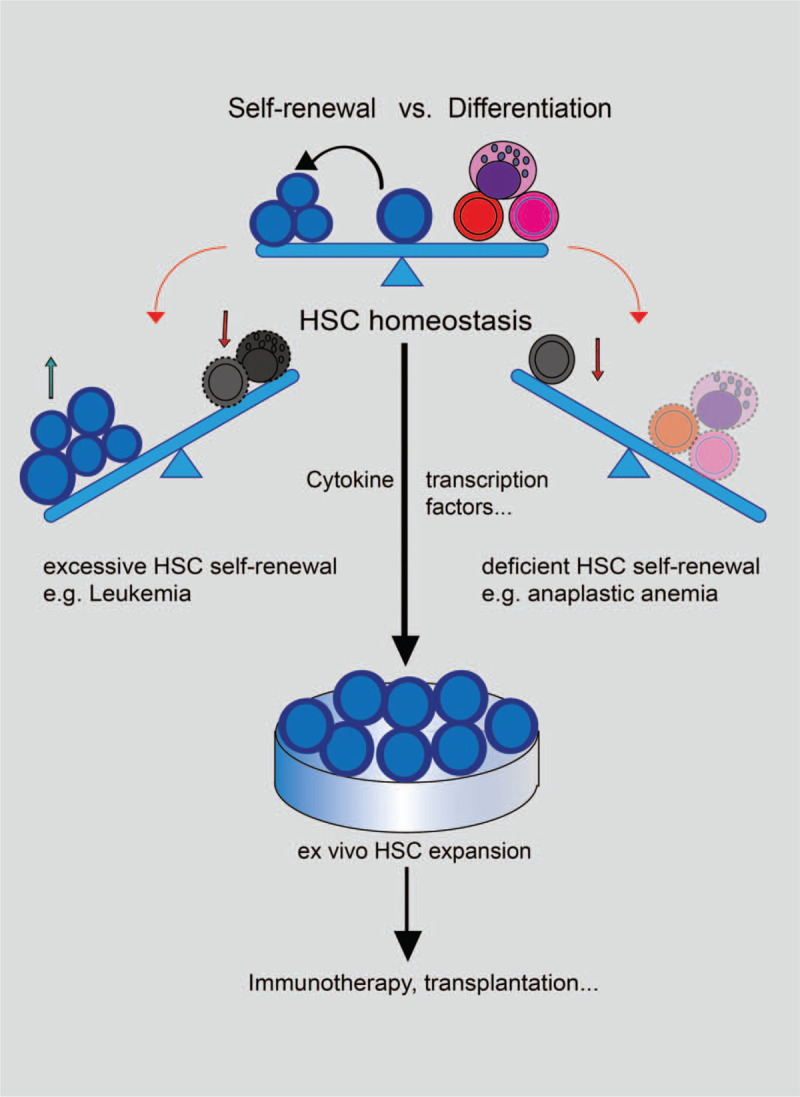
The model of HSC homeostasis. HSC homeostasis is the balance between HSC self-renewal and differentiation activity. The disrupted balance leads to hematopoietic malignancies. Excessive HSC self-renewal can result in lethal diseases like leukemia while deficient HSC self-renewal may cause the lack of differentiated cells and induce diseases like anaplastic anemia. Maintenance of HSC homeostasis can facilitate the ex vivo expansion of HSCs to achieve medical purposes like immunotherapy or transplantation.
Figure 2.
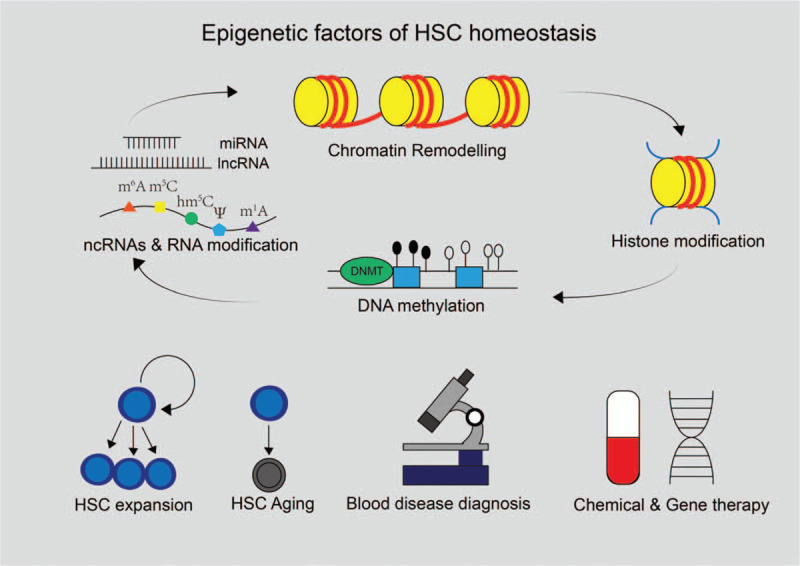
Epigenetic factors of HSC homeostasis and potential applications. Epigenetic factors including DNA methylation, histone modification, chromatin remodeling, non-coding RNAs, and RNA modification have been identified in recent decades. These factors are interactive and are essential for the regulation of HSC homeostasis. Understanding the regulatory mechanisms of these factors may unleash huge potential of HSCs in clinic such as blood disease diagnosis, developing chemical and gene therapies through expanding HSCs or slowing down their aging.
2. DNA METHYLATION
DNA methylation is the first investigated epigenetic marker, which plays important roles in gene imprinting, X chromosome inactivation, transposon inhibition, and developmental processes.5,6 In vertebrates, DNA methylation usually occurs on cytosine of CpG dinucleotides, which is predominantly established by DNA methyltransferases (DNMTs), and oxidized to 5-hydroxymethylcytosine (5hmC) by Ten-eleven translocation (TET) enzymes. DNMT1 participates in the maintenance of DNA methylation patterns after DNA replication while DNMT3A and DNMT3B are mainly responsible for de novo methylation. The regulatory mechanisms of DNA methylation on transcription shows diversity and complexity. Generally, methylation of cis-regulatory elements (such as promoters or enhancers) will inhibit the binding of some transcriptional factors and recruit methylation-binding proteins (such as MECP2, MBD) and co-regulatory factors to decrease chromatin accessibility, resulting in transcriptional inhibition.7,8 In mammals, most individual CpGs are methylated, but the CpG enrichment regions (CpG island), that usually locate near the promoters are generally unmethylated, and those near active enhancers are hypomethylated. Besides, imprinting control regions (ICRs) display allele-specific methylation, which leads to expression of imprinted genes in a parent-of-origin specific manner.6,9
Under physiological conditions, DNA methylation undergoes extensive changes during HSC differentiation into multipotent progenitor cells and lineage commitment cells. Along with advances in next-generation sequencing technology in recent years, researchers have built a genome-wide map of DNA methylation of hematopoiesis trajectory at single-base resolution.10–13 These dynamic changes are analyzed more comprehensively and deepen our understanding that DNA methylation maintains HSC functionality and abnormal methylation pattern leads to hematopoietic malignancies. Along with HSC differentiation, methylation and demethylation occur simultaneously in multiple regions. These differentially methylated regions (DMRs) significantly overrepresent in DNaseI hypersensitivity sites (DHSs), promoters, enhancers, and/or TF binding sites (TFBSs).10 DMRs frequently overlap with lineage-associated TFs and their binding sites, and correlate inversely with expression of these genes. For example, HSC-specific Hoxb cluster genes gain methylation on the DERARE region and show decreased expression during lineage commitment.11,12 DNA methylation maintains HSC homeostasis by regulating expression of non-coding RNAs. An ICR (H19-DMR) in the H19-Igf2 imprinted locus exhibits maternally-specific methylation, which results in H19 and H19-derived miR-675 expressing only on the maternal allele. Deletion of H19-DMR in LT-HSCs caused Igf2 upregulation and miR-675 downregulation, which further led to increased activation, proliferation, and eventual exhaustion of HSCs.14 Another imprinting control region located in the Dlk1-Gtl2 locus controls a miRNA mega-cluster expression. Dlk1-Gtl2 locus preserves functionality of LT-HSCs by inhibiting the PI3K-mTOR pathway and restricting mitochondrial metabolism.15
When HSCs differentiate into lineage commitment state, the methylation of stemness genes increases, while the methylation of lineage specific genes decreases,10 which suggests that abnormal activation or inactivation of demethylase or methylase may break the balance. Indeed, loss of function studies revealed the pivotal roles of DNA methylation enzymes in HSC homeostasis. Bröke and his colleagues demonstrated that constitutive DNA methylation is essential for HSC self-renewal activity and HSCs from mice with reduced DNMT1 activity can differentiate into myelo-erythroid, but not lymphoid, progenies.16 Conditional knockout of DNMT1 also caused defects of HSC self-renewal, niche retention, and improper differentiation of the myeloid lineage.17 Loss of DNMT3A resulted in HSC expansion in bone marrow and impaired differentiation over serial transplantation.18 DNMT3A-null HSCs do not show a decrease in overall DNA methylation levels and hypermethylated CpGs mainly enriched in substantial CpG island. This paradoxical hypermethylation is caused by DNMT3B and drives some DNMT3A-null HSCs differentiation.18,19 Conditional knockout of DNMT3B resulted in a similar but milder phenotype compared with DNMT3A. Combined loss of DNMT3A and DNMT3B can synergistically enhance HSC self-renewal and more severely block differentiation. Overactivation of β-catenin signaling induced by double DNMTs knockout partially explain differentiation blockade.19 Loss of TET2 leads to decreased genomic levels of 5hmC and an increase in the number of HSCs/HSPCs. TET2 haploinsufficiency contributes to hematopoietic transformation by increasing HSC self-renewal and extramedullary hematopoiesis.20–23
DNA methylation is essential for the maintenance of HSC homeostasis, and mutations in DNMTs or TET2 lead to deficiency of HSC differentiation. Thus, abnormal DNA methylation may involve in the occurrence of diseases such as leukemia. Indeed, there is a high mutation rate of DNMT3A in patients with myeloid malignancies including AML (17–34%), myelodysplastic syndromes (MDS, 3–8%), chronic myelomonocytic leukemia (CMML, 4%), myeloproliferative neoplasms (MPN, 10%), and immature adult T-cell acute lymphoblastic leukemia (T-ALL, 4%). A similar high mutation rate of TET2 was also observed in AML (12%), CMML (42%), and MPN (7.6%).24,25 Despite the opposite functions of DNMT3 and TET2, when DNMT3A and TET2 mutations occur simultaneously, they will accelerate the occurrence of malignant transformation.26
Till now, the detailed underlying mechanisms of DNMT3A or TET2 mutation in leukemia remain largely unclear. Several recent studies found that DNMT3A mutation is an early event of AML. For example, DNMT3A mutations were detected in both AML cells and T cells of a patient, suggesting that DNMT3A mutations may occur during the HSC phase, and that DNMT3A mutations may lead to normal HSC becoming a precursor state of AML (pre-leukemic HSC). If mutations occur in other genes, such as NPM1 or FLT3, it may lead to malignant transformation of HSCs into leukemia cells.27 Ebert and his coworkers found that some elderly people without blood-related malignant tumors also carry mutations such as DNMT3A and TET2. Although they can live normally, they are more likely to develop leukemia than normal people.28 Despite the high mutation rate of DNMT3A in patients, no malignant transformation was observed when performing in vivo serial transplantation assay of DNMT3A knockout HSCs in mice every 18 weeks.18 However, mice receiving transplantation of DNMT3A mutant HSCs, without serial transplantation, could recapitulate the types of hematologic diseases observed in patients harboring DNMT3A mutations.29,30 It is possible that the changes of microenvironment caused by the deletion of DNMT3A is an important factor leading to the occurrence of disease. A small number of mutations (e.g. NPM1, Kras, c-Kit) have also been observed in these DNMT3A mutation-induced diseases, and these mutations have also been reported in patients.29,30
The distribution of DNA methylation in HSCs has been well recognized, but the roles of DMRs during cell differentiation or malignant transformation remain to be explored. In the future, these tumor-specific methylation regions will facilitate our understanding and benefit clinical diagnosis, treatment, and prognosis.
3. HISTONE MODIFICATION
DNA double strands are compacted into nucleosomes, the basic chromatin units that then form complex chromosome architecture. Histones, another component of nucleosome, undergo diverse posttranslational modifications (also referred to as histone code) such as acetylation, methylation, ubiquitination, phosphorylation, etc. Histone modifications mainly determine the opening states and accessibility of specific genome regions, thus regulating gene expression levels. The most extensively studied histone modifications are acetylation, methylation, and ubiquitination, all of which play important roles in regulating HSC homeostasis. Histone acetylation is related to open chromatin state and more active transcription, and ubiquitination introduced by PRC1 complex, is usually considered as repressive mark. Histone methylation, however, reflects more complicated state depending on the numbers of methyl added and specific modified amino acids.
3.1. Histone acetylation
Histone acetylation leads to a more open state of chromatin, which therefore is more accessible for TFs. Genes involved in different processes of HSC homeostasis (self-renewal, differentiation) are differentially acetylated periodically, thus acetylation modulators are of great importance in this process. TRRAP (transformation/transcription domain-associated protein), cofactor of histone acetyltransferase, is reported to be implicated in the maintenance of HSCs, deletion of which led to bone marrow failure due to Myc dysregulation and p53-dependent apoptosis in hematopoietic progenitors.31 Besides, Moz (KAT6A), a histone acetyltransferase in the MYST family, is also involved in HSC homeostasis. Moz knockout led to HSC failure and embryonic lethality, but the lineage commitment of cells remained unaffected. C-Kit was downregulated in fetal liver and reconstitution deficiency upon transplantation was also observed in Moz-depleted mice.32,33 Oncogenic MOZ fusion proteins are commonly observed in AML patients, suggesting its critical roles in HSC homeostasis.34
On the other hand, acetyl moiety can be removed by histone deacetylases, which is of equal importance in HSC homeostasis. Heideman et al reported that defects of either histone deacetylase HDAC1 or HDAC2 could be compensated by the other, implying redundant activity of this partner. However, simultaneous deletion of HDAC1 and HDAC2 led to severe HSC defect, causing as anemia and cytopenia.35 Another deacetylase SIRT1 was also reported to be involved in HSC regulation. Knockout of SIRT1 resulted in defects in HSC self-renewal and myeloid-inclined differentiation. Further study revealed that longevity of TF FOXO3 is the key molecule mediating function of SIRT1 in HSC homeostasis regulation.36
3.2. Histone methylation
Effects of histone methylation on gene expression are methyl group number- and residual location-dependent. Three methylation states mark open chromosome and transcription activation: H3K4, H3K36, and H3K79, while K3K9, H3K27, and H4K20 often indicate transcription repression. PRC2 complex (polycomb repressive complex 2) is mainly responsible for repressive H3K27me3 modification and its role in HSC homeostasis regulation has been revealed in several studies. Majewski et al reported that heterozygous mutation in PRC2 components, Ezh2, Eed, or Suz12, promoted HSPC differentiation, and enhanced reconstitution ability in transplantation assay, implying that PRC2 functions as negative regulator of HSC activity and keeps them in quiescent state.37 However, overexpression of Ezh2 was found to be beneficial to LT-HSC reconstitution ability in transplantation assay while maintaining HSC stemness.38 Hidalgo et al reported that deletion of Ezh1, which is also one of the components of PRC2, significantly impaired HSC self-renewal ability, and induced a senescence-like phenotype.39 Therefore, roles of PRC2 components may differ among distinct contexts, and further studies are warranted to better illustrate their functions.
Other epigenetic regulators, which introduce active methylations and induce/enhance gene expression, is of equal importance in HSC homeostasis. For example, Dot1l (disruptor of telomere silencing 1-like), methylase of H3K79, can regulate expression of GATA2 and PU.1, important TFs during hematopoiesis and myelopoiesis, respectively. Knockout of Dot1l resulted in embryonic lethality due to deficiency in erythroid differentiation.40 Besides, MLL1 (mixed-lineage leukemia 1), component of H3K4 methylation complex, is involved in fetal hematopoiesis. Mll1 knockout mice exhibited dramatic decrease in fetal liver cellularity as well as LT-HSC and ST-HSC numbers.41
3.3. Histone ubiquitination
H2A is the main histone target of ubiquitination mediated by PRC1, mostly occurring at lysine119 residual. H2A-K119ub represents close chromatin state and gene silencing, and the ubiquitination state is controlled by counteracting ubiquitin ligating and erasing enzymes. PRC1 is a heterogeneous multi-subunit complex composed of Bmi1, RING1/2, Mel18, Rae28/MPH1, and M33/CBX2, which were all reported to affect HSC functionality. Ding and colleagues discovered that during embryonic stem cell differentiation, ectopic expression of Bmi1 promoted hematopoietic cell development by inducing GATA2, which is necessary for primitive hematopoiesis. In addition, PRC1 could also promote HSPC proliferation derived from ESCs through inhibition of P16INK4A/P19ARF expression.42 Another PRC1 component, Rae28, is also involved in early hematopoietic development. Depletion of Rae28 was lethal due to fetal liver HSC deficiency as reflected by colony forming and transplantation assays.43 However, knockout of Mel18, one PRC1 component, resulted in increased G0 phase HSCs and promoted self-renewal, suggesting that Mel18 functions as a negative regulator of HSC self-renewal.44 Nevertheless, further studies are needed to confirm whether function of PRC1 in HSC homeostasis is dependent on its H2A-ubiquitination activity.45
Over the past decade, mounting studies documented the crucial roles of histone modification in regulation of HSC homeostasis under patho/physiological conditions. Based on these findings, treatments targeting histone modifications are being developed against many kinds of hematological malignancies. On the other hand, it is still not clear how different epigenetic markers cooperate in regulating gene expression and further exploration is needed for a more thorough understanding of HSC homeostasis.
4. SPATIAL ORGANIZATION OF CHROMOSOMES
Chromosomes do not exist in a simple linear form, but in a three-dimensional structure of highly ordered folding in the nucleus. The spatial organization of chromosomes has a very important influence on transcription and cell-fate decisions. Especially, tissue-specific gene expression often requires interactions between promoters and tissue-specific activated enhancers during cell differentiation. Cohesin and the CCCTC-binding factor (CTCF) cooperate to form topologically associating domains (TADs) and loops, mediating the interaction of promoters and enhancers. TADs are separated by TAD boundary, which are often enriched with cohesin and CTCF. The frequency of interaction of intra-TAD is much greater than inter-TAD, which results in active enhancers that can only interact with genes in same TAD. If cohesin, CTCF mutation, or CTCF binding site deletion may cause TAD boundary disappear, it may lead to abnormal activation or repression of some genes.46
Cohesin and CTCF have high mutation rates in AML, suggesting that changes in chromatin structure play an important role in malignant transformation.47,48 This may be partially explained by the fact that depletion of cohesin subunit Rad21 enhanced HSC/HSPC self-renewal by activating ERG, GATA2, RUNX1, HOXA7, HOXA9, and increased resistance to myeloid differentiation induced by inflammatory stimuli.48–50 There are many other factors involved in regulating chromatin conformation, such as TFs, DNA methylation, histone modification, and lncRNA, which can directly or indirectly alter chromatin structure.
Largely limited by the rare number of HSCs, the chromosome conformation capture (3C) and 3C derived Hi-C approaches for exploring chromatin structure are difficult to carry out. Thus, few studies have been published on the regulation of chromatin structure in HSC homeostasis. Recently, whole genome promoters interaction maps of 17 human primary hematopoietic cell types were made by promoter capture Hi-C.51 These maps show that promoter interactions are highly cell type-specific and can reflect the lineage relationships of the hematopoietic tree. Interacting regions in the intergenic regions significantly enriched mutations, which have correlation with their target genes expression. Hu et al conducted multiple-enzyme Hi-C, which can use small cell numbers as input to explore chromosome conformation of each developmental stage from HSPC to lineage commitment of early T cells.52 They observed abrupt genome-wide changes of chromatin organization during the transition from double-negative stage 2 (DN2) to DN3, accompanying the T lineage commitment. Such gradual changes in chromatin conformation may provide a barrier to prevent cells from reversing or differentiating into other lineages.
In the future, single-cell Hi-C or micro-dose cell Hi-C can be expected to be applied in drawing a 3-D spatial structure map of the whole genome during HSC differentiation. Such an atlas will give us a further understanding about the role of chromatin spatial structure in HSC homeostasis.
5. NON-CODING RNAS
The ENCODE project revealed that human genome is extensively transcribed, while less than 5% of transcripts possess protein-coding potential, suggesting that there exists huge regulatory network composed of non-protein-coding transcripts in human cells, safeguarding regular physiological processes. The whole blood system in mammalians is supported by HSCs, and these cells generate all mature cells in the blood whose functions also need to be elegantly regulated. Both transcriptional and posttranscriptional regulatory mechanisms ensure proper cellular differentiation and function through modulation of cell death, proliferation, activation, and lineage commitment. Among the regulatory factors involved in these processes are non-coding RNAs whose pivotal roles in the hematopoietic system have been paid more and more attention in recent years.
Here we discuss functions of ncRNA in HSC homeostasis regulation by arbitrarily classifying them by length. miRNAs are short regulatory RNAs with length of 19–23 nt, and function by binding to 3′-UTR of targets base-pairingly, thus suppressing translation and/or mediating degradation of target mRNA. LncRNAs are transcripts longer than 200 nt without protein coding potential. Longer length confers lncRNAs more structure complexity, thus exhibiting more variety of regulatory mechanisms. Different from miRNAs, lncRNAs can interact with proteins, RNAs, or DNAs involved in diverse patho/physiological processes. Recently, with development in the next generation sequencing technologies, more and more ncRNAs were discovered and their importance and function mechanisms in hematopoiesis regulation are being extensively explored.
5.1. miRNAs
Through targeting various factors, miRNAs play an important role in HSC homeostasis, thus dysregulation of which may lead to serious hematological diseases. Besides, second generation sequencing-based profiling studies also show specific miRNA expression signatures in each hematological cell sub-population. O’Connell et al reported that miR-125a, miR-125b, miR-155, miR-99a, miR-126, miR-196b, miR-130a, etc are enriched in LT-HSCs. Among them, overexpression of miR-125b, miR-126, and miR-155 enhanced reconstitution ability of bone marrow cells, demonstrating their importance in maintaining HSC self-renewal. More importantly, some of these miRNAs were discovered to be dysregulated in hematological malignancies.53 For example, upregulation of miR-125b in bone marrow cells resulted in progressive myeloid leukemia.53 Similarly, miR-196b was reported to be overexpressed in most MLL (mixed lineage leukemia) patients.54
Furthermore, Guo et al reported the effect of knocking out Dicer, the master processor of miRNA production, in HSPC. Global downregulation of miRNAs induced apoptosis in HSCs and resulted in loss of functional HSCs. Besides, they also discovered that miR-125a plays a more important role in regulating HSC self-renewal, overexpression of which suppressed apoptosis in lineage negative cells.55 Bissels et al combined miRNA and mRNA profiling data and discovered series of miRNAs as well as their potential targets that may affect HSC stemness. Among them, miR-29a was found to help maintain HSC stemness by targeting molecules involved in Wnt signaling pathway.56 Similar studies also confirmed that overexpression of miR-29a led to enhanced self-renewal of mice HSCs. The authors also found that miR-29a was abnormally upregulated in AML patients, implying its role in HSC self-renewal.57 These works demonstrated the necessity and importance of proper miRNA expression in maintaining HSC homeostasis.
On the other hand, miRNAs are also involved in HSC differentiation. Tenedini et al reported that miR-299 was specifically up-regulated in megakaryoblast in vitro differentiated from human CD34+ hematopoietic progenitors, and ectopic expression of miR-299 in CD34+ cells promoted megakaryocytic-granulocytic differentiation.58 Felli et al discovered that miR-221 and miR-222 were involved in the regulation of erythropoiesis. Overexpression of miR-221 and miR-222 suppressed CD34+ progenitors proliferation while promoted erythropoietic differentiation, and authors further discovered this is accomplished at least partially by downregulating c-Kit.59 Besides, Fazi et al reported that miR-223 is of great importance during myeloid differentiation, and NFI-A and CEBP-α are the downstream targets mediating its function.60 MiR-144 and miR-451, induced by GATA1, contributed to erythrocytic differentiation of HSC,61,62 while miR-150, targeting c-Myb63 or NOTCH3,64 is involved in lymphoid differentiation.
Accumulating evidence show that miRNAs play pivotal roles in HSC homeostasis, and dysregulation of miRNAs are not only typical features of many hematological malignancies but also in some cases the leading causes. So, researches focusing on function and mechanisms will further extend our knowledge about HSC homeostasis and related diseases.
5.2. lncRNAs
Compared with miRNAs, lncRNAs are implicated in regulation of HSC homeostasis through more diverse mechanisms. H19 is among the first identified lncRNAs, which is highly expressed specifically in LT-HSCs while downregulated in progenitor cells. Functional study revealed that conditional knocking out of H19 in hematological cells resulted in reduced HSC number, and accordingly increased number of ST-HSC. Furthermore, reconstitution ability of bone marrow cells was also impaired due to H19 depletion.14,65 Besides, lncHSC-1 and lncHSC-2 are also implicated in HSC homeostasis regulation. Luo et al first discovered functions of lncHSC-1/2 in hematopoiesis, which are highly expressed but hardly detected in differentiated lineages. Knocking down of either lncRNA reduced HSC and progenitor numbers, meanwhile promoting differentiation into myeloid cells or T lymphocytes. Further study revealed that TF E2A is involved in lncHSC-2 functioning.66
There are also lncRNAs implicated in HSC differentiation towards downstream lineages. Myeloid cells, including granulocytes, monocytes, and macrophages, are differentiated from CMPs (common myeloid progenitors) and many lncRNAs are involved in these processes. LncRNA EGO (eosinophil granule ontogeny) was first reported by Wagner et al. As its name inferred, EGO is involved in eosinophil differentiation, and is highly expressed in mature eosinophils. Knocking down of EGO in human CD34+ progenitors suppressed neurotoxin production, and prevented eosinophil differentiation.67 Another lncRNA, HOTAIRM1 (HOX antisense intergenic RNA myeloid 1), is also implicated in myeloid differentiation. Located between HOXA1 and HOXA2, HOTAIRM1's expression is restricted to myeloid cells, and is upregulated during myeloid differentiation. Knocking down of HOTAIRM1 significantly suppressed CD11b and CD18 expression during myeloid differentiation.68 Other lncRNAs, such as lnc-DC, lnc-MC, are all reported to regulate myeloid differentiation through interacting with various molecues.69,70
Lymphocytes, including B cells, T cells, and NK cells, are all derived from CLPs (common lymphoid progenitors), and many lncRNAs can regulate this process. lncRNA NRON can suppress T cell activation by binding to NFAT and preventing its nuclear translocation, which is critical in T cell development and activation.71 Another well-studied lncRNA in hematopoiesis is GAS5 (growth arrest-specific 5), which was shown to suppress proliferation and finally induce apoptosis in T cells. Further studies demonstrated that GAS5 can inhibit target gene expression by interacting with GREs (glucocorticoid receptor response elements).72,73 Ranzani et al discovered the Th1 cell-specific lncRNA linc-MAF-4, which was proved to be critical during Th1 cell differentiation. Knocking down of linc-MAF-4 induced MAF expression and led to Th2 phenotype transition. Further study revealed that linc-MAF-4 can recruit PRC2 and EZH2 to MAF promoter, thus inhibiting its transcription through epigenetic modification.74
The average lifespan of human red blood cells is no more than 120 days, so everyday millions of them died and is replenished through erythropoiesis in bone marrow. Hu et al reported an erythroid-specific lncRNA-EPS that regulates erythropoiesis through affecting HSC differentiation and apoptosis. By transcriptional repression of Pycard, lncRNA-EPS was able to inhibit apoptosis and safeguard erythropoiesis, while knockdown EPS by shRNA inhibited HSC erythroid differentiation.75 Lodish et al reported another lncRNA EC7 that regulates erythropoiesis and functions as enhancer RNA. LncRNA-EC7 can affect expression of Band3 located nearby. The author also found that with the help of CTCF, lncRNA-EC7 could assist chromatin looping of this region and directly activate Band3 transcription.76 Other second generation sequencing-based profiling studies also revealed large amount of ncRNAs that are implicated in erythropoiesis. In Paralkar's work, erythroid lineages specific lncRNA were identified in mice and humans. As expected, lncRNAs are poorly conserved between species, and most of them are located in promoter regions. Besides, in-depth analysis discovered that many of those lncRNAs lay downstream of TFs crucial in erythropoiesis, such as GATA1 and TAL1. Erythroid differentiation suppression was observed after knocking down of some candidates, demonstrating their involvement in erythropoiesis.77
Thus, a number of ncRNAs have been shown to play specific roles in regulation of HSC homeostasis, but there are even larger amount of ncRNAs that have been cataloged but not fully studied. Thus, further exploration of the functions of these transcripts may shed new light on how HSCs and the whole blood system work.
6. RNA MODIFICATION
Recent data revealed particular importance of RNA modification in formation of hematological hierarchy system. In contrast to “epigenetic modification” acting on DNA sequences, RNA modifications or epitranscriptomic modification, are one step closer to terminal function readouts of genetic information, thus more directly affecting physiological conditions. RNA modification is also essential in mammalian hematopoiesis, dysregulation of which causes various hematological malignancies. Here we mainly discuss the roles of N6-methyladenosine (m6A) modification and A-to-I editing in HSC homeostasis.
6.1. m6A modification
Similar to epigenetic modifications, m6A marks are also dynamically regulated by “writers” and “erasers” according to physiological contexts. These epitranscriptomic codes are read out by “reader” proteins, such as YTHDFs and IGF2BPs, finally affecting stability and translation of target mRNAs. The importance of m6A for HSC homeostasis is being extensively studied in different models, and writers, erasers, and readers are all found to be pivotal regulators in HSC homeostasis.
Zhang et al reported that expression of METTL3, the methyltransferase of m6A writer complex, is restricted to the hematovascular regions and especially enriched in hemogenic endothelial cells during embryo development, implying importance of correct m6A modification for early stage hematopoiesis. The authors found that morpholino knockdown of METTL3 impaired endothelial-to-hemogenic transition (EHT) during embryogenesis and thus led to dramatical decrease of HSPC numbers. Further genome-wide m6A profiling discovered Notch1a as down-stream factors mediating roles of METTL3 in hematopoiesis. Notch1a plays a negative role in HSPC formation during EHT, and m6A marks introduced by METTL3 are predominantly decay signals recognized by YTHDF2. Loss of METTL3 led to decreased m6A modification on Notch1a mRNA and enhanced stability, thus suppressing EHT. Accordingly, knockout of YTHDF2 phenocopied defects of METTL3, which was able to be rescued by ectopical expression of wild-type YTHDF2 other than ones with mutation on the m6A binding domain.78 The critical roles of METTL3-m6A-notch1a-YTHDF2 axis in hematopoiesis was also discovered in mice models. Lv et al reported in mice model with conditional knockout of METTL3 in vascular system, that loss of METTL3 expression impaired ETH but did not affect definitive hematopoiesis in late stage of embryogenesis.79 In human CD34+ HSPCs derived from cord blood, overexpression of METTL3 promoted proliferation while blocking myeloid differentiation,80 and vice versa.81 Meantime, Lee et al reported that METTL3-dependent m6A modifications are required during HSC differentiation, and conditional knockout of METTL3 impaired myeloid differentiation of HSC while spared normal functioning myeloid lineage cells,82 bringing up novel insights about roles of METTL3-dependent m6A in HSC homeostasis. Furthermore, knockout of METTL3,83 or other m6A writer complex components such as METTL14,84 RBM15,85 impaired engraftment capacity of donor HSCs to different extents in various experimental settings, confirming critical roles of m6A writers in HSC self-renewal and differentiation.
On the other hand, FTO, an eraser of m6A, is also implicated in regulation of HSC homeostasis. Li et al reported that in AML patients with MLL rearrangements, FTO is upregulated, and ectopically expressed FTO promoted cell growth, colony formation in vitro, and accelerated leukemogenesis in vivo. In contrast, suppression of FTO by shRNA or Crispr-Cas9 has opposite effects. Given the pathogeny of AML, it is likely that upregulation of FTO disturbed HSC homeostasis and led to dysregulated proliferation of myeloid progenitors while blocking differentiation. Mechanistically, ASB2 and RARA were identified as the main downstream targets mediating function of upregulated FTO, implying critical roles of FTO in response to ATRA (all-trans-retinoic acid) regime.86 Besides, Elkashef et al reported that in AML patients with IDH1/2 mutation, D-2-hydroxyglutarate (D-2HG) abnormally accumulated and competitively inhibited FTO activity, leading to increased global m6A level. In this context, enhanced m6A modification, likely of specific target mRNAs, is favorable for aberrant leukemia cell expansion.87 Nevertheless, under conditions without IDH mutation, Su et al reported that R-2HG exhibits antileukemic activity by directly binding to and suppressing FTO function. In this study, researchers also identified MYC as the main downstream effector mediating function of R-2HG and FTO.88 Based on these studies, the roles of FTO in HSC homeostasis are diverse and context-dependent, especially on downstream specific targets, m6A modification of which are mostly affected.
Besides writers and erasers, m6A readers were also found to be critical for HSC homeostasis maintenance. Roles of YTHDF2 in HSC homeostasis and stem cell fate-decision are reported by our and other groups. We previously found that conditional knockout of mouse Ythdf2 increased numbers of HSCs with normal function. Besides, knockdown of human YTHDF2 led to more than 10-fold increase in the ex vivo expansion of hUCB HSCs.89 In a parallel study, Wang et al found that loss of YTHDF2-mediated m6A-dependent mRNA clearance facilitates hematopoietic stem cells regeneration by regulation of Wnt pathway.90 Interestingly, Paris et al also reported that loss of function of YTHDF2 selectively compromised AML cancer stem cells while enhanced normal HSC activity.91 Besides, YTH-domain containing protein IGF2BPs were reported to be novel m6A readers, mediating target mRNA decay by binding to GG(m6A)C motifs.92 Palanichamy et al reported that IGF2BP is specifically overexpressed in MLL-rearranged B-acute lymphoblastic leukemia (B-ALL), ectopic expression of which promoted proliferation of mice HSPC cells, skewing hematopoietic development towards B cell/myeloid lineage.93 The authors also found that IGF2BP functions through binding to target genes MYC and CDK6 at their 3′-UTR region, which are typical m6A hot spots.
In summary, writers, erasers, and readers of m6A are all engaged in regulation of HSC homeostasis by modifying m6A marks on essential mRNAs. Although it is recognized that RNA modification plays a critical role in gene-expression regulation and thus affecting HSC homeostasis, how RNA methylation controls cell-fate decisions in diverse physiologic settings warrants further investigation.
6.2. A-to-I editing
The deamination of adenosine to inosine, namely A-to-I editing, is another prevalent RNA modification in mammalian transcriptome, which is introduced by ADAR family proteins, including ADAR1/2/3. Inosine is then recognized as guanosine when the edited mRNA is translated, thus causing varying consequences depending on the affected sequences. The roles of A-to-I editing in HSC homeostasis regulation and hematopoiesis are under extensive investigation. Wang et al reported that heterogeneous loss of functional ADAR1 in mice was embryonic-lethal, and most ADAR1+/− chimeric embryos died before embryonic day 14 with defects in the hematopoietic system, demonstrating the importance of ADAR1 for embryonic erythropoiesis in the fetal liver.94 Hartner et al first reported that loss of ADAR1 in mice led to failure in fetal liver hematopoiesis and defects in erythroid lineage were most drastic.95 Then their group reported that ADAR1 is essential for maintenance of both fetal and adult hematopoietic stem cells, and loss of ADAR1 in HSCs induced global upregulation of type I and II interferon-inducible transcripts and rapid apoptosis, indicating critical roles of RNA editing in HSC homeostasis.94 Abrahamsson et al reported that in human CML CSCs, ADAR1 activation promotes glycogen synthase kinase 3β (GSK3B) mis-splicing, thus leading to accumulation of β-catenin, a self-renewal agonist. Thus, activation of ADAR1 may have a significant role in malignancies that have acquired aberrant stem cell self-renewal capacity.96 Feng et al reported that conditional knock out of ADAR1 specifically in HSCs resulted in a rapid hematopoiesis failure and reconstitution deficiency.97
These researches demonstrated the critical roles of ADARs-mediated RNA editing in regulation of HSC homeostasis. Given the diverse consequence of A-to-I editing, its roles in regulation of HSC homeostasis are also multifarious. Thus, specific downstream targets mediating functional effect of A-to-I editing as well as precise modified bases are among top priorities to be elucidated in the future.
7. CONCLUSION
In recent years, advances in second-generation sequencing technology have greatly expanded our understanding of the roles of epigenetic regulation in HSC homeostasis. However, bulk sequencing of cells with specific molecular markers is difficult to accurately identify the dynamic process of HSC from resting to differentiation. If high-throughput single-cell sequencing, such as single cell ChIP-seq and single cell Hi-C, can be used to explore the epigenetic changes in hematopoietic system, it will greatly facilitate our understanding of the mechanisms of HSC stemness maintenance and differentiation.
Abnormal epigenetic markers, ncRNAs, and RNA modification are involved in the occurrence and development of hematological malignancies. Although some inhibitors of epigenetic related enzymes, such as azacytidine and decitabine, have been approved by FDA to target leukemia and MDS, these inhibitors are only effective for some patients and prone to drug tolerance. In the future, if drugs can be designed for specific epigenetic markers, ncRNAs, or RNA modification, it will be expected to achieve better therapeutic effects.
ACKNOWLEDGMENTS
This work was supported by grants from the National Key R&D Program of China, Stem Cell and Translation Research (2018YFA0109300), Zhejiang Province Science Foundation for Distinguished Young Scholars (LR19H080001), and the National Natural Science Foundation of China (81870080).
REFERENCES
- [1].Siminovitch L, McCulloch EA, Till JE. The distribution of colony-forming cells among spleen colonies. J Cell Comp Physiol 1963;62:327–336. [DOI] [PubMed] [Google Scholar]
- [2].McCulloch EA, Till JE. Perspectives on the properties of stem cells. Nat Med 2005;11:1026–1028. [DOI] [PubMed] [Google Scholar]
- [3].Till JE, McCulloch EA. Hemopoietic stem cell differentiation. Biochim Biophys Acta 1980;605:431–459. [DOI] [PubMed] [Google Scholar]
- [4].Boulais PE, Frenette PS. Making sense of hematopoietic stem cell niches. Blood 2015;125:2621–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Luo C, Hajkova P, Ecker JR. Dynamic DNA methylation: in the right place at the right time. Science 2018;361:1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Skvortsova K, Iovino N, Bogdanovic O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol 2018;19:774–790. [DOI] [PubMed] [Google Scholar]
- [7].Du Q, Luu PL, Stirzaker C, Clark SJ. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics 2015;7:1051–1073. [DOI] [PubMed] [Google Scholar]
- [8].Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet 2016;17:551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stadler MB, Murr R, Bueger L, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011;480:490–495. [DOI] [PubMed] [Google Scholar]
- [10].Cabezas-Wallscheid N, Klimmeck D, Hansson J, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 2014;15:507–522. [DOI] [PubMed] [Google Scholar]
- [11].Qian P, De Kumar B, He XC, et al. Retinoid-sensitive epigenetic regulation of the Hoxb cluster maintains normal hematopoiesis and inhibits leukemogenesis. Cell Stem Cell 2018;22:740–754.e7. [DOI] [PubMed] [Google Scholar]
- [12].Farlik M, Halbritter F, Müller F, et al. DNA methylation dynamics of human hematopoietic stem cell differentiation. Cell Stem Cell 2016;19:808–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bock C, Beerman I, Lien WH, et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell 2012;47:633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Venkatraman A, He XC, Thorvaldsen JL, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 2013;500:345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Qian P, He XC, Paulson A, et al. The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell 2016;18:214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bröske AM, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 2009;41:1207–1215. [DOI] [PubMed] [Google Scholar]
- [17].Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 2009;5:442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 2011;44:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Challen GA, Sun D, Mayle A, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell 2014;15:350–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Quivoron C, Couronné L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011;20:25–38. [DOI] [PubMed] [Google Scholar]
- [21].Shide K, Kameda T, Shimoda H, et al. TET2 is essential for survival and hematopoietic stem cell homeostasis. Leukemia 2012;26:2216–2223. [DOI] [PubMed] [Google Scholar]
- [22].Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011;20:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ko M, Bandukwala HS, An J, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A 2011;108:14566–14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Roller A, Grossmann V, Bacher U, et al. Landmark analysis of DNMT3A mutations in hematological malignancies. Leukemia 2013;27:1573–1578. [DOI] [PubMed] [Google Scholar]
- [25].Abdel-Wahab O, Mullally A, Hedvat C, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 2009;114:144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang X, Su J, Jeong M, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet 2016;48:1014–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014;506:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mayle A, Yang L, Rodriguez B, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 2015;125:629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Celik H, Mallaney C, Kothari A, et al. Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation. Blood 2015;125:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Loizou JI, Oser G, Shukla V, et al. Histone acetyltransferase cofactor Trrap is essential for maintaining the hematopoietic stem/progenitor cell pool. J Immunol 2009;183:6422–6431. [DOI] [PubMed] [Google Scholar]
- [32].Katsumoto T, Aikawa Y, Iwama A, et al. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev 2006;20:1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Thomas T, Corcoran LM, Gugasyan R, et al. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev 2006;20:1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Katsumoto T, Yoshida N, Kitabayashi I. Roles of the histone acetyltransferase monocytic leukemia zinc finger protein in normal and malignant hematopoiesis. Cancer Sci 2008;99:1523–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Heideman MR, Lancini C, Proost N, Yanover E, Jacobs H, Dannenberg JH. Sin3a-associated Hdac1 and Hdac2 are essential for hematopoietic stem cell homeostasis and contribute differentially to hematopoiesis. Haematologica 2014;99:1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rimmelé P, Bigarella CL, Liang R, et al. Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Rep 2014;3:44–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Majewski IJ, Ritchie ME, Phipson B, et al. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood 2010;116:731–739. [DOI] [PubMed] [Google Scholar]
- [38].Kamminga LM, Bystrykh LV, de Boer A, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood 2006;107:2170–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hidalgo I, Herrera-Merchan A, Ligos JM, et al. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell 2012;11:649–662. [DOI] [PubMed] [Google Scholar]
- [40].Feng Y, Yang Y, Ortega MM, et al. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood 2010;116:4483–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].McMahon KA, Hiew SY, Hadjur S, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 2007;1:338–345. [DOI] [PubMed] [Google Scholar]
- [42].Ding X, Lin Q, Ensenat-Waser R, Rose-John S, Zenke M. Polycomb group protein Bmi1 promotes hematopoietic cell development from embryonic stem cells. Stem Cells Dev 2012;21:121–132. [DOI] [PubMed] [Google Scholar]
- [43].Ohta H, Sawada A, Kim JY, et al. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J Exp Med 2002;195:759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kajiume T, Ninomiya Y, Ishihara H, Kanno R, Kanno M. Polycomb group gene mel-18 modulates the self-renewal activity and cell cycle status of hematopoietic stem cells. Exp Hematol 2004;32:571–578. [DOI] [PubMed] [Google Scholar]
- [45].Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell 2004;118:409–418. [DOI] [PubMed] [Google Scholar]
- [46].Stadhouders R, Filion GJ, Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature 2019;569:345–354. [DOI] [PubMed] [Google Scholar]
- [47].Dolnik A, Engelmann JC, Scharfenberger-Schmeer M, et al. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood 2012;120:e83–e92. [DOI] [PubMed] [Google Scholar]
- [48].Cuartero S, Weiss FD, Dharmalingam G, et al. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol 2018;19:932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mazumdar C, Shen Y, Xavy S, et al. Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell 2015;17:675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fisher JB, Peterson J, Reimer M, et al. The cohesin subunit Rad21 is a negative regulator of hematopoietic self-renewal through epigenetic repression of Hoxa7 and Hoxa9. Leukemia 2017;31:712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Javierre BM, Burren OS, Wilder SP, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 2016;167:1369–1384.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hu G, Cui K, Fang D, et al. Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T cells. Immunity 2018;48:227–242.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].O’Connell RM, Chaudhuri AA, Rao DS, et al. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A 2010;107:14235–14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Popovic R, Riesbeck LE, Velu CS, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood 2009;113:3314–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Guo S, Lu J, Schlanger R, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci U S A 2010;107:14229–14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bissels U, Wild S, Tomiuk S, et al. Combined characterization of microRNA and mRNA profiles delineates early differentiation pathways of CD133+ and CD34+ hematopoietic stem and progenitor cells. Stem Cells 2011;29:847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Han YC, Park CY, Bhagat G, et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med 2010;207:475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tenedini E, Roncaglia E, Ferrari F, et al. Integrated analysis of microRNA and mRNA expression profiles in physiological myelopoiesis: role of hsa-mir-299-5p in CD34+ progenitor cells commitment. Cell Death Dis 2010;1:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Felli N, Fontana L, Pelosi E, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A 2005;102:18081–18086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 2005;123:819–831. [DOI] [PubMed] [Google Scholar]
- [61].Dore LC, Amigo JD, Dos Santos CO, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci U S A 2008;105:3333–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Papapetrou EP, Korkola JE, Sadelain M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells 2010;28:287–296. [DOI] [PubMed] [Google Scholar]
- [63].Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2016;165:1027. [DOI] [PubMed] [Google Scholar]
- [64].Ghisi M, Corradin A, Basso K, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood 2011;117:7053–7062. [DOI] [PubMed] [Google Scholar]
- [65].Berg JS, Lin KK, Sonnet C, et al. Imprinted genes that regulate early mammalian growth are coexpressed in somatic stem cells. PLoS One 2011;6:e26410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Luo M, Jeong M, Sun D, et al. Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell 2015;16:426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wagner LA, Christensen CJ, Dunn DM, et al. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood 2007;109:5191–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zhang X, Lian Z, Padden C, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009;113:2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wang P, Xue Y, Han Y, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014;344:310–313. [DOI] [PubMed] [Google Scholar]
- [70].Chen MT, Lin HS, Shen C, et al. PU.1-regulated long noncoding RNA lnc-MC controls human monocyte/macrophage differentiation through interaction with microRNA 199a-5p. Mol Cell Biol 2015;35:3212–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Imam H, Bano AS, Patel P, Holla P, Jameel S. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci Rep 2015;5:8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010;3:ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Williams GT, Mourtada-Maarabouni M, Farzaneh F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem Soc Trans 2011;39:482–486. [DOI] [PubMed] [Google Scholar]
- [74].Ranzani V, Rossetti G, Panzeri I, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol 2015;16:318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 2011;25:2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Alvarez-Dominguez JR, Hu W, Yuan B, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood 2014;123:570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Paralkar VR, Mishra T, Luan J, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood 2014;123:1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhang C, Chen Y, Sun B, et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature 2017;549:273–276. [DOI] [PubMed] [Google Scholar]
- [79].Lv J, Zhang Y, Gao S, et al. Endothelial-specific m6A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res 2018;28:249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Vu LP, Pickering BF, Cheng Y, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 2017;23:1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 2018;22:191–205.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lee H, Bao S, Qian Y, et al. Stage-specific requirement for Mettl3-dependent m6A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol 2019;21:700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017;552:126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Yao QJ, Sang L, Lin M, et al. Mettl3-Mettl14 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells. Cell Res 2018;28:952–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Niu C, Zhang J, Breslin P, et al. c-Myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood 2009;114:2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Xu K, Yang Y, Feng G-H, et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res 2017;27:1100–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Elkashef SM, Lin AP, Myers J, et al. IDH mutation, competitive inhibition of FTO, and RNA methylation. Cancer Cell 2017;31:619–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Su R, Dong L, Li C, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell 2018;172:90–105.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Li Z, Qian P, Shao W, et al. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res 2018;28:904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wang H, Zuo H, Liu J, et al. Loss of YTHDF2-mediated m6A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res 2018;28:1035–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Paris J, Morgan M, Campos J, et al. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 2019;25:137–148.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Huang H, Weng H, Sun W, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 2018;20:285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Palanichamy JK, Tran TM, Howard JM, et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J Clin Invest 2016;126:1495–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 2000;290:1765–1768. [DOI] [PubMed] [Google Scholar]
- [95].Hartner JC, Schmittwolf C, Kispert A, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 2004;279:4894–4902. [DOI] [PubMed] [Google Scholar]
- [96].Abrahamsson AE, Geron I, Gotlib J, et al. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc Natl Acad Sci U S A 2009;106:3925–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].XuFeng R, Boyer MJ, Shen H, et al. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc Natl Acad Sci U S A 2009;106:17763–17768. [DOI] [PMC free article] [PubMed] [Google Scholar]


