Abstract
The transcription of essentially the entire eukaryotic genome produces a huge amount of non-coding RNAs. Among them, long non-coding RNAs (lncRNAs) consist of a significant portion that widely exists across mammal genome, generating from high-throughput transcriptomic studies in the last decade. Although the functions of most lncRNAs remain to be further investigated, many of them have already been shown to play critical roles during normal development and disease conditions. Increasing evidence indicates that lncRNAs involve in versatile biological processes during erythroid proliferation and differentiation, including erythroid cell survival, heme metabolism, globin switching and regulation, erythroid enucleation, etc, via cis- or trans-mediated molecular mechanisms. In this review, we focus on recent advances regarding the functions and mechanisms of lncRNAs in normal erythropoiesis.
Keywords: Erythropoiesis, Globin regulation, Heme biosynthesis, Long non-coding RNAs
1. INTRODUCTION
Red blood cells (RBCs) enable to transport oxygen to all tissues over the body, which are generated in the bone marrow via a multistep process named erythropoiesis. During erythropoiesis, the earliest committed erythroid progenitors are burst-forming unit-erythroid (BFU-E) and colony-forming unit-erythroid (CFU-E). Then the CFU-E progenitors divide 3 to 5 times over 2 to 3 days as they differentiate and undergo many substantial changes, including a decrease in cell size, chromatin condensation, and hemoglobinization, leading up to enucleation and expulsion of extra organelles, ultimately turning into mature RBCs.1
Recently, with the advances of high throughput RNA-sequencing technology, it has been increasingly appreciated that there are substantial RNAs without protein-coding potential in the cells, termed non-coding RNAs (ncRNAs). Albeit >60% of the mammalian genome (∼2/3) is capable to be transcribed, protein-coding genes account for < 2% of these transcribed transcripts in the entire mammal genome.2 Such huge amounts of non-coding transcripts could be mainly classified as miRNAs, small interfering RNAs, housekeeping RNAs, and long non-coding RNAs (lncRNAs).3
LncRNAs are transcripts > 200 nucleotides in length that do not code for proteins. Based on their genomic location and their neighboring protein-coding gene(s), lncRNAs have been further classified into six subgroups: intergenic lncRNAs (lincRNAs), intronic overlapping lncRNAs (ilncRNAs), antisense lncRNAs (alncRNAs), enhancer lncRNAs (elncRNAs), sRNA-host lncRNAs (shlncRNAs), and Pseudogene lncRNAs (plncRNAs).4,5 Compared with protein-coding genes, lncRNAs, in general, process the characteristics such as relatively shorter transcript length with 2 to 3 exons, poor conservation across multiple species, lower expression abundance as well as developmentally spatiotemporal specificity.6,7 Mechanistically, lncRNAs could regulate its targets through diverse molecular mechanisms.8 First, lncRNAs can act as a ribonucleoprotein scaffold to modulate nuclear architecture and regulate gene expression.9 Secondly, LncRNAs also can be co-expressed with their neighbor protein-coding genes and regulate their expression.10 Thirdly, they can be involved in post-transcriptional RNA modifications, include splicing, editing, etc.11 In addition, a small subset of lncRNAs participate in signal transduction and thus control diverse biological processes.12 Since very few lncRNAs have been identified during erythroid disorders, in this review, we solely focus on the very recent advances of the functions and mechanisms of lncRNAs during physiological (normal) rather than pathological erythropoiesis.
1.1. LincEPS and LncRNA-Saf in erythroid cell survival
It has been well documented that lncRNAs serve as vital regulatory elements governing diverse cell differentiation processes, including erythropoiesis. Hu et al13 first explored lncRNA landscape during mouse erythropoiesis utilizing RNA-seq technology from purified mouse fetal liver erythroid progenitors BFU-Es, CFU-Es, and terminal differentiated Ter119+ erythroid cells. They identified > 400 putative lncRNAs that are expressed during mouse erythropoiesis. Among them, lincRNA-erythrocyte prosurvival (lincEPS) is highly induced in terminally differentiating erythroblasts. Its inhibition leads to apoptosis and severely impairs differentiation and subsequent enucleation of erythroblasts, while its ectopic expression protects erythroid progenitors from apoptosis, strongly suggesting a potent antiapoptotic activity (Table 1). Mechanistically, lincRNA-EPS diffuses throughout the nucleus to repress loci encoding common mediators of apoptotic in red cells, including the caspase-activating adaptor protein Pycard. In detail, lincRNA-EPS binds the promoters of its target genes, for example Pycard, and promotes nucleosome occupancy near their transcription start site, therefore repressing their expression, which tightly depends on its interacting protein HNRNPL.14 However, it still unclear how lincRNA-EPS reaches its targets, which awaits further investigation (Fig. 1B).
Table 1.
Long non-coding RNAs in normal erythropoiesis.
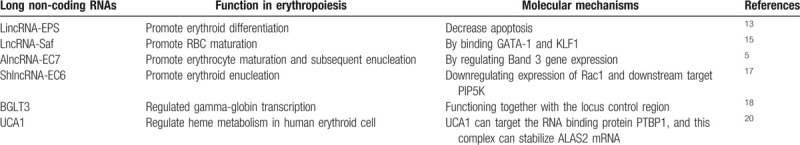
Figure 1.
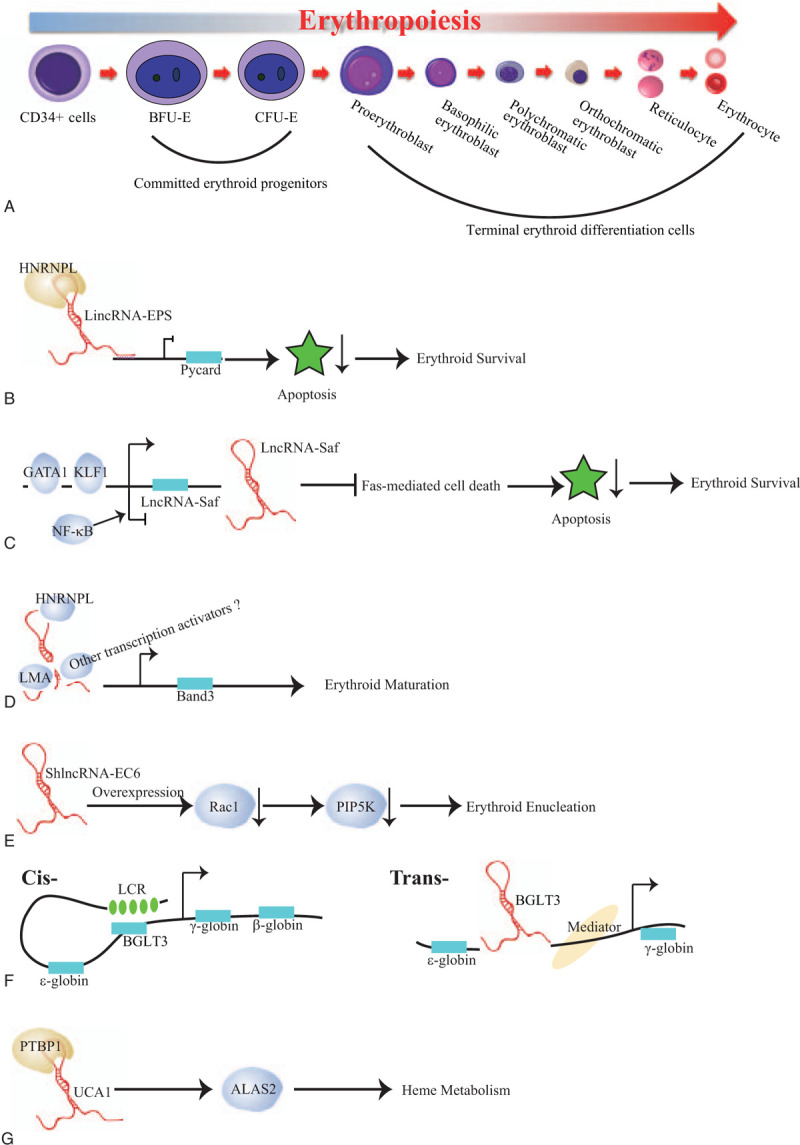
Erythropoiesis and possible modes of action of lncRNAs involved in erythropoiesis. (A) Erythropoiesis. The earliest committed erythroid progenitors are burst-forming unit-erythroid (BFU-E) and colony-forming unit-erythroid (CFU-E). During the terminal erythroid differentiation, the CFU-E progenitors divide 3 to 5 times and undergo many substantial changes, including a decrease in cell size, chromatin condensation, and hemoglobinization, leading up to enucleation and expulsion of extra organelles, ultimately turning into mature red blood cells. Here, Pro-E presents proerythroblast; baso-E presents basophilic erythroblast; Ploy-E presents polychromatic erythroblast; Ortho-E presents orthochromatic erythroblast; Reti presents reticulocyte; RBCs presents red blood cells. (B) LincRNA-EPS, interacting with the RNA-binding protein HNRNPL, binds to the promoter of Pycard to repress its transcription, executing a potent anti-apoptotic activity. (C) LncRNA Saf promotes erythroid cell survival via inhibiting the Fas-mediated cell death signals, which is transcriptionally upregulated by GATA1 and KLF1 while downregulated by NF-κB. (D) AlncRNA-EC7 induces erythroid maturation by enhancing Band 3 expression. Here, alncRNA-EC7 acts as a protein scaffold to carry on its enhancer function by binding to the chromatin attachment factor HNRNPU, the nuclear lamina component LMA and other unknown transcription coactivator(s). (E) ShlncRNA-EC6 promotes erythroid enucleation via regulating Rac1 and its downstream target PIP5K. (F) (left) Cis-regulatory mechanism: BGLT3 gene locus transcriptionally activates fetal γ-globin genes via facilitating chromatin looping between LCR and γ-globin promoters. (right) Tran-regulatory mechanism: albeit BGLT3 transcripts are dispensable for γ-globin/BGLT3 looping, it instead interacts with the Mediator complex, such as MED12 on chromatin to aid γ-globin transcriptional assembly. Here, LCR refers locus control region. (G) lncRNA UCA1 interacts with RNA binding protein PTBP1 to confer ALAS2 mRNA stability, which in turn regulates heme metabolism.
LncRNA Fas-antisense 1 (Fas AS1 or Saf) is upregulated at late stages of human erythroid maturation derived from human bone marrow CD34+ cells.15 Overexpression of this lincRNA in primary erythroid cells protects erythroblasts from Fas-mediated cell death signals. Its upstream regulators are the erythroid lineage specific transcription factors, GATA1 and KLF1, which bind to the promoter of lncRNA-Saf to active its transcription. In conversely, lncRNA-Saf is negatively regulated by NF-κB (Fig. 1C). Nonetheless, its downstream targets and the underlying molecular mechanisms in erythroid maturation are still largely unknown.
1.2. AlncRNA-EC7 and ShlncRNA-EC6 during erythroid differentiation
Alvarez-Dominguez et al also profiled lncRNAs in distinct differentiation stages of erythroid cells from mice fetal liver using RNA-seq.5 They rigorously filtered out 655 differentially expressed lncRNAs at E14.5 erythroid cells, including 132 previously unannotated lncRNAs. After integrating dynamic expressed lncRNAs with genome-wide surveys of chromatin states and transcription factor occupancy, they revealed that dynamic alteration of lncRNAs expression closely orchestrates with chromatin histone architecture and is targeted by key erythroid transcription factors, including GATA1, TAL1, and KLF1. They further performed the functional validation of 12 dynamically expressed lncRNAs and illustrated that majority of them are involved in erythroid differentiation and enucleation. For example, alncRNA-EC7 acts as an enhancer for SLC4A1, the gene encoding band 3, which is the primary anion exchanger of the erythrocyte, to modulate erythroid maturation. Mechanistically, alncRNA-EC7 executes its enhancer function in trans, serving as a scaffold via binding to the chromatin attachment factor HNRNPU, the nuclear lamina component LMA, other transcription coactivators (e.g., MYBBP1A) or corepressors (e.g., HSPA8) (Fig. 1D).16 Meanwhile, another lncRNA ShlncRNA-EC6, also called DLEU2, its down-regulation significantly disturbs erythroid enucleation in mouse fetal liver, mediated by upregulating Rac1 and its downstream targets PIP5K. Consistently, overexpression of Rac1 results in inducing PIP5K expression and then blocking erythroid enucleation (Fig. 1E).17 Additionally, transcribed from an enhancer co-bound by GATA1 and TAL1, ElncRNA-EC3 likely enhances KIF2A expression during human erythropoiesis, which is a kinesin motor involved in microtubule dynamics required for normal mitotic progression. However, KIP2A functions during erythropoiesis are yet to know.5
1.3. BGLT3 in globin regulation
The human β-globin locus comprises five coding genes, including the embryonic-specific ε (HBE1), fetal-restricted Gγ and Aγ (HBG2 and HBG1), and adult expressed δ and β (HBD and HBB) globin genes, which are sequentially activated during development. The intergenic region between the Aγ- and δ-globin genes contains a pseudogene (HBBP1) and a noncoding gene (BGLT3). BGLT3 is shown to specifically regulate fetal γ-globin expression.18 In erythroid cells BGLT3 co-transcribes with gamma-globin and primarily localizes in the nucleus. After combination of the CRISPR/Cas9 mediated whole gene body deletion, transcription start site deletion as well as antisense oligonucleotides (ASOs) mediated knockdown strategies, Ivaldi et al unraveled that BGLT3 locus and transcript have distinct functions. BGLT3 gene locus is exclusively involving in transcriptionally activation of fetal gamma globin genes, but not other types of globin genes, via chromatin looping. Whereas BGLT3 transcripts are dispensable for fetal gamma-globin/BGLT3 looping, it instead interacts with the Mediator complex, such as MED12 on chromatin to facilitate fetal γ-globin transcriptional assembly (Fig. 1 F). Since elevated fetal hemoglobin levels are capable to alleviate the syndrome of sickle cell disease or β-thalassemia in patients, HBBP1 and BGLT3 genes seem are two promising therapeutic targets for genome editing for those diseases.
1.4. UCA1 and heme biosynthesis
Shi et al has performed RNA sequencing (RNA-seq) at distinct differentiation stages derived from mobilized adult peripheral blood CD34+ cells and comprehensively identified 5326 potential long non-coding RNAs.19 After profiling the dynamically expressed lncRNAs during erythropoiesis, Liu and coworkers identified that lncRNA UCA1 regulates heme metabolism in human erythroid cells (Table 1).20 UCA1 expression peaks in proerythroblasts. Consistent with its expression pattern, diminished UCA1 expression blocked the erythroid differentiation at the proerythroblast stage due to the insufficient heme biosynthesis. Mechanistic analysis suggests that UCA1 interacts with RNA binding protein PTBP1 to confer ALAS2 mRNA stability, which is the rate limiting enzyme of heme biosynthesis (Fig. 1 G).20 This study revealed a new layer of lncRNA dependent, post-transcriptional regulation of ALAS2 mRNA, which might shed more lights on the heme disorders with currently unknown reasons.
1.5. Specie-, tissue-, and cellular-specificity of LncRNA
Despite a subset of lncRNAs are highly conserved, the majority of lncRNAs seem to be species- or lineage-specific. Paralkar and coworkers used RNA-seq to investigate tissue- and specie-specific lncRNAs during mouse and human erythropoiesis. In analyzing transcriptome of erythroblasts, megakaryocytes, and megakaryocyte-erythroid precursors, they identified 1109 potential lncRNAs expressed in mouse, while only 594 in humans.21 More importantly, they uncovered ∼15% of mouse lncRNAs are detected in humans or vice versa, reflecting dramatic species-specificity. In summary, these lncRNAs might help to shape the specie- and lineage-specific phenotypes.
1.6. Conclusions and future perspectives
The rapid development of high throughput transcriptome profiling techniques greatly aids the identification of lncRNAs, which has been exponentially increased for the past decade. Compared with the number of identified lncRNAs in erythroid development and differentiation, the lncRNAs with functional annotation are relatively few. Still, these studies raise interesting questions.
1. It is largely uncertain that the great number of lncRNAs identified during erythropoiesis is exclusively erythroid lineage specific or not. For example, although lncRNA UCA1 abundantly and dynamically expressed in erythroid cells,20 it is also highly expressed in spleen and heart.22 Since lncRNAs identified in prior studies were predominantly limited in hematopoietic cells/lineages, a broader comparison cross multiple tissues can answer this question soon.
2. It is intriguing to know whether the lncRNAs share the similar expression pattern possibly conduct resembling roles;
3. given that the majority of identified lncRNAs dynamically expressed during erythroid differentiation, it is yet to be known that the same lncRNA play distinct function at different differentiation stage;
4. although an increasing evidence indicates that lncRNAs play crucial roles during malignant hematopoiesis, the functions of lncRNAs in erythroid diseases have less been demonstrated. In future, it is of importance to continually address the function of these potential regulators to answer these questions. Mechanistically, nuclear localized lncRNAs are more frequently targeting genomic regions to modulate gene expression by recruiting chromatin modifying protein, whereas the cytoplasmic lncRNAs are more likely to interact with RNA-binding proteins to stabilize mRNA or participate in the translational regulation. Thus, combined biochemical and cell biology approaches are promising strategies to integrate the underlying molecular mechanisms of lncRNAs during erythroid development and differentiation, which will also provide valuable insights in various erythroid disorders.
ACKNOWLEDGEMENTS
This work was supported by the National Key Research and Development Program of China (2016YFA0102300 and 2017YFA0103102); CAMS Innovation Fund for Medical Sciences (2016-I2M-3-002, 2016-I2M-1-018 and 2017-I2M-1-015); National Natural Science Foundation of China (81870089 and 81700105); CAMS Medical Epigenetics Research Center (2018PT31033); Supported by the Fundamental Research Funds for the Central Universities (3332018157); State Key Laboratory of Experimental Hematology Research Grant (157-z18-07).
REFERENCES
- [1].Hattangadi SM, Wong P, Zhang L, Flygare J, Lodish HF. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood 2011;118(24):6258–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gallagher PG. Long noncoding RNAs in erythropoiesis. Blood 2014;123(4):465–466. [DOI] [PubMed] [Google Scholar]
- [3].Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet 2006;15 Spec No 1:R17–R29. [DOI] [PubMed] [Google Scholar]
- [4].Kulczynska K, Siatecka M. A regulatory function of long non-coding RNAs in red blood cell development. Acta Biochim Pol 2016;63(4):675–680. [DOI] [PubMed] [Google Scholar]
- [5].Alvarez-Dominguez JR, Hu W, Yuan B, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood 2014;123(4):570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489(7414):101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li F, Xiao Y, Huang F, et al. Spatiotemporal-specific lncRNAs in the brain, colon, liver and lung of macaque during development. Mol Biosyst 2015;11(12):3253–3263. [DOI] [PubMed] [Google Scholar]
- [8].Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17(1):47–62. [DOI] [PubMed] [Google Scholar]
- [9].Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013;20(3):300–307. [DOI] [PubMed] [Google Scholar]
- [10].Luo M, Jeong M, Sun D, et al. Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell 2015;16(4):426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang X, Lian Z, Padden C, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009;113(11):2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv 2017;3(9):eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 2011;25(24):2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Atianand MK, Hu W, Satpathy AT, et al. A Long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 2016;165(7):1672–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Villamizar O, Chambers CB, Mo YY, et al. Fas-antisense long noncoding RNA is differentially expressed during maturation of human erythrocytes and confers resistance to Fas-mediated cell death. Blood Cells Mol Dis 2016;58:57–66. [DOI] [PubMed] [Google Scholar]
- [16].Alvarez-Dominguez JR, Knoll M, Gromatzky AA, Lodish HF. The super-enhancer-derived alncRNA-EC7/Bloodlinc potentiates red blood cell development in trans. Cell Rep 2017;19(12):2503–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang C, Wu X, Shen F, Li Y, Zhang Y, Yu D. Shlnc-EC6 regulates murine erythroid enucleation by Rac1-PIP5K pathway. Dev Growth Differ 2015;57(6):466–473. [DOI] [PubMed] [Google Scholar]
- [18].Ivaldi MS, Diaz LF, Chakalova L, Lee J, Krivega I, Dean A. Fetal gamma-globin genes are regulated by the BGLT3 long noncoding RNA locus. Blood 2018;132(18):1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shi L, Lin YH, Sierant MC, et al. Developmental transcriptome analysis of human erythropoiesis. Hum Mol Genet 2014;23(17):4528–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu J, Li Y, Tong J, et al. Long non-coding RNA-dependent mechanism to regulate heme biosynthesis and erythrocyte development. Nat Commun 2018;9(1):4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Paralkar VR, Mishra T, Luan J, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood 2014;123(12):1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett 2008;582(13):1919–1927. [DOI] [PubMed] [Google Scholar]


