Abstract
We have previously identified, in Paenibacillus popilliae, a 708-bp sequence which has homology to the sequence of the enterococcal vanA gene. We have performed further studies revealing five genes encoding homologues of VanY, VanZ, VanH, VanA, and VanX in P. popilliae. The predicted amino acid sequences are similar to those in VanA vancomycin-resistant enterococci: 61% identity for VanY, 21% for VanZ, 74% for VanH, 77% for VanA, and 79% for VanX. The genes in P. popilliae may have been a precursor to or have had ancestral genes in common with vancomycin resistance genes in enterococci. The use of P. popilliae biopesticidal preparations in agricultural practice may have an impact on bacterial resistance in human pathogens.
In the United States, the percentage of nosocomial enterococcal infections caused by vancomycin-resistant enterococci (VRE) is increasing. This increase poses important problems, including the dearth of available antimicrobial therapy for these organisms and the possibility that vancomycin resistance genes can be transferred to other gram-positive bacteria, especially Staphylococcus aureus. Five glycopeptide resistance types in enterococci—VanA, VanB, VanC, VanD, and VanE—have been described and can be distinguished on the basis of the level and inducibility of resistance to vancomycin and teicoplanin, transferability of glycopeptide resistance, and presence of specific ligase genes. VanA and VanB types of glycopeptide resistance have been associated with outbreaks of VRE infections and are readily transferred from enterococci to other gram-positive organisms. One of the most worrisome examples of this is the transfer of high-level vancomycin resistance from enterococci to S. aureus in the laboratory (13). VanA vancomycin resistance has also been transferred in vitro by conjugation or transformation to Streptococcus sanguis, Lactococcus lactis, Streptococcus pyogenes, and Listeria monocytogenes (4, 16). In addition to laboratory experiments, the vanA gene has been found in vancomycin-resistant clinical isolates of Cellulomonas turbata, Arcanobacterium haemolyticum, and Bacillus circulans (8, 16) and the vanB gene has been found in a vancomycin-resistant isolate of Streptococcus bovis (17). Reduced susceptibility of S. aureus to vancomycin has recently been described in Japan and in the United States, although the mechanism of resistance to vancomycin in these isolates is distinct from that in VRE (19).
VanA-type glycopeptide resistance is characterized by acquired inducible resistance to both vancomycin and teicoplanin. It is mediated by Tn1546 or closely related elements which encode nine polypeptides assigned to groups with different functions: transposition functions; regulation of vancomycin resistance genes (VanR and VanS); synthesis of depsipeptide d-alanyl–d-lactate, which when incorporated into the pentapeptide peptidoglycan precursor forms a precursor to which vancomycin and teicoplanin bind with reduced affinity (VanH and VanA); and hydrolysis of precursors of normal peptidoglycan (VanX and VanY). The function of VanZ is unknown (2). The vanB and vanD gene clusters have homology to the vanA gene cluster but have been less well studied (5, 6, 14).
Vancomycin resistance present in nonenterococcal organisms may have been transferred to enterococci under the pressure of increased oral and parenteral vancomycin use in clinical practice and the use of glycopeptides (avoparcin and orienticin) in animal husbandry (3, 22). The source of these vancomycin resistance genes is unknown. It has recently been hypothesized that the source may be glycopeptide-producing organisms (11). Other environmental organisms may have been the more direct source. Paenibacillus (formerly Bacillus) popilliae, a vancomycin-resistant biopesticide (vancomycin MIC, 800 μg/ml; teicoplanin MIC, <1 μg/ml) (18), has been used in the United States for more than 50 years for suppression of Japanese beetle populations; P. popilliae causes milky disease of Japanese beetle larvae. We have previously identified, in P. popilliae, a 708-bp fragment which has homology to a portion of the enterococcal vanA gene (18). The putative ligase gene in P. popilliae has 77% nucleotide identity to the sequence of the vanA gene and was designated vanE (18). Since our original description, another vanE gene has been described (7); therefore, we have renamed the putative ligase gene in P. popilliae vanF. The purpose of this study was to determine whether vanY, vanZ, and vanX- and vanH-like genes are present in P. popilliae.
MATERIALS AND METHODS
P. popilliae ATCC 14706 was studied. DNA was extracted by using DNA-STAT (Tel-Test, Inc., Friendswood, Tex.). PCR amplification was performed as previously described (15). The PCR primers used included published primers designed to amplify the enterococcal vanH gene (12) and the P. popilliae vanF gene (18), newly designed primers designed (based on the published sequence of vanX) to amplify fragments of the P. popilliae vanXF gene (described herein), and newly designed primers based on sequences derived from restriction site PCR (Table 1). Restriction site PCR was used to extend the sequence in the 5′ and 3′ directions; restriction site PCR involves PCR using four separate universal primers which are representative of given restriction enzyme sites (restriction site primers) and a specific (first-stage) primer from one end of the known sequence (21). This is followed by nested PCR with the restriction site primers and an internal specific (second-stage) primer after which the product is sequenced by using a third internal specific primer (21). All sequences were confirmed in both the 5′-to-3′ and the 3′-to-5′ directions.
TABLE 1.
PCR primers used in this study
| Forward primer | Forward primer
|
Reverse primer | Reverse primer
|
Product size (bp) | ||
|---|---|---|---|---|---|---|
| Sequence | Location on sequence shown in Fig. 1 | Sequence | Location on sequence shown in Fig. 1 | |||
| BPOP-50F | TTTAATATTTACCCCACA | Upstream | BPOP-159R | TACTAACGCCATCCTCTT | 415 | 458 |
| BPOP-46F | AAGGGAATCTACTCTTGG | 221 | BPOP-158R | TTCAGGCGCTTTATCCAT | 576 | 356 |
| BPOP-40F | GACTTGGATGAGCAAAGCAG | 442 | BPOP-1308R | AGCGCAAGGTAGAACAGAAA | 1127 | 686 |
| BPOP-walk5′F | AATTCAATATGAACCTTGGCATATT | 654 | BPOP-4R | CACTAGCGCAAGGTAGATCA | 1131 | 478 |
| BPOP-1086F | TTGATGGAGTAATTGTGACC | 887 | BPOP-1909R | GGCAGAGCTTGTAATGG | 1726 | 840 |
| BPOP-298F | AAAGGTAAAGTTAGGAGACG | 951 | BPOP-147R | CCTAACGGCATAAACAGAAT | 1274 | 323 |
| BPOP4-F | TGATCTACCTTGCGCTAGTGT | 1112 | BPOP-433R | CATTGCGTTGGTTTCCGATA | 1747 | 636 |
| vanHEf | TCGCCACGCTTTGGTGTCATAC | 1685 | BPOPREV | AATTGCTTTCGCCGTCTC | 3416 | 1,732 |
| vanE-FOR | CTCGGCTGTAAAGGTCTT | 3426 | vanX-REV | AAAATAGSTATYGGGGTATGG | 4208 | 783 |
| BPOPX-2356F | GAGCGGCTTCGATTTTATGGA | 4030 | BPOPX-2739R | CCATGCTGGAACACGATGAGGT | 4434 | 405 |
| BPOP-3039F | AACGCCGTCGGAGTCCATTT | 4334 | BPOP-3670R | TATAGTTGCCGAATTTGTTTC | 4800 | 467 |
| BPOP-3508F | GGCGAAGAGGCTTAATATGC | 4616 | BPOP-walk3′R | AGGCACATGGCTCGTTGATTTAAT | 5025 | 410 |
| BPOP-4311F | TTGCTTCTATCGTTTTGTTA | 4965 | BPOP-5754R | CAGCTACTGATGAATTAGTG | 5545 | 581 |
| BPOP-5667F | TGAATGGTATCGCCGCAGTA | 5468 | BPOP-5970R | TCAGGTGAAATACCATAT | 5768 | 301 |
| BPOP-5251F | CCGCACAGCAACTATTTA | 5652 | BPOP-6010R | TCTGCACCTGCCATCAA | Downstream | 548 |
For sequencing, 6 μl of the PCR mix, 1 μl of a 1-U/μl concentration of shrimp alkaline phosphatase, and 1 μl of a 10-U/μl concentration of exonuclease I (United States Biochemical) were incubated at 37°C for 30 min followed by 80°C for 15 min. One microliter of dimethyl sulfoxide and 1 μl of a 3.2-μM sequencing primer were then added. The DNA sequence was determined in both the 5′-to-3′ and the 3′-to-5′ directions with a Taq dideoxy terminator cycle sequencing kit and a 373A DNA sequencer (Applied Biosystems, Foster City, Calif.) by using a series of internal sequencing primers that provided appropriate coverage of the van genes. The sequence data were analyzed with Sequencher 3.0 (Gene Codes Corp., Ann Arbor, Mich.).
Nucleotide sequence accession number.
The nucleotide sequence of the gene cluster of vanYF, vanZF, vanHF, vanF, and vanXF of P. popilliae ATCC 14706 has been submitted to GenBank and given accession no. AF155139.
RESULTS
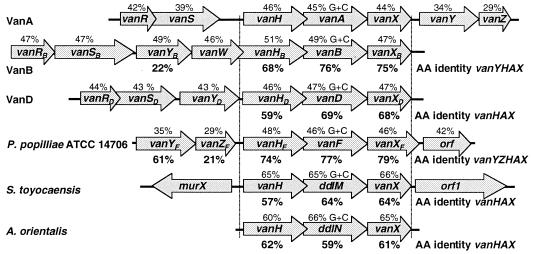
A total of 6,177 bp of P. popilliae DNA, encompassing vanY, vanZ, vanH, vanA, and vanX enterococcal gene homologues, was sequenced (Fig. 1). VanY and the putative P. popilliae VanY protein, VanYF, have 61% predicted amino acid identity (Fig. 2), although the vanY gene is 30 bp longer than the vanYF gene. VanYB and VanYF have 25% predicted amino acid identity, although vanYF is 72 bp longer than vanYB. VanZ and the putative P. popilliae VanZ protein, VanZF, in contrast, have only 21% predicted amino acid identity (Fig. 2), and the vanZF gene is 135 bp longer than the vanZ gene. vanH and the P. popilliae vanH gene homologue, vanHF, have 75% nucleotide identity and 74% predicted amino acid identity (Fig. 2). vanHB and vanHF have 69% nucleotide identity and 65% predicted amino acid identity, although vanHB has 3 more bp at the 5′ end than vanHF and vanH do. VanHD and VanHF have 60% predicted amino acid identity. vanA and vanF (the P. popilliae vanA gene homologue) have 77% nucleotide and predicted amino acid identity. vanB and vanF have 69% nucleotide identity and 67% predicted amino acid identity, although vanB has a 3-bp deletion, as compared to vanF (and vanA). VanD and VanF have 64% predicted amino acid identity. vanX and the P. popilliae vanX gene homologue, vanXF, have 80% nucleotide identity and 79% predicted amino acid identity, although vanXF has 63 additional bp at the 3′ end when compared to vanX. vanXB and vanXF have 74% nucleotide identity and 73% predicted amino acid identity, although vanXF has an additional 63 bp at the 3′ end when compared to vanXB. VanXD and VanXF have 69% predicted amino acid identity, although vanXF has 63 additional bp at the 3′ end when compared to vanX. The orientations of the vanHF, vanF, and vanXF genes are identical to the orientations found in VRE (Fig. 2). Downstream of this gene cluster is an open reading frame encoding a putative protein of unknown function which has 75% amino acid identity to the putative oxidoreductase in the inlA 5′ region in Listeria monocytogenes (9).
FIG. 1.
Sequence and structure of the vancomycin resistance gene cluster in P. popilliae. The putative proteins are shown under the nucleic acid sequence, and the proposed names are provided above the nucleic acid sequences.
FIG. 2.
Alignment of glycopeptide resistance gene clusters of VanA, VanB, and VanD VRE, P. popilliae (as described herein), and the glycopeptide-producing organisms Amycolotopsis orientalis and Streptomyces toyocaensis (1, 6, 7, 12, 15). The percent amino acid identity to vanY, vanZ, vanH, vanA, and vanX products are shown below the respective genes (below the arrows). The percent G+C content of each gene is shown above the genes (above the arrows).
DISCUSSION
We have detected a vancomycin resistance gene cluster in P. popilliae which is homologous to the vancomycin resistance gene cluster in VanA VRE. In addition to the considerable similarity in amino acid composition noted, the orientation and alignment of the vanHF, vanF, and vanXF genes are identical to the orientation and alignment of homologous genes in VanA VRE (except that vanXF has an additional 63 bp at the 3′ end) (Fig. 2). The 3′ extension of vanXF is not present in the genes encoding VanX and its homologues in VanA and VanB enterococci nor is it present in Streptomyces toyocaensis or Amycolotopsis orientalis (11). The signature overlap of the 5′ end of vanF with the 3′ end of vanH, as described by Marshall et al., has been identified (11). That the spatial arrangements of the vanHF, vanF, and vanXF genes are maintained suggests a common ancestry with vancomycin resistance genes in VanA and VanB VRE. vanY and vanZ gene homologues were also found. There are several implications of these findings. That P. popilliae possesses this gene cluster implies that P. popilliae has a glycopeptide resistance mechanism similar to that of VRE. The presence of the vanYF (putative carboxypeptidase) and the vanXF (putative d,d-dipeptidase) genes suggests that at some point during growth, P. popilliae switches from producing the conventional d-Ala-d-Ala peptidoglycan precursor terminus to producing a peptidoglycan precursor terminating in d-Ala-d-Lac. Although the presence of the vanZ gene has been associated with teicoplanin resistance, P. popilliae ATCC 14706 is teicoplanin susceptible (1).
Marshall et al. have hypothesized that the origin of clinically relevant vancomycin resistance lies within the glycopeptide-producing organisms (11). However, the amino acid identities identified by these authors between the glycopeptide-producing organisms Streptomyces toyocaensis and Amycolotopsis orientalis and VanA VRE were 57 to 62% for VanH enzymes, 59 to 64% for DdlM and DdlN enzymes (VanA homologues), and 61 to 63% for VanX enzymes, all substantially less than the identities we found between these genes in the vanA gene cluster and in P. popilliae (Fig. 2). Furthermore, the G+C contents of the P. popilliae vanHF, vanF, and vanXF genes are virtually identical to those of the homologous genes in VRE and significantly different from those of the glycopeptide resistance genes in Streptomyces toyocaensis and Amycolotopsis orientalis (Fig. 2). Therefore, the vancomycin resistance gene cluster in P. popilliae is more similar to that in VRE than are the gene clusters in Streptomyces toyocaensis and Amycolotopsis orientalis. Given that the G+C contents of the VRE vanH, vanA, and vanX genes are higher than those of the adjacent vanR, vanS, vanY, and vanZ genes, it is plausible that the vanH, vanA, and vanX genes have been mobilized as a unit from another source.
Recently, a new type of acquired glycopeptide resistance, termed VanE, has been described in Enterococcus faecalis (7). The partial sequence of VanE in E. faecalis BM4405 has 43% predicted amino acid identity to VanF in P. popilliae (7).
The P. popilliae isolate studied is an American Type Culture Collection type strain which was isolated from commercial spore dust and first described in the medical literature in 1961 (10). That the gene cluster present in P. popilliae has homology to the vanA (and vanB and vanD) gene cluster(s) suggests that it may have been a precursor to or have had a common ancestral origin with the vanA (and vanB and vanD) gene cluster(s) found in modern clinical isolates of enterococci. P. popilliae spores have been introduced into soil in the eastern United States as a biopesticidal powder since the early 1940s. An example of such a product, currently marketed in the United States, is Milky Spore (St. Gabriel Laboratories, Gainesville, Va.). Milky Spore is described by its producer as a product that does not affect humans or animals or contaminate well water. Once established in a lawn, Milky Spore is described as lasting 15 to 20 years. It has been suggested that spread of P. popilliae spores may have been accomplished by birds, insects, skunks, moles, and mice (20). Such widespread distribution of this organism may have provided the opportunity for its contact with enterococci. Furthermore, both enterococci and P. popilliae are able to survive for long periods in the environment; for example, we recently recovered viable P. popilliae from dried Japanese beetle hemolymph preserved on a microscope slide in 1945 (18). In the presence of the increasing use of oral and parenteral vancomycin in humans since the late 1970s for the treatment of Clostridium difficile and methicillin-resistant staphylococcal infections, respectively, and in the presence of glycopeptide usage in agriculture, the transfer of vancomycin resistance to enterococci has potentially been facilitated. Small amounts of P. popilliae produced in North America have been distributed in New Zealand and South America. Although we cannot prove that transfer of vancomycin resistance to enterococci occurred directly from P. popilliae, the evidence suggests that the use of biopesticidal preparations in agricultural practice may have an impact on bacterial resistance in human pathogens.
REFERENCES
- 1.Arthur M, Depardieu F, Molinas C, Reynolds P, Courvalin P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene. 1995;154:87–92. doi: 10.1016/0378-1119(94)00851-i. [DOI] [PubMed] [Google Scholar]
- 2.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bager F, Madsen M, Christensen J, Aarestrup F M. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev Vet Med. 1997;31:95–112. doi: 10.1016/s0167-5877(96)01119-1. [DOI] [PubMed] [Google Scholar]
- 4.Biavasco F, Giovanetti E, Miele A, Vignaroli C, Facinelli B, Varaldo P E. In vitro conjugative transfer of VanA vancomycin resistance between Enterococci and Listeriae of different species. Eur J Clin Microbiol Infect Dis. 1996;15:50–59. doi: 10.1007/BF01586185. [DOI] [PubMed] [Google Scholar]
- 5.Casadewall B, Courvalin P. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J Bacteriol. 1999;181:3644–3648. doi: 10.1128/jb.181.12.3644-3648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evers S, Courvalin P. Regulation of VanB-type vancomycin resistance gene expression by the VanS(B)-VanR (B) two-component regulatory system in Enterococcus faecalis V583. J Bacteriol. 1996;178:1302–1309. doi: 10.1128/jb.178.5.1302-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fines M, Perichon B, Reynolds P, Sahm D, Courvalin P. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob Agents Chemother. 1999;43:2161–2164. doi: 10.1128/aac.43.9.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontana R, Ligozzi M, Pedrotti C, Padovani E M, Cornaglia G. Vancomycin-resistant Bacillus circulans carrying the vanA gene responsible for vancomycin resistance in enterococci. Eur J Clin Microbiol Infect Dis. 1997;16:473–474. doi: 10.1007/BF02471915. [DOI] [PubMed] [Google Scholar]
- 9.Gaillard J L, Berche P, Frehel C, Gouin E, Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-n. [DOI] [PubMed] [Google Scholar]
- 10.Haynes W, St. Julian G, Shekelton M, Hall H, Tashiro H. Preservation of infectious milky disease bacteria by lyophilization. J Insect Pathol. 1961;3:55–61. [Google Scholar]
- 11.Marshall C G, Lessard I A, Park I, Wright G D. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob Agents Chemother. 1998;42:2215–2220. doi: 10.1128/aac.42.9.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miele A, Bandera M, Goldstein B P. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob Agents Chemother. 1995;39:1772–1778. doi: 10.1128/aac.39.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noble W C, Virani Z, Cree R G. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992;72:195–198. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- 14.Ostrowsky B E, Clark N C, Thauvin-Eliopoulos C, Venkataraman L, Samore M H, Tenover F C, Eliopoulos G M, Moellering R C, Gold H S. A cluster of VanD vancomycin-resistant Enterococcus faecium: molecular characterization and epidemiology. J Infect Dis. 1999;180:1177–1185. doi: 10.1086/315030. [DOI] [PubMed] [Google Scholar]
- 15.Patel R, Uhl J R, Kohner P, Hopkins M K, Steckelberg J M, Kline B, Cockerill F R. DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcus spp. isolates. Antimicrob Agents Chemother. 1998;42:202–205. doi: 10.1128/aac.42.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Power E G M, Abdulla Y H, Talsania H G, Spice W, Aathithan S, French G L. vanA genes in vancomycin-resistant isolates of Oerskovia turbata and Arcanobacterium (Corynebacterium) haemolyticum. J Antimicrob Chemother. 1995;36:595–606. doi: 10.1093/jac/36.4.595. [DOI] [PubMed] [Google Scholar]
- 17.Poyart C, Pierre C, Quesne G, Pron B, Berche P, Trieu-Cuot P. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob Agents Chemother. 1997;41:24–29. doi: 10.1128/aac.41.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rippere K, Patel R, Uhl J R, Piper K E, Steckelberg J M, Kline B C, Cockerill F R, Yousten A A. DNA sequence resembling vanA and vanB in the vancomycin-resistant biopesticide Bacillus popilliae. J Infect Dis. 1998;178:584–588. doi: 10.1086/517480. [DOI] [PubMed] [Google Scholar]
- 19.Smith T L, Pearson M L, Wilcox K R, Cruz C, Lancaster M V, Robinson-Dunn B, Tenover F C, Zervos M J, Band J D, White E, Jarvis W R. Emergence of vancomycin resistance in Staphylococcus aureus. N Engl J Med. 1999;340:493–501. doi: 10.1056/NEJM199902183400701. [DOI] [PubMed] [Google Scholar]
- 20.St. Julian G, Bulla L A. Milky disease. In: Cheng T C, editor. Current topics in comparative pathobiology. New York, N.Y: Academic Press, Inc.; 1973. pp. 57–87. [Google Scholar]
- 21.Weber K L, Bolander M E, Sarkar G. Rapid acquisition of unknown DNA sequence adjacent to a known segment by multiplex restriction site PCR. BioTechniques. 1998;25:415–419. doi: 10.2144/98253st02. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimura H, Ishimaru M, Endoh Y S, Suginaka M, Yamatani S. Isolation of glycopeptide-resistant enterococci from chickens in Japan. Antimicrob Agents Chemother. 1998;42:3333. doi: 10.1128/aac.42.12.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]