Abstract
ALKBH3, a demethylase responsible for demethylating N1-methyladenosine (m1A) in mRNA and N1-methyldeoxyadenosine in single-stranded DNA, plays an important role in DNA repair and cancer cell proliferation. However, its function in hematopoietic stem cells (HSCs) is unknown. In this study, we generated Alkbh3 knockout mice and observed that the deletion of Alkbh3 does not impair the reconstitution capacity of HSCs in both primary and secondary transplantation. Aged hematopoietic stem and progenitor cells exhibit increased expression of ALKBH3. Forced ALKBH3 rescued the differentiation skewing without affecting the reconstitution capacity of aged HSCs. In brief, our study for the first time investigated the functional role of ALKBH3 in hematopoietic system, and observed that ALKBH3 is dispensable for HSCs maintenance and differentiation, but overexpression of ALKBH3 rectified the differentiation skewing of aged HSCs.
Keywords: Aging, ALKBH3, Hematopoietic stem cell
1. INTRODUCTION
HSCs generate all lineage of blood cells and have the ability of self-renewal.1 During aging, the function of HSCs declines gradually, characterized by increased in HSCs number, decreased the ability to reconstitute blood system, and enhanced myeloid differentiation bias.2–5 Previous studies revealed that both the intrinsic and extrinsic factors contributed to HSCs maintenance and aging.6,7 However, the molecular mechanisms of HSCs self-renewal and aging are still not well understood.
RNA modification is regarded as a pivotal regulator of cellular fate. Previous studies identified over 100 different types of chemical modifications to cellular RNA.8 Among those modifications, N6-methyladenosine (m6A) is the most abundant type in human and mouse tissues, and it is well characterized in HSCs. A series of studies reveal that m6A signal and m6A writer protein METTL3/METTL14 are essential for hematopoiesis and HSCs maintenance.9–11 Furthermore, recent evidence shows that the depletion of an m6A reader protein YTHDF2 results in HSC expansion.12–14 Taken together, these studies indicated that the m6A pathway physiologically regulates HSCs function. Unlike m6A, N1-methyladenosine (m1A) is a less abundant modification whose physiological function has largely been ignored. In human cells, m1A is prevalent in cytoplasmic and mitochondrial tRNAs, catalyzed by TRMT6, TRMT61A, and TRMT61B, TRMT10C, respectively.15–17 In addition to tRNAs, m1A is also found in mammalian mRNA, mainly enriched in the 5′ UTR.18–20 Although limited evidence derived from cultured cell line indicated that m1A was a dynamic modification that responses to many cellular stresses, the in vivo function of this modification especially how it regulates HSC and HSC aging is largely unknown.
RNA modification could be reversed by demethylase. Most of the demethylases belong to the Alkb family which containing nine homologs (ALKBH1-8, FTO).21 FTO and ALKBH5 are responsible for removing m6A in mRNA and play a crucial role in human obesity and mouse fertility respectively.22,23 ALKBH1 is another Fe(II)- and α-ketoglutarate-dependent dioxygenases in the Alkb family, which mediates the demethylation of m1A in tRNA and has been shown to influence embryonic development.24,25 ALKBH3, initially identified as a DNA repair enzyme,26 has been demonstrated as an m1A demethylase in mRNA.19 Although previous studies reveal that ALKBH3 participated in cancer cell proliferation,26 the functional role of ALKBH3 in the hematopoietic system is largely unknown.
In this study, we generated the Alkbh3−/− mice and reveal that ALKBH3 is dispensable for normal hematopoiesis. The deficiency of ALKBH3 does not influence the self-renewal capacity of HSCs. The expression of ALKBH3 was up-regulated in aged hematopoietic stem and progenitor cells (HSPCs) while ALKBH3-overexpression does not affect the repopulation capacity of young or aged HSCs. Nevertheless, overexpression of ALKBH3 repressed aged HSCs differentiated into myeloid cells and promoted the T lymphoid differentiation. Taken together, our study found that ALKBH3 is dispensable for HSC maintenance and differentiation, but overexpression of ALKBH3 rectified the differentiation skewing of aged HSCs.
2. RESULTS AND DISCUSSION
2.1. ALKBH3 is dispensable for normal hematopoiesis
In order to investigate the function of ALKBH3 in the hematopoietic system, we generated the Alkbh3 knockout mice by deleting a region containing exon 3, exon 4, and exon 5 of the Alkbh3 gene using CRISPR-Cas9 technology (Fig. 1A). The deletion of ALKBH3 in hematopoietic stem and progenitor cells was confirmed by western blot analysis (Fig. 1B). The Alkbh3−/− mice developed normally and do not exhibit any spontaneous overt phenotypes. Consistent with a previous study, Alkbh3 knockout mice were viable and without overt phenotypes.27
FIGURE 1.
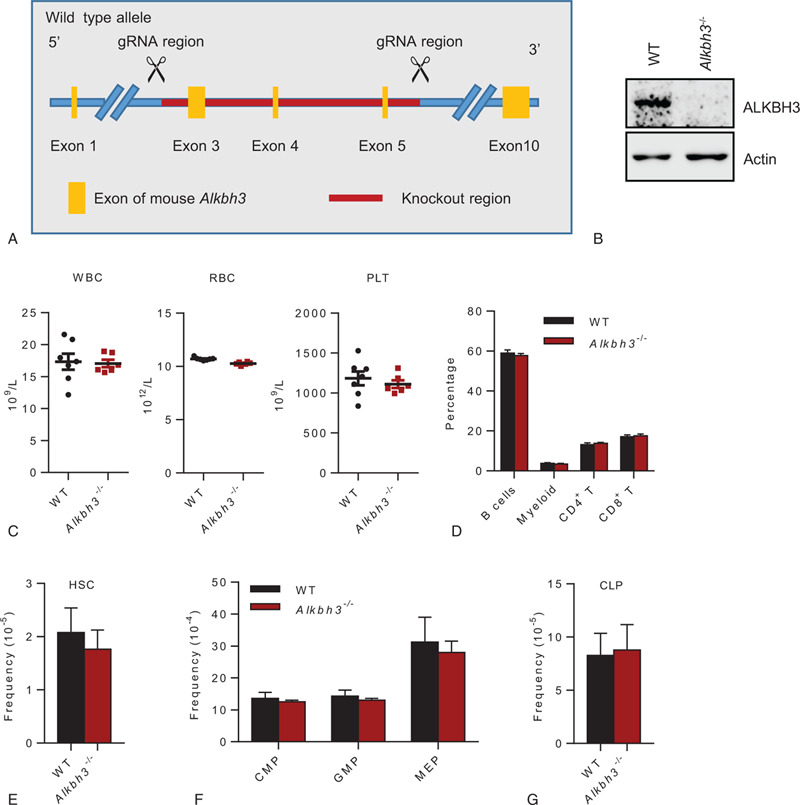
Alkbh3 is dispensable for steady-state hematopoiesis. (A) The schematic diagram showing the targeting strategy to generate Alkbh3 knockout mice. (B) Representative western blot showing the expression of ALKBH3 in WT and Alkbh3−/− hematopoietic stem and progenitor (c-Kit+) cells. (C) The scatter plots show the count of white blood cell (WBC), red blood cell (RBC) and platelet (PLT) between WT and Alkbh3−/− mice. N = 6–7 mice per group. Data are shown as mean ± SEM. (D) Six 2–4 mo old Alkbh3−/− and 7 age-matched WT mice were analyzed for myeloid, B and T cells. This histogram depicts the percentage of myeloid, B, CD4+ T and CD8+ T cells in peripheral blood. Data are shown as mean ± SEM. (E–G) Six 2–4 mo old Alkbh3−/− and 5 age-matched WT mice were analyzed for HSCs and progenitors. (E) This histogram depicts the frequency of HSCs (CD34− CD150+ KSL) between WT and Alkbh3−/− mice. (F) This histogram shows the frequency of CMPs (common myeloid progenitors), GMPs (granulocyte/macrophage progenitors), and MEPs (megakaryocytic/erythroid progenitors) between WT and Alkbh3−/− mice. (G) This histogram shows the frequency of CLPs (common lymphoid progenitors) between WT and Alkbh3−/− mice. Data are shown as mean ± SEM.
We next set out to investigate whether the deletion of Alkbh3 impairs normal hematopoiesis. First, we analyzed the blood cell count in peripheral blood from Alkbh3−/− mice or WT littermate and found no significant difference in white blood cell (WBC), red blood cell (RBC), and platelet (PLT) between WT and Alkbh3−/− mice (Fig. 1C). Next, we analyzed the percentage of B cells, myeloid cells, CD4+ T cells and CD8+ T cells in peripheral blood from Alkbh3−/− mice or WT littermate and observed no significant difference in these differentiated lineage cells between Alkbh3−/− and WT mice (Fig. 1D). Finally, we investigated HSCs and progenitor frequency in Alkbh3−/− or WT mice and found that Alkbh3 deficiency did not result in any significant changes in the percentage of HSCs, common myeloid progenitors (CMPs), granulocyte/macrophage progenitors (GMPs), megakaryocytic/erythroid progenitors (MEPs) and common lymphoid progenitors (CLPs) in bone marrow (Fig. 1E–G). Taken together, these results reveal that Alkbh3 is dispensable for steady-state hematopoiesis.
2.2. ALKBH3 loss does not impair the self-renewal capacity of HSCs
To further evaluate the influence of Alkbh3 deletion on the reconstitution capacity of HSCs, we performed competitive transplantation assays. In the beginning, 50 HSCs were freshly isolated from WT or Alkbh3−/− mice and transplanted together with 3 × 105 competitor cells into lethally irradiated recipients. The reconstitution capacity of Alkbh3−/− HSCs was similar to WT HSCs and no significant differences were observed in lineage distribution (Fig. 2A,B). We attempted to speculate that this phenomenon may be due to the excessive number of transplanted HSCs, thus masking the differences between individual cells. Accordingly, we reduced the number of transplanted HSCs to 20. However, we still have not found any significant difference in repopulation capacity and lineage distribution between WT and Alkbh3−/− HSCs (Fig. 2C,D). We next performed secondary transplantation assay using total bone marrow cells from primary recipients of 20 HSCs transplantation to evaluate the self-renewal capacity of Alkbh3 deficiency HSCs. No significant difference in reconstitution capacity and lineage distribution were observed between WT and Alkbh3−/− HSCs even in the secondary transplantation (Fig. 2E,F). Taken together, these results reveal that Alkbh3 loss does not impair the self-renewal capacity of HSCs.
FIGURE 2.
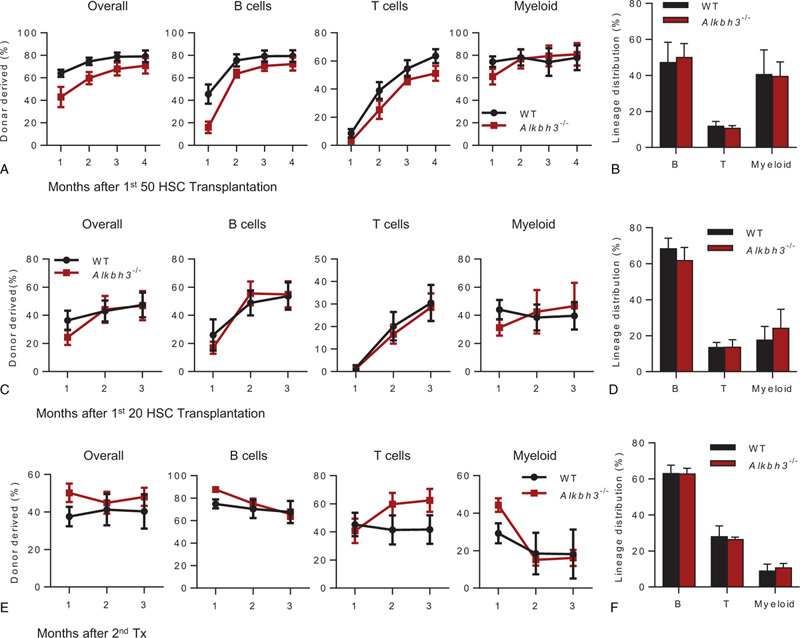
Alkbh3 deficiency does not impair the self-renewal capacity of HSCs. (A and B) Freshly isolated 50 HSCs from 4 mo old Alkbh3−/− mice or age-matched WT mice were transplanted into lethally irradiated recipients together with 3 × 105 competitor cells. Chimera in peripheral blood was checked every month until the fourth month. (A) This histogram displays the percentage of donor cell reconstitution in overall (CD45.2+), B (B220+), T (CD3+) and myeloid (Mac-1+) cell every month after transplantation. (B) This histogram displays the lineage distribution of donor-derived peripheral blood at the fourth month. N = 6 mice per group. Data are shown as mean ± SEM. (C and D) Freshly isolated 20 HSCs from 2 mo old Alkbh3−/− mice or age-matched WT mice were transplanted into lethally irradiated recipients together with 3 × 105 competitor cells. Chimera in peripheral blood was checked every month until the third month. (C) This histogram displays the percentage of donor cell reconstitution in overall (CD45.2+), B (B220+), T (CD3+) and myeloid (Mac-1+) cell every month after transplantation. (D) This histogram displays the lineage distribution of donor-derived peripheral blood at the fourth month. N = 6 mice per group. Data are shown as mean ± SEM. (E and F) Freshly isolated 20 HSCs from either 2 mo old Alkbh3−/− mice or age-matched WT mice were transplanted into lethally irradiated recipients together with 3 × 105 competitor cells. Three months after transplantation, two million bone marrow cells of primary recipients were transplanted into secondary recipients. Chimera in peripheral blood was checked every month until the third month. (E) This histogram displays the percentage of donor cell reconstitution in overall (CD45.2+), B (B220+), T (CD3+) and myeloid (Mac-1+) cell every month after transplantation. (F) This histogram displays the lineage distribution of donor-derived peripheral blood at the third month. N = 7 mice per group, data are shown as mean ± SEM.
2.3. The expression of ALKBH3 increased with aging and Alkbh3-overexpression rectified the differentiation skewing of aged HSCs
Since Alkbh3−/− mice have no overt phenotype in normal condition, we sought to investigate whether Alkbh3 take play a role in HSCs aging. By comparing the expression of ALKBH3 between young and aged HSCs-enriched populations, we found that ALKBH3 increased with aging (Fig. 3A,B). Aged HSC is commonly accompanied by accumulated DNA damage. Based on this, we aim to investigate whether the increase in ALKBH3 is caused by aging-associated DNA damage. Alkbh3 mRNA was measured in freshly sorted HSPCs from WT mice at 6 hours after 5 Gy whole-body X-ray irradiation (IR). Unexpectedly, we found that Alkbh3 expression was slightly decreased after X-ray irradiation. This result suggested that DNA damage was not the driving force of aging-elevated Alkbh3 expression (Fig. 3C).
FIGURE 3.
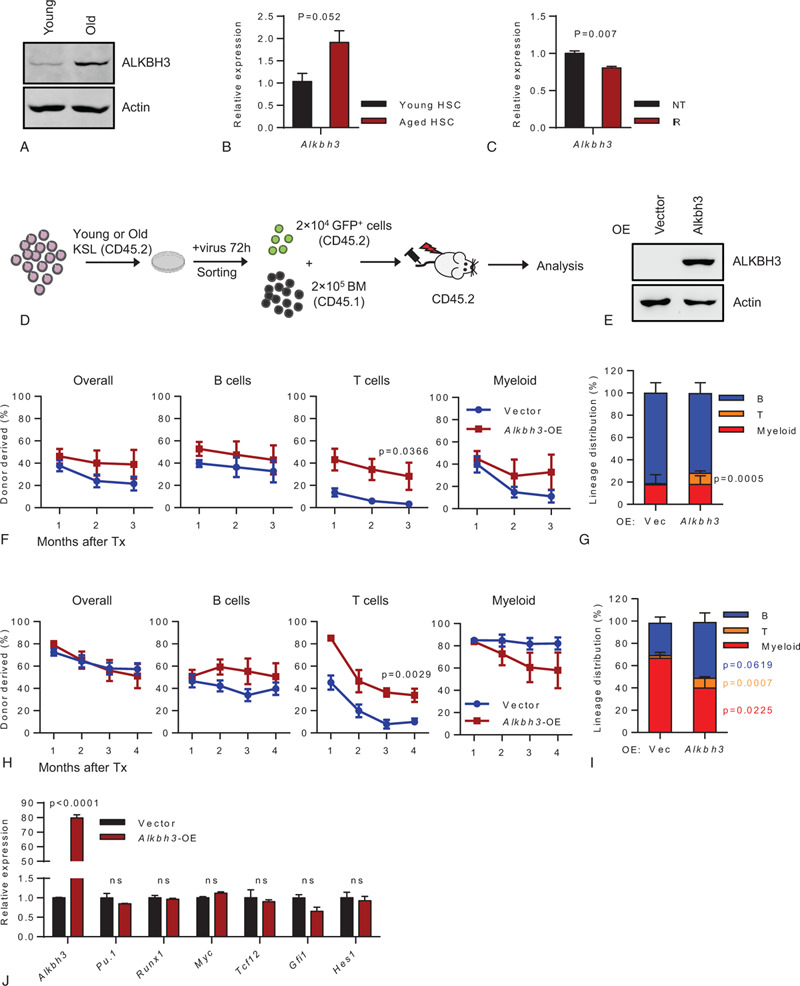
The expression of ALKBH3 increased with aging and Alkbh3-overexpression rectified the differentiation skewing of aged HSCs. (A) Representative western blot showing the expression of ALKBH3 in young (2 mo) and old (28 mo) hematopoietic stem and progenitor (c-Kit+) cells. (B) This histogram depicts the expression of ALKBH3 in young (2 mo) and old (28 mo) HSCs (CD34− CD150+ KSL). (C) This histogram depicts the expression of ALKBH3 in hematopoietic stem and progenitor (c-Kit+) cells in response to irradiation (5 Gy IR for 6 h). (D–I) Freshly isolated KSL cells from either (F, G) 2-mo-old or (H, I) 18-mo-old mice were infected by lentivirus carrying cDNA of Alkbh3, 72 h later, 2 × 104 GFP+ cells were sorted and injected into lethally irradiated recipients together with 2 × 105 competitor cells. Peripheral blood of recipient mice was evaluated every month until the third (F, G) or fourth (H, I) month. (D) Experimental design. (E) Representative western blot showing the expression of ALKBH3 in hematopoietic stem and progenitor (c-Kit+) cells after lentivirus infection. (F, H) These line plots depict the percentage of indicated donor-derived cells in peripheral blood of recipients carrying ectopic Alkbh3 or empty vector control. (G, I) This histogram displays the lineage distribution of donor-derived peripheral blood at the third month (G) or fourth month (I). (F, G) N = 6–8 mice per group, (H, I) N = 5–7 mice per group, data were shown as mean ± SEM. (J) This histogram exhibits the expression of Alkbh3, Pu.1, Runx1, Myc, Tcf12, Glf1, and Hes1 in hematopoietic stem and progenitor cells (KSL) in response to ALKBH3 overexpression.
Given that the expression of ALKBH3 is elevated in aged HSCs, we then sought to investigate the influence of Alkbh3-overexpression on HSCs function. We cloned the cDNA of Alkbh3 into a lentiviral vector and overexpressed Alkbh3 in HSCs freshly isolated from 2-month old mice. The infected HSCs were injected into lethally irradiated recipients together with 2 × 105 competitor cells (Fig. 3D,E). The results show that the reconstitution capacity of the Alkbh3-overexpressed HSCs was slightly higher than that of vector-overexpressed HSCs but the difference was not significant (Fig. 3F). Nevertheless, we found a significantly higher reconstitution capacity of the Alkbh3-overexpressed HSCs in T cells repopulation (Fig. 3F). Consistent with the advantage in T cells reconstitution, the result of lineage distribution revealed that Alkbh3-overexpressed HSCs exhibited T lymphoid lineage bias (Fig. 3G).
To further test whether overexpression of ALKBH3 in aged HSCs could ameliorate HSCs function, we overexpressed Alkbh3 in HSCs freshly isolated from 18-month old mice and injected the resulting cells into lethally irradiated recipients together with 2 × 105 competitor cells. Intriguingly, aged HSCs carrying empty vector shows higher reconstitution capacity than young vector-overexpressed HSCs (Fig. 3F,H). The possible reason is that our overexpression experiment used KSL (c-Kit+ Sca1+ Lineage−) cells as the donor (Fig. 3B), and the proportion of HSC in aged KSL cells is higher than that of young KSL cells, which leads to a higher rate of reconstruction of aged cells. This phenomenon has also been observed in our other studies. However, we have not found any difference in reconstitution capacity between Alkbh3-overexpressed HSCs and control (Fig. 3H). Similar to the results derived from young donor cells, aged Alkbh3-overexpressed HSCs exhibited significantly higher reconstitution capacity in T cells repopulation compared to control (Fig. 3H). Consistently, the result of lineage distribution revealed that aged Alkbh3-overexpressed HSCs exhibited a differentiation bias towards T lymphoid cells (Fig. 3I). Furthermore, aged Alkbh3-overexpressed HSCs generated more B lymphoid cells and did not exhibit the myeloid differentiation bias. Taken together, these results revealed that the expression of Alkbh3 increased with aging, and Alkbh3-overexpression rescued the differentiation skewing of aged HSCs.
To investigate the molecular mechanism of ALKBH3 in rectifying aged HSCs differentiation, we overexpressed Alkbh3 in HSCs freshly isolated from 2-month old mice and measured the expression of myelopoiesis and lymphopoiesis related genes in the resulting cells. However, we have not found any significant difference in the expression of Pu.1, Runx1, Myc, Tcf12, Gfi1, and Hes1 between control and Alkbh3-overexpressed HSCs (Fig. 3J). This result suggested that ALkbh3 rectified the differentiation skewing of aged HSCs may though a transcription-independent manner.
In this study, we provided experimental evidence that revealed ALKBH3 is dispensable for steady-state hematopoiesis and HSCs self-renewal, but forced ALKBH3 rectified the differentiation skewing of aged HSCs. We also observed that overexpression of ALKBH3 boosted T lymphoid differentiation both in young or aged HSCs. ALKBH3 has eight homologs in the Alkb family and most of them participated in DNA repair and RNA modification,21 so the function of ALKBH3 in hematopoiesis might be compensated by other homologs. Although the expression of Alkbh3 was elevated in aged HSPCs, overexpression of Alkbh3 did not result in a decline of HSC function, indicating that the increase of ALKBH3 may be the result or “by-product” of HSC aging, not the driven force.
Recent studies reported that the m6A eraser ALKBH5 promotes tumorigenesis in acute myeloid leukemia but not influences normal hematopoiesis.28,29 As the eraser of m1A, although ALKBH3 has no functional role in normal hematopoiesis, however, overexpression of ALKBH3 leads to rectify the differentiation skewing of aged HSCs, indicating that ALKBH3 may play a role in regulating HSCs differentiation. Given that ALKBH3 also overexpressed in multiple tumor cells,26 further research could shed light on the function of ALKBH3 in the tumorigenesis of leukemia.
3. METHODS
3.1. Mice
C57BL/6 mice (CD45.2), C57BL/6-SJL (CD45.1) mice were from the Jackson Laboratory. C57BL/6-Alkbh3tm1cyagen(Alkbh3−/−) mice were generated by Cyagen Biosciences. The recipient mice (CD45.1/2) were the first generation of C57BL/6 (CD45.2) and B6.SJL (CD45.1) mice. All mice were maintained in specific pathogen-free conditions. All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Tsinghua University.
3.2. Blood cell counts
Peripheral blood was collected from the tail vein and analyzed by the Auto Hematology Analyzer BC-5000 (MINDRAY).
3.3. Flow cytometric analysis and cell sorting
Bone marrow cells were collected by crushing the bones (femurs, tibias, and pelvis) from indicated donor mice. Cells were stained and sorted with following combination of surface markers: HSCs (CD34−CD150+ScaI+cKit+Lin−), CMPs (CD34+FcγR−Lin−Sca1−c-Kit+), GMPs (CD34+FcγR+Lin−Sca1−c-Kit+), MEPs (CD34−FcγR−Lin−Sca1−c-Kit+), CLPs (Lin−Sca1loc-KitloFlt3+IL7Rα+) B cells (B220+), T cells (CD3+) and myeloid cells (CD11b+).
For hematopoietic stem and progenitor cells (c-Kit+) enrichment, bone marrow cells were stained with the c-Kit-APC antibody followed by magnetic bead enrichment (Miltenyi Biotec). For HSCs sorting, c-Kit+ enriched cells were stained with combinations of the following surface markers: Sca1, c-Kit, CD150, CD34, and the lineage markers Gr-1, CD11b, Ter-119, B220, CD3, CD4, and CD8a. Dead cells were excluded by DAPI (Sigma-Aldrich, D8417) staining.
3.4. Transplantations and lineage tracking
HSCs (CD45.2) were freshly isolated and injected into lethally irradiated recipient mice (CD45.1/2) together with 3 × 105 competitor cells (CD45.1). Recipient mice were irradiated with a lethal dose (10Gy) of X-ray (RS-2000, Rad Source Technologies) before transplantation. In the secondary transplantation, 2 × 106 chimeric bone marrow cells from the primary recipients were injected into the secondary recipient mice. Recipient mice were administered with antibiotic water for 3 weeks after transplantation. Peripheral blood from the recipient were analyzed for donor-derived chimaerism (myeloid, B, and T cells) every month. The antibodies combination (CD3, B220, CD11b, CD45.1 CD45.2) were used to analyze peripheral blood chimaerism.
3.5. Lentivirus production and transduction
The mouse Alkbh3 cDNA was cloned into the pRRL-PPT-SF-newMCS-IRES2-EGFP vector. Lentivirus was packaged in 293T cells and concentrated by ultracentrifugation. KSL (c-Kit+ Sca1+ Lineage−) cells were sorted and cultured in SFEM medium (Stem Cell Technology, 09650) containing 20 μg/ml mSCF, 20 μg/ml mTPO and 1% penicillin/streptomycin in 96-well plate (∼1 × 105 per well). Lentivirus was added into KSL cells based on the titration result. 72 hours late, 2 × 104 GFP+ cells were sorted and injected into lethally irradiated recipients.
3.6. Western blot
c-Kit+ cells were collected and lysed with NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris–HCl, pH 8.0, 0.5% Nonidet P-40, containing phosphatase and protease inhibitors cocktail) on ice for 30 min, and then centrifuged at 4°C for 10 min. The supernatant was collected and mixed with a 2× Loading buffer. The resulting samples were boiled and then resolved on 10% SDS-PAGE. Membranes were blocked with 5% milk in TBST buffer and then probed with anti-ALKBH3 antibody (Millipore, 09-882).
3.7. Quantitative real-time PCR
Total RNA were isolated using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer's instruction. Total RNA were subjected to reverse transcription using PrimeScript RT reagent Kit (Takara). Acquired cDNA was analyzed by PowerUp™ SYBR™ Green mix (Applied Biosystems) with indicated primers on a QuantStudio-3 Real-time PCR System (Applied Biosystems). The PCR primers were as follows: Actin-F, 5′-GTGACGTTGACATCCGTAAAGA-3′; Actin-R, 5′-GCCGGACTCATCGTACTCC-3′. Alkbh3-F, 5′- GGAGGCTGGGCTACACCTA-3′; Alkbh3-R, 5′-CAAATTGTCGGTCACATTGCTG-3′. Pu.1-F, 5′-TGCGGAGAAATCCCAGTAGT-3′; Pu.1-R, 5′-CTCCATCGGATGACTTGGTT-3′. Runx1-F, 5′-CCGAGAACCCCGAAGACATC-3′; Runx1-R, 5′-GCAGTGGAGTGGTTCAAGGA-3′. Myc-F, 5′-ATGCCCCTCAACGTGAACTTC-3′; Myc-R, 5′-CGCAACATAGGATGGAGAGCA-3′. Tcf12-F, 5′-ATGTGCTACGAAACCATGCAG-3′; Tcf12-R, 5′-GCCATTGAGACTGACTGAATCTT-3′. Gfi1-F, 5′-CTATCACCAGCCGCGTTCTC-3′; Gfi1-R, 5′-AGCCTCGGTAAGCTGAGAGT-3′. Hes1-F, 5′-CCAGCCAGTGTCAACACGA-3′; Hes1-R, 5′-AATGCCGGGAGCTATCTTTCT-3′.
ACKNOWLEDGMENTS
This work was supported by grant numbers 2018YFA0800200, 2017YFA0104000, Z181100001818005 and 91849106 to J.W.W., from the National Key R&D Program of China or the Beijing Municipal Science & Technology Commission and the National Natural Science Foundation of China.
We thank the Beijing Advanced Innovation Center for Structural Biology and the Tsinghua-Peking Center for Life Sciences for facility and financial support.
REFERENCES
- [1].Crane GM, Jeffery E, Morrison SJ. Adult haematopoietic stem cell niches. Nat Rev Immunol 2017;17 (9):573–590. [DOI] [PubMed] [Google Scholar]
- [2].Wang J, Sun Q, Morita Y, et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 2012;148 (5):1001–1014. [DOI] [PubMed] [Google Scholar]
- [3].Wang J, Morita Y, Han B, Niemann S, Löffler B, Rudolph KL. Per2 induction limits lymphoid-biased haematopoietic stem cells and lymphopoiesis in the context of DNA damage and ageing. Nat Cell Biol 2016;18 (5):480–490. [DOI] [PubMed] [Google Scholar]
- [4].Wang J, Lu X, Sakk V, Klein CA, Rudolph KL. Senescence and apoptosis block hematopoietic activation of quiescent hematopoietic stem cells with short telomeres. Blood 2014;124 (22):3237–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].He H, Xu P, Zhang X, et al. Aging-induced IL27Ra signaling impairs hematopoietic stem cells. Blood 2020;136 (2):183–198. [DOI] [PubMed] [Google Scholar]
- [6].Ho Y-H, Méndez-Ferrer S. Microenvironmental contributions to hematopoietic stem cell aging. Haematologica 2020;105 (1):38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mejia-Ramirez E, Florian MC. Understanding intrinsic hematopoietic stem cell aging. Haematologica 2020;105 (1):22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell 2017;169 (7):1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang C, Chen Y, Sun B, et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature 2017;549 (7671):273–276. [DOI] [PubMed] [Google Scholar]
- [10].Vu LP, Pickering BF, Cheng Y, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nature Med 2017;23 (11):1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 2018;22 (2):191–205. e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li Z, Qian P, Shao W, et al. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res 2018;28 (9):904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang H, Zuo H, Liu J, et al. Loss of YTHDF2-mediated m6A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res 2018;28 (10):1035–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Paris J, Morgan M, Campos J, et al. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 2019;25 (1):137–148. e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chujo T, Suzuki T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA 2012;18 (12):2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 2005;11 (8):1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase-extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res 2012;40 (22):11583–11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016;530 (7591):441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li X, Xiong X, Wang K, et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat Chem Biol 2016;12 (5):311–316. [DOI] [PubMed] [Google Scholar]
- [20].Li X, Xiong X, Zhang M, et al. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell 2017;68 (5):993–1005. e1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. The AlkB family of Fe(II)/(α-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J Biol Chem 2015;290 (34):20734–20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011;7 (12):885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zheng G, Dahl John A, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013;49 (1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu F, Clark W, Luo G, et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell 2016;167 (3):816–828. e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ougland R, Lando D, Jonson I, et al. ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells 2012;30 (12):2672–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dango S, Mosammaparast N, Sowa Mathew E, et al. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol Cell 2011;44 (3):373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ringvoll J, Nordstrand LM, Vågb⊘ CB, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J 2006;25 (10):2189–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Wang J, Li Y, Wang P, et al. Leukemogenic chromatin alterations promote AML leukemia stem cells via a KDM4C-ALKBH5-AXL signaling axis. Cell Stem Cell. [DOI] [PubMed] [Google Scholar]
- [29]. Shen C, Sheng Y, Zhu AC, et al. RNA Demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]


