Abstract
PML nuclear bodies (NBs), which are increasingly recognized as the central hub of many cellular signaling events, are superassembled spherical complexes with diameters of 0.1–2 μm. Recent studies reveal that RING tetramerization and B1-box polymerization are key factors to the overall PML NBs assembly. The productive RBCC oligomerization allows subsequent PML biogenesis steps, including the PML auto-sumoylation and partners recruitment via SUMO–SIM interactions. In promyelocytic leukemia, the oncoprotein PML/RARα (P/R) inhibits PML NBs assembly and leads to a full-fledged leukemogenesis. In this review, we review the recent progress in PML and acute promyelocytic leukemia fields, highlighting the protein oligomerization as an important direction of future targeted therapy.
Keywords: Leukemogenesis, Oligomerization, PML nuclear body, PML/RARα
1. INTRODUCTION
The PML protein (also called TRIM19) is first observed in the 1960s.1,2PML gene is 53 kb in length and contains nine exons. Through alternatively splicing, at least seven PML isoforms can be observed.3 All of them share a conserved RBCC motif in N-terminus, in which R stands for RING, B for B1- and B2-box, and CC for coiled coil domain. It has been shown that these domains are indispensable for PML nuclear bodies (NBs) biogenesis.4–9 The C-terminus contributes to the specific functions of each PML isoform. For instance, the C-terminal portion of PML-IV (amid 361–633) is critical in p53 regulation.10 In comparison, the C-terminal domains of PML-II and PML-V may contribute to PML-NBs formation.11,12 The NBs-facilitated posttranslation modification is often recognized as the main function of this subcellular complex.13–17 In particular, PML sumoylation is highlighted in acute promyelocytic leukemia (APL) development and targeted therapy. Until today, three sumoylation sites (i.e. K65, K160, K490) are reported in PML. The PML auto-sumoylation and the sumo interaction motif (SIM) in its C-terminus can also contribute to the overall NBs assembly.18,19 In this review, we will look into the current understanding of PML NBs biogenesis pathway and their implication in APL pathogenesis and targeted therapy.
1.1. RBCC oligomerization is essential to PML NBs assembly
In the process of PML NBs assembly, the integrity of PML RBCC motif is thought essential.5,7–9 Amongst RBCC domain, the RING tetramerization might precede B1-driven oligomerization.9 Structural-dependent mutations on RING tetramerization and B1-box oligomerization abolish PML NBs formation.8,9 Many TRIMs could facilitate RING-binding E2 enzymes to their nascent substrates, and hence are often recognized as ubiquitin E3 ligases.20 TRIM RING dimerization is critical for ubiquitin E2–E3 interactions.21,22 In comparison, the PML RING tetramerization is required for the recruitment of SUMO-conjugating E2 UBC98 (Fig. 1), prompting the hypothesis that high-order assembly of RING domain might be the structural determinants that enable PML/TRIM19 into a sumoylation, but not ubiquitin ligase, E3.8 This is further supported by the recent observation of PML B1-box polymerization (n > 4).9 Li and coworkers have shown that PML B1-box could form dimer, tetramer, and much higher-order polymer via oligomeric sequence uniquely observed in PML but not other TRIMs (Fig. 1). Indeed, the B1-box oligomerization gives rise to an unexpected K160 lining/concentration that might explain why this lysine position is particularly prone to SUMO long chain. In previous studies,23 it is clear that CC domain can also contribute to RBCC oligomerization. In the recent study,9 via arsenic rescue experiment, a cooperative mechanism has been proposed among RING, B1/2-box, and CC domains. Future study should focus on the mechanism of how RING, B1/2-box, and CC might fold together, leading to the 2D–3D transition and ultimately to PML speckles formation.
Figure 1.
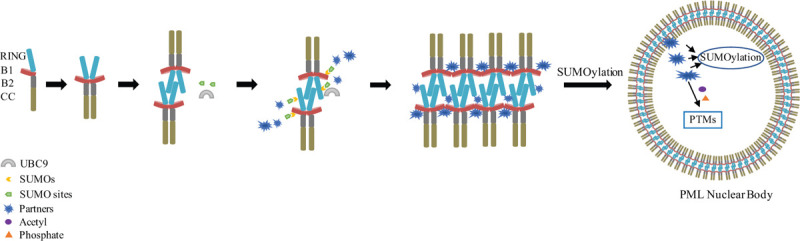
A model for PML nuclear bodies assembly. RBCC-mediated PML oligomerization constitutes the “shell” of PML NBs to recruit UBC9, and hence facilitates PML sumoylation. Sumoylated PML might allow the interaction of partner proteins that contain sumo-interaction-motif (SIM). All these partners’ recruitments ultimately enable the in situ sumoylation and other posttranslational modifications (PTMs).
1.2. PML NBs PTMs are involved in PML NBs assembly
PML often participates in various cellular signaling via posttranslational modification of itself and partners. Its best-known PTM function is sumoylation. PML could directly conjugate SUMOs via lysine residues, which endow their ability to interact with partner proteins equipped with a short hydrophobic sequence termed as SIM.24 Until today, three PML sumoylated sites, that is, K65/K160/K490, are reported. Furthermore, PML and its partners both contain SIM domain.24 The SUMO–SIM interaction might help the partner proteins sequestration within the NBs. Until today, it has been reported that PML could directly or indirectly interact with at least 120 proteins,25 including p53, DAXX, SP100, CBP, HDAC, and so on. Of note, the K160 site, which is not critical for NBs formation,26 is essential for partners recruitment.24 Furthermore, as demonstrated in P/R transgenic mice, K160 is critical for PML/RARα sumoylation and APL leukemogenesis.27
In addition to sumoylation, other PTM functions, including phosphorylation and acetylation, are also reported in PML. Like sumoylation, PML phosphorylation is catalyzed by kinases such as ATR, ATM, and CHK2 to assist in DNA damage process.28 PML acetylation is also reported to happen in the positions of K487 and K505 by the protein acetyltransferase involved in cell apoptosis and cell death regulation.29 Some anchored PML partners localized in the NBs core could also be modulated by other PTMs (Fig. 1). One of the best-known partners is the tumor suppressor p53. Therefore, PML is often recognized as a critical regulator of p53 activity and p53-mediated cellular processes, such as apoptosis, cell cycle arrest, and DNA repair and senescence, especially through the sequestration of Mdm2-dependent PTM of p53 and SIRT1-dependent deacetylation of p53.30,31
1.3. Enlightening in oligomerization and tumorigenesis
APL belongs to the M3 subtype of acute myeloid leukemia. The generation of PML/RARα (P/R) oncoprotein and PML NBs abolishment trigger APL pathogenesis.32,33 Primarily, the abnormal P/R expression prompts NBs disruption, leading to nuclear microspeckles formation and losing its pivot properties involved in tumor-suppressive processes, including DNA damage response, cellular apoptosis, senescence, and angiogenesis.25 In recent report, the B1-box-mediated P/R oligomerization is identified as a significant regulator in P/R-driven transactivation and leukemogenesis.9 Supportively, the PML RING–RING and CC–CC interactions are also a significant factor for differentiation arrest and transformation in vivo.8,9,34 All these results highlight the PML oligomerization as an important regulator in leukemogenesis. Furthermore, transcriptional repression due to DAXX recruitment of PML/RARα in a strictly K160-dependent manner and DNA binding factors (DBF)/CoR recruitment by RARα portion are recognized as key contributors in APL development.27,35 PML/RARα could recognize >3000 DNA binding sites via many DBF such as RXR and/or PU.1.35,36 The PML/RARα-DBF complex, which is regulated by multimerization, might cooperate with many corepressor complexes (CoR) such as histone deacetylases (HDACs), polycomb repressive complexes (PRCs), and DNA methyltransferases to exert oligomerization-driven transcriptional repression37–40 (Fig. 2).
Figure 2.
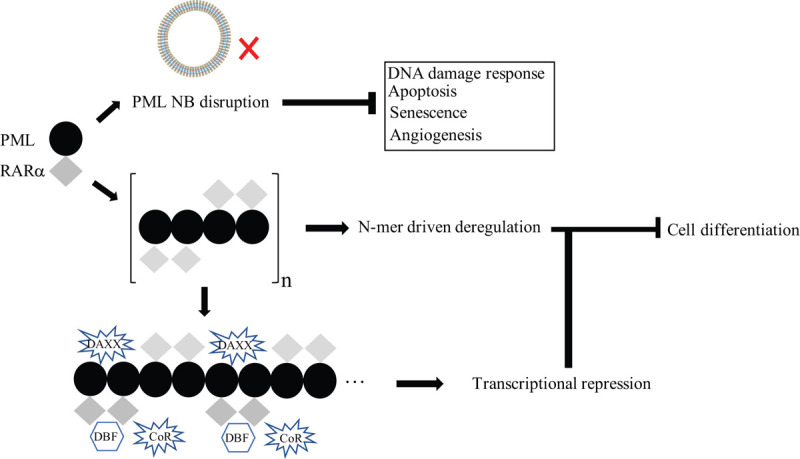
The scheme of PML/RARα-driven APL pathogenesis. The oncoprotein PML/RARα drives disruption of PML nuclear bodies while P/R-multimer exerts target genes in deregulation and recruits DBF (RXR, PU.1 and many others) to form hetero-multimers, which in turn might enhance CoR recruitment, leading to abnormal cell arrest.
Oligomerization-driven tumorigenesis is widely observed in other leukemias such as acute myeloid leukemia (AML), mixed-lineage leukemia (MLL), and acute lymphoblastic leukemia (ALL).41–45 In AML leukemia, the Nervy homology 2 (NHR2) domain in AML1/ETO is shown to form an alpha-helical tetramer. More importantly, NHR2 oligomerization is essential to AML1/ETO's oncogenic activity, that is, the inhibition of granulocyte differentiation.42 In MLL leukemia, it has been reported that the diverse oncoproteins, including MLL/GAS7, MLL/AF1p, MLL/GEPRIN, could form oligomerization via coiled coil domain. The disruption of MLL oligomerization precludes leukemogenic transformation.45 In ALL leukemia, the PAX5–PML is shown to repress PAX5 transactivity in heterodimer complex.41 Finally, the tetramerization of stat5, which is frequently associated with various leukemias, is also important for leukemogenesis. The stat5 mutants that target the tetramerization fail to trigger leukemias, reiterating the importance of oligomerization in oncogenic drivers.46
2. CONCLUSION AND PERSPECTIVES
RBCC-mediated oligomerization not only contributes to PML NBs assembly as a premise of the following PML sumoylation and partners recruitment via SUMO–SIM interaction but is also instrumental to APL pathogenesis. PML NBs disruption and the oligomerization of oncoproteins involved in various leukemias suggest that oligomerization could be recognized as an important tumorigenic regulator and valuable direction for future targeted intervention.
ACKNOWLEDGMENTS
This work was supported by research grants 81970132, 81770142, 81370620, 81570120, 31070645, 81800144, and 31800642 from National Natural Science Foundation of China, a research grant 20152504 from Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support, The Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institute of Higher Learning, a research grant 11JC1407200 from SMSTC, a research grant 12ZZ109 from SME, Program for New Century Excellent Talents in University (NCET-10–9571), Samuel Waxman Cancer Research Foundation.
Author Contributions: YL and GM planned, drafted the manuscript. YL, XM, and GM critically revised the manuscript. All authors read and approved the final manuscript.
REFERENCES
- [1].de Thé G, Rivière M, Bernhard W. Examen au microscope électronique de la tumeur VX2 du lapin domestique dérivée du papillome de Shope. Bull Ass Franç Cancer 1960;47:570–584. [PubMed] [Google Scholar]
- [2].Brasch K, Ochs RL. Nuclear bodies (NBs): a newly “rediscovered” organelle. Exp Cell Res 1992;202(2):211–223. [DOI] [PubMed] [Google Scholar]
- [3].Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 2007;8(12):1006–1016. [DOI] [PubMed] [Google Scholar]
- [4].Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene 2001;20(49):7223–7233. [DOI] [PubMed] [Google Scholar]
- [5].Boddy MN, Duprez E, Borden K, Freemont PS. Surface residue mutations of the PML ring finger domain alter the formation of nuclear matrix-associated PML bodies. J Cell Sci 1997;110(Part 18):2197–2205. [DOI] [PubMed] [Google Scholar]
- [6].Borden KLB, Lally JM, Martin SR, O’Reilly NJ, Solomon E, Freemont PS. In vivo and in vitro characterization of the B1 and B2 zinc-binding domains from the acute promyelocytic leukemia protoncoprotein PML. Proc Natl Acad Sci USA 1996;93:1601–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell 2006;24(3):331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang P, Benhenda S, Wu H, et al. RING tetramerization is required for nuclear body biogenesis and PML sumoylation. Nat Commun 2018;9(1):1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li Y, Ma X, Chen Z, et al. B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis. Nat Commun 2019;10(1):3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ivanschitz L, Takahashi Y, Jollivet F, Ayrault O, Le Bras M, de The H. PML IV/ARF interaction enhances p53 SUMO-1 conjugation, activation, and senescence. Proc Natl Acad Sci USA 2015;112(46):14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li C, Peng Q, Wan X, Sun H, Tang J. C-terminal motifs in promyelocytic leukemia protein isoforms critically regulate PML nuclear body formation. J Cell Sci 2017;130(20):3496–3506. [DOI] [PubMed] [Google Scholar]
- [12].Weidtkamp-Peters S, Lenser T, Negorev D, et al. Dynamics of component exchange at PML nuclear bodies. J Cell Sci 2008;121(Pt 16):2731–2743. [DOI] [PubMed] [Google Scholar]
- [13].Cheng X, Kao HY. Post-translational modifications of PML: consequences and implications. Front Oncol 2012;2:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lallemand-Breitenbach V, de Thé H. PML nuclear bodies. Cold Spring Harb Perspect Biol 2010;2:a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Scaglioni PP, Yung TM, Cai LF, et al. A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell 2006;126(2):269–283. [DOI] [PubMed] [Google Scholar]
- [16].Duprez E, Saurin AJ, Desterro JM, et al. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J Cell Sci 1999;112:381–393. [DOI] [PubMed] [Google Scholar]
- [17].Grignani F, de Matteis S, Nervi C, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 1998;391:815–818. [DOI] [PubMed] [Google Scholar]
- [18].Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 2006;281(23):16117–16127. [DOI] [PubMed] [Google Scholar]
- [19].Kamitani T, Kito K, Nguyen HP, Wada H, Fukuda-Kamitani T, Yeh ET. Identification of three major sentrinization sites in PML. J Biol Chem 1998;41:26675–26682. [DOI] [PubMed] [Google Scholar]
- [20].Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of 'single protein RING finger’ E3 ubiquitin ligases. Bioessays 2005;27(11):1147–1157. [DOI] [PubMed] [Google Scholar]
- [21].Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer 2011;11(11):792–804. [DOI] [PubMed] [Google Scholar]
- [22].Yudina Z, Roa A, Johnson R, et al. RING dimerization links higher-order assembly of TRIM5alpha to synthesis of K63-linked polyubiquitin. Cell Rep 2015;12(5):788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fagioli M, Alcalay M, Tomassoni L, et al. Cooperation between the Ring+B1-B2 and coiled-coil domains of PML is necessary for its effects on cell survival. Oncogene 1998;16:2905–2913. [DOI] [PubMed] [Google Scholar]
- [24].Sahin U, Ferhi O, Jeanne M, et al. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J Cell Biol 2014;204(6):931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Guan D, Kao HY. The function, regulation and therapeutic implications of the tumor suppressor protein, PML. Cell Biosci 2015;5:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lallemand-Breitenbach V, Zhu J, Puvion F, et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med 2001;193(12):1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhu J, Zhou J, Peres L, et al. A sumoylation site in PML/RARα is essential for leukemic transformation. Cancer Cell 2005;7(2):143–153. [DOI] [PubMed] [Google Scholar]
- [28].Dellaire G, Ching RW, Ahmed K, et al. Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J Cell Biol 2006;175(1):55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hayakawa F, Abe A, Kitabayashi I, Pandolfi PP, Naoe T. Acetylation of PML is involved in histone deacetylase inhibitor-mediated apoptosis. J Biol Chem 2008;283(36):24420–24425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol 2004;6(7):665–672. [DOI] [PubMed] [Google Scholar]
- [31].Guan D, Lim JH, Peng L, et al. Deacetylation of the tumor suppressor protein PML regulates hydrogen peroxide-induced cell death. Cell Death Dis 2014;5:e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood 2008;111(5):2505–2515. [DOI] [PubMed] [Google Scholar]
- [33].Voisset E, Moravcsik E, Stratford EW, et al. Pml nuclear body disruption cooperates in APL pathogenesis and impairs DNA damage repair pathways in mice. Blood 2018;131(6):636–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Occhionorelli M, Santoro F, Pallavicini I, et al. The self-association coiled-coil domain of PML is sufficient for the oncogenic conversion of the retinoic acid receptor (RAR) alpha. Leukemia 2011;25(5):814–820. [DOI] [PubMed] [Google Scholar]
- [35].Martens JH, Brinkman AB, Simmer F, et al. PML-RARalpha/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell 2010;17(2):173–185. [DOI] [PubMed] [Google Scholar]
- [36].Wang K, Wang P, Shi J, et al. PML/RARalpha targets promoter regions containing PU.1 consensus and RARE half sites in acute promyelocytic leukemia. Cancer Cell 2010;17(2):186–197. [DOI] [PubMed] [Google Scholar]
- [37].Grignani F, De Matteis S, Nervi C, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 1998;391(6669):815–818. [DOI] [PubMed] [Google Scholar]
- [38].Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 1998;391(6669):811–814. [DOI] [PubMed] [Google Scholar]
- [39].Villa R, Pasini D, Gutierrez A, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell 2007;11(6):513–525. [DOI] [PubMed] [Google Scholar]
- [40].Di Croce L, Raker VA, Corsaro M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 2002;295(5557):1079–1082. [DOI] [PubMed] [Google Scholar]
- [41].Kurahashi S, Hayakawa F, Miyata Y, et al. PAX5-PML acts as a dual dominant-negative form of both PAX5 and PML. Oncogene 2011;30(15):1822–1830. [DOI] [PubMed] [Google Scholar]
- [42].Liu Y, Cheney MD, Gaudet JJ, et al. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity. Cancer Cell 2006;9(4):249–260. [DOI] [PubMed] [Google Scholar]
- [43].So CW, Cleary ML. Dimerization: a versatile switch for oncogenesis. Blood 2004;104(4):919–922. [DOI] [PubMed] [Google Scholar]
- [44].Eguchi M, Eguchi-Ishimae M, Greaves M. The small oligomerization domain of gephyrin converts MLL to an oncogene. Blood 2004;103(10):3876–3882. [DOI] [PubMed] [Google Scholar]
- [45].So CW, Lin M, Ayton PM, Chen EH, Cleary ML. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell 2003;4(2):99–110. [DOI] [PubMed] [Google Scholar]
- [46].Moriggl R, Sexl V, Kenner L, et al. Stat5 tetramer formation is associated with leukemogenesis. Cancer Cell 2005;7(1):87–99. [DOI] [PubMed] [Google Scholar]


