Abstract
Erythropoiesis is a complex and sophisticated multi-stage process regulated by a variety of factors, including the transcription factor GATA1 and non-coding RNA. GATA1 is regarded as an essential transcriptional regulator promoting transcription of erythroid-specific genes—such as long non-coding RNAs (lncRNA). Here, we comprehensively screened lncRNAs that were potentially regulated by GATA1 in erythroid cells. We identified a novel lncRNA—PCED1B-AS1—and verified its role in promoting erythroid differentiation of K562 erythroid cells. We also predicted a model in which PCED1B-AS1 participates in erythroid differentiation via dynamic chromatin remodeling involving GATA1. The relationship between lncRNA and chromatin in the process of erythroid differentiation remains to be revealed, and in our study we have carried out preliminary explorations.
Keywords: Chromatin accessibility, Erythroid differentiation, Long non-coding RNA, PCED1B-AS1
1. INTRODUCTION
Hematopoietic stem cells (HSCs) initiate erythroid differentiation into common myeloid progenitor cells (CMPs), further differentiation into erythroblasts (EBs), and eventually maturation into red blood cells—a complex and dynamic process.1,2 HSCs have self-renewal and pluripotency differentiation abilities, and can differentiate into CMPs and lymphoid progenitor cells (CLPs).3 CMPs can be further differentiated into EBs, which approach the characteristics of mature red blood cells with morphological and structural changes, including a decrease in cell size and nuclear condensation.4 After denucleation, the cells form reticulocytes, which are released into the blood and eventually mature into red blood cells, playing the role of transporting oxygen.3,5
Erythroid differentiation is regulated by a variety of factors, including cytokines, signaling pathways, and transcription factors.3,6 In particular, a variety of transcription factors playing a significant role in erythroid differentiation have been discovered, such as GATA1—one of the important transcription factors during erythroid differentiation.7,8 The effects of alncRNA-EC3, alncRNA-EC7, and lincRNA-EPS on erythroid differentiation have been verified by many years of lncRNA research.9–11 A number of studies have shown that lncRNA has tissue specificity and plays an important role in stem cell differentiation and other biological process.12–14 Chromatin conformation also showed dynamic changes in erythroid differentiation,15,16 during the late stages of which the nucleus was concentrated and chromatin might change dramatically.17 However, the association between lncRNA in erythroid differentiation regulation and chromatin status remains to be studied.
In order to explore the association of lncRNA and chromatin status in erythroid differentiation, we first screened the lncRNAs 2 kb upstream and downstream with GATA1 binding sites based on ChIP-Seq in the ENCODE database, and further identified lncRNA candidates in the Blueprint database containing transcriptome and epigenome data relating to the hemotopoietic system. We found that lncRNA PCED1B-AS1 may have an effect on erythroid differentiation. Interestingly, we found that erythroid differentiation regulated by PCED1B-AS1 could be associated with GATA1 and chromatin remodeling in erythroblast cells.
2. METHODS
2.1. Cell culture
We cultured TF-1 cells in RPMI 1640 medium containing 10% fetal bovine serum, 1% penicillin and streptomycin, and 2 μg/ml–8 μg/ml granulocyte-macrophage colony stimulating factor. We cultured K562 cells in RPMI 1640 medium containing 10% fetal bovine serum, 1% penicillin, and streptomycin, and induced k562 erythroid differentiation with 50 μM hemin. We cultured 293T cells in DMEM containing 10% fetal calf serum, 1% penicillin and streptomycin. We isolated CD34+ cells from human cord blood using the EasySep Hu CB CD34 Pos Sel Kit II kit and tested its purity by flow cytometry. CD34+ cells were first cultured at 105 cells/ml for 6 days in SFEM II supplemented with 10% FBS, 10 ng/ml SCF, 1 ng/ml IL-3, and 3 IU/ml EPO at 37°C in 5% CO2, and then cultured for 8 more days in the above complete medium with the presence of 30% FBS and the absence of IL-3 and SCF. We cultured all of the above cells a constant temperature incubator at 37°C with 5% CO2. The subject has signed an informed consent form, and the sample collection was approved by the ethics committee of Beijing Institute of Genomics (BIG), CAS.
2.2. GATA1 knock down in TF-1 cells
We purchased three GATA1 siRNA sequences from Gemma gene company and diluted them to 20 μM with ddH2O (GATA1-siRNA-1: CAUUGCUCAACUGUAUGGATT, GATA1-siRNA-2: CCAAGCUUCGUGGAACUCUTT,GATA1-siRNA-3: GGUACUC AGUGCACCAACUTT). We added 0.8 × 106 TF-1 cells to each well of a six-well plates. We transferred cells of each well with 7.5 μl of 20 μM of siRNA via lipofectamine 2000 reagent, and cultured the cells in incubator for 48 h. We collected 1 × 106 cells and extracted total RNA using TRIzol. We detected knockdown efficiency by RT-PCR after reverse transcription.
2.3. Overexpression of PCED1B-AS1 in K562 cells
We cloned the sequence of PCED1B-AS1 into lentiviral plasmid pHAGE-fEF-1a-IRES-ZsGreen-2. We transfected the overexpressing recombinant construct into 293T cells, together with the packaging plasmids psPAX2 and pMD2.G with Lipofectamine 2000 regents, and collected the cell supernatant medium for 48 h and 72 h, respectively, as a virus stock solution. After filtration through a 0.45 μm filter, we concentrated the virus stock to 50× virus using 5× PEG8000. We added the concentrated virus solution and polybrene at a final concentration of 6 μg/ml to a medium in which we cultured K562 cells for infection, and isolated GFP-positive cells by flow cytometry and cultured them to obtain cells stably expressing PCED1B-AS1.
2.4. Quantitative real-time PCR
We extracted total RNA from the cells using the TRIzol Reagent kit, and reverse transcribed total RNA into cDNA using the PrimeScript RT reagent Kit with gDNA Eraser kit. We performed RT-PCR in a CFX96 Real-Time PCR detection system using Maxima SYBR Green/ROX qPCR Master Mix.
2.5. Flow cytometry
We collected 1 × 106 cells, washed them twice with 1 ml of 1× PBS, and then resuspended them in 100 μl of 1× PBS. We added 0.1 μl of phycoerythrin-conjugated anti-CD235a and 0.5 μl of allophycocyanin-conjugated anti-CD71 in a dark area, and incubated them in a refrigerator at 4°C for 10 min in the dark. We washed the cells twice with 1 ml of 1× PBS, and discarded the supernatant to obtain the cell pellet, which we resuspended in 500 μl of 1× PBS and detected on a FACSCalibur flow cytometer (BD Biosciences).
2.6. Benzidine staining
We dissolved 10 mg benzidine hydrochloride with 1 ml of 0.5 M acetic acid, and added 1 μl 30% hydrogen peroxide to 50 μl benzidine solution immediately before use. We collected 0.3 to 0.5 M cells in 100 μl 1× PBS for 2 washes, then resuspended the cells with 10 μl of 1× PBS. We added 1 μl benzidine solution to the cell suspension and incubated it for 5 min at room temperature. We spread the cells on a glass slide, and observed the staining results under a microscope after 30 min, counting the proportion of positive cells in three different fields.
2.7. ChIP-seq analysis of K562 cells in ENCODE
We downloaded the GATA1 ChIP-seq data in K562 erythroid cells from ENCODE.18 We screened for the GATA1 binding sites within 2 kb in the presence of the lncRNAs. The lncRNA is regarded to be potentially regulated by GATA1 in erythroid cells.
2.8. Chromatin immunoprecipitation
Appropriate 50 μl magnetic beads (Dynabeads M-280 Sheep anti-Rabbit IgG) were respectively mixed with 2.5 μg GATA1 antibody and IgG antibody, and rotated overnight at 4°C. A total of 107 K562 cells were cross-linked by 9% formaldehyde solution, and the chromatin was sheared to a fragment of 300 to 500 bp in size. The sheared chromatin was divided into two equal parts, in addition to 20 μl input and 20 μl to identify the ultrasound efficiency, and was added to the beads-GATA1-antibody and beads-IgG-antibody for immunoprecipitation overnight at 4°C. The DNA obtained by immunoprecipitation was purified by Agencourt AMPure XP Kit and detected by qPCR. Primers used are as follows:
Rank1-F: GCCATCTAGTCCCCAGACAA, Rank1-R: CCTCCAAGCTGTCCTTGTTC; Rank2-F: CAGTCCCACTGAGGAAAGGA, Rank2-R: AAGGAAGGGAGGTCTGTGGT; Rank3-F: ACCTGCTGGTTTCAAGTGGA, Rank3-R: CCACTGACGTCTTGATGGAA; Rank4-F: TCCGCTTCCTAAAGCAAGAA, Rank4-R: AGCTTGGGCTATAGGGGAAA.
2.9. Chromatin accessibility analysis
We downloaded the ATAC-seq raw data of HSC, CMP, and EB from the GSE74912 dataset.15 We based the data processing on a previous study.15 In brief, we trimmed reads by TrimGalore and aligned them to the human genome (GRCh37/hg19) using Bowtie 2.19 We identified peaks using MACS2 software.20
2.10. Motif analysis
We selected the peaks near the PCED1B-AS1 transcription start site of HSC, CMP, and EB, and performed motif analysis using Homer software with the findmotifsgenome.pl command module.
2.11. The potential regulation factor of PCED1B-AS1
We first identified the regulation factors of PCED1B-AS1 at 100 kb using the Cistrom Data Browser Tookit (http://dbtoolkit.cistrome.org/)21,22 and then retained the factors present in EB cells with a regulatory potential (RP) score greater than 0.5. We set regulation potential (RP) scores of >0.8, 0.6–0.8, and <0.6 as high, medium, and low, respectively.
2.12. Target gene prediction of PCED1B-AS1 and correlation analysis
We selected GATA1 as a potential regulation factor of PCED1B-AS1 based on the previous step analysis, which we also performed using the Cistrom Data Browser Tookit. We then obtained the top 10 putative targets of PCED1B-AS1 at the bottom of the results page according to public data. We also predicted target genes of PCED1B-AS1 using Starbase 3.0.23 We performed correlation analysis using the rcorr function in the Hmisc package and visualized it using the corrplot R package.
2.13. Statistical analysis
We performed each experiment three times or more, and tested the statistical difference between samples for significance using the Kruskal−Wallis test in R. We considered P ≤ .05 to be statistically significant.
3. RESULTS
3.1. LncRNA PCED1B-AS1 was identified as a potential regulator in promoting erythroid differentiation
In erythroid differentiation, GATA1 is an essential transcription factor involved in a considerable number of target genes that regulate the proliferation, differentiation, and survival of hematopoietic progenitors.24 Therefore, lncRNA regulated by GATA1 is more likely to play a role in erythropoiesis. To screen for potential GATA1-regulated lncRNAs, we first analyzed the ChIP-seq data from K562 erythroid cells in the ENCODE database and identified 107 lncRNAs with GATA1 binding sites within 2 kb of their genes.40 We extracted the expression data from these 107 lncRNAs in the following three stages—HSCs, CMPs, and EBs, considered to be representative stages in the process of erythropoiesis, and analyzed them for differences in expression.
Significant changes in the expression of lncRNAs might indicate that they have the potential to regulate erythroid differentiation. We utilized DESeq2 to analyze lncRNA expression in three stages of HSCs, CMPs, and EBs. With padj < .05 and |log2FoldChange|>1 as thresholds, we finally identified one significantly differentially expressed lncRNA—PCED1B-AS1—expressed in EBs at a far higher level than those of the other two stages, HSCs and CMPs (Fig. 1A). The variant of PCED1B-AS1 is ENST00000552269 that is the only transcript of PCED1B-AS1 screened in this study.
FIGURE 1.
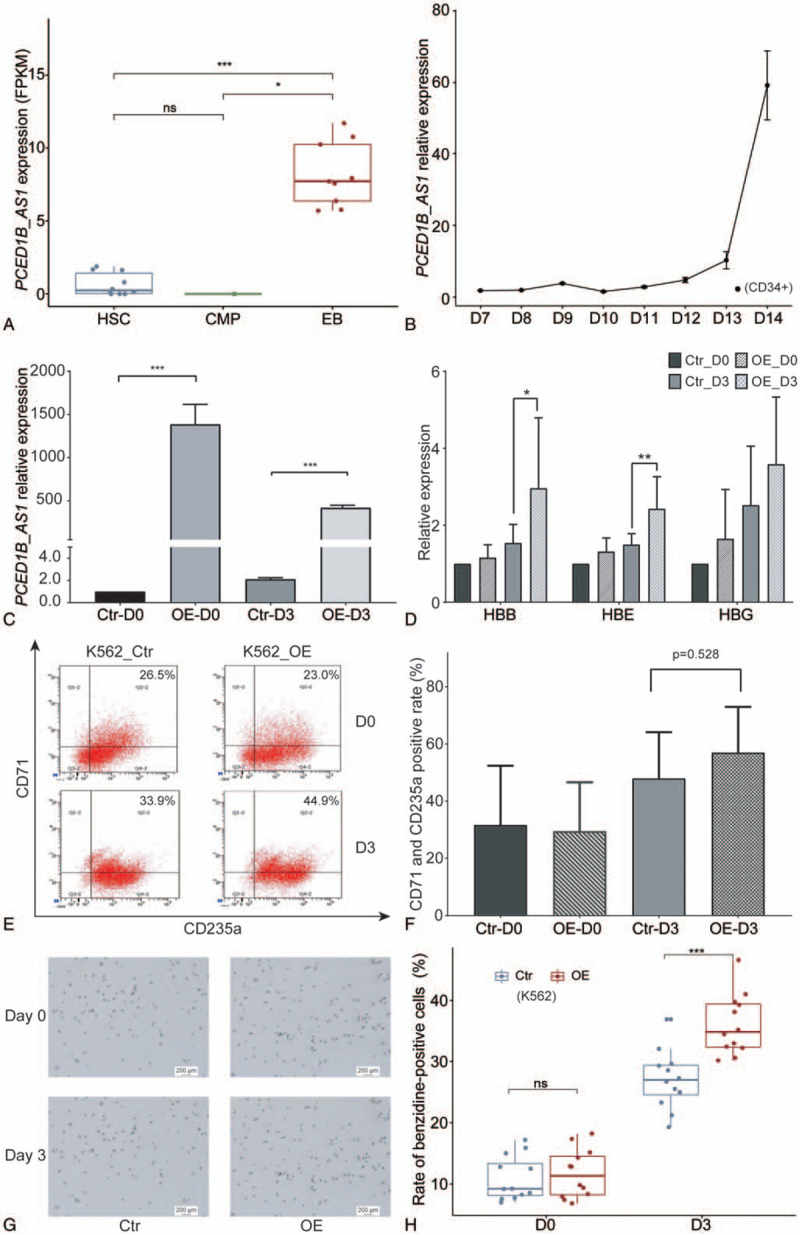
Identification and functional verification of lncRNA PCED1B-AS1. (A) Dynamic expression of lncRNA PCED1B-AS1 during erythroid differentiation in Blueprint database. (B) Relative expression of PCED1B-AS1 during late stage of erythroid differentiation of CD34+ cells. The horizontal axis represents the days after EPO induction. (C) The confirmation of PCED1B-AS1 overexpression in K562 cells by RT-PCR. D3 represents the cells induced with hemin for 3 days. (D) The analysis for globin gene expression changes before and after induction of PCED1B-AS1-overexpressed K562 cells compared with that in control cells. HBE, HBG and HBB are β-like genes that respectively represent ε (embryonic)-, γ (fetal)- and β (adult)-globin and are specifically expressed at different developmental stages during erythroid differentiation. (E) The expression changes of CD235a and CD71 was detected by flow cytometry before and after induction. The percentage in the upper right of each figure represents the proportion of CD235a+-CD71+ double positive cells. (F) Percentage of CD235a+-CD71+ double positive cells among the K562 cells before and after induction. Data are the means ± SD of three experimental replicates. N.S. = no significance between samples. (G,H) Benzidine-stained positive rate changes of K562 cells before and after induction with hemin. All relative mRNA expression was normalized to GAPDH. Statistical results were analyzed by student t-test and Kruskal−Wallis test, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, nsP > .05, ns: no significance. D0: Day0, D3: Day3. Ctr: Control, OE: overexpression of PCED1B-AS1.
Next, to further confirm the potential of PCED1B-AS1 in erythroid differentiation, we observed the endogenous expression pattern of PCED1B-AS1 during erythroid differentiation of cord blood-derived hematopoietic stem and progenitor cells (HSPCs), and from day 7 to day 15, the result showed a continuous upward expression trend of PCED1B-AS1 (Fig. 1B), suggesting its potential role in promoting erythroid differentiation.
3.2. PCED1B-AS1 promotes erythroid differentiation in K562 cells
To verify whether PCED1B-AS1 promotes erythroid differentiation, we conducted phenotypic analysis of K562 cells with PCED1B-AS1 overexpression (Fig. 1C) and control cells that were induced toward erythroid differentiation for 3 days by hemin. We first examined the expression changes in globin genes, finding a slight increase in the expression of ε- and β-globin before and after induction in the cells overexpressing PCED1B-AS1 compared with the control cells. However, the expression of γ-globin significantly increases after induction compared with the control cells (Fig. 1D). We analyzed the expression of cell surface markers CD235a and CD71 by flow cytometry. We found that the proportion of CD235a and CD71 double positive cells in the overexpression group was higher than that in the control cells after induction (Fig. 1F), but there was no significant difference. Benzidine staining results also showed the proportion of stained positive cells in total cells to be far higher than that of the control cells (Fig. 1G, H). These results demonstrated the role of PCED1B-AS1 in promoting erythroid differentiation of K562 cells.
3.3. GATA1-regulated PCED1B-AS1 coordinating with accessible chromatin participates in erythroid differentiation
To further explore the mechanism behind PCED1B-AS1 participation in erythroid differentiation, we analyzed the dynamic changes in chromatin accessibility around PCED1B-AS1 loci during erythroid differentiation. Interestingly, we found that the chromatin was tightly condensed at the PCED1B-AS1 promoter region in the EB compared with those at the HSCs and CMPs, while the dense structure of the nucleosome was newly opened at the distal region of the PCED1B-AS1 in EB, and the chromatin accessibility increased compared to that at other periods (Fig. 2A). Moreover, from the public ChIP-seq data and histone modification data of ENCODE, we observed strong GATA1 binding signals and H3K27ac signals at the distal region of PCED1B-AS1 (Fig. 2A). We verified the physical binding of GATA1 in that region by ChIP-qPCR (Fig. 2B). H3K27ac histone modification at the distal region of PCED1B-AS1 indicated the presence of an enhancer signal.
FIGURE 2.
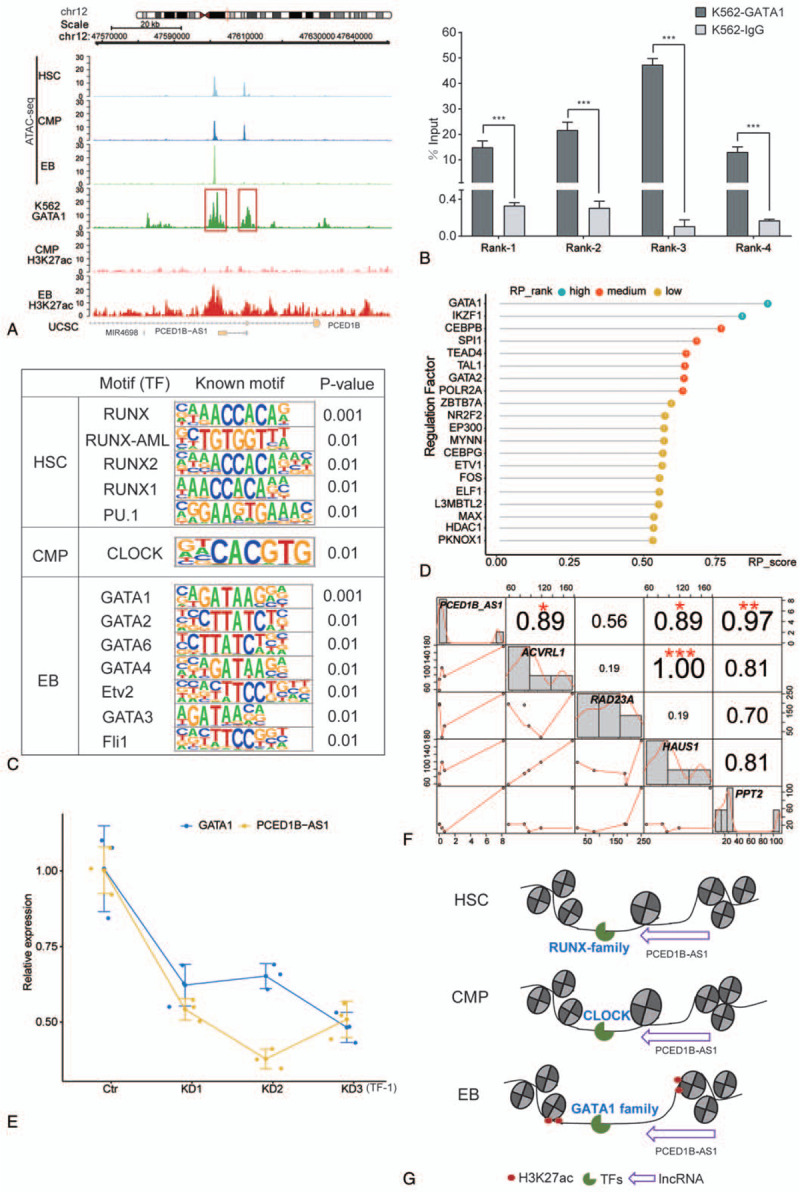
The association between lncRNA PCED1B-AS1 and chromatin accessibility during erythroid differentiation. (A) Dynamic profile of chromatin accessibility and histone modification of H3K27ac around PCED1B-AS1 loci during erythroid differentiation. (B) Confirmation of physical binding of GATA1 around PCED1B-AS1 loci by ChIP-qPCR in K562 cells. Rank1 and Rank2 are from the red box on the left side in (A) picture, and Rank3 and Rank4 are from the red box on the right side in (A). The bar graphs represent the average of percentage input (mean ± SD) from three independent ChIP experiments. (C) Known motif identification of peaks within 20 kb of PCED1B-AS1. (D) Regulation factors of PCED1B-AS1 within its 100 kb genomic regions in EB. (E) Relative expression of PCED1B-AS1 in TF-1 cells with GATA1 knockdown by siRNA. (F) Correlation between PCED1B-AS1 with its potential target genes. (G) Model of PCED1B-AS1 participates in erythroid differentiation via dynamic chromatin remodeling involving GATA1.
We then performed motif analysis of chromatin accessibility peaks around PCED1B-AS1 loci, including the promoter region and the distal region. We found that the transcription factor enriched in HSCs was mainly from the RUNX family (Fig. 2C), which play a significant role in hematopoiesis regulation—particularly in the important function of Runx1 in the generation of HSCs.25–27 The transcription factor CLOCK, a well known factor regulating circadian rhythms was enriched in CMPs (Fig. 2C).28,29 Transcription factors enriched at the EB stage belonged mainly to the GATA family, such as GATA1 (Fig. 2C), suggesting that accessibility of the chromatin was complete in preparation for the entrance of the GATA family TFs to regulate erythroid differentiation in EBs by PCED1B-AS1.
To further clarify which transcription factor may regulate PCED1B-AS1 at the distal region, we first used the published data of ChIP-seq (the protein factor and histone marker) and the chromatin accessibility (DNase-Seq and ATAC-seq) of EB cells to predict the potential regulatory factors within 100 kb upstream and downstream of PCED1B-AS1. The regulatory potential score is reflected by the standardized values of 0–1. We observed that GATA1 has the highest regulatory potential for PCED1B-AS1 (Fig. 2D). Moreover, we found that the expression of PCED1B-AS1 also decreased with GATA1 knockdown in the TF-1 cell line (Fig. 2E). These results suggested that—when chromatin was accessible—GATA1 regulated PCED1B-AS1 at the distal region of PCED1B-AS1.
In order to further explore the target genes regulated by PCED1B-AS1, we predicted the potential target genes of PCED1B-AS1 through Cistrom Data Browser and starBase 3.0 database and calculated the correlation analysis based on their expression. We found that PCED1B-AS1 has the strongest correlation with PPT2 (Fig. 2F). The palmitoyl protein thioesterase-2 (PPT2) gene encodes a lysosomal thioesterase homologous to PPT1.30 The correlation between PCED1B-AS1 and both HAUS1 and ACVRL1 is also high (Fig. 2F). HAUS1 is involved mainly in the assembly of mitotic spindles, which facilitates the maintenance of centrosome integrity and completion of cytokinesis. ACVRL1 is a type I receptor of TGF-beta family ligands BMP9/GDF2 and BMP10, and is an important regulator of normal vascular development.
In summary, we observed that enhanced chromatin accessibility in EBs allowed the transcription factor GATA1 and other GATA family members to bind to the distal region, thereby regulating the transcription of PCED1B-AS1, which in turn affected erythroid differentiation (Fig. 2G).
4. DISCUSSION
LncRNAs are abundant, large in number, and participate in many biological processes, including the process of erythropoiesis. Although several studies have identified multiple lncRNAs that function during erythroid differentiation, the function mechanism of lncRNA remains unclear. In our study, we screened antisense lncRNA PCED1B-AS1 from public ENCODE databases and Blueprint, and verified its role in promoting erythroid differentiation in the K562 cell model widely used for phenotypic analysis of the role played by unknown regulators in erythroid differentiation.
lncRNA is characterized by specificity at the cellular stage, a lower expression level than that of the coding gene,14 and a low conservation between species.31 Different lncRNAs play roles at specific cell stages during erythroid differentiation. For example, H19, highly expressed in long-term HSCs (LT-HSCs), regulates the maintenance and self-renewal of LT-HSCs via the Igf2 and Igf1r pathways,32 while lncRNA-EC6, highly expressed in red blood cells in the late stage of erythropoiesis, regulates the denucleation of mouse red blood cells through the RAC1/PIP5K signaling pathway.33 The expression level of PCED1B-AS1 gradually increases in the late stage of erythroid differentiation, indicating its important role in the late stage of red blood cell differentiation. The recent study of PCED1B-AS1 found that its expression was decreased in patients with active tuberculosis compared with that in healthy people, and that the apoptosis of monocytes was attenuated and autophagy was enhanced.34 Because, in the late stage of erythropoiesis, red blood cell islands are formed by macrophages and cells at different stages of terminal differentiation, and the role of macrophages is to help red blood cells denucleate by phagocytosis.35,36 Due to the rapidly increased expression of PCED1B-AS1 at day 14 of the culture, we suspect that PCED1B-AS1 may play a role in this process of denucleation.
The chromatin of eukaryotes is a high-level complex structure formed by dense compaction of nucleosomes. When the dense nucleosome structure is destroyed, the chromatin DNA can be approached by cis-regulatory elements such as a promoter, an enhancer, an insulator, a silencer, and a trans-acting factor. In the past, the study of lncRNA associated with erythroid differentiation was concerned mainly with the regulation of lncRNA by direct cis-regulation or trans-regulation with other factors,37 and few explore the changes of chromatin accessibility in this process. By inhibiting Fas-mediated cell death, lncNRA Fas-antisense 1 (Fas-AS1 or Saf) participates in the process of erythrocyte maturation, and binding sites of GATA1, KLF1, and NF-κB transcription factors were found in its promoter region. Moreover, in the early stage of EB expansion, NF-kB signaling inhibits the expression of Saf, followed by increased expression of Saf in the late stage of RBC maturation, and the concurrent elevation of GATA1 and KLF1 expression, indicating that Saf may be regulated by these transcription factors.38 This suggests that sequential actions of different transcription factors at different times on the same loci lead to changes in gene expression, which in turn regulate the differentiation process. In this study, we found that, at different stages of erythroid differentiation, chromatin accessibility changed in the vicinity of PCED1B-AS1 and was accompanied by successive binding of different transcription factors (RUNX/PU.1/GATA1). After knocking down GATA1 in TF-1 erythroid cells, we detected a decrease in the expression of PCED1B-AS1, indicating that GATA1 may be the factor that affects PCED1B-AS1, directly or indirectly, and further regulates erythroid differentiation.
We found there is higher correlation between PCED1B-AS1 with its potential target genes. For example, PPT2, one of candidate of α-Globin poly (C) binding proteins (αCPs) complex involved in posttranscriptional regulation, in addition to maintaining the stability of RNA, it also participates in translation control during erythroid differentiation.39
The link between lncRNA and chromatin has been reported in other research areas. LncRNA controls the chromatin structure and accessibility of genetic information through interaction with chromatin-modifying enzymes and nucleosome-remodeling factors.40,41 For instance, lncRNA TARID acts by coupling with GADD45A to direct DNA demethylation to specific loci in cancer cells to regulate gene expression.41,42 The relationship between lncRNA and chromatin in the process of erythroid differentiation remains to be revealed, and in our study we have carried out preliminary explorations.
ACKNOWLEDGMENTS
This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010602) and the National Natural Science Foundation of China (81670109, 81700097, 81870097, 81700116).
REFERENCES
- [1].Notta F, Zandi S, Takayama N, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 2016;351(6269):1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Baron MH, Vacaru A, Nieves J. Erythroid development in the mammalian embryo. Blood Cells Mol Dis 2013;51(4):213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dzierzak E, Philipsen S. Erythropoiesis: development and differentiation. Cold Spring Harb Perspect Med 2013;3(4):a011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tsiftsoglou AS, Vizirianakis IS, Strouboulis J. Erythropoiesis: Model systems, molecular regulators, and developmental programs. IUBMB Life 2009;61(8):800–830. [DOI] [PubMed] [Google Scholar]
- [5].Walkley CR. Erythropoiesis, anemia and the bone marrow microenvironment. Int J Hematol 2011;93(1):10–13. [DOI] [PubMed] [Google Scholar]
- [6].Hattangadi SM, Wong P, Zhang L, Flygare J, Lodish HF. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood 2019;118(24):6258–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wu W, Cheng Y, Keller CA, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res 2011;21(10):1659–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Onodera K, Takahashi S, Nishimura S, et al. GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc Natl Acad Sci U S A 1997;94(9):4487–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li W, Ren Y, Si Y, Wang F, Yu J. Long non-coding RNAs in hematopoietic regulation. Cell Regen (Lond) 2018;7(2):27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang X, Hu W. Long noncoding RNAs in hematopoiesis. F1000Research 2016;5:1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity 2015;42(5):792–804. [DOI] [PubMed] [Google Scholar]
- [12].Ding N, Xi J, Li Y, et al. Global transcriptome analysis for identification of interactions between coding and noncoding RNAs during human erythroid differentiation. Front Med 2016;10(3):297–310. [DOI] [PubMed] [Google Scholar]
- [13].Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17(1):47–62. [DOI] [PubMed] [Google Scholar]
- [14].Paralkar VR, Mishra T, Luan J, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood 2018;123(12):1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Corces MR, Chang HY, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 2016;48(10):1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Buenrostro JD, Corces MR, Lareau CA, et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 2018;173(6):1535–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ruan X, Chang K-H, Yan J, et al. Transcriptome dynamics during human erythroid differentiation and development. Genomics 2013;102((5–6)):431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bernstein BE, Brown M, Johnson DS, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9(9):R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wu Q, Brown M, Liu XS, et al. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res 2018;47((D1)):D729–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zang C, Brown M, Liu XS, et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res 2016;45((D1)):D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li JH, Liu S, Zhou H, Qu LH, Yang JH. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42((D1)):D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hasegawa A, Shimizu R. GATA1 activity governed by configurations of cis-acting elements. Front Oncol 2017;6:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cai X, Gaudet JJ, Mangan JK, et al. Runx1 loss minimally impacts long-term hematopoietic stem cells. PLoS One 2011;6(12):e28430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].de Bruijn M, Dzierzak E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood 2017;129(15):2061–2069. [DOI] [PubMed] [Google Scholar]
- [27].Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev 2005;19(19):2331–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Menet JS, Pescatore S, Rosbash M. CLOCK: BMAL1 is a pioneer-like transcription factor. Genes Dev 2014;28(1):8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rogers EH, Hunt JA, Pekovic-Vaughan V. Adult stem cell maintenance and tissue regeneration around the clock: do impaired stem cell clocks drive age-associated tissue degeneration? Biogerontology 2018;19(6):497–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gupta P, Soyombo AA, Shelton JM, et al. Disruption of PPT2 in mice causes an unusual lysosomal storage disorder with neurovisceral features. Proc Natl Acad Sci U S A 2003;100(21):12325–12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Alvarez-dominguez JR, Hu W, Yuan B, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood 2015;123(4):570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Venkatraman A, He XC, Thorvaldsen JL, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 2013;500(7462):345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang C, Wu X, Shen F, et al. Shlnc-EC6 regulates murine erythroid enucleation by Rac1-PIP5K pathway. Dev Growth Differ 2015;57(6):466–473. [DOI] [PubMed] [Google Scholar]
- [34].Li M, Cui J, Niu W, et al. Long non-coding PCED1B-AS1 regulates macrophage apoptosis and autophagy by sponging miR-155 in active tuberculosis. Biochem Biophys Res Commun 2019;509(3):803–809. [DOI] [PubMed] [Google Scholar]
- [35].Chasis JA, Mohandas N. Erythroblastic islands: Niches for erythropoiesis. Blood 2008;112(3):470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li W, Wang Y, Zhao H, et al. Identification and transcriptome analysis of erythroblastic island macrophages. Blood 2019; 2019000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kulczyńska K, Siatecka M. A regulatory function of long non-coding RNAs in red blood cell development. Acta Biochim Pol 2016;63(4):675–680. [DOI] [PubMed] [Google Scholar]
- [38].Villamizar O, Chambers CB, Mo YY, et al. Fas-antisense long noncoding RNA is differentially expressed during maturation of human erythrocytes and confers resistance to Fas-mediated cell death. Blood Cells Mol Dis 2016;58:57–66. [DOI] [PubMed] [Google Scholar]
- [39].Waggoner SA, Liebhaber SA. Identification of mRNAs associated with alphaCP2-containing RNP complexes. Mol Cell Biol 2003;23(19):7055–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol 2015;12(10):1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li X, Fu X-D. Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat Rev Genet 2019;20(9):503–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Arab K, Park YJ, Lindroth AM, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 2014;55(4):604–614. [DOI] [PubMed] [Google Scholar]


