Abstract
DNA methylation is one of the most important components of epigenetics, which plays essential roles in maintaining genome stability and regulating gene expression. In recent years, DNA methylation measuring methods have been continuously optimized. Combined with next generation sequencing technologies, these approaches have enabled the detection of genome-wide cytosine methylation at single-base resolution. In this paper, we review the development of 5-methylcytosine and its oxidized derivatives measuring methods, and recent advancement of single-cell epigenome sequencing technologies, offering more referable information for the selection and optimization of DNA methylation sequencing technologies and related research.
Keywords: 5-Methylcytosine derivatives, DNA methylation, Next-generation sequencing, Single-cell sequencing
1. INTRODUCTION
DNA methylation refers to the process of adding a methyl group to the fifth carbon atom of cytosine covalently, under the catalysis of DNA methyltransferase (DNMTs) with S-adenosyl-methionine (SAM) as the methyl donor. This process mainly occurs at the CpG or CpHpG (H = A, T, C) sites of DNA.1 In 1950s, scientists discovered this phenomenon in the DNA of calf thymus.2 In 1975, Holliday et al first proposed that DNA methylation plays an important role in epigenetic modification during vertebrate development.3 As one of the epigenetic markers, DNA methylation is widely believed to be reversible and inheritable.4
In eukaryotes, CpG sites usually exist in 2 forms: one is randomly dispersed in DNA sequences, the other is highly aggregated, called CpG islands (CGI). In normal tissues, around 80% of the dispersive CpG sites are methylated, while CGIs are often unmethylated (except for some special regions or genes, such as genes located on the inactivated X chromosome and imprinted genes). CGIs are often located near the transcriptional regulatory regions and overlapped with the transcription start sites (TSS).5
1.1. Functions of DNA methylation
In transcriptional regulatory regions of the genome, DNA methylation levels controls gene expression: DNA hypermethylation can inactivate genes, while demethylation can induce gene reactivation.6 DNA methylation can affect the interaction between DNA and proteins, block the binding of transcription factors and reduce the level of gene expression7; it can also destabilize nucleosomes and change DNA conformation, resulting in chromatin structure remodeling and transcriptional repression states.8 Moreover, studies have shown that DNA methylation involved in the regulation of gene expression is also distributed in control regions outside TSS, repetitive DNA sequences and gene bodies.9 DNA methylation helps maintain the stability of imprinted genes and the genome as well. However, abnormal DNA methylation may cause a variety of diseases, such as tumors, vascular diseases, neurological diseases and so on. In the process of tumorigenesis, genomic hypomethylation and regional hypermethylation will activate proto-oncogenes and inactivate tumor suppressor genes and DNA repair genes, leading to the occurrence of tumors eventually.10
During the hierarchical differentiation as described in Waddington's epigenetic landscape,11 taking hematopoiesis as an example, the bifurcations from stem cells to lineage-committed progenitors on this road are similar to the cell fate decision, all of which are inseparable from the rigid and subtle regulation by epigenetics.12 DNA methylation plays a vital role in establishment and maintenance of cell identity, lineage commitment, transcription factor expression, as well as the regulation of mammalian embryonic development and other biological processes.13
1.2. DNA methylation related enzymes and 5mC derivatives
In eukaryotic genomes, DNA methylation modification related enzymes mainly include DNMT and ten-eleven translocation (TET) families. DNA methylation is mainly catalyzed by a series of highly conserved DNMTs, including DNMT3A, DNMT3B for regulating de novo DNA methylation, DNMT1 for maintaining methylation status,14 The TET family, containing TET1, TET2 and TET3, catalyzes the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), all of which are key intermediates in the DNA demethylation processes.15 Studies have shown that 5hmC is a stable epigenetic modification whose quantity is far less than 5mC in adult mammals.16 The distribution of 5hmC is tissue-specific: it is abundant in embryonic stem cells and nervous systems, but significantly reduced in many types of cancer cells.17 It has been revealed that 5hmC was highly concentrated in exons and near TSS in mouse embryonic stem cells, suggesting its potential impact on regulating transcriptional activation.18
In short, the acquisition of the above-mentioned DNA methylation related information requires corresponding methylation measuring methods. In the past decades, methylation detection technologies have made great progress. Till now, the widely used next generation sequencing (NGS)-based methylation measuring methods have achieved quantitative detection of genome-wide methylation and generated a large amount of sequence information. The revolution of DNA methylation sequencing technologies has offered us a more comprehensive understanding of biological growth, development, health, and disease.
2. DNA METHYLATION MEASURING METHODS
2.1. Development of DNA methylation sequencing technologies
As an early DNA methylation detection technique, liquid chromatograph (LC) was used to quantitatively detect the global DNA methylation levels.19 Combined with fluorescence, ultraviolet (UV) and mass spectrometry (MS), the sensitivity of LC for DNA methylation was greatly enhanced. However, due to their inability to obtain detailed methylation information at specific sites, these methods gradually faded out. Later, electrophoresis-based techniques enabled the detection of DNA methylated regions and specific sites on a genomic scale. Methylation-specific PCR (MSP) is a classic method for detecting DNA methylation quickly and qualitatively. It used 2 pairs of primers, corresponding to the methylated and unmethylated DNA sequences respectively, to conduct PCR amplification on the bisulfite-converted DNA sequences and then analyzed DNA methylation by gel electrophoresis.20 Besides, In the 1990s, restriction landmark genomic scanning (RLGS) adopted methylation sensitive restriction enzymes (MRE) to perform multiple endonuclease digestion, followed by isotope labeling, 2-dimensional electrophoresis, and scanning analysis of genomic DNA (gDNA). Then, sites on the scanned maps were compared with related DNA databases to gain the DNA methylation information.21 However, because of the high labor intensity, technical requirements and relatively low resolution, electrophoresis-based techniques were gradually replaced by microarray hybridization techniques with better resolution and throughput. Combined with endonuclease digestion—immunoprecipitation—and bisulfite conversion-based methods respectively, microarray hybridization techniques have contributed to a broader understanding of DNA methylation patterns.22
Currently, high-throughput sequencing technologies are undergoing rapid development with a tendency to replace microarrays gradually. In 1984, based on Sanger sequencing technologies, researchers proposed to measure DNA methylation levels according to the reactivity difference between unmethylated and methylated cytosines with hydrazine (N2H4).23 Since 2005, by converting the release of pyrophosphate (PPi) into light signals during the synthesis, pyrosequencing has increased several orders of magnitude per run based on sequencing-by-synthesis (SBS) techniques.24 The advanced third-generation sequencing technologies hybridize single molecule fluorescence and nanopores to achieve single-molecule real-time (SMRT) sequencing.25 However, due to the technical instability and high error rates of SMRT sequencing at the moment, the second-generation sequencing technologies still occupy the dominant position.
Since PCR amplification cannot retain DNA methylation information, currently, most sequencing-based DNA methylation detection methods rely on various pretreatments of DNA samples. Depending on different pretreatment methods, NGS-based DNA methylation technologies can be mainly divided into 3 categories: endonuclease digestion, affinity enrichment, and base conversion. This paper mainly introduces the principles, applications, advantages, and disadvantages of these methods; and further discusses the DNA methylation detection methods related to 5mC derivatives, and single-cell multi-omics.
2.2. Endonuclease digestion-based methods
Before high-throughput sequencing technologies were available, MREs were used to gain methylation information about certain specific sites in combination with electrophoresis or PCR. The endonuclease digestion methods mainly utilize MREs, insensitive to DNA methylation sites, to cut DNA double-strands into fragments. Then, NGS is applied to clarify the DNA methylation status.26 Classical enzyme pairs available in these methods mainly include MspI and HpaII, a group of isoschizomers with different CpG methylation sensitivity, where MspI recognizes CCGG sites, and HpaII identifies whether these sequences are methylated.27
In 2010, aimed at exploring the function of intragenic DNA methylation, Maunakea et al developed methylation sensitive restriction enzyme sequencing (MRE-seq) (Fig. 1), which could cover approximately 30% of the genome.28,29 Combining 3 endonucleases (HpaII, Hin6I and AciI), MRE-seq inferred the DNA methylation status from the inverse relationship between methylation of MRE targets and MRE-seq readouts. In this work, authors also proposed methylated DNA immunoprecipitation and sequencing (MeDIP-seq) as an appropriate supplement to MRE-seq. Combination operation with MeDIP-seq and MRE-seq generated high-resolution methylome maps of human brain frontal cortex grey matter.
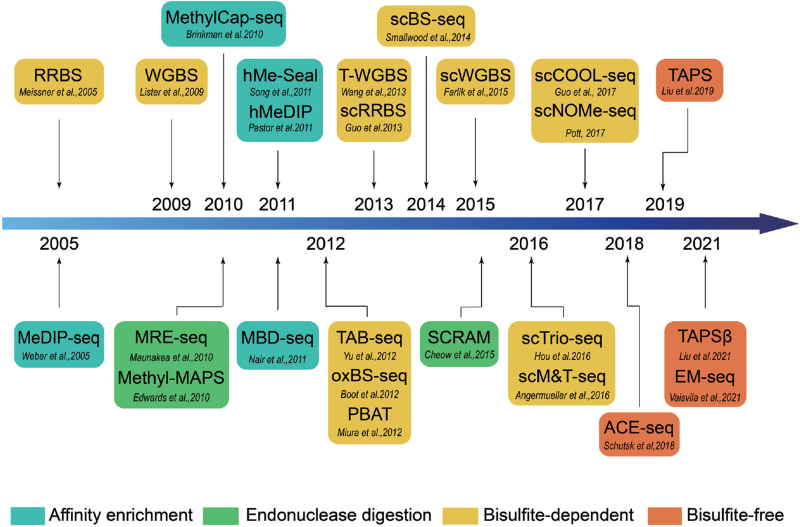
Figure 1.
Evolution of NGS-based techniques applied to DNA methylation profiling. Different background colors indicate different categories of DNA methylation measuring methods (bottom). ACE-seq: APOBEC-coupled epigenetic sequencing, EM-seq = Enzymatic Methyl-seq, hMeDIP = hydroxymethylated DNA immunoprecipitation, hMe-Seal = 5hmC-selective chemical labeling method, MBD-seq = methyl-CpG-binding domain protein sequencing, MeDIP-seq = methylated DNA immunoprecipitation sequencing, Methyl-MAPS = methylation mapping analysis by paired-end sequencing, MRE-seq = methylation sensitive restriction enzyme sequencing, oxBS-seq: oxidative bisulfite sequencing, PBAT = post-bisulfite adaptor tagging, RRBS = reduced representation bisulfite sequencing, scBS-seq = single cell bisulfite sequencing, scCOOL-seq = single-cell Chromatin Overall Omic-scale Landscape Sequencing, scM&T-seq = single-cell methylome and transcriptome sequencing, scNOMe-seq = single-cell Nucleosome Occupancy and Methylome Sequencing, SCRAM = single-cell restriction analysis of methylation, scRRBS = single-cell reduced representation bisulfite sequencing, scTrio-seq = single-cell triple omics sequencing, scWGBS = single cell whole genome bisulfite sequencing, TAB-seq = Tet-assisted bisulfite sequencing, TAPS = TET-assisted pyridine borane sequencing, TAPSβ = TAPS with β-glucosyltransferase (β-GT) blocking, T-WGBS = Tagmentation based WGBS, WGBS = whole-genome bisulfite sequencing.
MRE-seq provides estimates of DNA methylation at single base resolution, but its dependence on different kinds of endonucleases limits the genomic coverage (Table 1). Researchers attempted to improve the coverage of CpG sites by increasing the number of endonucleases. Methyl-MAPS (methylation mapping analysis by paired-end sequencing) adopted 5 MREs and a methylation-dependent endonuclease McrBC complex to divide the genome into methylated and unmethylated fragments. The methylated compartment of the genome was isolated by limit digest with 5 MREs, while the unmethylated compartment was isolated by digestion with the McrBC complex. Researchers could then evaluate the DNA methylation status by analyzing the resistance or sensitivity of CpG sites to McrBC complex and MREs,30 This method provides high coverage of DNA methylation status at single-copies and repeated sequences with relatively low cost.
Table 1.
Comparison of different DNA Methylation measuring methods.
| Base conversion | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Category | Endonuclease digestion | Affinity enrichment | Bisulfite-dependent | Bisulfite-free | |||
| Methods | MRE-seq | MeDIP-seq | MBD-seq | RRBS | WGBS | TAPS | EM-seq |
| Resolution | Single-base | 150–300 bp | Single-base | ||||
| Coverage | – | +/– | + + | ||||
| DNA input | + | + | + + | – | |||
| Cost | – | – | + + | – | |||
| Application | Site-specific studies | Low-resolution, large-scale studies | High resolution, whole-genome studies | ||||
| Pros | Cost-effective, user-friendly | High sensitivity, low cost | Detect whole-genome methylation at single-base | ||||
| Cons | Enzyme-dependent methylation detection; incomplete digestion | Low resolution; biased towards highly methylated regions | DNA degradation; high cost; reduced sequence complexity | Unsuitable for single cells; Large investment | |||
EM-seq = Enzymatic Methyl-seq, MBD = methyl-CpG-binding domain protein, MeDIP = methylated DNA immunoprecipitation, MRE = methylation sensitive restriction enzyme, RRBS = reduced representation bisulfite sequencing, -seq = sequencingg, TAPS = TET-assisted pyridine borane sequencing, WGBS = whole-genome bisulfite sequencin.
Coverage, DNA input and cost in table 1 are divided into 5 levels, followed by + + > + > +/– > – > – –.
Another disadvantage of the endonuclease digestion-based technologies is the incomplete digestion of MREs, which may lead to false positive results. Considering that the fragments produced by endonuclease digestion may still contain some important methylation information, bisulfite conversion followed by MRE treatment (MREBS) added a bisulfite conversion step to detect the methyl groups at CpG sites within DNA fragments after MRE treatment. This approach enables a significant increase in the coverage of genome-wide DNA methylation.31
2.3. Affinity enrichment-based methods
Given the related shortcomings of endonuclease digestion-based methods, in 2005, Weber et al established methylated DNA immune precipitation (MeDIP), that isolated and detected CpG sites based on the interaction between proteins and methylated DNA.32 MeDIP captured single-strand methylated DNA fragments with 5mC specific antibodies. By changing specific antibodies, MeDIP could detect different DNA methylation intermediates, like 5hmC.33 Subsequently, researchers combined MeDIP with NGS, named MeDIP-seq, which could achieve an approximate 100 to 300 bp resolution at genomic level.
In addition to MeDIP-seq, Methyl-CpG-binding domain sequencing (MBD-seq) is another DNA methylation detection method based on affinity enrichment.34 The MBD domain contained in MBD protein family is capable of binding to the single symmetrically methylated CpG dinucleotides, which is essential for the regulation of epigenome. In MBD-seq, Methyl-CpG Binding Domain Protein 2 (MBD2) was used to capture CpG sites on double-stranded DNA (dsDNA). After enrichment of methylated DNA with different CpG densities through a stepwise elution method, high-throughput sequencing was performed with prepared libraries on NGS platforms. The resolution of this method depends on the size of the sonicated DNA fragments. Methylated DNA capture by affinity purification (MethylCap-seq) utilizes the MBD domain of Methyl-CpG Binding Protein 2 (MeCP2) to capture methylated DNA, which can attain higher sequence coverage with lower CpG densities.35
Compared with the single-stranded DNA (ssDNA) methylated fragments captured by MeDIP-seq, dsDNA methylated regions enriched by MBD-seq and MethylCap-seq are more conducive to subsequent library construction. However, methylated-CpG-binding proteins are unable to cover unmethylated CpG sites, which limits their access to CGIs (hypomethylation) and other related information. Furthermore, neither of the above methods can attain a single-base resolution of genome. Nowadays, MBD-seq has been widely employed in the studies of genome-wide DNA methylation due to its low cost, high sensitivity, and strong specificity.
2.4. Base conversion-based methods
2.4.1. Bisulfite-dependent base conversion
In the 1990s, Hayatsu et al discovered that unmethylated cytosine residues and methylated cytosines in denatured gDNA exhibited different reactivity to bisulfite treatment: the former was deaminated and converted to thymine faster than the latter. This phenomenon transformed the epigenetic differences into changes of DNA sequences, which promoted another revolution in DNA methylation detection methods.36 Sequencing technologies, especially the current popular NGS techniques, possess good compatibility with bisulfite-treated DNA samples. Hence, the DNA methylation measuring methods based on bisulfite treatment occupy a dominant position in DNA methylome profiling.
In 2005, Reduced representation bisulfite sequencing (RRBS), developed by Meissner et al, employed endonuclease MspI to the genome to generate DNA fragments for adaptor ligation. Then, the amplification products were purified and sequenced after bisulfite treatment.37 RRBS is mainly suitable for the detection of representative high-density DNA methylation regions. Despite the small number of readouts produced, this method is able to achieve a broader coverage of CGIs, promoters, and enhancer regions with less DNA inputs (generally 10–300ng). Nevertheless, due to the application of restriction enzymes, RRBS could not cover all CpG sites in the genome. Overall, RRBS is a time-saving, cost-effective and accurate DNA methylation detection method.
Whole genome bisulfite sequencing (WGBS) detects the whole-genome DNA methylation at a single base resolution. In this protocol, DNA was fragmented, end-repaired and ligated to the adapters first. After bisulfite treatment, the unmethylated C bases were converted to U bases and then transformed into T bases by PCR amplification, distinguished from the methylated C bases (Table 2). Finally, the PCR products were subjected to high-throughput sequencing.38 In 2008, Cokus and his colleagues first applied this method to obtain the DNA methylation landscape of Arabidopsis thaliana.39 Nowadays, WGBS has become the “gold standard” for DNA methylation detection because of its single-base resolution, less time consumption, high throughput, and genomic coverage. However, WGBS is most expensive and requires a large amount of DNA inputs (minimum 200–500 ng, maximum 5 μg). In addition, the acidic and thermal environment during the bisulfite conversion may lead to DNA degradation, resulting in quantities of DNA loss. Besides, the complexity of the DNA sequences is reduced after bisulfite treatment, which increases the redundancy in sequencing process. These problems were shared by series of methods based on bisulfite conversion.
Table 2.
Comparison of the conversion of C, 5mC, 5hmC in different DNA methylation profiling methods.
| Base | WGBS | TAPS | TAPSβ | EM-seq | ACE-seq | TAB-seq | oxBS-seq |
|---|---|---|---|---|---|---|---|
| C | T | C | C | T | T | T | T |
| 5mC | C | T | T | C | T | T | C |
| 5hmC | C | T | C | C | C | C | T |
ACE-seq = APOBEC-coupled epigenetic sequencing, EM-seq = Enzymatic Methyl-seq, oxBS-seq = oxidative bisulfite sequencing, TAB-seq = Tet-assisted bisulfite sequencing, TAPS = TET-assisted pyridine borane sequencing, TAPSβ = TAPS with β-glucosyltransferase (β-GT) blocking, WGBS = whole-genome bisulfite sequencing.
To circumvent these limitations, after many attempts, researchers developed post-bisulfite adaptor tagging (PBAT) and Tagmentation based WGBS (T-WGBS) which have become the main alternative to WGBS. T-WGBS fragmented gDNA by the active Tn5 transposase, and then added sequencing adapters to the amplification products. This improvement brought the required amount of DNA inputs below 20 ng.40 PBAT was a PCR-free WGBS method which put the bisulfite treatment step ahead of adapter ligation, avoiding bisulfite-induced degradation of the DNA adapter templates. It could generate a large quantity of unamplified reads just from a small amount of DNA inputs.41 Compared with WGBS, PBAT does not need to utilize Covaris for fragmentation, which can simplify purification steps, shorten operating time, and lower product loss, but it is necessary to consider issues such as the low efficiency of adapter ligation of ssDNA fragments.
2.4.2. Bisulfite-free base conversion
Given the shortcomings of bisulfite treatment such as massive DNA degradation and sequence complexity reduction, scientists have developed a variety of bisulfite-free DNA methylation detection methods. Among them, the electrochemical oxidation-based assays depend on the direct oxidation of different DNA bases on various electrodes to distinguish C from 5mC. In 2010, Wang et al developed a choline chloride monolayer supported multiwalled carbon nanotubes film modified glassy carbon electrode (MWNTs/Ch/GCE), which could clearly identify all purine and pyrimidine bases of adenine (A), guanine (G), thymine (T), C and 5mC.42 In addition, since the C5–C6 double bond of pyrimidine bases of 5mC can be oxidized more easily than that of cytosine, many chemical labelling-based methods take advantage of this phenomenon to distinguish between cytosine and 5mC. In 2006, Okamoto et al employed osmium complexes to oxidize 5mC. Combined with bipyridine derivative functionalized fluorescent dyes labelling 5mC in DNA, they could further detect the 5mC through fluorescence spectra, fluorescence resonance energy transfer (FRET), and an electrochemical assay.43 However, these methods described above cannot detect the DNA methylation profiling of the whole genome. Besides, they are not compatible with NGS which limits their applications.
In 2019, Liu et al developed a mild DNA methylation detection method, named TET-assisted pyridine borane sequencing (TAPS). This method is independent of bisulfite conversion, and has achieved whole-genome detection of methylated cytosines at a single-base resolution. In this protocol, 5hmC and 5mC were oxidized to 5caC by TET, and then reduced to dihydrouridine (DHU) by pyridine borane. The subsequent PCR amplification converted DHU to T bases, thereby realizing the conversion from C to T. Combined with high-throughput sequencing, TAPS could quantitatively detect the DNA methylation levels of the whole genome.44 As a reductant, pyridine borane is inactive against the unmethylated cytosines. In 2021, based on TAPS, TAPS with β-glucosyltransferase blocking (TAPSβ) employed β-glucosyltransferase (β-GT) to glycosylate 5hmC to protect it from TET oxidation and pyridine borane reduction, thereby achieving 5mC specific sequencing. TAPSβ is a bisulfite-free, single-base resolution method with high sensitivity and specificity.45 It has overcome the disadvantages of bisulfite conversion, and can retain DNA fragments over 10 kilobases while improving the sequencing quality, comparison rate and average genome coverage. Since this method has just been proposed and not widely applied to academic research and clinical diagnosis, more detailed tests are required for the construction of specific chemical processes and the usage of reagent components. Besides, TAPSβ has not been adopted to single-cell analysis yet, which is the direction of further research.
In addition, Enzymatic Methyl-seq (EM-seq) developed by Vaisvila et al is another bisulfite-free method.46 It first employed TET2 and T4-βGT to convert 5mC and 5hmC into substrates that cannot be deaminated by APOBEC3A (A3A). Then A3A catalyzed the deamination of unmodified cytosines to U bases which were then converted into T bases by PCR amplification. Combined with high-throughput sequencing, EM-seq has realized the detection of DNA methylation levels of the whole genome. A3A is a DNA deaminase with high activity and a particular proficiency for 5mC deamination. It can distinguish different cytosine modification states effectively, and then preform deaminization through enzymatic reaction instead of chemical reaction, thereby greatly reducing the amount of DNA loss.47 APOBEC-coupled epigenetic sequencing (ACE-seq) applied β-GT to glycosylate 5hmC to protect it from the deamination of A3A, which could localize 5hmC at a single-base resolution.48 Comparing the sequencing results of DNA samples processed by EM-seq and ACE-seq, we can figure out the distribution of 5mC at single-base resolution specifically. The DNA libraries processed by EM-seq were superior to the bisulfite treated libraries in terms of coverage, repeatability, sensitivity, and base composition. Meanwhile, the requirement of DNA inputs for EM-seq could be as low as 100pg, which offered a new way for the research and clinical applications. However, A3A modification has severely reduced the sequence complexity, which will lead to low mapping rates, poor base quality, and uneven genome coverage.
3. 5mC DERIVATIVES RELATED DNA METHYLATION DETECTION METHODS
5hmC, 5fC and 5caC are obtained after the oxidation of 5mC by TET proteins. Identifying these DNA modifications provides important clues for studying DNA demethylation and its potential functions in gene expression.49 Since 5mC and 5hmC are recognized as C bases, whereas 5fC, 5caC, and unmodified C bases are converted into T bases after bisulfite treatment, the bisulfite-dependent DNA methylation methods cannot distinguish 5mC from 5hmC.
Hydroxymethylated DNA immuneprecipitation (hMeDIP) adopted specific antibodies to carry out the specific detection of 5hmC, providing a cost-effective strategy for understanding the whole-genome distribution of 5hmC, but it cannot achieve the quantitative detection of 5hmC. Tet-assisted bisulfite sequencing (TAB-seq) used β-GT to catalyze the glycosylation of 5hmC while converting 5mC to 5caC by TET-mediated oxidation, and then coupled with bisulfite treatment and DNA sequencing to obtain the methylation status of 5hmC sites.50 In 2011, hMe-Seal (5hmC-selective chemical labeling method) employed β-GT of T4 phage to transfer the designed azide-containing glucose, which can be chemically modified by biotin, to the hydroxyl group of 5hmC. Then 5hmC was pulled down by the tight binding between biotin and streptavidin to attain the methylation detection of DNA fragments.51
Potassium perrhenate (KRuO4) can selectively oxidize 5hmC to 5fC. The oxidative bisulfite sequencing (oxBS-seq) took advantage of this phenomenon to oxidize 5hmC to 5fC while keeping 5mC unchanged. Comparing the methylation levels of the oxidized and unoxidized samples enabled us to distinguish 5hmC from 5mC at single-base resolution. Since oxidative conditions may result in serious degradation and damage of gDNA, this method needs a relatively large amount of DNA inputs.52
In recent years, the detection methods of DNA methylation derivatives have extended to single-cell areas. The scAba-seq, established in 2016, applied restriction enzyme AbaSI to identify the glycosylated 5hmC sites. Afterwards, the digested gDNA were used for an end-repair reaction and ligated to adapters, and then the linear amplification was performed by in vitro transcription, greatly increasing the throughput of single-cell analysis.53
4. SINGLE-CELL DNA METHYLATION SEQUENCING TECHNOLOGIES
Generally, cells in different cell types have different epigenetic characteristics, while cells within a given cell type also exhibit inherent epigenetic heterogeneity due to the inaccurate definition of cellular identity and their differences in cell cycle as well as microenvironment. In recent years, with the rapid development of single-cell transcriptome sequencing technologies, single-cell epigenome sequencing technologies have been emerging rapidly, and studies of epigenetics have undergone the transition from bulk cells to single cells.54
In 2013, based on RRBS mentioned above, Guo et al established single-cell reduced representation bisulfite sequencing (scRRBS). By concentrating the reaction steps into a small tube before PCR amplification, this method minimized the gDNA loss in single cells caused by purification, which is suitable for the detection of high-density DNA methylated regions.55 Single cell whole genome bisulfite sequencing (scWGBS) and single cell bisulfite sequencing (scBS-seq) were 2 PABT-based methods. scWGBS lysed cells through bisulfite treatment directly, and then used the labeled random hexamer primers that can bind to the 3’ end of the DNA fragments to extend the DNA strands.56 In terms of scBS-seq, after bisulfite conversion, 5 rounds of DNA pre-amplification were added to increase the number of labeled DNA strands, which could generate more DNA copies and increase the genomic coverage.57 Compared with scBS-seq, scWGBS does not require any DNA pre-amplification, which can reduce reagent costs, operation time and amplification biases, but the complexity of the DNA libraries generated by this method is relatively low. scBS-seq has higher genomic coverage than scWGBS, which can accurately measure up to 48.4% of CpG sites. Other methods reported recently, like single-nucleus methyl-cytosine sequencing58 (snmC-seq) and single-cell combinatorial indexing for methylation analysis59 (sci-MET), could further improve the throughput of single-cell methylation sequencing.
In 2015, single-cell restriction analysis of methylation (SCRAM), developed by Cheow et al, combined single-cell MSRE digestion with multiplex PCR analyses to detect the methylation status of multiple CpG sites in single cells, avoiding the massive degradation of DNA molecules during the bisulfite conversion. Within a relatively short time (<2 days), this method could reliably and accurately finish the detection of DNA methylation status in single cells at a low cost.60 Nevertheless, as it was based on targeted amplification, multiple PCR primers were required to obtain each CpG site. Therefore, SCRAM was not suitable for genome-wide DNA methylation detection. Since only CpG sites distributed in the enzyme recognition regions can be detected at single-base resolution, the coverage of SCRAM is lower than other single cell DNA methylation detection methods based on bisulfite conversion.
With the rapid advances of single-cell sequencing technologies, single cell multi-omics sequencing methods have come into reality, which provide a unique opportunity to study the associations among different layers of omics directly. While exploring gDNA methylation, researchers can directly observe the relationships between different omics (genome, epigenome, and transcriptome) within an individual cell. In these methods, firstly, gDNA and mRNA within a single cell are separated, and then genome and transcriptome sequencing are performed simultaneously. Combined with scBS-seq, single-cell methylome and transcriptome sequencing (scM&T-seq) used biotinylated oligo-dT primers conjugated to the magnetic beads to separate mRNA and gDNA for single cell profiling respectively.61 Single-cell triple omics sequencing (scTrio-seq) slightly lysed cells first, released cytoplasmic RNA while keeping the nucleus intact, and then the nucleus is recovered by centrifugation to separate gDNA and mRNA.62 Subsequently, combined with scRRBS, it could detect the transcriptome and epigenome levels within an individual cell. Since cancer cells show strong heterogeneity in these 3 omics, multi-omics methods, like scTrio-seq, are especially crucial for cancer research.
Additionally, many methods can also analyze the chromatin accessibility while studying DNA methylation status: single-cell Nucleosome Occupancy and Methylome Sequencing (scNOMe-seq) can not only determine the DNA methylation status in single cells, but also locate the nucleosome footprint on high resolution63; single-cell Chromatin Overall Omic-scale Landscape Sequencing (scCOOL-seq) combines NOMe-seq with PBAT to estimate gDNA methylation, chromatin open states and copy number variations (CNVs).64 At present, as the latest and most cutting-edge analysis methods, single-cell epigenome sequencing technologies have relatively low coverage and throughput, and there is still much room for improvement.
5. PERSPECTIVES
The above-mentioned NGS-based DNA methylation detection methods have their own advantages and disadvantages respectively. In order to choose a suitable DNA methylation detection method, factors such as quantity and quality of DNA samples, required resolution and coverage, accuracy and reproducibility of the methods need to be taken into consideration.
The endonuclease digestion methods depend on endonucleases which limits their coverage of the genome. The affinity enrichment methods are cost-effective with low requirements for DNA purity and integrity; however, they are not suitable for low methylation detection and unable to achieve single-base resolution. Compared with the former 2 methods, the bisulfite conversion methods improve the sensitivity, coverage, and resolution of DNA methylation detection. Nevertheless, the coverage, sensitivity, and accuracy of sequencing might also be compromised by severe DNA degradation caused by excessive reaction conditions, low efficiency of adapter ligation, and loss of DNA fragments resulted from subsequent PCR amplification biases. Bisulfite-free DNA methylation detection methods, like TAPS, can better circumvent these shortcomings above, but require significant investment in the development and application of new analysis tools for data analysis.
Single-cell sequencing technologies can eliminate the heterogeneity of cells within the same cell type; single-cell multi-omics sequencing methods reveal the connection of different omics and provide a universal and fundamental solution for studying epigenetic regulation such as DNA methylation. However, the amount of DNA provided by individual cells is relatively low, and single cell epigenome sequencing technologies have relatively low coverage, accuracy, and throughput. Furthermore, there is also a problem of the loss of spatial information. Additionally, how to fully exploit the information in the large number of omics data has become a new challenge in the investigation of DNA methylation. In short, there is still much room for improvement in DNA methylation measuring methods. We anticipate that more high-quality DNA methylation detection technologies and analysis methods will be developed in the future, which will pave the way for in-depth epigenetic research.
ACKNOWLEDGMENT
We thank the insightful suggestions from the Zhu lab members.
REFERENCES
- [1].Schoofs T, Muller-Tidow C. DNA methylation as a pathogenic event and as a therapeutic target in AML. Cancer Treat Rev 2011;37 (Suppl 1):S13–S18. doi: 10.1016/j.ctrv.2011.04.013. [DOI] [PubMed] [Google Scholar]
- [2].Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem 1948;175 (1):315–332. [PubMed] [Google Scholar]
- [3].Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science (New York, NY) 1975;187 (4173):226–232. [PubMed] [Google Scholar]
- [4].Gertz J, Varley KE, Reddy TE, et al. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet 2011;7 (8):e1002228. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13 (7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- [6].Schübeler D. Function and information content of DNA methylation. Nature 2015;517 (7534):321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- [7].Suelves M, Carrió E, Núñez-Álvarez Y, Peinado MA. DNA methylation dynamics in cellular commitment and differentiation. Brief Funct Genomics 2016;15 (6):443–453. doi: 10.1093/bfgp/elw017. [DOI] [PubMed] [Google Scholar]
- [8].Fournier A, Sasai N, Nakao M, Defossez PA. The role of methyl-binding proteins in chromatin organization and epigenome maintenance. Brief Funct Genomics 2012;11 (3):251–264. doi: 10.1093/bfgp/elr040. [DOI] [PubMed] [Google Scholar]
- [9].Teif VB, Beshnova DA, Vainshtein Y, et al. Nucleosome repositioning links DNA (de)methylation and differential CTCF binding during stem cell development. Genome Res 2014;24 (8):1285–1295. doi: 10.1101/gr.164418.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 2011;11 (10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007;128 (4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- [12].Guirguis AA, Liddicoat BJ, Dawson MA. The old and the new: DNA and RNA methylation in normal and malignant hematopoiesis. Exp Hematol 2020;90:1–11. doi: 10.1016/j.exphem.2020.09.193. [DOI] [PubMed] [Google Scholar]
- [13].Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 2008;135 (7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Edwards JR, Yarychkivska O, Boulard M, Bestor TH. DNA methylation and DNA methyltransferases. Epigenetics Chromatin 2017;10 (1):23. doi: 10.1186/s13072-017-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science (New York, NY) 2009;324 (5929):930–935. doi: 10.1126/science. 1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer's disease human brain. Neurobiol Aging 2014;35 (6):1334–1344. doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- [17].Pastor WA, Pape UJ, Huang Y, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 2011;473 (7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pfeifer GP, Kadam S, Jin S-G. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin 2013;6 (1):10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Randt C, Linscheid M. Analysis of 5-methyl-deoxycytidine in DNA by micro-HPLC. Fresenius’ Z Anal Chem 1988;331 (3):459–463. doi: 10.1007/BF00481927. [Google Scholar]
- [20].Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996;93 (18):9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hatada I, Hayashizaki Y, Hirotsune S, Komatsubara H, Mukai T. A genomic scanning method for higher organisms using restriction sites as landmarks. Proc Natl Acad Sci U S A 1991;88 (21):9523. doi: 10.1073/pnas.88.21.9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yan PS, Perry MR, Laux DE, Asare AL, Caldwell CW, Huang TH. CpG island arrays: an application toward deciphering epigenetic signatures of breast cancer. Clin Cancer Res 2000;6 (4):1432–1438. [PubMed] [Google Scholar]
- [23].Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A 1984;81 (7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc 2007;2 (9):2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- [25].Clark TA, Spittle KE, Turner SW, Korlach J. Direct detection and sequencing of damaged DNA bases. Genome Integr 2011;2:10. doi: 10.1186/2041-9414-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bird AP, Southern EM. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol 1978;118 (1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- [27].Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 2009;27 (4):361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Clermont P-L, Parolia A, Liu HH, Helgason CD. DNA methylation at enhancer regions: novel avenues for epigenetic biomarker development. Front Biosci-Landmark 2016;21 (2):430–446. doi: 10.2741/4399. [DOI] [PubMed] [Google Scholar]
- [29].Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010;466 (7303):253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Edwards JR, O’Donnell AH, Rollins RA, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res 2010;20 (7):972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bonora G, Rubbi L, Morselli M, et al. DNA methylation estimation using methylation-sensitive restriction enzyme bisulfite sequencing (MREBS). PLoS One 2019;14 (4):e0214368. doi: 10.1371/journal.pone.0214368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 2005;37 (8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- [33].Tan L, Xiong L, Xu W, et al. Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method. Nucleic Acids Res 2013;41 (7):e84. doi: 10.1093/nar/gkt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nair SS, Coolen MW, Stirzaker C, et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 2011;6 (1):34–44. doi: 10.4161/epi.6.1.13313. [DOI] [PubMed] [Google Scholar]
- [35].Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG. Whole-genome DNA methylation profiling using MethylCap-seq. Methods 2010;52 (3):232–236. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- [36].Hayatsu H. Discovery of bisulfite-mediated cytosine conversion to uracil, the key reaction for DNA methylation analysis—a personal account. Proc Jpn Acad Ser B Phys Biol Sci 2008;84 (8):321–330. doi: 10.2183/pjab.84.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 2005;33 (18):5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009;462 (7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008;452 (7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang Q, Gu L, Adey A, et al. Tagmentation-based whole-genome bisulfite sequencing. Nat Protoc 2013;8 (10):2022–2032. doi: 10.1038/nprot.2013.118. [DOI] [PubMed] [Google Scholar]
- [41].Miura F, Enomoto Y, Dairiki R, Ito T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res 2012;40 (17):e136–e1136. doi: 10.1093/nar/gks454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang P, Mai Z, Dai Z, Zou X. Investigation of DNA methylation by direct electrocatalytic oxidation. Chem Commun 2010;46 (41):7781–7783. doi: 10.1039/C0CC00983K. [DOI] [PubMed] [Google Scholar]
- [43].Okamoto A, Tainaka K, Kamei T. Sequence-selective osmium oxidation of DNA: efficient distinction between 5-methylcytosine and cytosine. Org Biomol Chem 2006;4 (9):1638–1640. doi: 10.1039/B600401F. [DOI] [PubMed] [Google Scholar]
- [44].Liu Y, Siejka-Zielinska P, Velikova G, et al. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat Biotechnol 2019;37 (4):424–429. doi: 10.1038/s41587-019-0041-2. [DOI] [PubMed] [Google Scholar]
- [45].Liu Y, Hu Z, Cheng J, et al. Subtraction-free and bisulfite-free specific sequencing of 5-methylcytosine and its oxidized derivatives at base resolution. Nat Commun 2021;12 (1):618. doi: 10.1038/s41467-021-20920-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vaisvila R, Ponnaluri VKC, Sun Z, et al. Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA. Genome Res 2021;31 (7):1280–1289. doi: 10.1101/gr.266551.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Carpenter MA, Li M, Rathore A, et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem 2012;287 (41):34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Schutsky EK, DeNizio JE, Hu P, et al. Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat Biotechnol 2018;36:1083–1090. doi: 10.1038/nbt.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev 2015;263 (1):6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yu M, Hon GC, Szulwach KE, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012;149 (6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Song C-X, Szulwach KE, Fu Y, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol 2011;29 (1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Booth MJ, Branco MR, Ficz G, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science (New York, NY) 2012;336 (6083):934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- [53].Mooijman D, Dey SS, Boisset J-C, Crosetto N, van Oudenaarden A. Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nat Biotechnol 2016;34 (8):852–856. doi: 10.1038/nbt.3598. [DOI] [PubMed] [Google Scholar]
- [54].Wen L, Tang F. Single cell epigenome sequencing technologies. Mol Aspects Med 2018;59:62–69. doi: 10.1016/j.mam.2017.09.002. [DOI] [PubMed] [Google Scholar]
- [55].Guo H, Zhu P, Guo F, et al. Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat Protocols 2015;10 (5):645–659. doi: 10.1038/nprot.2015.039. [DOI] [PubMed] [Google Scholar]
- [56].Farlik M, Sheffield NC, Nuzzo A, et al. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep 2015;10 (8):1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Smallwood SA, Lee HJ, Angermueller C, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 2014;11 (8):817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Luo C, Keown CL, Kurihara L, et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science (New York, NY) 2017;357 (6351):600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Mulqueen RM, Pokholok D, Norberg S, et al. Scalable and efficient single-cell DNA methylation sequencing by combinatorial indexing. BioRxiv 2017;157230. [Google Scholar]
- [60].Cheow LF, Quake SR, Burkholder WF, Messerschmidt DM. Multiplexed locus-specific analysis of DNA methylation in single cells. Nat Protoc 2015;10 (4):619–631. doi: 10.1038/nprot.2015.041. [DOI] [PubMed] [Google Scholar]
- [61].Angermueller C, Lee HJ, Reik W, Stegle O. DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol 2017;18 (1):67. doi: 10.1186/s13059-017-1189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hou Y, Guo H, Cao C, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res 2016;26 (3):304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Pott S. Simultaneous measurement of chromatin accessibility, DNA methylation, and nucleosome phasing in single cells. eLife 2017;6:e23203. doi: 10.7554/eLife.23203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Guo F, Li L, Li J, et al. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res 2017;27 (8):967–988. doi: 10.1038/cr.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]